ABSTRACT
Hydrogen sulfide (H2S) is a versatile signaling molecule that regulates multiple physiological processes in plants, including growth and development, immunity, and stress response as well. Signaling triggered by H2S is proposed to occur via persulfidation, an oxidative post-translational modification (PTM) of cysteine residues (–SH) to persulfides (–SSH). Notwithstanding the growing body of information for the plant persulfidation proteome, the gap between the molecular mechanism of H2S and physiological functions of protein persulfidation remains large. In this mini-review, we discussed the specific regulatory mechanism of persulfidation on guard cell abscisic acid (ABA) signaling and the possible link of persulfidation, sulfenylation, and S-nitrosylation within the framework of redox-based regulation.
KEYWORDS: Hydrogen sulfide, persulfidation, ABA signaling, sulfenylation
Hydrogen sulfide (H2S) has been recognized as an important bio-active signaling molecule in both animal and plant, and is as important as nitric oxide (NO), carbon monoxide (CO), and hydrogen peroxide (H2O2).1,2 A growing body of reports have demonstrated that H2S is involved in various biological processes during the whole lifespan, including seed dormancy/germination, root growth, development, defense responses to biotic and abiotic stresses.3-7 The exogenously administration of H2S confers to the positive effects in regulating plants adaptation to environmental stress and growth.8-10Despite numerous studies highlighting the importance of H2S-based signaling transduction, its mechanism of action is not fully understood.9 Current knowledge suggested that the primary signaling mechanism of H2S work through a new oxidative posttranslational modification (PTM), called persulfidation, which convert cysteine thiols (–SH) into persulfides (–SSH).12,13 This PTM on target proteins lead to structural and functional changes. Recently, persulfidation proteome revealed that a considerable amount of proteins which involved a wide range of biological functions are sensitive to persulfidation.14,15 The proteomes greatly extend our knowledge of the function of persulfidation and provide potential candidate targets for further investigation. In this review, we summarize and discuss the features of persulfidation in regulating guard cell abscisic acid (ABA) signaling and the potential crosstalk of persulfidation, sulfenylation, and S-nitrosylation.
Persulfidation regulates guard cell ABA signaling
ABA as an important phytohormone involves in the regulation of diverse plant stress resistance and developmental processes. ABA is also the central regulator that triggers a complex signaling network to regulate stomatal movement.16 ABA could bind with PYR/RCAR (PYRABACTINRESISTANCE/REGULATORY COMPONENT OF ABA RECEPTOR) receptors. Binding of ABA to these receptors enhances their interaction with and inhibition of clade A protein phosphatases (PP2Cs).17 PP2Cs inhibition enables the activation of SnRK2/OST1 (Open Stomata 1) protein kinases. These kinases phosphorylate and activate targets, in concert with ROS and Ca2+ together with Ca2+-dependent protein kinases (CDPKs) activate ion channels that mediate stomatal closure.18
The participation of H2S in stomatal closure has also been widely reported.19,20 L-cysteine desulfhydrase (DES1) has been characterized as the major production of endogenous cytosolic H2S.2,21 ABA cannot induce stomatal closure of des1 mutants, while this effect is restored by the application of exogenous H2S.22 Recent study showed that the guard cell itself could be able to produce H2S, which is involved in the ABA-induced stomatal closure.23 Accordingly, a cluster of proteins including protein kinases and phosphatases, which involved in guard cell ABA signaling, was found in the persulfidation proteome (Table 1).24 Recent studies demonstrated that H2S regulates guard cell ABA signaling pathways through persulfidation of specific targets. For example, H2S produced by DES1 positively regulates ABA signaling by persulfidation of SNF1-RELATED PROTEIN KINASE2.6 (SnRK2.6)/open stomata (OST1) in guard cell, which is essential for the control of stomatal closure.25 The persulfidation on Cys131 and Cys137 of SnRK2.6 promotes its phosphorylating activity and the interaction with ABA response element-binding factor 2 (ABF2), and further activate the downstream genes expression. It should be mentioned that SnRK2.6/OST1 is also S-nitrosylated by nitric oxide (NO) at Cys137, lead to the inhibition of its activity, and further negatively regulating guard cell ABA signaling.26 H2S is required for the ABA-induced NO production, and acts as upstream of NO in ABA-dependent stomatal closure.22 It can be deduced that NO forms a feedback loop to fine-tune guard cell ABA/H2S signal by nitrosylating SnRK2.6/OST1. Coincidentally, our recent results revealed that persulfidation of DES1 and NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG PROTEIN D (RBOHD) fine-tunes guard cell ABA signaling.27 ABA triggers the persulfidation of DES1 itself at Cys44 and Cys205 and increase DES1 activity to produce sustainable H2S, which could regard as an amplification of H2S signaling. Enhanced H2S production by DES1 further persulfides RBOHD at Cys825 and Cys 890 to facilitate the rapid induction of ROS and stomatal closure. Interestingly, both persulfidation on DES1 and RBOHD were redox dependent, which could be further oxidized by over-accumulated ROS and inhibit its activity. Thus, these processes form a negative feedback loop through H2S and ROS-mediated modification that fine-tunes guard cell redox homeostasis and ABA signaling. Besides that, our study also implied that the crosstalk between H2S and ROS are essential for the rapid response to environmental changes. Considering the versatile biological function of DES/H2S and the universality of persulfidation modification in plant, it would be reasonable to believe that persulfidation may function in multiple ways in regulating not only stomatal movement, but also other plant developmental and stress-responsive processes.1,28
Table 1.
The list of persulfidated proteins which involved in guard cell ABA signaling.24
| Protein description | AGI accession |
|---|---|
| Abscisic acid receptor (PYL1) | AT5G46790 |
| Calcium-dependent protein kinase 3 (CPK3) | AT4G23650 |
| Calcium-dependent protein kinase 5(CPK5) | AT4G35310 |
| Calcium-dependent protein kinase 6(CPK6) | AT2G17290 |
| Calmodulin-domain protein kinase 9 (CPK9) | AT3G20410 |
| SNF1-related protein kinases 2.1 (SnRK2.1) | AT5G08590 |
| SNF1-related protein kinases 2.2 (SnRK2.2) | AT3G50500 |
| SNF1-related protein kinases 2.4 (SnRK2.4) | AT1G10940 |
| SNF1-related protein kinases 2.6 (SnRK2.6/OST1) | AT4G33950 |
| SNF1-related protein kinases 2.10 (SnRK2.10) | AT1G60940 |
| Protein phosphatase 1 (PP1) | AT1G50370 |
| Protein phosphatase 2A subunit A2 (PP2AA2) | AT3G25800 |
| Protein phosphatase 2A subunit A3 (PP2AA3) | AT1G13320 |
| Serine/threonine-protein phosphatase catalytic subunit (PP2A-3) | AT3G58500 |
| Serine/threonine-protein phosphatase catalytic subunit (PP2A5) | AT1G69960 |
| Serine/threonine protein phosphatase 2A subunit B (PP2AB2) | AT1G17720 |
| Probable protein phosphatase 2C 20 (PP2C 20) | AT2G20630 |
| Probable protein phosphatase 2C 21 (PP2C 21) | AT2G25070 |
| Probable protein phosphatase 2C 26 (PP2C 26) | AT2G30170 |
| Probable protein phosphatase 2C 28 (PP2C 28) | AT2G34740 |
| Probable protein phosphatase 2C 39 (PP2C 39) | AT3G15260 |
| Probable protein phosphatase 2C 58 (PP2C 58) | AT4G28400 |
| Probable protein phosphatase 2C 62 (PP2C 62) | AT4G33500 |
| Probable protein phosphatase 2C 76 (PP2C 76) | AT5G53140 |
| Probable protein phosphatase 2C 80 (PP2C 80) | AT5G66720 |
Redox-based cysteine post-translational modifications
The crosstalk with other signal molecules, including NO and H2O2, is important for biological function of H2S.1,22,29 The protein thiols are the important intermediates that integrate the interaction network, since they can be modified by all of the signal molecules–mediated PTMs.7 The cysteine thiols can be easily oxidized by reactive oxygen species (ROS) to form sulfenic acid (SOH), which usually results in enzymatic inactivation. This reversible modification is proposed to functions as an intermediate for further redox modifications, or overoxidation to sulfinic (SO2H) and sulfonic (SO3H) acids when ROS exposure proceeds.30,31 Overoxidation to sulfinic or sulfonic acid is irreversible and usually leads to permanent functional loss and protein degradation. It should be noted that H2S cannot directly react with cysteine thiols to form persulfides, precondition of oxidation step is required for persulfidation. Reactions of H2S with either disulfides or sulfenic acids yield persulfides.32 Persulfides also can be oxidized to perthiosulfenic acids (–SSOH) and further perthiosulfinic (–SSO2H), and perthiosulfonic (–SSO3H) acid, due to its high reactivity to ROS.12,13 However, this oxidized status could be reduced by thioredoxins or glutaredoxins, which would recycle it back to its thiol form. Hence, it is more plausible that protein persulfidation is an evolutionarily conserved modification that act as a protective mechanism to prevent irreversible thiol overoxidation under oxidative stress.12 It could be confirmed by the results from animal. By using a dimedone-based probe, the authors found the persulfides are mainly produced from the reaction of sulfenic acids with H2S. More importantly, temporal dynamics of persulfidation perfectly matches the sulfenylation, with the latter preceding the former.33
In plants, persulfidation proteome showed that at least 5% of the whole proteome in Arabidopsis is persulfidated, and H2S is involved in regulation of the redox status of cysteine residues of proteins involved in diverse biological processes.24 A comparison of the persulfidation and sulfenylation proteome were performed for the investigation of its potential crosstalk in plants based on two latest studies.24,31 Four hundred thirty-seven proteins were identified in both two modifications (Figure 1(a)), illustrating the overlap between these two PTMs. GO biological process analysis indicated that these proteins are mainly involved in stress responses, oxidation–reduction process, glycolytic process and tricarboxylic acid cycle (Figure 1(b)). Interestingly, several antioxidant enzymes and redox regulatory protein, including glutathione S-transferase (GSTU5 and GSTT1), cytosolic L-ascorbate peroxidase (cAPX1 and cAPXT), thioredoxin, and thioredoxin-like protein were found. As cysteine-rich proteins, antioxidant enzymes such as, APX, catalase (CAT), glutathione peroxidase (GPX), and peroxiredoxin are susceptible to oxidation and thereby function suppressed, especially for APX and peroxiredoxin, which often use thiol-based mechanisms to decompose H2O2.34,35 Nevertheless, the function of persulfidation and sulfenylation are usually opposite. For example, sulfenylation at Cys32 causes cAPX1 inactivation,36 while persulfidation at Cys32 significantly increase the activity of cAPX1.14 Glyceraldehyde-3-phosphate dehydrogenase (GAPC1) was also found persulfidated and sulfenylated at the same cysteine residue, while its activity was enhanced by persulfidation, and inhibited by sulfenylation.14,37 It further indicated that persulfidation function as an efficient mechanism to rescue antioxidant capacity by resolving sulfenylation and enhance activity of thiol-based antioxidant enzyme/proteins in response to oxidative damage.
Figure 1.
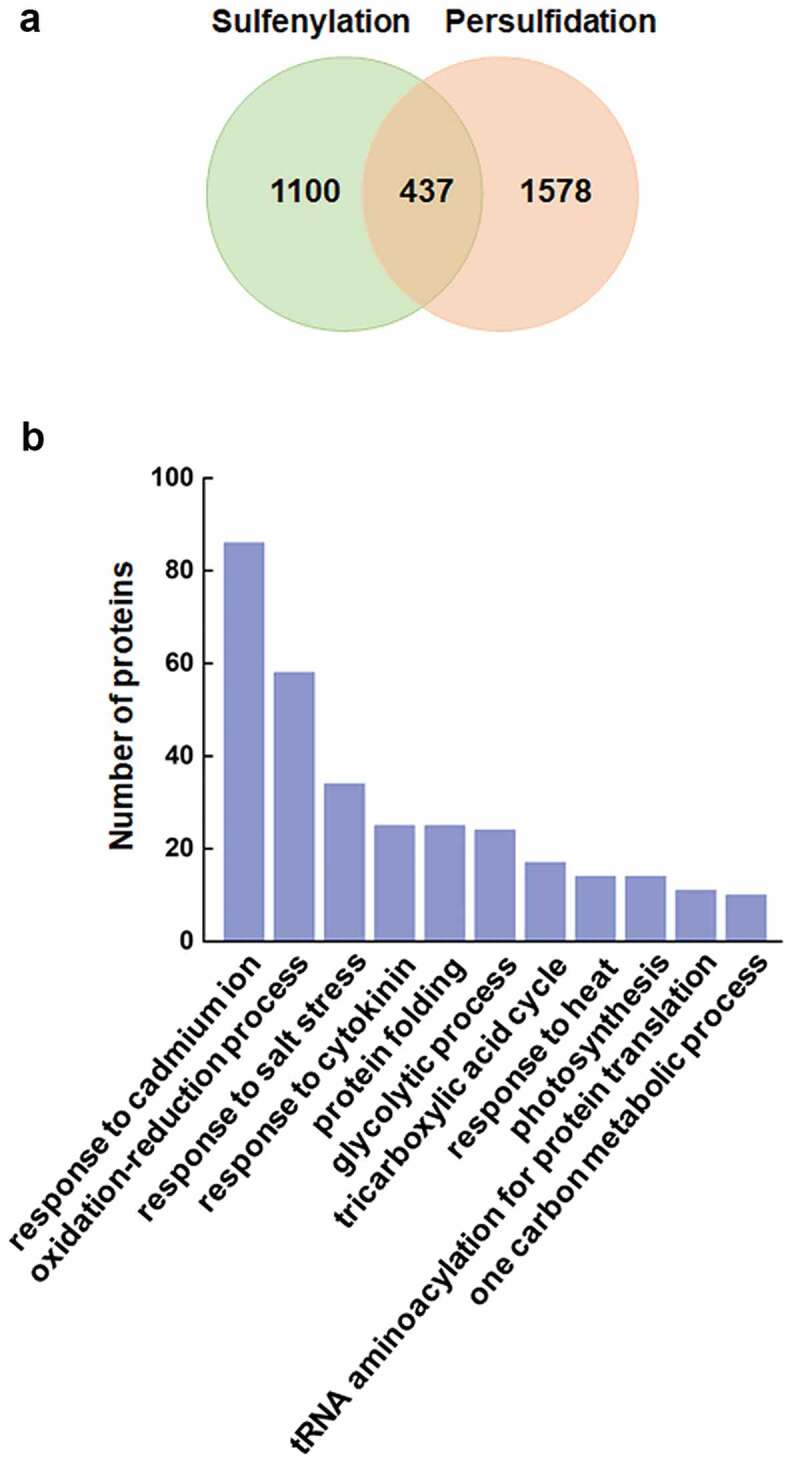
Comparison of sulfenylation and persulfidation proteome in Arabidopsis. Venn diagram shows the sulfenylated31 and persulfidated24 proteins identified from recent studies (a). GO biological process analysis shows the 11 most enriched terms from the 437 overlapped proteins (b).
Interestingly, the largest nitrosylation proteome identified 927 endogenously S-nitrosylated proteins in Arabidopsis.38 Six hundred thirty-nine of which are susceptible to being persulfidated.7 Although targets of these two PTMs share high coverage scale, the function vary from protein to protein. For example, different from the opposite effect on the activity of SnRK2.6/OST1, both persulfidation and S-nitrosylation at Cys32 significantly enhance the activity of cAPX1.39,40 S-nitrosylation abolishes the catalytic activity of GAPC, whereas the persulfidation increase its activity.14,41 It indicated that these proteins may finely regulate by different PTMs in response to different stress conditions.
Taken together, these proteomic data provide valuable tools for further investigation of function of these oxidative translational modifications and the crosstalk regulation among protein sulfenylation, persulfidation, and S-nitrosylation.
Concluding remarks
A considerable number of articles highlight the important roles of H2S in numerous biological process in plant. Benefit from the continuous improved persulfidation proteome, a number of proteins that undergo persulfidation have been characterized. However, the base of targets is too broad and the function validation will be laborious. Further effort should focus on the comparative proteomic analyses under different developmental process or environmental changes. Additionally, the extent of interaction of persulfidation and sulfenylation on redox regulation and how these two modifications shift on cysteine residues in response to environmental changes need further investigation. Most importantly, the crosstalk among these signal molecules are complicate in response to environment changes and the physiological evidence sometimes are controversial, the research on the regulatory mechanism and biological functions of the specific targets of these PTMs will shed more light on the regulatory role of H2 in plant cells.
Funding Statement
The work was supported by grants from the Chinese Natural Science Foundation [31670255], the Fundamental Research Funds for the Central Universities [KYZ201859], the Natural Science Foundation of Jiangsu Province [BK20161447].
Acknowledgments
We are grateful to Dr. Frantisek Baluska for kindly inviting this review.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Corpas FJ, González-Gordo S, Cañas A, Palma JM.. Nitric oxide and hydrogen sulfide in plants: which comes first? J Exp Bot. 2019;70(17):1–5. doi: 10.1093/jxb/erz031. [DOI] [PubMed] [Google Scholar]
- 2.Gotor C, García I, Á A, Laureano-Marín AM, Arenas-Alfonseca L, Jurado-Flores A, Moreno I, Romero LC.. Signaling by hydrogen sulfide and cyanide through post-translational modification. J Exp Bot. 2019;70:4251–4265. doi: 10.1093/jxb/erz225. [DOI] [PubMed] [Google Scholar]
- 3.Xie Y, Lai D, Mao Y, Zhang W, Shen W, Guan R. Molecular cloning, characterization, and expression analysis of a novel gene encoding L-cysteine desulfhydrase from Brassica napus. Mol Biotech. 2013;54(3):737–746. doi: 10.1007/s12033-012-9621-9. [DOI] [PubMed] [Google Scholar]
- 4.Xie Y, Zhang C, Lai D, Sun Y, Samma MK, Zhang J, Shen W. Hydrogen sulfide delays GA-triggered programmed cell death in wheat aleurone layers by the modulation of glutathione homeostasis and heme oxygenase-1 expression. J Plant Physiol. 2014;171:53–62. doi: 10.1016/j.jplph.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Guo H, Xiao T, Zhou H, Xie Y, Shen W. Hydrogen sulfide: a versatile regulator of environmental stress in plants. Acta Physiol Plant. 2016;38(1):16. doi: 10.1007/s11738-015-2038-x. [DOI] [Google Scholar]
- 6.Guo H, Zhou H, Zhang J, Guan W, Xu S, Shen W, Xu G, Xie Y, Foyer CH. L‐cysteine desulfhydrase‐related H2S production is involved in OsSE5‐promoted ammonium tolerance in roots of Oryza sativa. Plant Cell Environ. 2017;40:1777–1790. doi: 10.1111/pce.12982. [DOI] [PubMed] [Google Scholar]
- 7.Angeles A, Á A, Gotor C, Romero LC. Hydrogen sulfide signaling in plants: emerging roles of protein persulfidation. Front Plant Sci. 2018;9:1369. doi: 10.3389/fpls.2018.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Ye YK, Wang SH, Luo JP, Tang J, Ma DF. Hydrogen sulfide counteracts chlorophyll loss in sweetpotato seedling leaves and alleviates oxidative damage against osmotic stress. Plant Growth Regu. 2009;58:243–250. doi: 10.1007/s10725-009-9372-1. [DOI] [Google Scholar]
- 9.Jin Z, Xue S, Luo Y, Tian B, Fang H, Li H, Pei Y. Hydrogen sulfide interacting with abscisic acid in stomatal regulation responses to drought stress in Arabidopsis. Plant Physiol Biochem. 2013;62:41–46. doi: 10.1016/j.plaphy.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Jin Z, Wang Z, Ma Q, Sun L, Zhang L, Liu Z, Liu D, Hao X, Pei Y. Hydrogen sulfide mediates ion fluxes inducing stomatal closure in response to drought stress in Arabidopsis thaliana. Plant Soil. 2017;419(1–2):141–152. doi: 10.1007/s11104-017-3335-5. [DOI] [Google Scholar]
- 11.Yamasaki H, Cohen MF. Biological consilience of hydrogen sulfide and nitric oxide in plants: gases of primordial earth linking plant, microbial and animal physiologies. Nitric Oxide. 2016;55:91–100. doi: 10.1016/j.niox.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Filipovic MR, Jovanović VM. More than just an intermediate: hydrogen sulfide signalling in plants. J Exp Bot. 2017;68(17):4733–4736. doi: 10.1093/jxb/erx352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipovic MR, Zivanovic J, Alvarez B, Banerjee R. Chemical biology of H2S signaling through persulfidation. Chem Rev. 2018;118(3):1253–1337. doi: 10.1021/acs.chemrev.7b00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angeles A, Á A, Serna A, Gotor C, Romero LC. S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol. 2015;168(1):334–342. doi: 10.1104/pp.15.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angeles A, Á A, Schneider M, Scheibe R, Gotor C, Romero LC. Hydrogen sulfide regulates the cytosolic/nuclear partitioning of glyceraldehyde-3-phosphate dehydrogenase by enhancing its nuclear localization. Plant Cell Physiol. 2017;58(6):983–992. doi: 10.1093/pcp/pcx056. [DOI] [PubMed] [Google Scholar]
- 16.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2019;324:1064–1068. [DOI] [PubMed] [Google Scholar]
- 18.Hauser F, Li Z, Waadt R, Schroeder JI. SnapShot: abscisic acid signaling. Cell. 2017;171(7):1708. doi: 10.1016/j.cell.2017.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Mata C, Lamattina L. Hydrogen sulphide, a novel gasotransmitter involved in guard cell signalling. New Phytol. 2010;188(4):977–984. doi: 10.1111/j.1469-8137.2010.03465.x. [DOI] [PubMed] [Google Scholar]
- 20.Papanatsiou M, Scuffi D, Blatt MR, García-Mata C. Hydrogen sulfide regulates inward-rectifying K+ channels in conjunction with stomatal closure. Plant Physiol. 2015;168(1):29–35. doi: 10.1104/pp.114.256057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Álvarez C, Calo L, Romero LC, García I, Gotor C. An O-acetylserine (thiol) lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol. 2010;152(2):656–669. doi: 10.1104/pp.109.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scuffi D, Álvarez C, Laspina N, Gotor C, Lamattina L, García-Mata C. Hydrogen sulfide generated by L-cysteine desulfhydrase acts upstream of nitric oxide to modulate abscisic acid-dependent stomatal closure. Plant Physiol. 2014;166(4):2065–2076. doi: 10.1104/pp.114.245373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Zhou M, Ge Z, Shen J, Zhou C, Gotor C, Romero LC, Duan X, Liu X, Wu D, et al. Abscisic acid‐triggered guard cell L-cysteine desulfhydrase function and in situ hydrogen sulfide production contributes to heme oxygenase-modulated stomatal closure. Plant Cell Environ. 2019. doi: 10.1111/pce.13685. [DOI] [PubMed] [Google Scholar]
- 24.Á A, Benito JM, Gotor C, Romero LC. Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J Exp Bot. 2017;68(17):4915–4927. doi: 10.1093/jxb/erx294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Jia H, Wang X, Shi C, Wang X, Ma P, Wang J, Ren M, Li J. Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2. 6 in guard cells. Mol Plant. 2020. doi: 10.1016/j.molp.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Wang P, Du Y, Hou YJ, Zhao Y, Hsu CC, Yuan F, Zhu X, Tao WA, Song CP, Zhu JK. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Pro Natl Acad Sci U S A. USA. 2015;112:613–618. doi: 10.1073/pnas.1423481112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen J, Zhang J, Zhou M, Zhou H, Cui B, Gotor C, Romero LC, Fu L, Yang J, C H F, et al. Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell. 2020. doi: 10.1105/tpc.19.00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamasaki H, Ogura MP, Kingjoe KA, Cohen MF. d-cysteine-induced rapid root abscission in the Water Fern Azolla Pinnata: implications for the linkage between d-amino acid and reactive sulfur species (RSS) in plant environmental responses. Antioxidants. 2019;8(9):411. doi: 10.3390/antiox8090411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei Y, Chen H, Shen W, Shen W, Huang L. Hydrogen peroxide is involved in hydrogen sulfide-induced lateral root formation in tomato seedlings. BMC Plant Biol. 2017;17(1):62. doi: 10.1186/s12870-017-1110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L N S, Dominguez-Solis JR, Allary AL, Buchanan BB, Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Pro Natl Acad Sci U S A. 2016;103:9732–9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Willems P, Wei B, Tian C, Ferreira RB, Bodra N, Gache S, Wahni K, Liu K, Vertommen D, et al. Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Pro Natl Acad Sci U S A. 2019;116(42):21256–21261. doi: 10.1073/pnas.1906768116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuevasanta E, Lange M, Bonanata J, Coitiño EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J Biol Chem. 2015;290(45):26866–26880. doi: 10.1074/jbc.M115.672816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zivanovic J, Kouroussis E, Kohl JB, Adhikari B, Bursac B, Schott-Roux S, Petrovic D, Miljkovic JL, Thomas-Lopez D, Jung Y, et al. Selective persulfide detection reveals evolutionarily conserved antiaging effects of S-sulfhydration. Cell Metab. 2019;30(6):1152–1170. doi: 10.1016/j.cmet.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111(4):471–481. doi: 10.1016/S0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 35.Calabrese G, Peker E, Amponsah PS, Hoehne MN, Riemer T, Mai M, Bienert GP, Deponte M, Morgan B, Riemer J. Hyperoxidation of mitochondrial peroxiredoxin limits H2O2-induced cell death in yeast. Embo J. 2019;38(18):e101552. doi: 10.15252/embj.2019101552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitajima S, Kurioka M, Yoshimoto T, Shindo M, Kanaori K, Tajima K, Oda K. A cysteine residue near the propionate side chain of heme is the radical site in ascorbate peroxidase. Febs J. 2008;275(3):470–480. doi: 10.1111/j.1742-4658.2007.06214.x. [DOI] [PubMed] [Google Scholar]
- 37.Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P. Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci. 2013;4:450. doi: 10.3389/fpls.2013.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Wang Y, Zuo J. Site-specifc nitrosoproteomic identifcation of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2015;167:1731–1746. doi: 10.1104/pp.15.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Begara-Morales JC, Sánchez-Calvo B, Chaki M, Valderrama R, Mata-Pérez C, López-Jaramillo J, Padilla MN, Carreras A, Corpas FJ, Barroso JB. Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J Exp Bot. 2014;65:527–538. doi: 10.1093/jxb/ert396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Mu J, Chen L, Feng J, Hu J, Li L, Zhou JM, Zuo J. S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 2015;167:1604–1615. doi: 10.1104/pp.114.255216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindermayr C, Saalbach G, Durner J. Proteomic identification of S-Nitrosylated proteins in Arabidopsis. Plant Physiol. 2015;137(3):921–930. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]


