ABSTRACT
Leaf development is a complex process and factors such as size, shape, curvature, compounding, and texture determine the final leaf morphology. MicroRNA160 is one of the crucial players that has been shown to regulate lamina formation and compounding in tomato. In this study, we show that miR160 also regulates leaf curvature in potato. miR160 targets a group of Auxin Response Factors – StARF10, StARF16, and StARF17 – that are proposed to function majorly as repressors of auxin signaling. We observed that overexpression of miR160 (miR160-OE) results in decrease in the levels of these ARFs along with hypersensitivity to exogenous auxin treatment, whereas knockdown of miR160 (miR160-KD) causes increased ARF levels and auxin hyposensitivity. The leaves of miR160-OE plants have a high positive curvature, but of miR160-KD plants are flattened compared to wildtype. A prolonged activation of cell cycle – as indicated by increased levels of StCYCLIND3;2 – in the center region of miR160-OE leaves appears to have caused this positive curvature. However, a comparable StTCP4 activity at both center and margin regions of miR160-KD leaves could be the cause for its flattened leaf phenotype. In summary, we show that miR160 plays an important role in regulating leaf curvature in potato plants.
KEYWORDS: Auxin, leaf development, MicroRNA160, potato
Introduction
The leaves of most plants have evolved into a flat surface to efficiently capture sunlight for photosynthesis.1 A leaf initiates at the flank of the shoot apical meristem as an incipient primordium. The primordium transforms into a flat leaf by undergoing multiple rounds of tightly controlled cell divisions and maturation along three different axis of growth i.e proximo-distal, medio-lateral and adaxial-abaxial.2 Additional complexity to leaf shape is contributed by different types of margins and its invaginations leading to simple, lobed or compound leaves3.
Auxin is intimately linked to several aspects of leaf development.4 Mutations in the components of auxin signal transduction have been shown to affect leaf development.5 Canonical auxin signaling pathway involves repression of the ARF (Auxin Response Factor) transcription factors by the AUX/IAA repressors under low auxin conditions. When auxin levels increase, AUX/IAA repressors are degraded by the proteasomal pathway and ARFs are free to initiate the transcription of auxin responsive genes.6
ARFs have been broadly grouped into three classes based on phylogenetic analysis7 and invitro transcriptional activity8,9 as class A activator ARFs and class B and C repressor ARFs. This categorization is not straightforward and it appears that the transcriptional activity of ARFs is highly context dependent and cell specific.10–12 Nevertheless, the current model of auxin pathway only incorporates the functions of activator ARFs and its interaction with AUX/IAA repressors.13,14 The function of repressor ARFs in this pathway is still not clear. It is predicted that repressor ARFs may compete with activator for binding sites on the promoter of auxin responsive target genes.10,14
In tomato, the C-ARFs, SlARF10, 16 and 17 have been shown to play a crucial role in leaf compounding and lamina formation.15–17 C-ARFs are post-transcriptionally regulated by a conserved microRNA, miR160. Transgenic tomato plants that either have reduced levels of miR16015 or increased levels of miR160-resistant SlARF1016 develop narrow lamina, suggesting that miR160 promotes lamina formation and SlARF10 represses it. Additionally, overexpression of C-ARFs was shown to increase leaf complexity in tomato (compounding), while overexpression of miR160 resulted in the development of simplified leaves.17 Plants with mutation in the protein AUX/IAA9 or ENTIRE also produce simplified leaves. It is proposed that repression of auxin signaling by both C-ARFs and ENTIRE is required for proper leaf development in tomato.17
In our previous study,18 we observed that overexpression of miR160 results in formation of highly downward curved leaves in potato. Though, this observation is not reported in the close relative tomato, previous studies in Arabidopsis have hinted an altered leaf curvature in plants overexpressing miR160-resistant versions of class C ARFs.19–21 These studies, however, did not elaborate the mechanistic basis for the altered leaf curvature. In the present report, we have attempted to investigate the underlying reason for the altered leaf curvature using miR160 overexpression (OE) and knockdown (KD) plants in potato. Our morphometric analysis revealed that leaves of miR160-OE plants have high degree of downward curvature, whereas leaves of miR160-KD plants are flattened compared to wildtype leaves. Based on the expression patterns of the cell proliferation marker, StCYCD3;2, and the cell differentiation marker, StTCP4, we propose that the medio-lateral patterning during leaf development could be affected in miR160 transgenic plants. miR160-OE plants have reduced C-ARF levels and are hypersensitive to exogenous auxin treatment, whereas miR160-KD plants have high expression of C-ARF and are hyposensitive. Hence, miR160 appears to play a crucial role in medio-lateral patterning and development of flattened leaf surface in potato.
Results
Leaf curvature is altered in potato plants misexpressing miR160
During our previous study with miR160-OE and miR160-KD plants,18 we observed that overexpression of miR160 results in leaves with high positive curvature (downward curve) (Figure 1(a)). A detailed look at the leaves from two independent overexpression lines (Supplementary Figure1 and Figure 1(b,c)) strengthened our hypothesis that miR160 could be regulating leaf curvature. To quantify the curvature phenotype, we estimated the longitudinal and transverse curvature indices (CIL and CIT) (Liu et al., 2010) of the miR160-OE and -KD plants (Figure 1(d)). A curvature index of zero suggests that the leaf surface is completely flat. We observed that wildtype (WT) and vector control (VC) leaves were slightly downward curved with both CIL and CIT around 0.2 (Figure 1(e,f)). But, the curvature indices were drastically high for leaves from both the miR160-OE independent lines compared to WT and VC (Figure 1(e,f)). Interestingly, this quantification also revealed that the leaves from miR160-KD-1 plants had a lower CIT than WT, suggesting that its leaf surface had flattened compared to WT (figure 1(f)). At this point, we do not have an explanation of why the expression levels are not proportional to the curvature phenotypes observed in the two miR160-OE lines. We also noticed additional morphogenetic changes in the leaves of miR160 transgenic lines compared to WT and chose to characterize the lines miR160-OE-1 and miR160-KD-1 in a greater detail in the rest of this study because of their extreme phenotypes.
Figure 1.
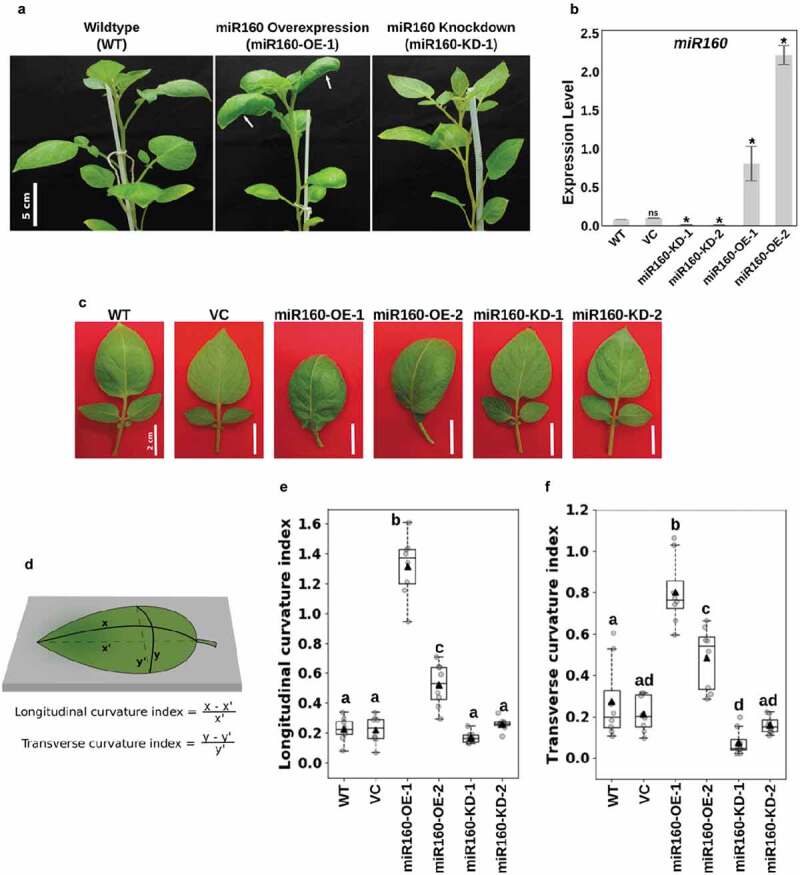
Misregulation of miR160 results in altered leaf curvature. (a) Photographs of 6-week old wildtype (WT), miR160-OE and miR160-KD plants. White arrows in the middle panel show curved leaves of miR160-OE plants. (b) Quantitative real-time PCR-based expression analysis of miR160 in the leaf tissues of WT, vector control (VC), and two independent lines each of miR160 overexpression and knockdown plants. Bars indicate mean ± SD from four biological replicates. Statistically significant differences compared to WT samples were estimated using Student’s unpaired t-test (p < .05, two-tailed), ns indicates not significant. (c) A closer look at the individual leaves of miR160-OE and miR160-KD independent lines. (d) An illustration of the method used to calculate longitudinal and transverse curvature indices, which was originally used by Liu et al, 2010. (e-f) The longitudinal (e) and transverse (f) curvature indices of leaves emerging from the 5th-node of 6-week old plants. Data are represented as boxplots with gray-filled circles representing data points from eight individual plants and black filled triangles representing mean of the data points. Statistically significant differences were estimated using one-way ANOVA followed by a Tukey HSD test (p < .05).
Multiple leaf characteristics are affected in miR160-OE and miR160-KD plants
In an array of leaves ordered from the top of the plant to the bottom (L1 to L12, 6-weeks old) (Figure 2(a)), we could clearly observe the differences in shape and structure of leaves from WT, miR160-OE-1 and miR160-KD-1 plants. To flatten a curved leaf, incisions are required to be made at the leaf margin (Figure 2(b)). Corroborating our measurements of the curvature indices (Figure 1(d,e)), we observed that leaves from miR160-OE-1 plants required several more incisions to flatten than the WT leaves (Figure 2(a), numbers on top of each leaf) revealing their highly curved nature; conversely, leaves from miR160-KD-1 plants required fewer incisions suggesting that their curvature has decreased compared to WT leaves (Figure 2(a)). We observed similar patterns in leaves obtained from older plants (10-weeks old) (Supplementary Figure 2) suggesting that the phenotype manifests early in leaf development.
Figure 2.
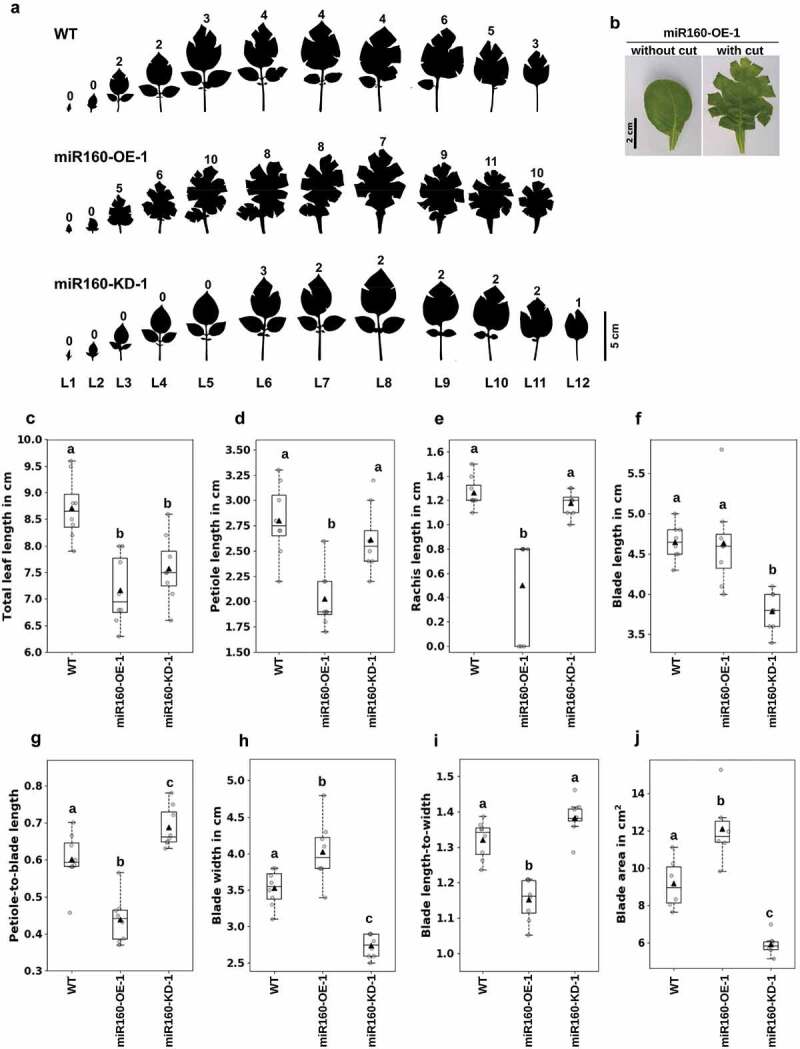
Both overexpression and knockdown of miR160 affect multiple leaf characteristics. (a) Silhouettes of all the leaves collected from 6-week old plant arranged in the order of youngest (L1) to oldest (L12). Cuts were made along the margin of the leaves to flatten it. The numeral on top of each leaf indicates the number of cuts that was required to flatten the leaf: miR160-OE leaves required more cuts and miR160-KD leaves required less cuts than WT. (b) A leaf from the miR160-OE-1 plants before and after the cuts was made at its margin. (c-j) Various morphometric parameters, as indicated along the y-axis, were measured using the leaf emerging from the 5th-node of 6-week old plants (L5). Each boxplot is prepared using data from eight individual plants (indicated by gray-filled circles), except blade area for which six individual plants were used. The black-filled triangle represents the mean of the data. Statistically significant differences were estimated using one-way ANOVA followed by a Tukey HSD test (p < .05). This experiment was performed three times with similar results.
To understand how the leaf architecture had changed in our transgenic plants, we estimated various leaf morphometric parameters of the terminal leaf arising from the fifth node (L5 in Figure 2(a)). We observed that the total leaf length (the sum of the lengths of blade, rachis and petiole) had reduced in both miR160-OE-1 and miR160-KD-1 plants compared to the WT (Figure 2(c)). However, different factors had contributed to the reduction in leaf length in each of these plants. In miR160-OE-1 leaves, petiole and rachis length were reduced as shown in Figure 2(d,e), but the blade length was comparable to WT leaves (Figure 2(f)). Whereas, in miR160-KD-1 leaves, the petiole and rachis length were similar to WT, but the blade length was smaller Figure 2(d–f).
We considered the presence of rachis as a surrogate for leaf compounding. All the leaves of miR160-KD-1 plants had rachis and showed compounding similar to WT leaves (Figure 2(a,e)). But, miR160-OE-1 leaves had a range of rachis length (some did not have rachis and some had a rachis length smaller than WT), suggesting that their compounding was not similar to WT leaves (Figure 2(a,e)). We also observed an ectopic growth of blade on the petiole and rachis of miR160-OE leaves indicating that miR160 promotes blade formation (Supplementary Figure 3). miR160-OE-1 plants had a lower ratio of petiole-to-blade length owing to their reduced petiole length, and miR160-KD-1 plants had higher ratio owing to their reduced blade length (Figure 2(g)).
We also observed that the blade width had increased in miR160-OE-1 leaves, and decreased in miR160-KD-1 leaves compared to WT (Figure 2(h)). The blade length-to-width ratio provides insights about the shape of the leaf: a ratio near 1.0 suggests a circular shape. For the WT leaves, as the length was greater than the width, a mean ratio of ~1.3 was observed (Figure 2(i)). For miR160-OE-1 leaves, though the blade length was not altered, the blade width had increased compared to WT (figure 2(f,h)). This increased the blade (lamina) area (Figure 2(j)), but reduced the blade length-to-width ratio to 1.15 (Figure 2(i)), suggesting that the shape of miR160-OE-1 leaves was approaching circularity. For miR160-KD-1 leaves, both blade length and width had decreased (Figure 2(f,h)); this reduced the blade area (Figure 2(j)), but the ratio remained comparable to WT leaves (Figure 2(i)). All these analyzes clearly demonstrated that the leaf architecture is modified in both miR160-OE-1 and miR160-KD-1 plants.
miR160 targets a group of auxin response factors for degradation
In our previous study, we had validated ARF10 and ARF16 as targets of miR160 in Solanum chacoense plants.18 In the present study, we were able to validate that StARF17 also undergoes miR160-guided cleavage along with StARF10 and StARF16 in Solanum tuberosum L. cv. Désirée (Figure 3(a)). As expected for a true target, the expressions of StARF10 and StARF17 were reduced in the leaves of miR160-OE-1 plants and elevated in miR160-KD-1 plants compared to WT (Figure 3(b,d)). But, expression of StARF16 did not follow a similar pattern (Figure 3(c)). A tissue-level expression analysis revealed that the highest expression for StARF10 and StARF17 was in petiole and leaf samples, respectively (Figure 3(e), red arrows), hinting toward their potential role in leaf development, unlike StARF16, which had its highest expression in the carpels (Figure 3(e)).
Figure 3.
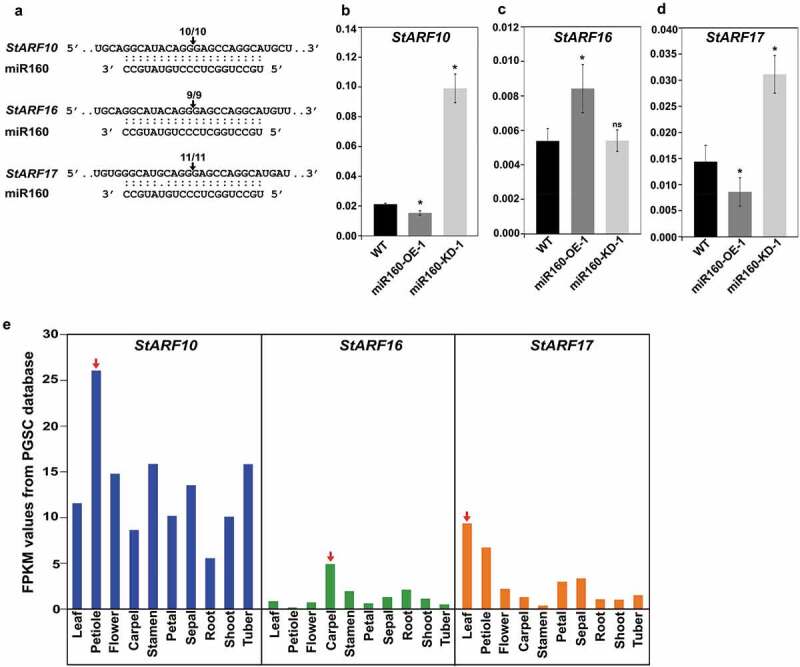
miR160 targets the class C auxin response factors (ARF) in potato. (a) Cleavage site mapping assay revealed that StARF10, StARF16 and StARF17 undergo miR160-guided cleavage. The arrow and the numbers indicate the site at which miR160 cleaves the ARFs and the frequency with which we were able to recover cleaved targets using RLM-RACE assay. (b-d) qRT-PCR-based expression analysis of StARF10 (b), StARF16 (c) and StARF17 (d) in the leaves of WT, miR160-OE-1 and miR160-KD-1 leaf samples. Bars indicate mean ± SD from four biological replicates. Statistically significant differences compared to WT samples were estimated using Student’s unpaired t-test (p < .05, two-tailed), ns indicates not significant. The expression analysis was performed twice with similar results. (e) The expression profile of StARF10, StARF16 and StARF17 transcripts across various tissue types as analyzed by the RNA-seq data set from the PGSC database. Red arrow indicates the tissue with the highest expression for each transcript.
miR160-OE-1 plants are hypersensitive to exogenous auxin treatment, whereas miR160-KD-1 plants are hyposensitive
Both miR160 and its target ARFs are known to function in auxin signaling.19,20 Hence, we determined the auxin responsiveness of our miR160-transgenic plants. For this purpose, we sub-cultured WT, miR160-OE-1, and miR160-KD-1 plants on the Murashige and Skoog media containing different concentrations (0.3 and 1 μM) of the synthetic auxin 2,4-Dichlorophenoxyacetic acid (2,4-D). After 12 days of growth, we quantified the length of newly emerged roots. It is well known that treatment of plants with increasing concentrations of exogenous auxin progressively decreases root length.22 Similarly, in our experiments, we observed that all the plant types showed reduction in root length with increased concentrations of auxin (Figure 4(a,b)). But, the root length reduced drastically in miR160-OE-1 plants even at a lower concentration of auxin (0.3 μM). This suggested that miR160-OE plants were hypersensitive to auxin. On the other hand, miR160-KD-1 plants responded to auxin similar to WT. However, when we compared the percentage reduction of root length between untreated and 1.0 μM auxin-treated plants, we observed a 84% reduction in WT, 96% in miR160-OE-1, and only 75% in miR160-KD-1 plants (Figure 4(c)). This analysis indicated that miR160-KD-1 plants were hyposensitive to auxin treatment.
Figure 4.
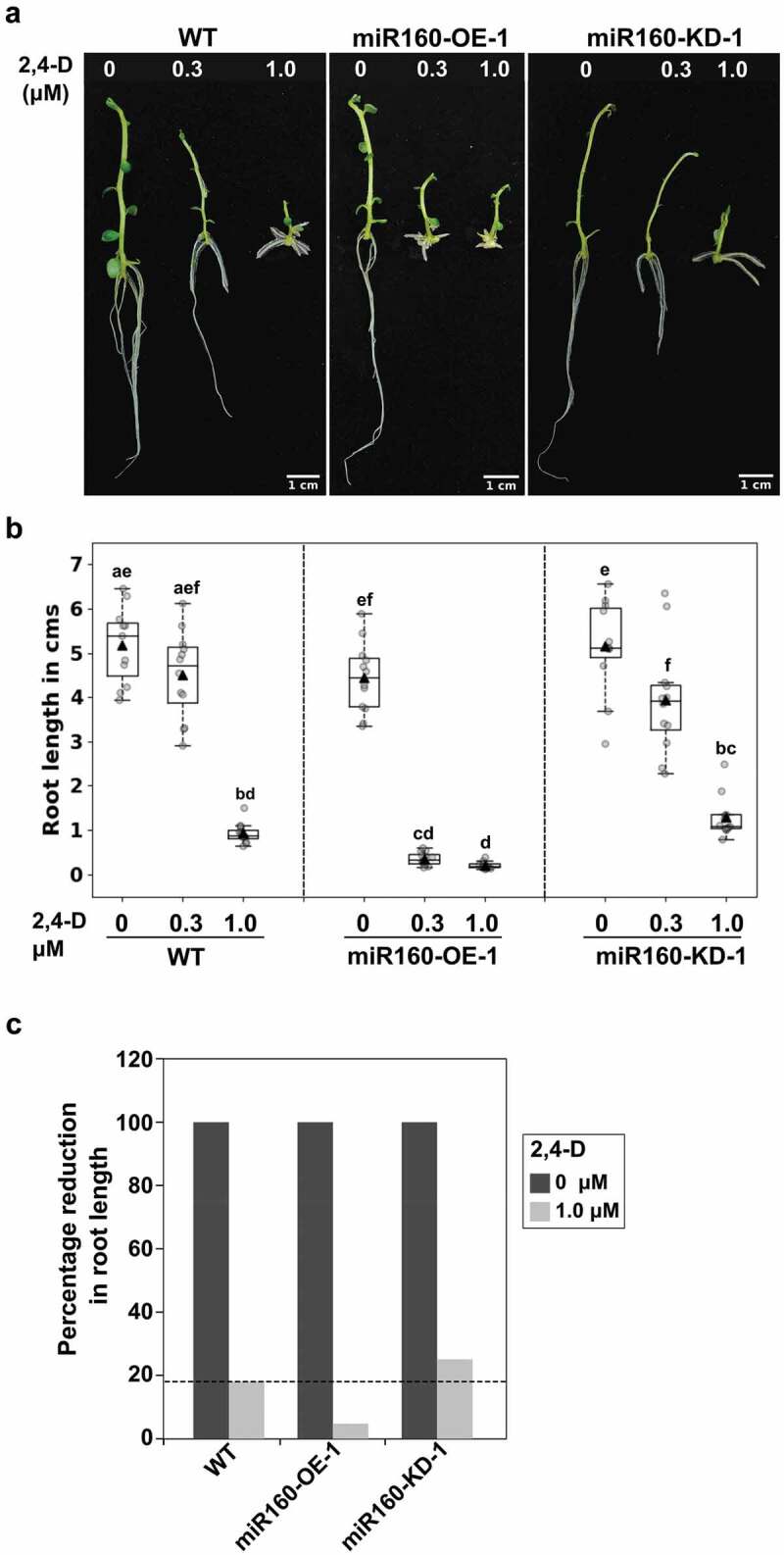
miR160-OE plants are auxin hypersensitive and miR160-KD plants are hyposensitive. (a) Roots of plants grown without or with (0.3 or 1.0 μM) auxin (2,4-D) for 12 days. (b) Quantification of root length after exogenous auxin treatment. Boxplots were prepared using data from 12 individual plants in each treatment. Grey-filled circles are data from individual plants and black-filled triangle is the mean of the data points. Drastic reduction in root length in miR160-OE-1 plants at 0.3 μM auxin treatment shows that miR160-OE plants are hypersensitive to auxin. Statistically significant differences were estimated using one-way ANOVA followed by a Tukey HSD test (p < .05). (c) Percentage reduction in root length in 1.0 μM treatment compared to untreated control shows that miR160-KD-1 plants are hyposensitive to auxin. This experiment was performed twice with similar results.
StCYCLIND3;2 and StTCP4 expressions are altered in the leaves of miR160 transgenic plants
To further understand the cause of the altered curvature, we analyzed the expressions of StCYCD3;2 (involved in cell proliferation)23 and StTCP4 (involved in cell differentiation)24 in the center and margin region (i.e. along the medio-lateral axis) of leaves of different lengths from WT, miR160-OE-1 and miR160-KD-1 plants (Figure 5(A,b)). Our quantitative-PCR analysis revealed that StCYCD3;2 levels decreased in all the plant-types as the leaves matured (Figure 5(a)). In WT leaves, both center and margin regions expressed comparable levels of StCYCD3;2 (Figure 5(a)). In miR160-OE-1 plants, ~0.6 and ~1.0 cm leaves had higher expression of StCYCD3;2 in the center compared to the margin region. miR160-KD-1 leaves had higher StCYCD3;2 expression in the center region of ~0.6 cm leaves, but in the leaves of other lengths its levels were comparable to the margin region (Figure 5(a)). These results suggested that the prolonged activity of StCYCD3;2 in the center of miR160-OE leaves might have resulted in the positively curved phenotype.
Figure 5.
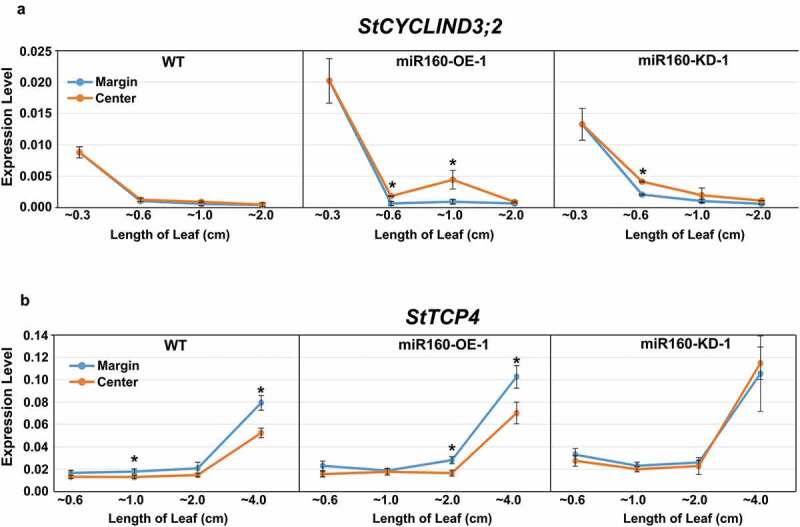
Expression of StCYCD3;2 and StTCP4 are altered in miR160 transgenic plants. (a-b) qRT-PCR-based expression analysis of StCYCD3;2 (a) and StTCP4 (b) in the center and margin regions of leaves of different lengths from WT, miR160-OE-1 and miR160-KD-1 plants. Values indicate mean ± SD from at least two biological replicates. Statistically significant differences were estimated using Student’s unpaired t-test (p < .05, two-tailed).
We observed that StTCP4 expression increased as the leaves matured (Figure 5(b)). In WT plants, the StTCP4 expression was higher in the margin regions of ~1.0 and ~4.0 cm leaves when compared to the center. Similarly, in miR160-OE-1 plants, ~2.0 and ~4.0 cm leaves had higher expression of StTCP4 in the margin than in the center (Figure 5(b)). In miR160-KD-1 leaves, we could not observe any difference in StTCP4 expression between the margin and center regions. These results suggest that equivalent expression of StTCP4 in both center and margin regions of miR160-KD-1 leaves might have resulted in their increased flatness.
Discussion
The mechanism of leaf development is an extremely controlled process. The leaf primordia initiates from the periphery of shoot apical meristem where auxin maxima is established, followed by initiation of growth along different axes (i.e. the proximo-distal, the adaxial-abaxial and the medio-lateral).2 The final leaf shape and structure depends on the rate of cell proliferation and differentiation across these different axes of growth. miR160 and its target class C ARFs have been shown to regulate lamina growth and leaf complexity in tomato.15–17 However, it was not known if miR160 could regulate any other aspects of leaf development. In the present study, we show that miR160 also governs leaf curvature in potato.
miR160 and its targets are crucial regulators of final leaf shape in potato
We observed that when miR160 levels are elevated (miR160-OE), leaves develop with high positive curvature, but when miR160 levels are reduced (miR160-KD), the leaves attain a more flattened surface compared to WT. Such observations are not reported in its close relative tomato. But, in Arabidopsis transgenic plants expressing the miR160-resistant versions of ARF17,19 ARF16,20 and ARF1021 displayed upward curling of the leaf margins, suggesting negative curvature that corroborated with our observations on miR160-KD plants. The overexpression of miR160 in Arabidopsis, however, did not affect the curvature.21 A (positively or negatively) curved leaf is formed when the growth rates are altered severely along medio-lateral axis.25 Cyclins and CINCINNATA-like TCPs (CIN-TCP) can serve as markers for leaf cells that are undergoing proliferation and differentiation along the medio-lateral axis, respectively.23,24 In Antirrhinum, CIN-TCP is expressed highly at the margin26 and a mutation in this gene results in negatively curved leaves with excessive growth of the margin.25 Similarly in Arabidopsis, reduced TCP activity causes excessive growth of margin, and increased TCP activity results in plants with smaller and flattened leaves compared to WT.27 Leaf differentiation is delayed when the activity of Lanceolate (a TCP family member) is lost in tomato.28 The equivalent StTCP4 levels observed at both center and margin regions in our miR160-KD mature leaves (Figure 5(b)) suggest that cells along the medio-lateral axis might be differentiating at the similar rate. This observation also correlates with the flattened blade surface and reduced blade area of the miR160-KD leaves (Figure 2(a)).
Positive curvature in leaves develops when cells at the center proliferate more than ones at the margin.29 In Arabidopsis, mutations in two genes encoding the DNA-binding protein PEAPOD result in the formation of positively curved leaves with increased blade growth; this occurs due to prolonged proliferative phase of dispersed meristematic cells.30 This is similar to our observation on miR160-OE leaves, which are also positively curved (Figure 1) and have increased blade area (Figure 2). A recent study showed that a JA signaling related protein NOVEL INTERACTOR OF JAZ (NINJA) – whose mutation also results in the formation of positively curved leaves, but without the excess growth of blade – physically interacts with PEAPOD to directly repress CYCD3;2 expression.31 Leaves of ninja mutants have a highly convex cell cycle arrest front with increased expression of CYCLINS at the central region of leaves. The high positive curvature and the increased blade area of our miR160-OE leaves also appear to have resulted from the prolonged StCYCD3;2 expression at the central region (Figure 5(a)). It is possible that cell cycle arrest front in miR160-OE leaves have also increased in convexity compared to WT leaves. All these observations indicate that miR160 controls leaf curvature in potato by modulating the activity of StTCP4 and StCYCLIND3;2.
In tomato, miR160-targeted C-ARFs are proposed to promote leaflet separation by inhibiting blade formation between leaflets;17 C-ARFs function in a parallel pathway alongside the AUX/IAA9 protein, ENTIRE, to achieve this. In potato also C-ARFs appear to restrict blade formation as the blade area was increased in miR160-OE leaves and reduced in miR160-KD leaves (Figure 2(j)). The leaflet separation was certainly affected in miR160-OE leaves as they developed rachis and petiole with reduced lengths (Figure 2). In fact, we observed ectopic blade formation on the rachis and petioles on miR160-OE leaves (Supplementary Fig. 3). The impact of C-ARFs on leaflet separation consequently affects leaf complexity: overexpression of miR160 or downregulation of its targets results in formation of simple leaves and the vice versa results in the formation of complex leaves in tomato.17 In Arabidopsis, downregulation of miR16032 or overexpression of miR160-resistant targets19,20 introduced serrations at the leaf margins, indicating that loss of miR160 activity can induce complexity even in a simple leaf species. Similarly in potato, miR160-OE plants developed simple leaves and miR160-KD plants developed leaves with smaller blade and longer petiole, which might be a subtle manifestation of increased leaf complexity (Figure 2). It is proposed that different expression levels of C-ARFs results in variable degree of leaf complexity in tomato.33 Hence, the leaf compounding phenotypes of our potato plants could be attributed to the extent of miR160 overexpression or knockdown.
miR160-targeted C-ARFs modulate auxin sensitivity
We have validated that StARF10, StARF16, and StARF17 undergo miR160-guided cleavage in potato (Figure 3). The levels of StARF10 were drastically altered upon miR160-OE and miR160-KD plants, followed by the levels of StARF17, suggesting that StARF10 is the major target of miR160 in potato. ARF10 was also suggested to be the major target in tomato.33 Even though we could detect miR160-targeted cleavage products of StARF16 through RLM-RACE technique, the expression of StARF16 was not complementary to that of miR160 in our transgenic plants. This atypical miR160-ARF16 expression pattern was also reported in tomato15 suggesting a complex regulation of ARF16 expression by factors other than miR160.
As mentioned earlier, the function of ARFs as a transcriptional activator or a repressor appears to be highly context dependent.10 Previous reports have suggested the role of class C-ARFs as both repressor17 and activator.34 In case of leaf development in potato and tomato, however, C-ARFs appear to function as repressors of auxin signaling. In tomato, the expression domain of an auxin responsive promoter DR5 (driving the VENUS reporter gene) had expanded in plants overexpressing miR160 and reduced in plants overexpressing miR160-resistant versions of SlARF10 and SlARF17.17 Additionally, the repressive effect of SlARF10 was enhanced when fused with a repressive domain (SRDX) and compromised when fused with an activation domain (VP16).17 Our auxin treatment assays in potato also suggest that miR160-targeted ARFs function as repressors of auxin signaling, specifically by modulating the sensitivity to auxin response. miR160-OE plants were hypersensitive to exogenous auxin treatment, and miR160-KD plants were hyposensitive (Figure 4). Soybean plants that overexpressed miR160 were also shown to be hypersensitive to auxin35 corroborating our results. These observations suggest that altered C-ARF levels affect the threshold at which auxin responses occur: in miR160-OE plants this threshold is brought down hence plants are hypersensitive, and in miR160-KD plants the threshold is raised up and the plants become hyposensitive.
Overall, we conclude that miR160 controls the final leaf shape and structure in potato by governing leaf curvature in addition to compounding and blade growth. There are still several gaps in our understanding of this developmental process. Some examples are: (1) How does miR160 interact with other leaf development pathways (such as TCP and PEAPOD)? (2) Do C-ARFs control the expression of Cyclins and TCPs? And (3) What is the relationship between the levels of miR160 in a cell and its auxin sensitivity? We propose that detailed studies of C-ARFs have the potential to reveal these answers.
Materials and methods
Plant materials and growth conditions
Solanum tuberosum L. cv. Désirée plants were used for all the experiments. The miR160 overexpression and knockdown lines were same as described in our previous study.18 For the sake of clarity in the current study, we had renamed our miR160 overexpressor lines as miR160-OE-1 and miR160-OE-2 (pre160-L17-D1 and pre160-L17-C1, respectively in our previous study), and miR160 knockdown lines as miR160-KD-1 and miR160-KD-2 (eTM160-L24-2 and eTM160-26, respectively in our previous study). All plants were grown in vitro in soild Muroshige and Skoog (MS) medium for two-three weeks, after which they were transferred to soil mixture (equal parts of soil, perlite and cocopit) and grown in greenhouse. The in vitro growth conditions were maintained as 16:8 hr light:dark cycle with a temperature of 24°C. In greenhouse, plants were maintained at 22°C under a 16:8 hr light:dark cycle.
Morphometric analysis
All the morphometric analyses were performed on 6–7 weeks old soil-grown plants. Mature terminal leaves at the fifth node were considered for all the measurements. The curvature indices, petiole-to-blade length and blade length-to-width ratio were analyzed following methods described previously.21 Presence of rachis was considered as a surrogate for the presence of leaf compounding. This experiment was performed three times with similar results.
Cleavage site mapping assay
To validate StARF10, StARF16 and StARF17 as true targets of miR160 in Solanum tuberosum L. cv. Désirée plants, modified 5ʹ-RNA ligase mediated rapid amplification of cDNA ends (RLM RACE) technique was performed as described previously.18 Primers used for this analysis are provided in Supplementary Table S1.
Reverse transcriptase-quantitative PCR (RT-qPCR)
One microgram of total RNA was used for oligo(dT) or miRNA-specific cDNA preparation with Superscript-IV Reverse Transcriptase (Invitrogen). qRT-PCR reactions were set up using TB Green Premix Ex Taq II (Tli Rnase H Plus) (Takara) and were carried out in Biorad CFX96 instrument. microRNA RT-qPCR was carried following a previously published protocol.36 Data were analyzed using 2-∆Ct method. GAPDH and U6 were used as normalization genes for analysis of mRNA and microRNA expression, respectively. Primer details are provided in Supplementary Table S1.
Effect of auxin on root development
Apical nodes from 2 to 3 weeks old in vitro grown plants were subcultured onto solid MS media with or without the synthetic auxin 2,4-Dichlorophenoxyacetic acid (2,4-D) and allowed to grow for 12 days. After this period, plants were removed from the media and imaged using a smartphone camera. The quantification of root length was performed using ImageJ image analysis software.37 This experiment was performed twice and both yielded similar results.
Analysis of leaf center and margin
To determine StCYCLIND3;2 and StTCP4 expression levels, center and margin regions were carefully dissected as described previously38 from the leaves of different lengths (~0.3, ~0.6, ~1.0, ~2.0 and ~4.0 cm). The leaves of length ~0.3 cm were taken as a whole since dissection into center and margin was not possible. RNA was isolated from these samples and RT-qPCR was performed as described above.
Histology of the petiole
Petiole from mature leaves at the fourth and fifth nodes of a 8-week old soil-grown plants were used for histological analysis. Ten micrometer thin paraffin-embedded sections were prepared as described earlier by Bhogale et al., 2014.39 Images were taken using CarlZeiss PALMRobo microscope. To obtain the complete image of a petiole, multiple images were manually stitched together using Inkscape (a free vector graphics software)
Accession numbers
U6: X60506, StGAPDH: PGSC0003DMT400044944, StARF10: PGSC0003DMT400020874, StARF16: PGSC0003DMT400062489, StARF17: XM_006364896/PGSC0003DMT400071449, StTCP4: PGSC0003DMT400024329, StCYCD3;2: PGSC0003DMT400066557
Funding Statement
This work was supported by Indian Institute of Science Education and Research (IISER) Pune and the research associate fellowship to BN was from Council of Scientific and Industrial Research (CSIR) Government of India .
Acknowledgments
We thank Prof. Xiu-Jie Wang for ath-eTM160 constructs using which miR160-KD plants were generated, Dr. Rita Ulloa for sharing with us Solanum tuberosum cv. Désirée plants, Mr. Harpreet Singh Kalsi for help with generation of transgenic potato plants, Mr. Nitish Lahigude for help in plant maintenance, and all the members of molecular plant biology lab at IISER Pune for useful insights and discussions on the project.
Disclosures
The authors have no conflicts of interest to declare.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Niinemets Ü, Sack L.. Structural determinants of leaf light-harvesting capacity and photosynthetic potentials. Progress in Botany. 2006;1–10. [Google Scholar]
- 2.Moon J, Hake S.. How a leaf gets its shape. Curr Opin Plant Biol. [Internet]. 2011;14:24–30. doi: 10.1016/j.pbi.2010.08.012. . [DOI] [PubMed] [Google Scholar]
- 3.Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P. A conserved molecular framework for compound leaf development. Science (80-). 2008;322(5909):1835–1839. doi: 10.1126/science.1166168. [DOI] [PubMed] [Google Scholar]
- 4.Xiong Y, Jiao Y. The diverse roles of auxin in regulating leaf development. Plants. 2019;8(7):243. doi: 10.3390/plants8070243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shwartz I, Levy M, Ori N, Bar M. Hormones in tomato leaf development. Dev Biol. [Internet]. 2016;419:132–142. doi: 10.1016/j.ydbio.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Leyser O. Auxin signaling. Plant Physiol [Internet] 2018; 176:465–479. Available from: http://www.plantphysiol.org/lookup/doi/10.1104/pp.17.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finet C, Berne-Dedieu A, Scutt CP, Marlétaz F. Evolution of the ARF gene family in land plants: old domains, new tricks. Mol Biol Evol. 2013;30(1):45–56. doi: 10.1093/molbev/mss220. [DOI] [PubMed] [Google Scholar]
- 8.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin response factors. Proc Natl Acad Sci. 1999;96(10):5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. [Internet] 2003; 15(2):533–543. Available from: http://www.nature.com/articles/nplants2016190%5Cnhttp://www.plantphysiol.org/cgi/doi/10.1104/pp.106.079509%5Cnpapers3://publication/doi/10.1038/nature08122%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4145118&tool=pmcentrez&rendertype=abst [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Bianco M, Kepinski S. Context, specificity, and self-organization in auxin response. Cold Spring Harb Perspect Biol. [Internet] 2011; 3(1):a001578. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21047914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brackmann K, Qi J, Gebert M, Jouannet V, Schlamp T, Grünwald K, Wallner ES, Novikova DD, Levitsky VG, Agustí J, et al. Spatial specificity of auxin responses coordinates wood formation. Nat Commun. [Internet] 2018; 9(1). Available from: 10.1038/s41467-018-03256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang JY, He SB, Li L, Yang HQ. Auxin inhibits stomatal development through MONOPTEROS repression of a mobile peptide gene STOMAGEN in mesophyll. Proc Natl Acad Sci U S A. 2014;111(29):E3015–E3023. doi: 10.1073/pnas.1400542111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piya S, Shrestha SK, Binder B, Neal Stewart C, Hewezi T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci. 2014;5:1–9. doi: 10.3389/fpls.2014.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernoux T, Brunoud G, Farcot E, Morin V, Van Den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damodharan S, Zhao D, Arazi T. A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 2016;86:458–471. doi: 10.1111/tpj.13127. [DOI] [PubMed] [Google Scholar]
- 16.Hendelman A, Buxdorf K, Stav R, Kravchik M, Arazi T. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Mol Biol. 2012;10(6):561–576. doi: 10.1007/s11103-012-9883-4. [DOI] [PubMed] [Google Scholar]
- 17.Ben-Gera H, Dafna A, Alvarez JP, Bar M, Mauerer M, Ori N. Auxin-mediated lamina growth in tomato leaves is restricted by two parallel mechanisms. Plant J. 2016;86:443–457. doi: 10.1111/tpj.13188. [DOI] [PubMed] [Google Scholar]
- 18.Natarajan B, Kalsi HS, Godbole P, Malankar N, Thiagarayaselvam A, Siddappa S, Thulasiram HV, Chakrabarti SK, Banerjee AK. MiRNA160 is associated with local defense and systemic acquired resistance against Phytophthora infestans infection in potato. J Exp Bot. 2018;69(8):2023–2036. doi: 10.1093/jxb/ery025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17(5):1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Wang L, Mao Y, Cai W, Xue H, Chen X. Control of root cap formation by MicroRNA-targeted auxin response factors in arabidopsis. Plant Cell. 2005;17(8):2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, Jia L, Mao Y, He Y. Classification and quantification of leaf curvature. J Exp Bot. 2010;61(10):2757–2767. doi: 10.1093/jxb/erq111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonner J, Koepfli JB. The inhibition of root growth by auxins. Am J Bot. [Internet] 1939; 26:557–566. Available from: http://www.jstor.org/stable/2436583 [Google Scholar]
- 23.Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci U S A. 2007;104(36):14537–14542. doi: 10.1073/pnas.0704166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Challa KR, Rath M, Nath U. The CIN-TCP transcription factors promote commitment to differentiation in Arabidopsis leaf pavement cells via both auxin-dependent and independent pathways. PLoS Genet. 2019;15(2):1–30. doi: 10.1371/journal.pgen.1007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nath U, Crawford BCW, Carpenter R, Coen E. Genetic control of surface curvature. Science (80-). 2003;299(5611):1404–1407. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- 26.Das Gupta M, Aggarwal P, Nath U. CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 2014;204(4):901–912. doi: 10.1111/nph.12963. [DOI] [PubMed] [Google Scholar]
- 27.Koyama T, Sato F, Ohme-Takagi M. Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 2017;175(2):874–885. doi: 10.1104/pp.17.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet. 2007;39(6):787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- 29.Sarvepalli K, Nath U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 2011;67:595–607. doi: 10.1111/j.1365-313X.2011.04616.x. [DOI] [PubMed] [Google Scholar]
- 30.White DWR. PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci USA. 2006;103(35):13238–13243. doi: 10.1073/pnas.0604349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baekelandt A, Pauwels L, Wang Z, Li N, De Milde L, Natran A, Vermeersch M, Li Y, Goossens A, Inzé D, et al. Arabidopsis leaf flatness is regulated by PPD2 and NINJA through repression of CYCLIN D3 genes. Plant Physiol. 2018;178(1):217–232. doi: 10.1104/pp.18.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. [Internet] 2010. [cited 2015 April16]; 6:e1001031. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2908682&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damodharan S, Corem S, Gupta SK, Arazi T. Tuning of SlARF10A dosage by sly-miR160a is critical for auxin-mediated compound leaf and flower development. Plant J. 2018;96(4):855–868. doi: 10.1111/tpj.14073. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Tian L, Sun MX, Huang XY, Zhu J, Guan YF, Jia QS, Yang ZN. AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiol. 2013;162(2):720–731. doi: 10.1104/pp.113.214940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner M, Nizampatnam NR, Baron M, Coppin S, Damodaran S, Adhikari S, Arunachalam SP, Yu O, Subramanian S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013;162(4):2042–2055. doi: 10.1104/pp.113.220699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3(1):1–12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. [Internet]. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bresso EG, Chorostecki U, Rodriguez RE, Palatnik JF, Schommer C. Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 2018;176(2):1694–1708. doi: 10.1104/pp.17.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhogale S, Mahajan AS, Natarajan B, Rajabhoj M, Thulasiram HV, Banerjee AK. MicroRNA156: a potential graft-transmissible MicroRNA that modulates plant architecture and tuberization in solanum tuberosum ssp andigena. Plant Physiol. 2014;164:1011–1027. doi: 10.1104/pp.113.230714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


