Abstract
Background
Circular RNAs (circRNAs) are research hotspots in the network of noncoding RNAs in numerous tumours. The purpose of our study was to evaluate the clinicopathological, prognostic and diagnostic value of circRNAs in colorectal cancer.
Methods
The PubMed, Cochrane Library, and Web of Science online databases were searched for relevant studies before May 15, 2019. Pooled hazard ratios (HRs) and odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to assess the association between circRNAs expression, and overall survival (OS) and clinical parameters. Pooled sensitivity, specificity, and the area under the curve (AUC) were employed to assess the diagnostic value of circRNAs.
Results
A total of 19 studies were enrolled in this meta-analysis, with 11 on clinicopathological parameters, 8 on prognosis and 7 on diagnosis. For clinicopathological and prognostic value, elevated expression of oncogenic circRNAs was correlated with poor clinical parameters (tumor size: OR = 1.769, 95% CI: 1.097–2.852; differentiation grade: OR = 1.743, 95% CI: 1.032–2.946; TNM stage: OR = 3.320, 95% CI: 1.529–7.207; T classification: OR = 3.410, 95% CI: 2.088–5.567; lymph node metastasis: OR = 3.357, 95% CI: 2.160–5.215; distal metastasis: OR = 4.338, 95% CI: 2.503–7.520) and worse prognosis (HR = 2.29, 95% CI: 1.50–3.52). However, elevated expression of tumor-suppressor circRNAs was correlated with better clinical parameters (differentiation grade: OR = 0.453, 95% CI: 0.261–0.787; T classification: OR = 0.553, 95% CI: 0.328–0.934; distal metastasis: OR = 0.196, 95% CI: 0.077–0.498) and favorable prognosis (HR = 0.37, 95% CI: 0.22–0.64). For diagnostic value, the pooled sensitivity, specificity, and AUC were 0.82 (95% CI, 0.75–0.88), 0.72 (95% CI, 0.66–0.78), and 0.82 (95% CI, 0.78–0.85), respectively.
Conclusions
These results indicate that circRNAs may be potential biomarkers for the diagnosis and prognosis of colorectal cancer.
Keywords: Circular RNA, Colorectal cancer, Diagnosis, Prognosis
Background
Circular RNAs (circRNAs), consisting of a circular configuration through a typical 5′ to 3′-phosphodiester bonds, are a novel class of endogenous noncoding RNAs [1–3]. CircRNAs play a special role as molecular markers in many human diseases including tumors, due to their conservation, abundance and tissue specificity [4]. In addition, circRNAs can be classified into four categories: exon circRNAs, intron circRNAs, exon-intron circRNAs, and intergenic circRNAs [5]. Different types of circRNAs have distinct functions, including interacting with RNA binding proteins, regulating the stability of the mRNAs, regulating gene transcription, sponging microRNAs and participating in translation [5–7]. However, the underlying mechanisms and functions of circRNAs remain uncertain.
Extensive studies have indicated that circRNAs play a major role in tumorigenesis, the development of cardiovascular diseases, and the pathogenesis of neurodegenerative diseases [8]. However, the differential expression of circRNAs and their definite functions are still not totally clear in colorectal cancer (CRC). Colorectal cancer is among the most common malignancies of the digestive system and the fourth leading cause of cancer-related death worldwide [9]. Although considerable progress has been made in the diagnosis and treatment of this disease, the prognosis of CRC patients is still poor, due to the delay in early diagnosis and the high frequency of metastasis and recurrence [10]. In this study, we performed a meta-analysis and a comprehensive search of all relevant literature to summarize the diagnostic, prognostic, and clinical significance of circRNAs in CRC.
Methods
Data search strategy
The PubMed, Cochrane Library, and Web of Science online databases were searched for studies on circRNA research that were published in English before May 15, 2019. The following search strategy was applied: (1) “circRNA” or “circular RNA” and (2) “colorectal cancer” or “colorectal carcinoma” or “colorectal tumour” or “CRC”. Two researchers (JPY and DMG) assessed the title, abstract and full text to identify the appropriate articles. Other researchers (XXL), together with two researchers (JPY and DMG) were involved in the data extraction. Any disagreements were settled by a third researcher (JTC). Then, the data were extracted from the selected articles and populated it into a table.
Inclusion and exclusion criteria
This study used the following criteria when selecting articles. Studies that met the following inclusion criteria were included in the meta-analysis: (1) patients with a pathological diagnosis of CRC; (2) cohort study or case-control study; and (3) studies that detected the circRNA expression level and provided information on the clinicopathological features and prognosis of patients. Studies were excluded if the following excluded criteria were met: (1) studies irrelevant to CRC or circRNAs; (2) data similar to that in prior studies; (3) case reports, letters, animal experiments, reviews, conference reports and meta-analysis; and (4) insufficient data.
Data extraction and quality assessment
All relevant studies were independently screened by two researchers (JPY and DMG) and the following data were extracted from eligible studies: (1) first author, publication year, type of cancer and circRNA, sample size and detection method of circRNA; (2) the role of circRNAs, follow-up time; (3) diagnostic sensitivity and specificity of circRNAs; and (4) clinicopathological features with age, gender, tumour size, tumor location, differentiation grade, TNM stage, T classification, lymph node metastasis, distal metastasis [11]. The Newcastle-Ottawa Scale (NOS) [12] was adopted for the quality assessment of the studies by two independent researchers (JPY and DMG). A third investigator (XXL) discussed any differences. A study with a score ≥ 7 was considered of high quality.
Statistical analysis
Statistical analysis was conducted using STATA software (version 14). Pooled ORs and 95% CIs were used to explore the association between circRNAs expression and clinicopathological features. HRs and 95% CIs were used to assess the prognostic value of circRNAs. The number of true positive (TP), false positive (FP), false negative (FN) and true negative (TN) were calculated and finally the pooled sensitivity, specificity and AUC were obtained to assess the diagnostic value of circRNAs. The chi-square test were used to evaluate heterogeneity. When the I2 value was < 50%, no observable heterogeneity was suggested and a fixed effects model was used [13]; otherwise, a random effects model was utilized. Sensitivity analysis was performed to explore the source of heterogeneity. Qualitative analysis of publication bias was conducted using funnel plots and quantitative analysis was conducted using Begg and Egger’s tests.
Results
Search results
As shown in Fig. 1, 83 relevant studies were obtained from several databases. After abstract reviews, 46 studies were obtained for further full-text reviews. Then, 27 articles were excluded for the following reasons: 5 were not about circRNAs or CRC, 10 did not report relevant results, 3 were review articles, 1 was animal data, and 8 had insufficient data. In summary, there were 19 studies [14–32] included in this study, with a total of 1307 patients, including 11 on clinicopathological features, 8 on prognosis and 7 on diagnosis.
Fig. 1.
Flowchart of trial selection
Study characteristics
The basic information of studies are showed in Table 1 and Table 2. All studies were published between 2015 and 2019. The follow-up time of patients ranged from 57 months to 123 months and the number of samples ranged from 40 to 204. As shown in Tables 1, 6 circRNAs were identified as tumour promoters, and 2 circRNAs were identified as tumour suppressors. As shown in Tables 2, 7 articles with AUC, sensitivity and specificity were included for the diagnosis analysis. The included studies were of high quality (See Supplementary Table 1, Additional File 1).
Table 1.
Basic features of studies for prognosis analysis
| CircRNA expression |
||||||||
|---|---|---|---|---|---|---|---|---|
| Study | Year | CircRNA | Cancer Type | High | Low | Detection Method | Regulation | Follow-up (months) |
| Zeng et al. [27] | 2018 | circHIPK3 | CRC | 89 | 89 | qRT-PCR | Upregulated | 91 |
| Fang et al. [14] | 2018 | circ_100290 | CRC | 24 | 20 | qRT-PCR | Upregulated | 59 |
| Weng et al. [31] | 2017 | circCiRS7 | CRC | 89 | 76 | qRT-PCR | Upregulated | 123 |
| Wang et al. [25] | 2019 | circPVT1 | CRC | 32 | 32 | qRT-PCR | Upregulated | 58 |
| Jin et al. [17] | 2018 | circ_0136666 | CRC | 26 | 26 | qRT-PCR | Upregulated | 60 |
| Wang et al. [26] | 2018 | circ_0071589 | CRC | 20 | 20 | qRT-PCR | Upregulated | 58 |
| Li et al. [18] | 2018 | circ_0000711 | CRC | 50 | 51 | qRT-PCR | Downregulated | 60 |
| Wang et al. [23] | 2018 | circ_0014717 | CRC | 23 | 23 | qRT-PCR | Downregulated | 57 |
CRC Colorectal cancer; qRT-PCR Quantitative real time polymerase chain reaction
Table 2.
Basic features of studies for diagnosis analysis
| Sample size | Diagnosis power | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Study | Year | CircRNA | Cancer Type | case | control | Method | Regulation | Sen. | Spe. | AUC. |
| Ji et al. [16] | 2018 | circ_0001649 | CRC | 64 | 64 | qRT-PCR | downregulated | 0.828 | 0.781 | 0.857 |
| Li et al. [19] | 2018 | circITGA7 | CRC | 69 | 48 | qRT-PCR | downregulated | 0.928 | 0.667 | 0.879 |
| Wang et al. [24] | 2017 | circ_0000567 | CRC | 102 | 102 | qRT-PCR | downregulated | 0.833 | 0.765 | 0.865 |
| Zhuo et al. [28] | 2017 | circ_0003906 | CRC | 122 | 40 | qRT-PCR | downregulated | 0.803 | 0.725 | 0.818 |
| Ruan et al. [22] | 2019 | circ_0002138 | CRC | 35 | 35 | qRT-PCR | downregulated | 0.629 | 0.743 | 0.725 |
| Wang et al. [32] | 2015 | circ_001988 | CRC | 31 | 31 | qRT-PCR | downregulated | 0.680 | 0.730 | 0.788 |
| Li et al. [18] | 2018 | circ_0000711 | CRC | 101 | 101 | qRT-PCR | downregulated | 0.910 | 0.58 | 0.810 |
AUC Area under the ROC curve; qRT-PCR Quantitative real-time polymerase chain reaction; Sen Sensitivity; Spe. Specificity; CRC Colorectal cancer
Clinicopathological parameters
The associations between circRNAs and the clinical parameters are shown in Table 3. Up-regulation of oncogenic circRNAs was closely associated with unfavorable clinical features (tumor size: OR = 1.769, 95% CI: 1.097–2.852; differentiation grade: OR = 1.743, 95% CI: 1.032–2.946; TNM stage: OR = 3.320, 95% CI: 1.529–7.207; T classification: OR = 3.410, 95% CI: 2.088–5.567; lymph node metastasis: OR = 3.357, 95% CI: 2.160–5.215; distal metastasis: OR = 4.338, 95% CI: 2.503–7.520). Additionally, down-regulation of tumor-suppressor circRNAs was closely associated with favorable clinical parameters (differentiation grade: OR = 0.453, 95% CI: 0.261–0.787; T classification: OR = 0.553, 95% CI: 0.328–0.934; distal metastasis: OR = 0.196, 95% CI: 0.077–0.498). However, there was no difference between oncogenic circRNAs expression and other clinical parameters such as age, gender, and tumor location.
Table 3.
Clinical Parameters of circRNAs in CRC
| Tumor promoter | Tumor Suppressor | |||||
|---|---|---|---|---|---|---|
| OR | 95%CI | P | OR | 95%CI | P | |
| Age (older/younger) | 1.078 | 0.737–1.577 | 0.698 | 0.589 | 0.241–1.437 | 0.224 |
| Gender (M/W) | 1.114 | 0.757–1.639 | 0.968 | 0.805 | 0.491–1.320 | 0.390 |
| Tumor size (larger/smaller) | 1.769 | 1.097–2.852 | 0.019 | 0.658 | 0.382–1.132 | 0.131 |
| Tumor location (rectum/colon) | 0.888 | 0.572–1.380 | 0.598 | 0.902 | 0.480–1.694 | 0.748 |
|
Differentiation grade (poor/well & moderate) |
1.743 | 1.032–2.946 | 0.038 | 0.453 | 0.261–0.787 | 0.005 |
| TNM stage (III + IV/I + II) | 3.320 | 1.529–7.207 | 0.002 | 0.442 | 0.187–1.042 | 0.062 |
|
T classification (T3 + T4/T1 + T2) |
3.410 | 2.088–5.567 | 0.000 | 0.533 | 0.328–0.934 | 0.027 |
| Lymph node metastasis (Y/N) | 3.357 | 2.160–5.215 | 0.000 | 0.389 | 0.116–1.307 | 0.127 |
| Distant metastasis (Y/N) | 4.338 | 2.503–7.520 | 0.000 | 0.196 | 0.077–0.498 | 0.001 |
CI Confidence interval; M Men; N No; W Women; Y Yes; OR Odds ratio. The results are in bold if p < 0.05
Overall survival
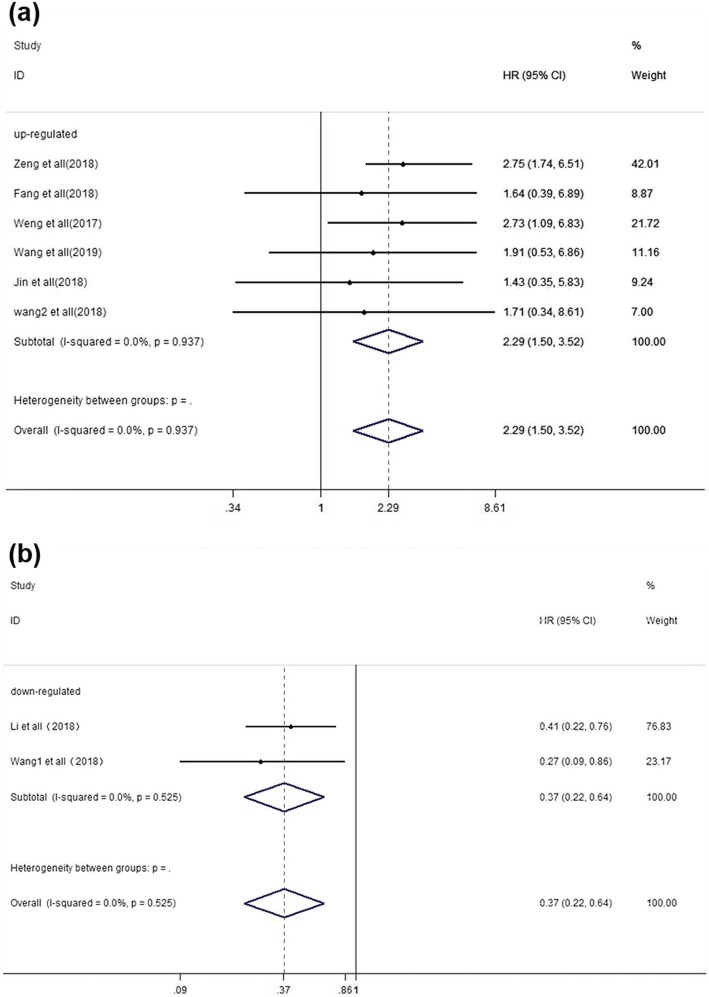
Up-regulation of oncogenic circRNAs was notably associated with worse prognosis (HR = 2.29, 95% Cl: 1.50–3.52, p < 0.001, Fig. 2 a), and a fixed-effects model was utilized as no heterogeneity was found (I2 = 0.0%, p = 0.937). In addition, down-regulation of tumour-suppressor circRNAs was associated with better prognosis (HR = 0.37, 95% Cl: 0.22–0.64, p < 0.001, Fig. 2 b), and a fixed-effects model was applied because of no heterogeneity between studies (I2 = 0.0%, p = 0.525).
Fig. 2.
Forest plots for the association between circRNAs and overall survival (OS) in colorectal cancer (CRC). a oncogenic circRNAs; b tumor suppressor circRNAs
Diagnosis analysis
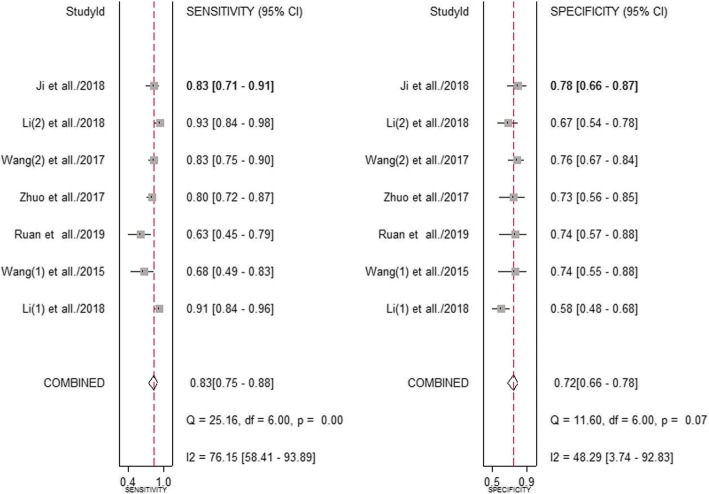
To further evaluate the diagnostic value of circRNAs, the pooled sensitivity and specificity were calculated, and the results were shown in Fig. 3. And a random-effects model was utilized because of high heterogeneity between studies (I2 = 76.15% and I2 = 48.29%). The pooled results showed a sensitivity of 0.83 (95% CI: 0.75–0.88) and a specificity of 0.72 (95% CI: 0.66–0.78). In addition, the summary receiver operator characteristic (SROC) curve analysis indicated AUC of 0.82 (95% CI 0.78–0.85, Fig. 4). Taken together, these results suggested that circRNAs have a good diagnostic accuracy for CRC.
Fig. 3.
Forest plots for the pooled sensitivity and specificity of circRNAs
Fig. 4.

SROC curve in the diagnostic analysis
Publication bias and sensitivity analysis
No evidence of publication bias were identified from the funnel plot by qualitative analysis (See Supplementary Fig. 1, Additional File 2). In quantitative analysis, there was no obvious publication bias by Begg’s (p = 0.213, See Supplementary Fig. 2, Additional File 2) and Egger’s test (p = 0.722, See Supplementary Fig. 3, Additional File 2). Furthermore, Deek’s funnel plot asymmetry test [33] was performed to assess the publication bias among studies for diagnosis analysis, and the result showed no obvious publication bias was found (p = 0.07, See Supplementary Fig. 4, Additional File 2). Sensitivity analysis indicated the pooled results were stable in our studies (See Supplementary Fig. 5, Additional File 2).
Discussion
Recently, many studies have focused on the significant role of circRNAs, whereas no relevant meta-analyses on circRNA expression in CRC have been performed. A total of 1307 cancer patients from 19 eligible studies were collected and analyzed in this study, including 7 on diagnosis, 8 on prognosis, and 11 on clinicopathological features. For diagnostic value, the summarized results revealed AUC of 0.82, with a sensitivity of 83% and a specificity of 72%. For clinical and prognostic value, abnormal expression of circRNAs were closely associated with clinical parameters and prognosis.
Our current study observed a significant relationship between abnormal circRNA expression and its diagnostic value in CRC patients. As aberrant expression of circRNAs in different tumor tissue can be easily detected, measurements can be performed conveniently and economically. Coupled with the structural stability of circRNAs, circRNAs are considered as potential biomarkers for the diagnosis of CRC patients. Although sensitivity analysis showed no significant heterogeneity, more pertinent investigations are warranted to corroborate our findings.
In previous meta-analyses, only five meta-analyses [34–38] detected an association between the circRNAs and carcinoma. However, in the studies of Wang et al. [34], Chen et al. [35] and Li et al. [36], only one study was included to investigate the relationship between the circRNAs and CRC. Li et al. [37] and Ding et al. [38] assessed the diagnostic value of circRNAs for human cancers, in which five articles were included to investigate the diagnostic value of circRNAs in CRC, whereas they failed to discuss the role of circRNAs in CRC patients. In the present study, we collected all the relevant articles published to date and performed a meta-analysis including 19 articles with 1307 CRC patients. Furthermore, we evaluated the prognostic and diagnostic value of circRNA expression in CRC patients. Nonetheless, further large-scale studies are needed to confirm these results.
However, several limitations must be considered when interpreting the conclusions of this meta-analysis. First, since all patients included in the article were from China, this reduced the applicability of the results across different ethnicities and regions. Moreover, there was a limited number of articles for a subgroup analysis. Furthermore, a relatively small number of patients was included in this meta-analysis, so larger-scale studies would be necessary to verify the obtained results. Finally, several studies did not provide HRs with their 95% CIs in the article, so we needed to extract them from the Kaplan-Meier survival curve.
Conclusions
In summary, our study demonstrated a crucial relationship between the aberrant expression of circRNAs and clinicopathological, prognostic, and diagnostic value in CRC patients. Furthermore, circRNAs may be promising biomarkers and treatment targets for colorectal cancer.
Supplementary information
Additional file 1: Table S1. Quality assessment of included studies (Newcastle-Ottawa Scale).
Additional file 2: Figure S1. Funnel plot for the evaluation of publication bias. Figure S2. Begg’s funnel plot for the evaluation of publication bias. Figure S3. Egger’s funnel plot for the evaluation of publication bias. Figure S4. Deeks’ funnel plot asymmetry test for the evaluation of publication bias. Figure S5. Sensitivity analysis to assess the stability of results.
Acknowledgments
Not applicable.
Abbreviations
- OR
Odds ratios
- 95% CI
95% Confidence interval
- HR
Hazard ratio
- OS
Overall survival
- circRNAs
Circular RNAs
- CRC
Colorectal cancer
- SROC
The summary receiver operator characteristic curve
- AUC
The area under the curve
Authors’ contributions
JTC and XXL conceived and designed the study. JPY, DMG, XXL and JTC performed data assessment. JPY and DMG analyzed the data and wrote the manuscript. All authors reviewed the paper. All authors have read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
All data analyzed during this study are included in this article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jinpeng Yuan and Dongming Guo equally contributed as first author.
Contributor Information
Xinxin Li, Email: savageli23@163.com.
Juntian Chen, Email: 13809846668@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12885-020-06932-z.
References
- 1.Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. EMBO J. 2013;32(7):923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Yuhai, Alexandrov Peter, Jaber Vivian, Lukiw Walter. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7) Genes. 2016;7(12):116. doi: 10.3390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 10.Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17(4):268. doi: 10.1038/nrc.2017.24. [DOI] [PubMed] [Google Scholar]
- 11.Huang X, Zhang W, Shao Z. Prognostic and diagnostic significance of circRNAs expression in lung cancer. J Cell Physiol. 2019;234(10):18459–18465. doi: 10.1002/jcp.28481. [DOI] [PubMed] [Google Scholar]
- 12.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 13.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang G, Ye BL, Hu BR, Ruan XJ, Shi YX. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2018;504(1):184–189. doi: 10.1016/j.bbrc.2018.08.152. [DOI] [PubMed] [Google Scholar]
- 15.Guo JN, Li J, Zhu CL, Feng WT, Shao JX, Wan L, Huang MD, He JD. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi: 10.2147/OTT.S123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji W, Qiu C, Wang M, Mao N, Wu S, Dai Y. Hsa_circ_0001649: a circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497(1):122–126. doi: 10.1016/j.bbrc.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol. 2019;234(5):7247–7256. doi: 10.1002/jcp.27482. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Ni S, Zhou C, Ye M. The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer. Cancer Manag Res. 2018;10:2777–2784. doi: 10.2147/CMAR.S172388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Wang J, Zhang C, Lin C, Zhang J, Zhang W, Zhang W, Lu Y, Zheng L, Li X. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246(2):166–179. doi: 10.1002/path.5125. [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Liu S, Xiang Y, Qu X, Xie Y, Zhang X. Bioinformatic analysis of circular RNA-associated ceRNA network associated with hepatocellular carcinoma. Biomed Res Int. 2019;2019:8308694. doi: 10.1155/2019/8308694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):325. doi: 10.1186/s13046-018-1006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruan H, Deng X, Dong L, Yang D, Xu Y, Peng H, Guan M. Circular RNA circ_0002138 is down-regulated and suppresses cell proliferation in colorectal cancer. Biomed Pharmacother. 2019;111:1022–1028. doi: 10.1016/j.biopha.2018.12.150. [DOI] [PubMed] [Google Scholar]
- 23.Wang F, Wang J, Cao X, Xu L, Chen L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed Pharmacother. 2018;98:775–782. doi: 10.1016/j.biopha.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Li X, Lu L, He L, Hu H, Xu Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal. 2018;32(5):e22379. doi: 10.1002/jcla.22379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Su M, Xiang B, Zhao K, Qin B. Circular RNA PVT1 promotes metastasis via miR-145 sponging in CRC. Biochem Biophys Res Commun. 2019;512(4):716–722. doi: 10.1016/j.bbrc.2019.03.121. [DOI] [PubMed] [Google Scholar]
- 26.Yong W, Zhuoqi X, Baocheng W, Dongsheng Z, Chuan Z, Yueming S. Hsa_circ_0071589 promotes carcinogenesis via the miR-600/EZH2 axis in colorectal cancer. Biomed Pharmacother. 2018;102:1188–1194. doi: 10.1016/j.biopha.2018.03.085. [DOI] [PubMed] [Google Scholar]
- 27.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Zhuo F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–5193. doi: 10.2147/OTT.S147378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, Xu J, Zhao J, Wang X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(1):118–126. doi: 10.26355/eurrev_201801_14108. [DOI] [PubMed] [Google Scholar]
- 30.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A. Circular RNA ciRS-7-a promising prognostic biomarker and a potential therapeutic target in colorectal Cancer. Clin Cancer Res. 2017;23(14):3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, Yan Y, Jia B, Liu H, Li S, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 33.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58(9):882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Wang M, Yang Y, Xu J, Bai W, Ren X, Wu H. CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer. 2018;18(1):303. doi: 10.1186/s12885-018-4213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Zhang L, Han G, Zuo X, Zhang Y, Zhu Q, Wu J, Wang X. A meta-analysis of the diagnostic accuracy of circular RNAs in digestive system malignancy. Cell Physiol Biochem. 2018;45(3):962–972. doi: 10.1159/000487291. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Li H, Lv X, Yang Z, Gao M, Bi Y, Zhang Z, Wang S, Cui Z, Zhou B, et al. Diagnostic performance of circular RNAs in human cancers: a systematic review and meta-analysis. Mol Genet Genomic Med. 2019;7(7):e00749. doi: 10.1002/mgg3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Zeng X, He J, Gui Y, Zhao S, Chen H, Sun Q, Jia N, Yuan H. Circular RNA as a biomarker for cancer: a systematic meta-analysis. Oncol Lett. 2018;16(3):4078–4084. doi: 10.3892/ol.2018.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding HX, Lv Z, Yuan Y, Xu Q. The expression of circRNAs as a promising biomarker in the diagnosis and prognosis of human cancers: a systematic review and meta-analysis. Oncotarget. 2018;9(14):11824–11836. doi: 10.18632/oncotarget.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Quality assessment of included studies (Newcastle-Ottawa Scale).
Additional file 2: Figure S1. Funnel plot for the evaluation of publication bias. Figure S2. Begg’s funnel plot for the evaluation of publication bias. Figure S3. Egger’s funnel plot for the evaluation of publication bias. Figure S4. Deeks’ funnel plot asymmetry test for the evaluation of publication bias. Figure S5. Sensitivity analysis to assess the stability of results.
Data Availability Statement
All data analyzed during this study are included in this article.