Abstract
Background
Culicoides obsoletus is an abundant and widely distributed Holarctic biting midge species, involved in the transmission of bluetongue virus (BTV) and Schmallenberg virus (SBV) to wild and domestic ruminants. Females of this vector species are often reported jointly with two morphologically very close species, C. scoticus and C. montanus, forming the Obsoletus/Scoticus Complex. Recently, cryptic diversity within C. obsoletus was reported in geographically distant sites. Clear delineation of species and characterization of genetic variability is mandatory to revise their taxonomic status and assess the vector role of each taxonomic entity. Our objectives were to characterize and map the cryptic diversity within the Obsoletus/Scoticus Complex.
Methods
Portion of the cox1 mitochondrial gene of 3763 individuals belonging to the Obsoletus/Scoticus Complex was sequenced. Populations from 20 countries along a Palaearctic Mediterranean transect covering Scandinavia to Canary islands (North to South) and Canary islands to Turkey (West to East) were included. Genetic diversity based on cox1 barcoding was supported by 16S rDNA mitochondrial gene sequences and a gene coding for ribosomal 28S rDNA. Species delimitation using a multi-marker methodology was used to revise the current taxonomic scheme of the Obsoletus/Scoticus Complex.
Results
Our analysis showed the existence of three phylogenetic clades (C. obsoletus clade O2, C. obsoletus clade dark and one not yet named and identified) within C. obsoletus. These analyses also revealed two intra-specific clades within C. scoticus and raised questions about the taxonomic status of C. montanus.
Conclusions
To our knowledge, our study provides the first genetic characterization of the Obsoletus/Scoticus Complex on a large geographical scale and allows a revision of the current taxonomic classification for an important group of vector species of livestock viruses in the Palaearctic region.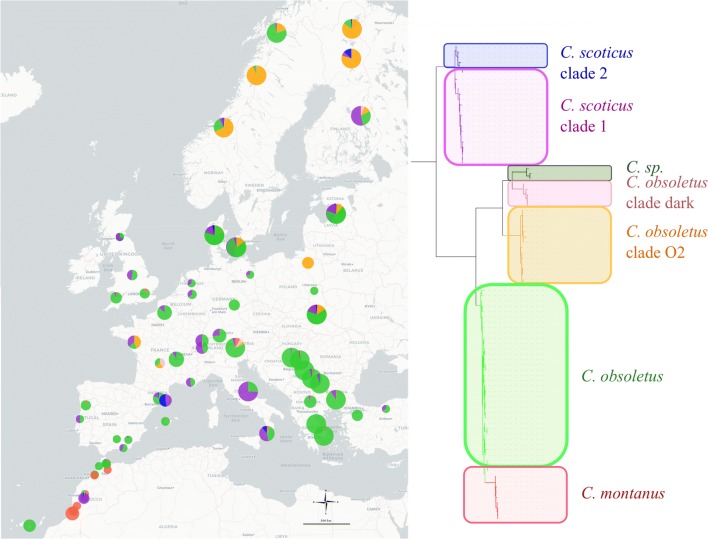
Keywords: Culicoides spp., Cryptic species, Phylogeny, Taxonomy, Species delimitation, Palaearctic Region, Biting midge
Background
In 2006, northern Europe faced massive outbreaks of bluetongue disease (BTV), a Culicoides-borne viral infection which affects wild and domestic ruminants. This was followed by the emergence of Schmallenberg virus (SBV) in 2011, another Culicoides-borne virus, which also caused important economic losses for farmers of domestic ruminants [1]. Both, the emergence and massive spread of these diseases over the Palaearctic region raised questions about the vector competence of native Palaearctic biting midges members of the genus Culicoides [2, 3]. Quickly following these epizootics, studies confirmed Culicoides species of the subgenus Avaritia as the most likely vector species and particularly those of the Obsoletus group (see below) [3–5]. Culicoides obsoletus, C. scoticus, C. dewulfi and C. chiopterus are reported to be implicated in BTV and SBV transmission in Europe based on virus detection or isolation in field-collected populations [6]. Laboratory experimental infections have also confirmed the vector competence of C. obsoletus and C. scoticus, highlighting species variations in their competence level [7].
The literature defines the Obsoletus Group as a group of species with a similar morphology, especially for the characters commonly used for the identification of these insect vectors, namely wing spot pattern (poorly defined spotted wings and a second radial cell with a light spot) and distinctive male genitalia [8–10]. Adults of species in this Group are known to be abundant, widespread across central and northern Europe, and are characterized by long seasonal occurrence [11, 12]. At present, the group is an artificial taxonomic entity still poorly defined, with no real consensus on the included species, with variable internal groupings and naming. Indeed, the world catalogue of Culicoides does not account for levels below subgenus and does not identify species complexes as groups [13]. At present, the Obsoletus Group is composed of nine valid species: C. obsoletus (Meigen), 1818; C. sinanoensis Tokunaga, 1937; C. scoticus Downes & Kettle, 1952; C. montanus Shakirzjanova, 1962; C. gornostaevae Mirzaeva, 1984; C. abchazicus Dzhafarov, 1964; C. filicinus Gornostaeva & Gachegova, 1972; C. alachua Jamnback & Wirth, 1963; and C. sanguisuga (Coquillet, 1901). The latter two are the only species belonging to this group exclusively present in the Nearctic region, while the others, are sympatric in the Palaearctic region [8]. Culicoides obsoletus is considered Holarctic because it is present in both Nearctic and Palaearctic regions [8]. Combined studies of the geometric morphometry of wings, coupled with molecular analysis have excluded C. chiopterus and C. dewulfi from the Obsoletus Group, despite previously being considered as part of the Group, based only on morphological features [9, 10, 14–16]. Life-cycles and trophic behaviors for most of the species in the Obsoletus Group are not well described or vary greatly. For instance, ecological niches of C. chiopterus and C. obsoletus are suspected to be different, although these two species are phylogenetically very close within the subgenus Avaritia. Culicoides obsoletus is a widespread generalist species and occupies a wide range of larval habitats such as forest litter, silage residue, tree holes or manure [17]. Culicoides obsoletus shows opportunistic trophic preferences and is able to take blood meals on various hosts species (man, sheep, goat, cow, horse or rodent) and, occasionally, on birds [18]. Culicoides chiopterus is a more specialist species, found engorged almost exclusively on cattle blood with larvae associated with cattle dung [19, 20].
In addition to the group, Meiswinkel et al. [21], defined the Obsoletus Complex to close taxa with very similar female adult morphology, including C. obsoletus, C. montanus and C. scoticus. Several authors have recently reported the existence of cryptic diversity within C. obsoletus, namely the C. obsoletus clade ‘O2’ and C. obsoletus clade ‘O3’ in Sweden and Switzerland [15], and the C. obsoletus clade ‘dark’ in the Netherlands [21, 22]. We will use the term “Obsoletus/Scoticus Complex” here to refer to the cryptic species C. obsoletus, C. scoticus and C. montanus as well as all previously described operational taxonomic units in the literature (see above). The terminology “Obsoletus/Scoticus Complex” is written according to the rules defined by Harbach [23] for infrasubgeneric categories within the genus Anopheles. Considering that sympatric cryptic species may exhibit different vector competence and may confound epidemiological investigations, it is mandatory to assess the intra- and interspecific diversity within the Obsoletus/Scoticus Complex.
Given the difficulty of separating these species based on morphological identification, various molecular markers have been used to overcome specific identification problems, including 16S ribosomal DNA [24, 25], 28S ribosomal DNA [26], cytochrome oxidase b (cytb), the internal transcribed spacer region 1 (ITS1) [27] and ITS2 rDNA [28]. However, the DNA region primarily used to infer phylogenetic relationships in this complex has been the mitochondrial cytochrome c oxidase subunit 1 (cox1) [29]. Most of the diversity within the Obsoletus/Scoticus Complex has been identified using cox1 [29–33]. Despite the diversity of markers used to characterize the phylogeny of the complex, few studies have used a multi-marker approach [34]. This study, therefore, will integrate a multi-marker approach in order to strengthen genetic reconstruction of the Obsoletus/Scoticus Complex.
The Obsoletus/Scoticus Complex, as currently presented in the literature, is confused and needs taxonomic revision. We characterized and mapped the genetic diversity of the Obsoletus/Scoticus Complex along a Palaearctic-Mediterranean transect covering Scandinavia to Canary islands (North to South) and Canary islands to Turkey (West to East). Our main objectives were to identify and describe the cryptic diversity observed within the Obsoletus/Scoticus Complex over a wide geographical area using molecular analyses and to question the taxonomic status of some newly described clades. In order to achieve these objectives, we conducted a molecular analysis which combined multi-marker sequencing, phylogenetic analyses and species delimitation to explore the genetic diversity of the Obsoletus/Scoticus Complex in the western European portion of the Palaearctic region.
Methods
Culicoides capture and morphological identification
Biting midges were collected at 68 sites located in 20 countries in the western European portion of the Palaearctic region, between 2009 and 2017, using national surveillance networks for Culicoides populations or local collections (Additional file 1: Table S1). Collections were made overnight with Onderstepoort Veterinary Institute (OVI) light traps set at farms near horses, cattle or sheep and all insects were stored in 70% ethanol. Morphological identification to the species level of adult Culicoides spp. was performed under a binocular microscope using the available identification keys [35, 36].
DNA extraction, amplification and sequencing
DNA was extracted from a total of 3883 adult females belonging to the Obsoletus/Scoticus Complex using the NucleoSpin® DNA kit RapidLyse (Macherey-Nagel, Duren, Germany), following the manufacturer’s instructions. An additional step was added, before extraction, for all individuals (specimens were ground in 50 μl of 1× PBS buffer). DNA samples are available upon request. Fragments of cox1 were amplified for the 3883 individuals. After sequence cleaning, 3763 sequences of cox1 were obtained [dataset cox1 (1)].Fragments of 16S and 28S rDNA were amplified on individuals chosen to be representative of the entire species diversity resulting from cox1 [dataset cox1 (2)] to reinforce mitochondrial gene sequences. All primer sequences as well as the information relating to them are present in Additional file 2: Table S2. PCR’s were performed in a 25 μl reaction volume. The PCR mix contained 1× Qiagen buffer, 1 mM MgCl2, 0.25 mM of each dNTP, 0.2 μM of each primer, 1.25 U Qiagen Taq Polymerase and 0.7 ng/μl genomic DNA for all genes. PCR programs included one-step of 5 cycles before a second step with 35 cycles for 16S rDNA and 28S rDNA. PCR amplification conditions were: an initial denaturation step at 94 °C for 5 min followed by 5 cycles of 94 °C for 30 s; 45 °C for cox1, 42 °C for 16S rDNA or 55 °C for 28S rDNA for 40 s; 72 °C for 1min; 35 cycles of 94 °C for 30 s; 51 °C for cox1, 55 °C for 16S rDNA or 50 °C for 28S rDNA for 30 s; 72 °C for 1 min; and a final extension step at 72 °C for 10 min. For each amplification reaction, negative controls were carried out. The PCR products were visualized on 1.5% agarose gels with a GelRed® Nucleic Acid Gel Stain, staining after migration of 90 min at 130 V by electrophoresis for quality control. After purifications, carried out by the sequencing service provider, the remaining 20 μl were sequenced with the same forward primers used for PCR (https://www.genewiz.com).
Sequence analyses
The reference sequences of cox1 used to identify individuals to species are available in Additional file 3: Table S3. A total of 3763 cox1 sequences (Additional file 4: Table S4) from female adults morphologically identified as belonging to the Obsoletus/Scoticus Complex were obtained after deletion of short and poor-quality sequences. The cox1 alignment was used to identify all Culicoides to species- or clade-level within the complex using the reference sequences. Among the reference sequences used to specifically assign our Culicoides samples, some sequences previously identified as C. obsoletus O1 and O3 [15] were included in our analysis in order to cover the diversity of the clades described in the literature within the Obsoletus/Scoticus Complex. After comparison with other sequences, it appears that the sequences named C. obsoletus O1 were actually C. obsoletus and that C. obsoletus O3 belongs to C. obsoletus clade dark. For this purpose, cox1 sequences were aligned with reference sequences [dataset cox1 (1)]. A phylogenetic tree based on maximum likelihood method allowed designation of a species name to each sequence if the latter belonged to a monophyletic clade with strong support (bootstrap < 900) that included a reference sequence. Thus, Culicoides were sequenced for markers 16S rDNA and 28S rDNA to support the phylogenetic reconstruction of the complex. All cox1 [dataset cox1 (2)], 16S rDNA and 28S rDNA sequences were independently aligned with the MUSCLE [37] algorithm available in the software GENEIOUS v.6.0.5 (Biomatters, http://www.geneious.com). Genetic diversity indices, haplotype and nucleotide diversity were evaluated using DNASP v.5.10 [38]. Alignments with gaps were cleaned using the software Gblocks 0.91b [39]. To assess genetic distance between clades and species within, barcoding gap bar chart using R software was performed with ggplot2 [40] and ggthemes packages. Intra- and interspecific genetic differences based on the Kimura 2-Parameter (K2P) distance model [41, 42] were calculated with MEGAX [43]. In order to map the specific diversity of the Obsoletus Group, the R software version 3.6.0 was used with the Leaflet version 2.0.2 and shiny packages version 1.4.0.
Phylogenetic inferences
Phylogenetic trees were constructed for the three markers using maximum-likelihood (ML) and Bayesian inference (BI). Bayesian inference analyses were conducted on MrBayes version 3.2.6, with tree sampling every 1000 generations in order to calculate posterior probabilities (PP) and 10 million generations. Optimal sequence evolutionary models for each analysis were obtained with Bayesian information criterion (BIC) using jModelTest. Maximum-likelihood analyses were conducted on PhyML 3.0. The ML analyses were conducted with the best model selected using 1000 bootstrap replicates for each dataset to investigate the level of support at each node, with starting tree determined by BioNJ analysis. After independent analysis of each gene, alignment of cox1 and 16S rDNA were concatenated, and analyzed following ML and BI methods.
Species delimitation methods
Two species delimitation methods were applied. The first method was a Bayesian implementation of classical GMYC method, Bayesian General Mixed Yule Coalescent (bGMYC) [44]. The single-locus ultrametric gene trees used for bGMYC methods were created with BEAST 1.8.0 [45] under a strict clock model, a Yule Process Tree Model of speciation, and a random starting tree. This analysis was carried out with default prior distribution, without outgroups and with 10 million generations sampled every 1000 cycles with HKY + G substitution model for 16S rDNA, and with HKY + I for 28S rDNA [46]. The software TREEANNOTATOR v1.8.2 was used to find Ultrametric maximum clade credibility (MCC). Single-threshold GMYC analyses were conducted with splits package in R.
The second species delimitation method used was the Bayesian Poisson Tree Processes (bPTP) through the web server PTP (http://species.h-its.org/ptp/) [47] with 100,000 MCMC generations and a thinning parameter of 100 on a maximum likelihood phylogenetic tree constructed with cox1 and 16S rDNA genes concatenated with C. dewulfi as outgroup.
Results
Molecular analysis
In total, 3763 sequences were obtained for cox1, 95 for 16S rDNA and 95 for 28S rDNA (Table 1). All sequences were deposited in GenBank (Additional file 4: Table S4). No stop codons, insertions or deletions were found in any of the cox1 sequences, indicating functional mitochondrial products.
Table 1.
Sequence statistics for four gene fragments used to reconstruct the phylogeny of the Obsoletus/Scoticus complex
| Dataset | n | Length (bp) | S | C+G (%) | h | Hd (SD) | π (SD) | Nucleotide model (under BIC) | Implemented model (BI) |
|---|---|---|---|---|---|---|---|---|---|
| cox1 (1) | 3763 | 512–627 | 141 | 34.5 | 228 | 0.890 (0.003) | 0.06299 (0.00473) | ||
| cox1 (2) | 95 | 528–623 | 146 | 33.7 | 77 | 0.994 (0.003) | 0.0921 (0.00489) | TPM2uf+I+G | nst = 6; rates = invgamma |
| 16S rDNA | 95 | 263 | 51 | 15.8 | 13 | 0.851 (0.02) | 0.04791 (0.00446) | HKY+G | nst = 2; rates = gamma |
| 28S rDNA | 95 | 576 | 25 | 39.8 | 24 | 0.911 (0.015) | 0.00753 (0.00099) | HKY+I | nst = 2; rates = propinv |
| Concatened genes | 95 | 731 | 197 | 32.8 | 90 | 0.998 (0.002) | 0.04594 (0.00297) | TPM2uf+I+G | nst = 6; rates = invgamma |
Abbreviations: n, number of individuals; h, number of haplotypes; Hd, haplotype (gene) diversity; π, nucleotide diversity; S, number of polymorphic sites; SD, standard deviation; bp, base pairs; BIC, Bayesian information criterion; BI, Bayesian inference
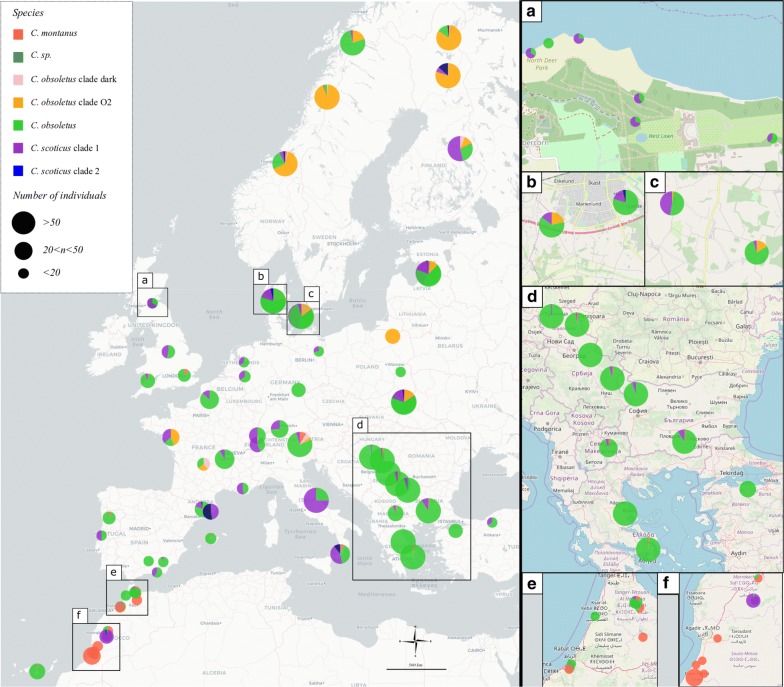
Within the selected samples present in our data set, the most abundant species was C. obsoletus, with 2416 individuals sampled, representing 68% of all Culicoides caught (Fig. 1). Species diversity within the complex varied according to the latitudes of the sampling sites (Fig. 1). The most sampled species in northern Europe (Norway and Finland) was the C. obsoletus clade O2 with 62% (162 individuals) and 58% (147 individuals) of this clade, respectively, in each country. For eastern Europe (Latvia, Poland, Serbia, Bulgaria, Macedonia, Greece and Turkey) the most sampled species was C. obsoletus, representing, for example, 99% (162 individuals) of all Culicoides sampled in Greece. However, a population in Poland (Wronka) appears to be an exception with 100% (35 individuals) of C. obsoletus clade O2. Three individuals, belonging to a phylogenetic clade unidentifiable by our reference sequences close to C. obsoletus dark, were reported from Latvia. The latter sequences are identical to sequences present in the BOLD database (accession numbers: GMGRC1056-13, GMGRC1000-13, GMGRD2587-13) of Culicoides collected in Bavaria, Germany. Finally, western and central Europe (Portugal, Spain, UK, France, Italy, Netherlands, Germany, Switzerland and Denmark) had the higher species diversity of all the species found in the Palaearctic transect. The most sampled species within this area was C. obsoletus with 913 individuals, representing 64% of the Culicoides sampled. However, unlike in eastern Europe, C. scoticus clade 1 was also found in significant numbers with 386 individuals, or 27% of the samples. Culicoides obsoletus clade dark was rarely reported in Europe with only 26 individuals found in France, Denmark, Finland, Italy, Latvia, Norway and Switzerland. Culicoides montanus was found in relatively high proportion in Morocco with 54% (80 individuals) of samples, whereas it was much rarer and more sporadic in European countries.
Fig. 1.
Map of biting midges sampling sites represented by the number of individuals per clade within the Obsoletus/Scoticus Complex. The different clades identified are shown in different colours. The size of circles on Europe map correspond to the number of individuals per clade within the cryptic species. The numbered maps on the right correspond to magnifications of some study areas. The sites of sampling too close and thus the pie charts this superimposed are to be taken into account in the numbered magnifications and not on the main map. Magnifications: a Scotland sample sites; b, c Denmark sample sites; d Balkans sample sites; e, f Morroco sample sites
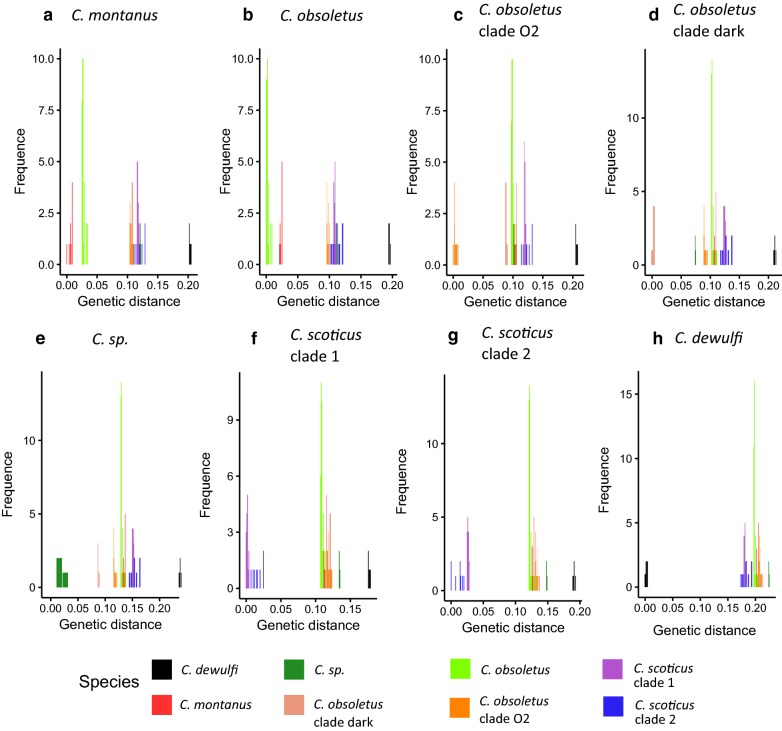
A total of 228 different cox1 haplotypes were identified. Haplotype diversity varied from 0.829 for C. obsoletus clade O2 to 0.517 for C. scoticus clade 1 and C. obsoletus clade dark (Table 2). In accordance with mitochondrial genes in insects [48], the GC composition ranged from 16% for 16S rDNA alignment to 39.8% for 28S rDNA, with a strong AT bias (Table 2). Culicoides obsoletus showed very high intraspecific diversity with 94 different haplotypes, representing nearly 43% of the total haplotypic diversity encountered in this study. The maximum interspecific genetic distance (Fig. 2) was reached between C. dewulfi and other taxonomic units, with a minimum of 17% of genetic distance between this outgroup and all other members inside the complex. Culicoides dewulfi is used here as an outgroup. Genetic distances of the same level as the other intraspecific distances were observed between C. scoticus clade 1 and C. scoticus clade 2, and between C. obsoletus and C. montanus, with a maximum of 2% and 4%, respectively. Similar interspecific genetic distances, were observed between all the other clades within the complex, with a minimum of 8% distance. All species had mean intraspecific distances of less than 1%, other than C. scoticus clade 2.
Table 2.
Genetic diversity indices for mitochondrial cox1 gene segment of Culicoides spp. in the Obsoletus/Scoticus Complex
| Species | n | h | Hd (SD) | π (SD) | S | C+G (%) |
|---|---|---|---|---|---|---|
| C. montanus | 106 | 13 | 0.642 (0.039) | 0.00726 (0.00317) | 66 | 0.334 |
| C. obsoletus | 2416 | 106 | 0.773 (0.005) | 0.00450 (nd) | 72 | 0.324 |
| C. obsoletus clade O2 | 512 | 38 | 0.829 (0.012) | 0.00447 (nd) | 53 | 0.310 |
| C. obsoletus clade dark | 26 | 6 | 0.517 (0.113) | 0.00191 (0.00058) | 7 | 0.315 |
| Culicoides sp. | 3 | 2 | 0.667 (0.314) | 0.00749 (0.00353) | 6 | 0.331 |
| C. scoticus clade 1 | 645 | 61 | 0.562 (0.023) | 0.00463 (nd) | 93 | 0.347 |
| C. scoticus clade 2 | 55 | 12 | 0.753 (0.046) | 0.00499 (0.00093) | 16 | 0.328 |
Abbreviations: n, number of individuals; h, number of haplotypes; Hd, haplotype (gene) diversity; π, nucleotide diversity; nd, not determined; S, number of polymorphic sites; SD, standard deviation
Fig. 2.
Relative distribution of interspecific divergence and intraspecific variation in cox1 for all cryptic species within the Obsoletus/Scoticus Complex
Among the 228 unique cox1 haplotypes, 95 were selected to represent the specific diversity of the complex. Using the same individuals, two alignments of 95 sequences of 16S rDNA (Additional file 5: Figure S1.) and 28S rDNA were constructed. In order to compare tree topology and to concatenate markers, a second cox1 alignment of the 95 sequences was performed (Additional file 6: Figure S2.). The cox1 dataset had a greater haplotype diversity than 16S rDNA and 28S rDNA datasets, with 0.994 vs 0.851 and 0.911 respectively. 28S rDNA was more monomorphic than cox1 and 16S rDNA, with 25, 146 and 51 polymorphic sites, respectively.
Phylogenetic analysis
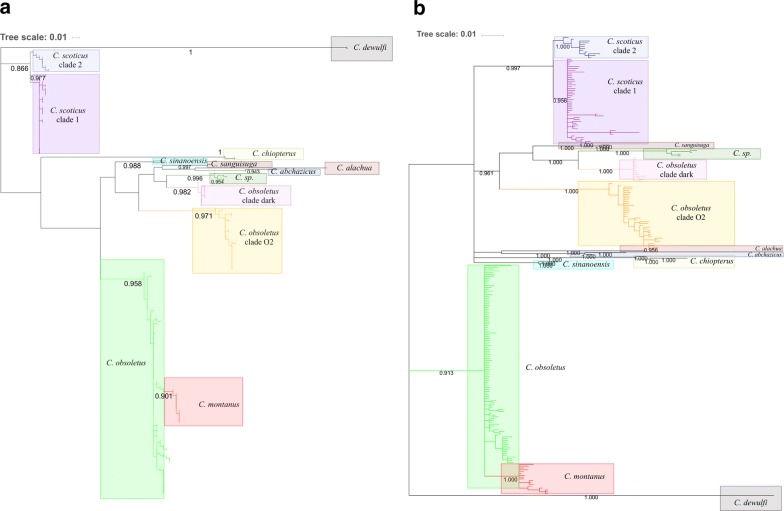
Information on the alignments used for the construction of phylogenetic trees is provided in Table 1. Species with confirmed taxonomic validity (C. montanus, C. obsoletus and C. scoticus) and cryptic taxa (C. obsoletus clade O2, C. obsoletus clade dark) highlighted by previous studies were strongly supported (bootstrap > 90%) (Fig. 3). Culicoides obsoletus clade O2 constituted a monophyletic clade with strong support (bootstrap > 90%). Culicoides obsoletus and C. montanus formed a monophyletic clade. Culicoides scoticus, another species considered valid, showed two phylogenetic clades, i.e. C. scoticus clade 1 and C. scoticus clade 2. A monophyletic clade close to C. obsoletus clade dark, was strongly supported by bootstrap values. Topologies of phylogenetics trees constructed via maximum likelihood (Fig. 3a) and Bayesian inference (BI) analyses (Fig. 3b) were congruent for alignment of full haplotype diversity.
Fig. 3.
Maximum likelihood (a) and Bayesian inference (b) phylogenetic tree using cox1 representing the haplotypic diversity within the Obsoletus/Scoticus Complex at the European scale. Values at the nodes represent bootstrap (a) and posterior probability (b) values greater than 900 for the most important nodes (1000 replicates)
Species delimitation
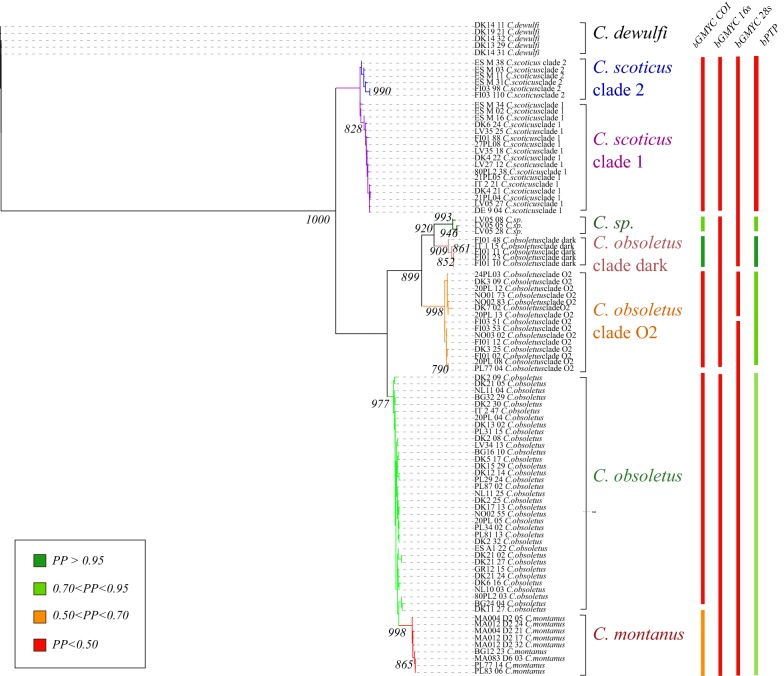
Using the cox1 dataset and bGMYC method for species delimitation (Fig. 4), 6 molecular operational taxonomic units (MOTUs) were observed: C. montanus, C. obsoletus, C. scoticus, C. obsoletus clade O2, C. obsoletus clade dark and Culicoides sp. within the Obsoletus/Scoticus Complex. Based on the 16S rDNA dataset analysed with the same delimitation method, three MOTUs were characterised including (i) two clades within C. scoticus; (ii) Culicoides sp., C. obsoletus clade dark and C. obsoletus clade O2; and (iii) C. obsoletus and C. montanus. Species delimitation with the 28S rDNA and 16S rDNA dataset had a lower resolution compared to cox1. The 28S rDNA and 16S rDNA datasets were invaluable in identifying cryptic diversity within the Obsoletus/Scoticus Complex. Indeed, the tree generated with the 28S rDNA dataset showed very low polymorphisms and resolution signal (Additional file 7: Figure S3.). The bPTP method was conducted on concatenated dataset of cox1 and 16S rDNA genes. The best statistical support for species delimitation was for concatenated dataset with bPTP. MOTUs found with the concatenated alignment and cox1 were the same, except for the delimitation of C. montanus and C. obsoletus. Indeed, using molecular delineation based on bPTP analysis, MOTUs were observed for C. scoticus, C. obsoletus clade O2, C. obsoletus clade dark, and Culicoides sp. Using the bPTP method we were not able to distinguish C. obsoletus and C. montanus as well as C. scoticus clades 1 and 2, which were not classified as separate species by either method.
Fig. 4.
Maximum likelihood phylogenetic tree using concatenated genes (cox1; 16S rDNA) representing species delimitation and molecular relationships within the Obsoletus/Scoticus Complex
Discussion
Our study investigated the genetic diversity of the Obsoletus/Scoticus Complex at the European level (68 sampling sites across 20 Palaearctic countries with 3763 cox1 sequences). For the first time, the sample selection covers the whole known western Palaearctic distribution area of these species. The complementary use of the mitochondrial 16S rDNA and nuclear 28S rDNA genes confirms the important level of cryptic diversity found within the Obsoletus/Scoticus Complex. Indeed species delimitation methods allowed us to delineate five MOTUs and: (i) to provide evidence of the taxonomic validity of C. obsoletus clade O2 and C. obsoletus clade dark (ii) to identify individuals belonging to a species not yet described or not present into the databases; and (iii) to question the taxonomic status of C. montanus.
Species assignment at the European scale showed variation in the distribution of the cryptic diversity of Obsoletus/Scoticus Complex. This result confirmed a previous study by Möhlmann et al. [49], who found a strong latitudinal effect on the relative abundance of species of the Obsoletus/Scoticus Complex. However, the previous study was carried out with few individuals from a relatively small number of countries and sampling sites [49]. This contrasts with our study, the first to be conducted at a European scale with a large data set sufficient to provide a more precise idea of the cryptic diversity within the Obsoletus/Scoticus Complex. The latitudinal variation in the relative abundance of the different cryptic species in the Obsoletus/Scoticus Complex, could be due to a wide range of factors like different ecological niches, or differences in the availability of hosts and breeding sites [50]. For example, a study conducted in Italy showed that C. scoticus collection sites were dominated by areas of natural vegetation or forest, at medium altitudes, preferably in wilder and more pristine environments [51]. However, the heterogeneity of Culicoides collection dates may also explain these variations in specific diversity within the complex. For instance, a study conducted in Sweden, found a seasonal variation in Culicoides community structure [52]. The omnipresence of C. obsoletus makes it the dominant species in Europe, confirming its status as a generalist species, which tolerates a wide range of eco-climatic conditions. The dominant species also varies according to geographical location. For example, C. obsoletus clade O2 is the most sampled species in Nordic countries while C. montanus prevails in Morocco. France, Italy and Spain appear to have the highest specific diversity of Culicoides belonging to the Obsoletus/Scoticus Complex. These three countries bring together all the cryptic diversity known so far, except the new, unidentified, Culicoides taxon that has only been found in Latvia regarding our dataset. This could be due to the significant diversity of ecological niches as well as the high density of hosts in these countries and this species diversity variation is in line with general patterns of latitudinal increase in species richness [53].
Our phylogenetic analysis allowed us to define seven well supported phylogenetic clades. Some of them correspond to species with taxonomic validity (C. montanus, C. obsoletus and C. scoticus), some more recently described phylogenetic clades (C. obsoletus clade dark and C. obsoletus clade O2) [52, 54] and some clades never described before (Culicoides sp. and C. scoticus clade 2).
Without taking into account C. scoticus, the phylogenetic reconstruction produced herein, confirmed the presence of two divergent groups; one consisting of C. obsoletus and C. montanus and the other of C. obsoletus clade dark, C. obsoletus clade O2 and a clade not yet described in the literature. We were unable to identify these sequences due to the absence of reference sequences identified at the species level in the sequence databases. However, three sequences, from specimens collected in Bavaria in Germany, present in the BOLD database are identical. Further sampling and sequencing of Culicoides from eastern Europe are necessary in order to associate morphological features with this new cryptic species. The large number of Culicoides processed during this study made it difficult to use non-destructive DNA extraction techniques. Non-destructive techniques would be necessary to couple morphological criteria with genetic analysis, in order to identify this species.
We also described a second clade phylogenetically very close to C. scoticus, C. scoticus clade 2 [55]. However, given the small genetic distance observed between these two clades, they can be considered as intraspecific variation within C. scoticus.
According to this study, C. obsoletus clade dark appears as a true cryptic species, with high phylogenetic support. Meiswinkel et al. [21] hypothesized that C. obsoletus clade dark could be C. gornostaevae Mirzaeva, 1984, but C. gornostaevae was significantly larger and had a distribution restricted to the boreal zone of Siberia. However, this species has been recently reported from Norway, Poland and Sweden [56] but the lack of C. gornostaevae reference sequences in publicly available databases prevented comparisons with C. obsoletus clade dark.
Culicoides obsoletus clade O2 was also strongly supported by phylogenetic and species delimitation analysis, and thus could be considered a cryptic species within the Obsoletus/Scoticus Complex. However, our sample showed a high abundance of C. obsoletus clade O2 mainly at high latitudes, whereas it had initially been identified for the first time further south in the Swiss Alps [54, 56, 57] and France.
Little is known about the ecology of C. montanus. We found this species over a large geographical area, from Morocco to a few individuals in Norway. In spite of this, we have identified only a very small number of C. montanus except in Morocco where this species constitutes the majority. This latter result is in accordance with the fact that sites in Italy, where C. montanus is predominant are characterised by a high land surface temperature, higher than the values registered in the C. obsoletus and C. scoticus sites [51]. Culicoides obsoletus and C. scoticus are considered to be sibling species because of diagnostic female morphological characters, which are difficult to observe or overlap [16], as opposed to the morphological identification of males which is simpler [58]. If the genetic analyses by Pages & Sarto [58] confirm that both C. obsoletus and C. scoticus are distinct species, the question remains open for C. montanus. Previous phylogenetic studies based on cox1 indeed showed limited genetic distance between C. obsoletus and C. montanus and in studies based on ITS sequences C. montanus always appears in one of the subclades of C. obsoletus [59]. In our multi-marker phylogenetic tree, C. montanus and C. obsoletus formed a monophyletic clade. In addition, the genetic distance between C. obsoletus and C. montanus was of the same order of magnitude as some intraspecific distances.
The number of putative species defined within the Obsoletus/Scoticus Complex varied depending on the molecular markers and species delimitation methods used (Fig. 4), particularly pertaining to the status of C. obsoletus and C. montanus. Unlike the other methods, the bGMYc method, based on cox1, distinguished these two species. This can be explained by the fact that some parameters of the analysis (i.e. priors), like differences in population size or speciation rates, can bias the GMYC method by overestimating the number of species [46, 60–62]. Insufficient sampling, high gene flow or a recent speciation event are also likely explanations for the differences in results between phylogenetic trees and species delimitation results [63, 64]. Although subject to the same constraints, it has been shown that the bPTP method is significantly more robust [65]. Moreover, methods based on multiple loci improve discovery, resolution and stability of species delimitation [66, 67]. Furthermore, studies have shown that species delineation in insects is more appropriate with multilocus species delimitation methods [68–71]. These arguments allowed us to validate the species delimitation scheme produced with the bPTP method. This analysis coupled with the low level of genetic distances observed between C. montanus and C. obsoletus led us to question the taxonomic status of C. montanus. This could be the beginning of a speciation process of C. montanus within the clade of C. obsoletus. Hovewer, C. montanus was originally described from Kazakhstan and present in central Asia [57]. Therefore, examination of individuals sampled in this geographical area could strengthen our conclusions.
In the light of our conclusions, in-depth morphological analyses with the deposition of reference individuals will have to be carried out in order to decide whether or not to definitively rule out the taxonomic status of the cryptic species making up this complex. Indeed, although adult females are not morphologically distinguishable on a routine basis, males are easier to identify using their genitalia and pupal differences and can provide evidence of morphological differences between the species.
Conclusions
This study provides clarification of the distribution pattern of species belonging to the Obsoletus/Scoticus Complex, using a dataset based on samples from western Palaeartic and Mediterranean transect. Strong variations in latitudinal cryptic species diversity was observed. This study clarifies the phylogenetic relationships between species belonging to the Obsoletus/Scoticus Complex. We identified and validated five MOTUs, C. obsoletus, C. scoticus, C. obsoletus clade O2, C. obsoletus clade dark and a MOTUs corresponding to an unidentified species. The latter three species have not been formally described but our results confirm that they should be considered as species in their own right. More detailed studies of their morphology and ecology are needed to provide more detailed descriptions of these species. Furthermore, our results raise questions concerning the taxonomic status of C. montanus, which was previously considered as a taxonomically valid species.
Supplementary information
Additional file 1: Table S1. Information on adult female Culicoides sampling sites and results of specific assignation using cox1.
Additional file 2: Table S2 Primers used for PCRs and sequencing in this study.
Additional file 3: Table S3. Reference sequences used for specific assignation.
Additional file 4: Table S4. Information on all Culicoides sequenced and GenBank accession numbers.
Additional file 5: Figure S1. Maximum likelihood phylogenetic tree using 16S rDNA. Values at the nodes represent bootstrap values greater than 800 for the most important nodes (1000 replicates).
Additional file 6: Figure S2. Maximum likelihood phylogenetic tree using cox1. Values at the nodes represent bootstrap values greater than 800 for the most important nodes (1000 replicates).
Additional file 7: Figure S3. Maximum likelihood phylogenetic tree using 28S rDNA. Values at the nodes represent bootstrap values greater than 800 for the most important nodes (1000 replicates).
Acknowledgements
The authors are grateful to all European partners who assisted in Culicoides sampling and shipping. We acknowledge Bruno Mathieu for his friendly cooperation and for allowing us access to use some of his sequences as references for our study.
Abbreviations
- BTV
bluetongue virus
- SBV
Schmallenberg virus
- cytb
cytochrome oxidase b
- ITS1
internal transcribed spacer region 1
- ITS2
internal transcribed spacer region 2
- cox1
cytochrome c oxidase subunit 1
- N
number of individuals
- h
number of haplotypes
- Hd
haplotype (gene) diversity
- π
nucleotide diversity
- nd
not determined
- S
number of polymorphic sites
- bGMYC
Bayesian General Mixed Yule Coalescent
- MOTUs
molecular operational taxonomic units
- bPTP
Bayesian Poisson Tree Processes
Authors’ contributions
AM, KH and CG designed the study. AIJ, AD, AC, BVP, DWR, DS, DW, DPu, DPe, EV, FJ, HK, IPF, JL, JN, JMP, JS, KRS, KK, CLC, ML, MBo, MG, MBi, ME, MR, MO, MAMC, RB, REP, SC, ST, SB, SS, SMS, ZO, ZS and ZV provided samples of Culicoides collected in their respective countries. IR, MD contributed the identification of Culicoides. AM, LG, LP and LT performed DNA extraction, amplification, quality control and sequencing. BM provided some sequences as references for our study. AM analysed the data. KH and CG contributed to the manuscript written first by AM. All authors read and approved on the final manuscript.
Funding
This study was partially funded by the EU grant H2020-727393 PALE-Blu and by the VectorNet project (OC/EFSA/AHAW/2013/02-FWC1) funded by the European Centre for Disease Prevention and Control (ECDC) and the European Food Safety Authority (EFSA). The contents of this publication are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission, of the ECDC or of the EFSA.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional files. The newly generated sequences were submitted in the GenBank database under the accession numbers MT170026-MT173788.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
All of the authors participated equally by providing samples
Contributor Information
Antoine Mignotte, Email: antoine.mignotte@cirad.fr.
Claire Garros, Email: claire.garros@cirad.fr.
Laetitia Gardès, Email: laetitia.gardes@cirad.fr.
Thomas Balenghien, Email: thomas.balenghien@cirad.fr.
Maxime Duhayon, Email: maxime.duhayon@cirad.fr.
Ignace Rakotoarivony, Email: ignace.rakotoarivony@cirad.fr.
Laura Tabourin, Email: laura.tabourin34@gmail.com.
Léa Poujol, Email: proleapoujol@gmail.com.
Bruno Mathieu, Email: bmathieu@unistra.fr.
Adolfo Ibañez-Justicia, Email: a.ibanezjusticia@nvwa.nl.
Ahmet Deniz, Email: ahmetdeniz@tarim.gov.tr.
Aleksandar Cvetkovikj, Email: acvetkovic@fvm.ukim.edu.mk.
Bethan V. Purse, Email: beth@ceh.ac.uk
David W. Ramilo, Email: dwrramilo@hotmail.com
Despoina Stougiou, Email: dimdesk@otenet.gr.
Doreen Werner, Email: Doreen.Werner@zalf.de.
Dubravka Pudar, Email: dubravkapudar@gmail.com.
Dušan Petrić, Email: dusanp@polj.uns.ac.rs.
Eva Veronesi, Email: eva.veronesi@uzh.ch.
Frans Jacobs, Email: F.H.H.Jacobs@nvwa.nl.
Helge Kampen, Email: Helge.Kampen@fli.de.
Isabel Pereira da Fonseca, Email: ifonseca@fmv.ulisboa.pt.
Javier Lucientes, Email: jlucien@unizar.es.
Javier Navarro, Email: ajavier.navarro@juntadeandalucia.es.
Josue Martinez de la Puente, Email: jmp@ebd.csic.es.
Jovana Stefanovska, Email: jstefanovska@fvm.ukim.edu.mk.
Kate R. Searle, Email: katrle@ceh.ac.uk
Khalid Khallaayoune, Email: k.khallaayoune@iav.ac.ma.
C. Lorna Culverwell, Email: lorna.culverwell@helsinki.fi.
Magdalena Larska, Email: m.larska@piwet.pulawy.pl.
Maria Bourquia, Email: bourquia.maria@gmail.com.
Maria Goffredo, Email: m.goffredo@izs.it.
Marina Bisia, Email: marina_ilisia@msn.com.
Marion England, Email: marion.england@pirbright.ac.uk.
Matthew Robin, Email: mr1964@liv.ac.uk.
Michela Quaglia, Email: michelaquaglia@yahoo.it.
Miguel Ángel Miranda-Chueca, Email: ma.miranda@uib.es.
René Bødker, Email: rebo@sund.ku.dk.
Rosa Estrada-Peña, Email: estrada@unizar.es.
Simon Carpenter, Email: simon.carpenter@pirbright.ac.uk.
Simona Tchakarova, Email: simona.tchakarova@hotmail.co.uk.
Sofia Boutsini, Email: sboutsini@yahoo.gr.
Ståle Sviland, Email: stale.sviland@vetinst.no.
Stefanie M. Schäfer, Email: smsc@ceh.ac.uk
Zanda Ozoliņa, Email: zanda.ozolina@bior.lv.
Zanda Segliņa, Email: zanda.seglina@bior.lv.
Zati Vatansever, Email: zativet@gmail.com.
Karine Huber, Email: karine.huber@cirad.fr.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-020-04114-1.
References
- 1.Carpenter S, Groschup MH, Garros C, Felippe-Bauer ML, Purse BV. Culicoides biting midges, arboviruses and public health in Europe. Antiviral Res. 2013;100:102–113. doi: 10.1016/j.antiviral.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 2.Wilson AJ, Mellor PS. Bluetongue in Europe: past, present and future. Phil Trans R Soc Lond B. 2009;364:2669–2681. doi: 10.1098/rstb.2009.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter S, Wilson A, Mellor PS. Culicoides and the emergence of bluetongue virus in northern Europe. Trends Microbiol. 2009;17:172–178. doi: 10.1016/j.tim.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann B, Eschbaumer M, Beer M. Real-time quantitative reverse transcription-PCR assays specifically detecting bluetongue virus serotypes 1, 6, and 8. J Clin Microbiol. 2009;47:2992–2994. doi: 10.1128/JCM.00599-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savini G, MacLachlan NJ, Sanchez-Vizcaino JM, Zientara S. Vaccines against bluetongue in Europe. Comp Immunol Microbiol Infect Dis. 2008;31:101–120. doi: 10.1016/j.cimid.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter S, Lunt HL, Arav D, Venter GJ, Mellor PS. Oral susceptibility to bluetongue virus of Culicoides (Diptera: Ceratopogonidae) from the United Kingdom. J Med Entomol. 2006;43:73–78. doi: 10.1093/jmedent/43.1.73. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter S, Mcarthur C, Selby R, Ward R, Nolan DV, Luntz AJ, et al. Experimental infection studies of UK Culicoides species midges with bluetongue virus serotypes 8 and 9. Vet Rec. 2008;163:589–592. doi: 10.1136/vr.163.20.589. [DOI] [PubMed] [Google Scholar]
- 8.Mathieu B. Les espèces de Culicoides du sous-genre Avaritia (Diptera: Ceratopogonidae) dans le monde: révision systématique et taxonomique des espèces d’intérêt dans la transmission dʼOrbivirus. École doctorale Sciences de la vie et de la santé Université de Strasbourg; 2011.
- 9.Meiswinkel R, Gomulski LM, Delecolle JC, Goffredo M, Gasperi G. The taxonomy of Culicoides vector complexes—unfinished business. Vet Ital. 2004;40:151–159. [PubMed] [Google Scholar]
- 10.Hajd Henni L, Sauvage F, Ninio C, Depaquit J, Augot D. Wing geometry as a tool for discrimination of Obsoletus group (Diptera: Ceratopogonidae: Culicoides) in France. Infect Genet Evol. 2014;21:110–117. doi: 10.1016/j.meegid.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Mehlhorn H, Walldorf V, Klimpel S, Jahn B, Jaeger F, Eschweiler J, et al. First occurrence of Culicoides obsoletus-transmitted bluetongue virus epidemic in central Europe. Parasitol Res. 2007;101:219–228. doi: 10.1007/s00436-007-0519-6. [DOI] [PubMed] [Google Scholar]
- 12.Savini G, Goffredo M, Monaco F, Di Gennaro A, Cafiero MA, Baldi L, et al. Bluetongue virus isolations from midges belonging to the Obsoletus complex (Culicoides, Diptera: Ceratopogonidae) in Italy. Vet Rec. 2005;157:133–139. doi: 10.1136/vr.157.5.133. [DOI] [PubMed] [Google Scholar]
- 13.Borkent A. World species of biting midges (Diptera: Ceratopogonidae) Salmon Arm: American Museum of Natural History, and Instituto Nacional de Biodiversidad; 2016. [Google Scholar]
- 14.Gomulski LM, Meiswinkel R, Delecolle JC, Goffredo M, Gasperi G. Phylogenetic relationships of the subgenus Avaritia Fox, 1955 including Culicoides obsoletus (Diptera, Ceratopogonidae) in Italy based on internal transcribed spacer 2 ribosomal DNA sequences. Syst Entomol. 2005;30:619–631. [Google Scholar]
- 15.Ander M, Troell K, Chirico J. Barcoding of biting midges in the genus Culicoides: a tool for species determination. Med Vet Entomol. 2013;27:323–331. doi: 10.1111/j.1365-2915.2012.01050.x. [DOI] [PubMed] [Google Scholar]
- 16.Kluiters G, Pages N, Carpenter S, Gardes L, Guis H, Baylis M, et al. Morphometric discrimination of two sympatric sibling species in the Palaearctic region, Culicoides obsoletus Meigen and C. scoticus Downes & Kettle (Diptera: Ceratopogonidae), vectors of bluetongue and Schmallenberg viruses. Parasit Vectors. 2016;9:262. [DOI] [PMC free article] [PubMed]
- 17.Ninio C, Augot D, Delecolle JC, Dufour B, Depaquit J. Contribution to the knowledge of Culicoides (Diptera: Ceratopogonidae) host preferences in France. Parasitol Res. 2011;108:657–663. doi: 10.1007/s00436-010-2110-9. [DOI] [PubMed] [Google Scholar]
- 18.Martínez-de la Puente Josué, Figuerola Jordi, Soriguer Ramón. Fur or feather? Feeding preferences of species of Culicoides biting midges in Europe. Trends in Parasitology. 2015;31(1):16–22. doi: 10.1016/j.pt.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Garros C, Gardes L, Allene X, Rakotoarivony I, Viennet E, Rossi S, et al. Adaptation of a species-specific multiplex PCR assay for the identification of blood meal source in Culicoides (Ceratopogonidae: Diptera): applications on Palaearctic biting midge species, vectors of Orbiviruses. Infect Genet Evol. 2011;11:1103–1110. doi: 10.1016/j.meegid.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Kettle DS, Lawson JWH. The early stages of British biting midges Culicoides Latreille (Diptera: Ceratopogonidae) and allied Genera. Bull Entomol Res. 2009;43:421–467. [Google Scholar]
- 21.Meiswinkel R, De Bree F, Bossers-De Vries R, Elbers AR. An unrecognized species of the Culicoides obsoletus complex feeding on livestock in The Netherlands. Vet Parasitol. 2015;207:324–328. doi: 10.1016/j.vetpar.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 22.Elbers AR, Meiswinkel R. Culicoides (Diptera: Ceratopogonidae) host preferences and biting rates in the Netherlands: comparing cattle, sheep and the black-light suction trap. Vet Parasitol. 2014;205:330–337. doi: 10.1016/j.vetpar.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Harbach RE. Review of the internal classification of the genus Anopheles (Diptera, Culicidae) - the foundation for comparative systematics and phylogenetic research. Bull Entomol Res. 1994;84:331–342. [Google Scholar]
- 24.Debeila TJ. Characterisatoin of selected Culicoides population in South Africa using genetic markers. PhD Thesis, Faculty of Veterinary Science, University of Pretoria; 2010.
- 25.Tabachnick W. Culicoides and the global epidemiology of bluetongue virus infection. Vet Ital. 2004;40:144–150. [PubMed] [Google Scholar]
- 26.Bakhoum MT, Labuschagne K, Huber K, Fall M, Mathieu B, Venter G, et al. Phylogenetic relationships and molecular delimitation of Culicoides Latreille (Diptera: Ceratopogonidae) species in the Afrotropical region: interest for the subgenus Avaritia. Syst Entomol. 2018;43:355–371. [Google Scholar]
- 27.Cêtre-Sossah Catherine, Baldet Thierry, Delécolle Jean-Claude, Mathieu Bruno, Perrin Aurélie, Grillet Colette, Albina Emmanuel. Molecular detection ofCulicoidesspp. andCulicoides imicola, the principal vector of bluetongue (BT) and African horse sickness (AHS) in Africa and Europe. Veterinary Research. 2004;35(3):325–337. doi: 10.1051/vetres:2004015. [DOI] [PubMed] [Google Scholar]
- 28.Gomulski LM, Meiswinkel R, Delecolle JC, Goffredo M, Gasperi G. Phylogeny of the subgenus Culicoides and related species in Italy, inferred from internal transcribed spacer 2 ribosomal DNA sequences. Med Vet Entomol. 2006;20:229–238. doi: 10.1111/j.1365-2915.2006.00620.x. [DOI] [PubMed] [Google Scholar]
- 29.Nolan DV, Carpenter S, Barber J, Mellor PS, Dallas JF, Mordue Luntz AJ, et al. Rapid diagnostic PCR assays for members of the Culicoides obsoletus and Culicoides pulicaris species complexes, implicated vectors of bluetongue virus in Europe. Vet Microbiol. 2007;124:82–94. doi: 10.1016/j.vetmic.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 30.Augot D, Sauvage F, Jouet D, Simphal E, Veuille M, Couloux A, et al. Discrimination of Culicoides obsoletus and Culicoides scoticus, potential bluetongue vectors, by morphometrical and mitochondrial cytochrome oxidase subunit I analysis. Infect Genet Evol. 2010;10:629–637. doi: 10.1016/j.meegid.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Pages N, Munoz-Munoz F, Talavera S, Sarto V, Lorca C, Nunez JI. Identification of cryptic species of Culicoides (Diptera: Ceratopogonidae) in the subgenus Culicoides and development of species-specific PCR assays based on barcode regions. Vet Parasitol. 2009;165:298–310. doi: 10.1016/j.vetpar.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 32.Schwenkenbecher JM, Mordue AJ, Piertney SB. Phylogenetic analysis indicates that Culicoides dewulfi should not be considered part of the Culicoides obsoletus complex. Bull Entomol Res. 2009;99:371–375. doi: 10.1017/S0007485308006391. [DOI] [PubMed] [Google Scholar]
- 33.Sarto i Monteys V., Ventura D., Pages N., Aranda C., Escosa R. Expansion of Culicoides imicola, the main bluetongue virus vector in Europe, into Catalonia, Spain. Veterinary Record. 2005;156(13):415–417. doi: 10.1136/vr.156.13.415. [DOI] [PubMed] [Google Scholar]
- 34.Harrup LE, Bellis GA, Balenghien T, Garros C. Culicoides Latreille (Diptera: Ceratopogonidae) taxonomy: current challenges and future directions. Infect Genet Evol. 2015;30:249–266. doi: 10.1016/j.meegid.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delecolle JC. Nouvelle contribution à lʼétude systématique et iconographique des espèces du genre Culicoides (Diptéra: Cératopogonidae) du Nord-Est de la France. PhD Thesis, University of Strasbourg, Strasbourg; 1985.
- 36.Campbell JA, Pelham-Clinton EC. Taxonomic review of the British species of Culicoides Latreille (Diptera, Ceratopogonidae) Proc R Entomol Soc. 1960;67:181–302. [Google Scholar]
- 37.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 39.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 40.Wickham H. ggplot2: Elegant graphics for data analysis. J R Stat Soc A Stat. 2009;174:245–250. [Google Scholar]
- 41.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 42.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid NM, Carstens BC. Phylogenetic estimation error can decrease the accuracy of species delimitation: a Bayesian implementation of the general mixed Yule-coalescent model. BMC Evol Biol. 2012;12:196. doi: 10.1186/1471-2148-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond Alexei J., Suchard Marc A., Xie Dong, Rambaut Andrew. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujisawa T, Barraclough TG. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol. 2013;62:707–724. doi: 10.1093/sysbio/syt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crozier RH, Crozier YC. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Möhlmann TWR, Bekendam AM, van Kemenade I, Wennergren U, Favia G, Takken W, et al. Latitudinal diversity of biting midge species within the Obsoletus group across three habitats in Europe. Med Vet Entomol. 2019;33:420–426. doi: 10.1111/mve.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goffredo M, Meiswinkel R, Federici V, Di Nicola F, Mancini G, Ippoliti C, et al. The ‛Culicoides obsoletus groupʼ in Italy: relative abundance, geographic range, and role as vector for bluetongue virus. Vet Ital. 2016;52:235–241. doi: 10.12834/VetIt.35.100.1. [DOI] [PubMed] [Google Scholar]
- 51.Ippoliti C, Goffredo M, Salini R, Bocci M, Quaglia M, Federici V, et al. Eco-climatic indicators for three Culicoides species of the Obsoletus complex in Italy. Vet Ital. 2016;52:213–222. doi: 10.12834/VetIt.1150.6293.1. [DOI] [PubMed] [Google Scholar]
- 52.Ander M, Meiswinkel R, Chirico J. Seasonal dynamics of biting midges (Diptera: Ceratopogonidae: Culicoides), the potential vectors of bluetongue virus, in Sweden. Vet Parasitol. 2012;184:59–67. doi: 10.1016/j.vetpar.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 53.Allen AP, Gillooly JF. Assessing latitudinal gradients in speciation rates and biodiversity at the global scale. Ecol Lett. 2006;9:947–954. doi: 10.1111/j.1461-0248.2006.00946.x. [DOI] [PubMed] [Google Scholar]
- 54.Wenk CE, Kaufmann C, Schaffner F, Mathis A. Molecular characterization of Swiss Ceratopogonidae (Diptera) and evaluation of real-time PCR assays for the identification of Culicoides biting midges. Vet Parasitol. 2012;184:258–266. doi: 10.1016/j.vetpar.2011.08.034. [DOI] [PubMed] [Google Scholar]
- 55.Augot D, Mathieu B, Hadj-Henni L, Barriel V, Zapata Mena S, Smolis S, et al. Molecular phylogeny of 42 species of Culicoides (Diptera, Ceratopogonidae) from three continents. Parasite. 2017;24:23. doi: 10.1051/parasite/2017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirkeby C, Dominiak P. Culicoides (Avaritia) gornostaevae Mirzaeva, 1984 (Diptera: Ceratopogonidae)—a possible vector species of the Obsoletus group new to the European fauna. Parasit Vectors. 2014;7:445. doi: 10.1186/1756-3305-7-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mathieu B, Delecolle JC, Garros C, Balenghien T, Setier-Rio ML, Candolfi E, et al. Simultaneous quantification of the relative abundance of species complex members: application to Culicoides obsoletus and Culicoides scoticus (Diptera: Ceratopogonidae), potential vectors of bluetongue virus. Vet Parasitol. 2011;182:297–306. doi: 10.1016/j.vetpar.2011.05.052. [DOI] [PubMed] [Google Scholar]
- 58.Pages N, Sarto IVM. Differentiation of Culicoides obsoletus and Culicoides scoticus (Diptera: Ceratopogonidae) based on mitochondrial cytochrome oxidase subunit I. J Med Entomol. 2005;42:1026–1034. doi: 10.1093/jmedent/42.6.1026. [DOI] [PubMed] [Google Scholar]
- 59.Kiehl Ernst, Walldorf Volker, Klimpel Sven, Al-Quraishy Saleh, Mehlhorn Heinz. The European vectors of Bluetongue virus: are there species complexes, single species or races in Culicoides obsoletus and C. pulicaris detectable by sequencing ITS-1, ITS-2 and 18S-rDNA? Parasitology Research. 2009;105(2):331–336. doi: 10.1007/s00436-009-1414-0. [DOI] [PubMed] [Google Scholar]
- 60.Esselstyn JA, Evans BJ, Sedlock JL, Anwarali Khan FA, Heaney LR. Single-locus species delimitation: a test of the mixed Yule-coalescent model, with an empirical application to Philippine round-leaf bats. Proc R Entomol Soc. 2012;279:3678–3686. doi: 10.1098/rspb.2012.0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talavera G, Dinca V, Vila R. Factors affecting species delimitations with the GMYC model: insights from a butterfly survey. Methods Ecol Evol. 2013;4:1101–1110. [Google Scholar]
- 62.Dellicour S, Flot JF. Delimiting species-poor data sets using single molecular markers: a study of barcode gaps, haplowebs and GMYC. Syst Biol. 2015;64:900–908. doi: 10.1093/sysbio/syu130. [DOI] [PubMed] [Google Scholar]
- 63.Rosenberg NA. The probability of topological concordance of gene trees and species trees. Theor Popul Biol. 2002;61:225–247. doi: 10.1006/tpbi.2001.1568. [DOI] [PubMed] [Google Scholar]
- 64.Nichols R. Gene trees and species trees are not the same. Trends Ecol Evol. 2001;16:358–364. doi: 10.1016/s0169-5347(01)02203-0. [DOI] [PubMed] [Google Scholar]
- 65.Pentinsaari M, Vos R, Mutanen M. Algorithmic single-locus species delimitation: effects of sampling effort, variation and nonmonophyly in four methods and 1870 species of beetles. Mol Ecol Resour. 2017;17:393–404. doi: 10.1111/1755-0998.12557. [DOI] [PubMed] [Google Scholar]
- 66.Fujita MK, Leache AD, Burbrink FT, McGuire JA, Moritz C. Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol Evol. 2012;27:480–488. doi: 10.1016/j.tree.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 67.Leache AD, Fujita MK. Bayesian species delimitation in West African forest geckos (Hemidactylus fasciatus) Proc R Entomol Soc. 2010;277:3071–3077. doi: 10.1098/rspb.2010.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boykin LM, Schutze MK, Krosch MN, Chomič A, Chapman TA, Englezou A, et al. Multi-gene phylogenetic analysis of south-east Asian pest members of the Bactrocera dorsalis species complex (Diptera: Tephritidae) does not support current taxonomy. J Appl Entomol. 2014;138:235–253. [Google Scholar]
- 69.Dinca V, Lukhtanov VA, Talavera G, Vila R. Unexpected layers of cryptic diversity in wood white Leptidea butterflies. Nat Commun. 2011;2:324. doi: 10.1038/ncomms1329. [DOI] [PubMed] [Google Scholar]
- 70.Hsieh CH, Ko CC, Chung CH, Wang HY. Multilocus approach to clarify species status and the divergence history of the Bemisia tabaci (Hemiptera: Aleyrodidae) species complex. Mol Phylogenet Evol. 2014;76:172–180. doi: 10.1016/j.ympev.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 71.Song J-H, Ahn K-J. Species delimitation in the Aleochara fucicola species complex (Coleoptera: Staphylinidae: Aleocharinae) and its phylogenetic relationships. Zool Scr. 2014;43:629–640. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Information on adult female Culicoides sampling sites and results of specific assignation using cox1.
Additional file 2: Table S2 Primers used for PCRs and sequencing in this study.
Additional file 3: Table S3. Reference sequences used for specific assignation.
Additional file 4: Table S4. Information on all Culicoides sequenced and GenBank accession numbers.
Additional file 5: Figure S1. Maximum likelihood phylogenetic tree using 16S rDNA. Values at the nodes represent bootstrap values greater than 800 for the most important nodes (1000 replicates).
Additional file 6: Figure S2. Maximum likelihood phylogenetic tree using cox1. Values at the nodes represent bootstrap values greater than 800 for the most important nodes (1000 replicates).
Additional file 7: Figure S3. Maximum likelihood phylogenetic tree using 28S rDNA. Values at the nodes represent bootstrap values greater than 800 for the most important nodes (1000 replicates).
Data Availability Statement
All data generated or analysed during this study are included in this published article and its additional files. The newly generated sequences were submitted in the GenBank database under the accession numbers MT170026-MT173788.