Abstract
Efficient strategies are required to implement comprehensive suspect screening methods using high-resolution mass spectrometry within environmental monitoring campaigns. In this study, both liquid and gas chromatography time-of-flight mass spectrometry (LC-QTOF-MS and GC-QTOF-MS) were used to screen for >5,000 target and suspect compounds in the Sacramento-San Joaquin River Delta in Northern California. LC-QTOF-MS data were acquired in All-Ions fragmentation mode in both positive and negative electrospray ionization (ESI). LC suspects were identified using two accurate mass LC-QTOF-MS/MS libraries containing pesticides, pharmaceuticals and other environmental contaminants and a custom exact mass database with predicted transformation products (TPs). The additional fragment information from the All-Ions acquisition improved the confirmation of the compound identity; with a low false positive rate (9%). Overall, 25 targets, 73 suspects and 5 TPs were detected. GC-QTOF-MS extracts were run in negative chemical ionization (NCI) for 21 targets (mainly pyrethroids) at sub-ng/L levels. For suspect screening, extracts were re-run in electron ionization (EI) mode with a retention time locked method using a GC-QTOF-MS pesticide library (containing exact mass fragments and retention times). Sixteen targets and 42 suspects were detected, of which 12 and 17, respectively, were not identified by LC-ESI-QTOF-MS. The results highlight the importance of analyzing water samples using multiple separation techniques and in multiple ionization modes to obtain a comprehensive chemical contaminant profile. The investigated river delta experiences significant pesticide inputs, leading to environmentally critical concentrations during rain events.
Introduction
The investigation of micropollutants in waste water, surface water and drinking water is an important component of water quality assessments1, 2. Classical monitoring approaches consist of screening for a defined number of target compounds. However, it has been shown that with a targeted approach investigating a few compounds, the exposure and risk of pollutants towards aquatic organisms can be significantly underestimated compared to more comprehensive screenings3, 4. With the use of high-resolution mass spectrometry (HRMS) it is possible to go beyond target analysis5–8. The field of suspect and non-target screening, primarily using liquid chromatography (LC)-electrospray ionization (ESI)-HRMS, is currently expanding, especially for emerging contaminants in water. Efficient and practical approaches with quick confirmation of compound identities are, however, still needed.
Suspect screening employs compound databases containing chemical formulas, accurate monoisotopic masses and isotope patterns, and, in some instances, MS/MS spectra5. This enables users to presumptively identify compounds without the need for procuring analytical reference standards. It has proven to be an efficient and successful approach for detecting expected and unexpected compounds in the water9–13. Schymanski et al. (2014)14 proposed a system for communicating confidence in unknown assignments depending on the amount of information available. It ranges from level 1 (confirmed structure by reference standard), level 2 (probable structure by library spectrum match or diagnostic evidence), level 3 (tentative candidates by plausible sub-structure or chemical class), level 4 (unequivocal molecular formula by isotope pattern match) to level 5 (exact mass only). This system is widely accepted by the environmental non-target community6 and is used here to describe the findings.
If the molecular formula is the only a priori information about the compound in a suspect screening11, it can initially only be identified with a confidence level 4, because all isomers have the same exact mass and isotope pattern. As MS/MS libraries become increasingly available from open sources (e.g., NORMAN MassBank15) and vendors (e.g., Agilent Technologies Personal Compound Database and Library, PCDL), additional fragment information should be considered when doing suspect screening16.
MS/MS information can be acquired by either data-dependent acquisition (DDA, isolating precursor masses of compounds in the suspect list or using preset intensity triggers) or data-independent fragmentation (DIA, fragmenting all ions or ions between certain mass ranges independent of a suspect list or MS data). DIA with a constant, wide mass window is also known as broadband DIA17 or All-Ions fragmentation. DDA provides very specific MS/MS spectra which is very helpful in identifying unknown chemicals from a non-target screening, but scan speed will not be high enough to trigger all MS/MS scans in large suspect lists. DIA can become very complex due to co-eluting chemicals in an environmental matrix, and it is difficult to reconstruct an individual MS/MS spectrum. However, DIA gives additional confidence in confirmation of a suspect compound with known MS/MS fragments, when the chromatographic co-elution of library fragments with the molecular ion in the MS full scan is monitored. A compound with matching isotope pattern and at least one co-eluting fragment can be considered as level 2 identification14.
For compounds missing from MS/MS libraries, such as predicted transformation products, suspect screening is limited by necessity to the molecular formula. Although a larger effort is necessary in the subsequent identification, findings of novel relevant TPs are important.
While several studies have identified numerous non-target compounds in water using LC-ESI-HRMS9–13, this approach does not provide a comprehensive picture of chemical pollution. Specific compound classes of environmental relevance such as pyrethroids cannot be analyzed by this method. Therefore, GC-MS is a necessary complementary method for more non-polar compounds. As the fragmentation pattern in electron ionization (EI) mode is highly reproducible between instruments, reliable unit mass library spectra have been assembled for over 200,000 compounds (NIST 14)18. Because GC-HRMS instruments are relatively new, only a limited number of exact mass libraries are currently available19 (e.g., Agilent GC/Q-TOF – Pesticide PCDL). If available, the more specific accurate mass fragments should reduce the number of false positives in a library search20. With such a library, a suspect screening analogous to the one in LC-HRMS can be carried out. An additional advantage of GC is that retention times (RTs) are easier to compare. Thus, RT indexing (relative RTs between different methods) or even RT locking (adapting a method from an existing method to have matching RTs) allows confirmation of compound identity with high certainty.
This study presents a holistic approach for screening over 5,000 micropollutants in surface water including both LC-QTOF-MS and GC-QTOF-MS platforms using a combined target and suspect screening workflow to produce comprehensive chemical contaminant profiles. Two new approaches - i) LC-QTOF-MS suspect screening using All-Ions acquisition and curated accurate mass MS/MS libraries and ii) GC-QTOF-MS suspect screening using a RT locked method and an accurate mass fragment library - are validated at environmental concentrations. To our knowledge, this is the first study to combine these methods to assess surface water quality. The screening was applied in a large storm-driven field study conducted in a sensitive habitat of the Sacramento-San Joaquin River Delta in Northern California.
Materials and Methods
Study Site and Sampling
Sampling was carried out at six locations throughout the Cache-Slough-Complex, located in the Sacramento-San Joaquin River Delta in Northern California during two rain events in winter 2016 predicted to have over 3 cm of precipitation (January 4 – 8, and March 4 – 9, respectively, see SI-1). The main input of point-source micropollutants as well as diffuse pollutants is expected to be via Ulatis Creek because of the discharge of a large waste water treatment plant (WWTP, 100,000 population equivalents) from the Vacaville urban area, and significant agricultural activity in the upstream catchment. During rain events, runoff from urban and agricultural areas is expected to increase the concentrations of pollutants with diffuse sources, while pollutants emitted by point sources, like municipal wastewater facilities with sanitary sewers, are expected to remain steady or decline. A transect of five locations (Ulatis Creek at Brown Road (UB) and Cache Slough locations C1-C4) was sampled to track pollutant dynamics. One reference site, Liberty Island (LI), which is separated from the transect and expected to have low micropollutant loading, was also sampled. Two 1 L grab samples − one for LC-MS and one for GC-MS − were taken in the middle of the river/wetland at roughly 30 cm depth during four and five days in the January and March events, respectively (1 sample before, 2–3 samples during and 1 sample after each rain event, SI-1). Three samples were not taken for logistical reasons resulting in a total of 51 samples. All samples were cooled during transport and stored in the dark at 4 °C until extraction.
Chemicals and Solvents
For the target analysis, 32 LC-MS amenable pesticides and 21 GC-MS amenable pesticides were selected (see SI-2). Five compounds were measurable on both instruments. Targets were chosen: (i) to include high use compounds in Solano County, CA at the time the methods were established (California DPR, 201221) and (ii) to represent pesticides from different classes and with different physico-chemical properties (see SI-2). For the LC-MS measurements, 11 internal standards were used; for the GC-MS measurements, two surrogates and one internal standard were used (see SI-2). All solvents were high purity (methanol, ethyl acetate, hexane, acetone, dichloromethane from Fisher Scientific, acetonitrile from Burdick and Jackson); ultra-pure water was supplied by an in-house deionized water system (MilliQ Millipore).
Extraction and Analytical Method for LC-QTOF-MS
Surface water samples were extracted for polar and semi-polar micropollutant analysis using a method developed by Kern et al. (2009). In brief, surface water (1 L) was filtered through a GF/F filter (0.45μm), the pH was adjusted to 6.5–7, and 200 ng of internal standard mix was added. Samples were passed over a multilayered cartridge containing Oasis HLB (Waters, Massachusetts, USA), Strata XAW, Strata XCW (both Phenomenex, Munich, Germany) and Isolute ENV+ (Biotage, Uppsala, Sweden), to enrich neutral, cationic and anionic species with a broad range of Kow values (see Fig. 1). Cartridges were dried for one hour; elution was performed with 6 mL ethyl acetate/methanol 50:50 with 0.5% ammonia, followed by 3 mL ethyl acetate/methanol 50:50 with 1.7% formic acid, and finally by 2 mL methanol. Extracts were evaporated to 0.2 mL with nitrogen using a Turbovap (Biotage) and reconstituted to 1 mL using ultra-pure water. A calibration curve consisting of ten points between 0.1 – 250 ng/mL was prepared in ultra-pure water/methanol (80:20) and spiked with the same amount of internal standards as the samples.
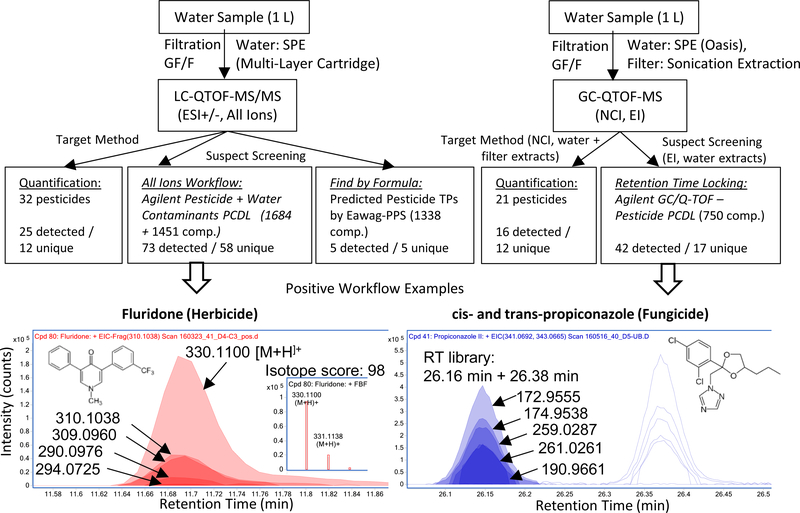
Figure 1.
Top: Flowchart of the extraction and data evaluation method. “Unique” compounds were only detected on either LC-QTOF-MS or GC-QTOF-MS, not on both instruments. TP: transformation product. Bottom: Example of two identified compounds in real environmental samples by the two suspect screening methods. Left: LC-QTOF-MS All-Ions workflow. Shown is an overlay plot of the exact mass of the [M+H]+ and the four main fragments of fluridone from the spectral library. Inset: comparison of theoretical and measured isotope pattern. Right: GC-QTOF-MS retention time locking workflow. Shown is an overlay plot of the five main fragments of cis-/trans-propiconazole in EI mode together with the library retention time (RT) information.
LC-QTOF-MS (Agilent 1260 Infinity HPLC coupled to an Agilent 6530 QTOF-MS with a Zorbax Eclipse Plus C18 column; 100 mm, 2.5 mm, 1.8 μm, Agilent Technologies, Inc.) analysis was performed by injecting 40 μL of extract with the following mobile phases used in a 23 min run at a flow rate of 0.35 mL/min: positive ionization mode: A) deionized water plus 0.1% formic acid, B) acetonitrile plus 0.1% formic acid; negative ionization mode: A) ultra-pure water plus 1mM ammonium fluoride, B) acetonitrile (see SI-3.1 for details). The instrument was run in the 2 GHz, extended dynamic range mode at 4 spectra/second. Acquisition was done in All-Ions fragmentation mode using collision energies (CE) of 0, 10, 20, and 40 eV, i.e., all ions with m/z 50−1,050 were fragmented in the collision cell with the corresponding CE. CE=0 means no fragmentation and is equal to a full MS scan. MS settings (gas flows, gas temperatures, etc.) were optimized separately in positive and negative ionization modes (see SI-3.1) using the 32 target pesticides.
Extraction and Analytical Method for GC-QTOF-MS
For non-polar compounds, the surface water samples were extracted based on a method developed by Hladik et al. (2009)22 who analyzed over 60 pesticides and TPs from multiple compound classes. Surface water (1 L) was filtered through a GF/F filter, filtrate was spiked with two surrogates and passed over an Oasis HLB cartridge (Waters). The cartridges were dried for one hour and eluted with 10 mL of ethyl acetate. A bottle rinse (3 × 4 mL dichloromethane) was used to recover pyrethroids sorbed to the glass wall in post-filtration samples22. The resulting extracts were combined and reduced to 0.2 mL. The filters containing suspended sediment were spiked with surrogates, sonication extracted with hexane/acetone (1:1; 2 × 20 mL), and the extracts were reduced to 0.2 mL without further cleanup. Water and filter extracts were measured individually. Dibromooctafluorobisphenol (DBOFB, 10 ng) was spiked as an internal standard to all samples. A calibration curve consisting of ten points between 0.1 – 250 ng/mL was prepared in ethyl acetate, spiking the same amount of surrogates and internal standard as the samples.
GC-QTOF-MS analysis (Agilent 7890B GC coupled to an Agilent QTOF/MS 7200B with a HP-5MS 30 m × 0.25 mm, 0.25 μm column, Agilent Technologies, Inc.) was conducted once in negative chemical ionization (NCI) mode using methane as collision gas and a second time in electron ionization (EI) mode (Fig. 1). NCI mode was used to quantify all 21 targets since NCI is much more sensitive for pyrethroids and other halogenated compounds than EI23. The filter extracts were only run in NCI mode to quantify the very non-polar target pyrethroids, which are expected to have the highest particle bound fractions. The optimized analytical parameters for NCI and additional analytical details are found in SI-3.2.
EI mode was used for screening the Agilent GC/Q-TOF – Pesticide PCDL20 containing 750 pesticides with exact mass EI fragments and retention times. The chromatographic parameters for the GC-EI-MS method were adapted from the Agilent method (SI-3.2). Using these settings, the measured RTs matched with the library RT within 0.5 min. To get the measured RT even closer to the library RT, retention time locking20 was implemented via five injections of the same standard, one at the original helium flow rate and four with −20%, −10%, +10%, and +20% of the selected helium flow rate. The retention time of chlorpyrifos (library RT 19.993 min in the 40 min run) was used to optimize helium flow by a regression curve of the multiple injections. Retention time locking provided RTs for targets within 0.2 min of their library RTs.
Target Quantification
Target compounds (SI-2) were quantified using Agilent MassHunter Quantitative Analysis software (B.07). For LC-QTOF-MS, the [M+H]+ or [M-H]- were used as quantifier (exact mass window ±10 ppm) and two main MS/MS fragments (taken from an existing library spectra) measured in the All-Ions scans were used as qualifiers. For GC-QTOF-MS, the main NCI fragment was used as quantifier and two additional fragments were used as qualifiers. For method validation and quality control, pre-spiked (before extraction), post-spiked (before injection) and procedural blank (extracted in ultra-pure water) samples, in triplicate, were used (see SI-4).
Suspect Screening using All-Ions Workflow on LC-QTOF-MS
Suspect screening employed the Agilent MassHunter Qualitative Analysis (B.07) software by applying the Find by Formula workflow in ESI+ and ESI- mode (SI-5.1 provides details). The Agilent Pesticide PCDL containing 1684 pesticides and transformation products (914 with MS/MS spectra) and the Agilent Water Contaminants PCDL containing 1451 compounds (1157 with spectra) were used as suspect lists (Fig. 1). [M+H]+ and [M+Na]+ in the positive mode as well as [M-H]- and [M+F]- in the negative mode were searched at m/z ±10 ppm and an isotope score (including exact mass deviation of monoisotopic m/z, abundance deviation and exact mass difference of isotopes versus theoretical pattern) of >70 was selected as threshold. The threshold value was selected as on optimum between false negatives and false positives (see results). For compounds without MS/MS fragments in the library, the workflow stopped here. For compounds with MS/MS fragments, the software automatically searched the five main fragments from the library in the All-Ions scans (CE 10, 20, 40). If one or more library fragments were present and co-eluting with the precursor mass, the compound was automatically flagged as qualified. All automatically detected compounds that had more than two detections in the 51 samples with intensities at least five times higher than in the blank were manually inspected for peak shape, signal-to-noise ratio and plausibility of the qualified fragments. If possible, a reference standard was purchased for the tentatively identified compounds for full confirmation and retrospective quantification.
Suspect Screening with RT Locked Method on GC-QTOF-MS
Suspect screening for GC-EI-QTOF-MS employed Agilent MassHunter Qualitative Analysis software using a Find by Formula workflow similar to the LC-QTOF-MS workflow. The Agilent GC/Q-TOF – Pesticide PCDL containing 750 pesticides with exact mass fragments and retention times was used (Fig. 1). In contrast to the LC-QTOF-MS workflow, the molecular ion was set as optional, a retention time tolerance of ± 0.2 min was included and the minimum number of qualified fragments was two (see SI-5.2). After manual inspection of the automatically detected compounds, reference standards were purchased for complete identification and for retrospective quantification.
Extended Pesticide Transformation Product Screening
To expand the search for transformation products (TPs) beyond those present in the databases mentioned above, an extensive TP screening for pesticides was conducted (Fig. 1). The batch-mode of the Eawag Pathway Prediction System (EAWAG-PPS24) was used to generate a list of 1409 TPs (SMILES codes) from 76 pesticides detected in this study using three recursion steps. The structures were evaluated for their theoretical ionization in ESI11 and 71 were eliminated. The molecular formulas of the remaining 1338 structures were added into a custom database and all 51 LC-QTOF-MS water samples were screened using the Find by Formula workflow in MassHunter Qualitative Analysis in ESI+ and ESI- (see SI-5.1 for parameters). As no MS/MS spectra were available for these compounds, only the exact mass and the isotope score (threshold 70) were used as criteria. Manual inspection was performed as described above for all compounds with more than five detections in the 51 samples and intensities more than five times above the blank. Additionally, at least one detection needed an isotope score >85 to eliminate compounds with consistently low intensities. Retention time plausibility was evaluated by comparing measured RTs for suspects to their predicted RTs using a correlation of logDow (pH 4 in ESI+, and pH 7 in ESI-, ChemAxon Jchem for Excel) and RT for target compounds. RT differences over 4 min were considered as not plausible.
For the remaining plausible candidates, the samples with the highest abundances were re-run in targeted MS/MS mode (CE 20), isolating the [M+H]+ or [M-H]- mass to obtain MS/MS spectra, which were imported into Agilent Molecular Structure Correlator (MSC, B.07.). MSC searches a selected database (e.g., Chemspider, Pubchem, or a custom PCDL containing molecular structures) for all compounds with the same exact mass as the isolated mass. In-silico fragments of all possible compounds are then compared with the measured MS/MS spectra. As output, it lists all measured fragments that can be explained by each structure and calculates a score based on a weighted match. For the purpose of this study, a custom PCDL containing the molecular structures of all remaining plausible TPs was made and MSC calculated the likelihood that the in-silico fragments of the compounds explain the measured MS/MS spectra. The identification was also supported by predicting MS/MS spectra of the plausible TPs using CFM-ID (http://cfmid.wishartlab.com/predict)25 by importing the SMILES codes into the software. If the candidate had plausible fragments, the compounds were considered as confirmed with a confidence level 3.14 If a library spectrum or reference standard was available, the level of confidence could be reduced to 2 or 1, respectively.
Priority Compounds
In 51 samples, compounds were prioritized by number of detections, maximum measured concentration (Max MEC) and maximum risk quotient (Max RQ, see SI-6). Max RQ was calculated by dividing Max MEC by the lowest available acute toxicity value for each compound. If available, the sensitive toxicity concentration (STC) as defined by Nowell et al. (2014)26 for three organism groups (fish, cladocerans and benthic invertebrates) was used as a toxicity value. The STC represents the 5th percentile of a wide range of data and is therefore highly robust towards outliers. For all other compounds, the lowest acute EC50 value (48 h – 96 h) from standard test species exposures (fish, invertebrates, nonvascular plants) as reported in the EPA ECOTOX database (https://cfpub.epa.gov/ecotox) was used.
Results and Discussion
Validation of Target Analysis (LC-QTOF-MS and GC-QTOF-MS)
From the 32 LC-QTOF-MS targets, all achieved absolute recoveries >70%, 26 had accuracies between 70–130%, 30 had precisions (standard deviation of triplicates) <10%, and 31 achieved low method detection limits (MDL) <10 ng/L (see SI-4.1). In spite of having an isotope-labelled internal standard for only one third of the compounds, accuracies were generally good and therefore, quantification is reliable. Detection limits are comparable to Moschet et al. (2013)11 who used the same extraction method but a different instrument for analysis. This shows that the extraction, separation and detection method is suitable to successfully detect pesticides with a broad range of physico-chemical properties (e.g., logKow: −3.3 to 6.2) from all pesticide types (herbicides, fungicides, insecticides).
From the 21 GC-NCI-QTOF-MS targets, 17 achieved absolute recoveries >70% in the water extracts, 15 absolute recoveries >70% in the filter extracts, 19 had accuracies between 70–130%, all 21 had precisions <10%, and 18 achieved very low MDLs <1 ng/L (see SI-4.2). The extremely low MDLs of non-polar pesticides in both dissolved and particle bound fractions are clearly below the EC50 values for H. azteca lab cultures27 and are comparable to the lowest reported MDLs in literature22, 23, 28.
Suspect Screening using All-Ions workflow on LC-QTOF-MS
The LC-MS target pesticides were used to validate the performance of the suspect screening using the All-Ions fragmentation workflow. Targets with more than one detection (19) in the 51 environmental samples were listed in the PCDLs; 15 of these were automatically found by the suspect screening; while four were not (cyprodinil, imidacloprid, propanil, thiamethoxame). These four compounds had maximum intensities of 2,000 in the samples. At this low intensity, isotope scores can fall below the cutoff value (<70) because their isotopes are either not present or had increased mass error or relative abundance deviation.
The fragment confirmation in the All-Ions workflow did not increase the false negative rate, i.e. compounds were not missed because of a missing fragment if a peak with matching isotope score was present. This is because the intensity of the main fragment in the high energy scans (CE 10, 20, 40 eV) was usually similar to or only slightly lower than the intensity of the monoisotopic ion mass in the MS full scan. In addition, the parameter settings to qualify a peak were chosen to be deliberately loose (1 fragment needed) because some compounds only have one usable fragment even when multiple CE scans are available. These compounds would be missed if the settings were more stringent.
Overall, this procedure was efficient because the number of software generated hits was manageable and false negative suspect identifications were primarily associated with low intensity detections. It is clear that an automated suspect screening yields higher detection limits than a manually evaluated target approach.11 Namely, the screening of the 51 water samples by the two Agilent PCDLs containing >2000 compounds automatically detected and qualified 83 compounds in positive mode and 39 in negative mode (with criteria: detections in at least two samples and intensities at least five times higher than in the blank). The manual inspection procedure described above reduced this number to 70 plausible candidates. These were considered as identification with confidence level 214. For example, the herbicide fluridone was detected in 39 samples with high isotope scores >90 and three to four qualified fragments that were co-eluting with the [M+H]+ mass (Fig. 1). From these 70 compounds, 64 reference standards could be purchased and 58 were confirmed by matching retention time as well as matching MS/MS spectra (see SI-6). This resulted in a false positive rate of 9% based on the software filters for mass accuracy, isotope pattern and fragment confirmation selected for this study. This is a low number considering that with an all ion fragmentation approach a large number of co-eluting peaks can occur in complex matrices. The six compounds for which no reference standard was available were reported as tentatively identified with confidence level 2.
Compounds in the two PCDLs for which no MS/MS spectra were available (770 in the Agilent Pesticide PCDL and 294 in the Agilent Water Contaminants PCDL) were screened by the Find by Formula workflow, too. Here, only the isotope score cutoff was considered and the peaks were manually inspected for peak shape and signal-to-noise ratio. Fifteen candidates remained after manual inspection and a reference standard was purchased for ten compounds. For the other five compounds the samples were re-run in a targeted MS/MS approach and the fragments were evaluated (analog to TP screening, see method section). Nine compounds could be confirmed by a reference standard, one rejected by a reference standard and five rejected due to implausible fragments. As expected, a higher false positive rate was obtained when only the molecular formula information was available compared to the All-Ions workflow using MS/MS fragments.
Suspect Screening Using Retention Time Locked Method on GC-QTOF-MS
Screening the 51 water extracts measured by GC-EI-QTOF-MS using the Agilent GC/Q-TOF – Pesticide PCDL (750 pesticides) with a retention time locked acquisition method resulted in the detection of 84 software generated hits (criteria: more than two detections and intensities higher than five times the blank). Again, the criterion for the number of confirmed fragments (2) was deliberately chosen to be conservative. The manual inspection eliminated 39 compounds with bad peak shape or because one important fragment from the library spectrum was missing in the measurement. From the remaining 45 compounds, 4 were targets of the GC-NCI-QTOF-MS method, 24 were already found on LC-QTOF-MS by either target or suspect screening approaches described above, and 17 compounds were uniquely detected by GC-EI-QTOF-MS (see Fig. 1 and SI-6). Because at least two co-eluting accurate mass fragments and the retention time had to match the library, the confidence of the identification is very high with this approach. For 39 of the 45 compounds, reference standards could be obtained and as expected, all were positively confirmed. The remaining six compounds were reported as tentatively identified with confidence level 2. One positive example is the fungicide propiconazole (cis- and trans- isomers), which was detected in 38 out of 51 samples with at least four matching fragments and retention time deviations of 0.01 min from the library retention time (Fig. 1). Both cis- and trans- isomers were confirmed with RT using the library.
Extended Transformation Product Screening
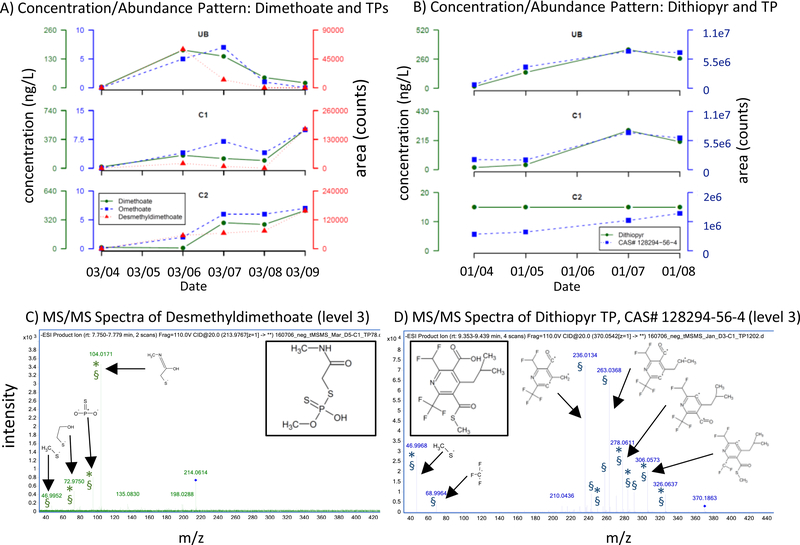
The screening of the 51 samples with 1338 predicted theoretically ionizable pesticide TPs resulted in 33 and 77 software generated hits in positive and negative ionization modes, respectively (detections in more than five samples with intensities higher than five times that in the blank). Manual inspection for peak shape and signal-to-noise ratio, as well as further evaluations such as RT plausibility and consideration of whether the detected compound is theoretically ionizable in the selected mode eliminated most compounds leaving only 13 and 20 plausible compounds in positive and negative modes, respectively. In a further step toward confirmation of the TPs, the abundance pattern of the 33 compounds in the 51 samples was plotted and compared with the concentration pattern of their potential parent compounds. Six compounds in positive mode and ten in negative mode (two of them detected in both modes) thereby showed a pattern that is expected from a compound introduced by a runoff event and was very similar to the pattern of the parent compound (see Fig. 2 and SI-7). The other seven and ten compounds had an undefined abundance pattern and were therefore eliminated from the candidate list. The similarity between the abundance patterns of the 14 tentatively identified TPs and their parent compounds suggests that these TPs were most likely formed at the source (i.e., prior to or coincident with discharge).
Figure 2.
Top: Concentration/area pattern in three locations (Ulatis Creek, UB, Cache Slough C1 and C2) of A) the insecticide dimethoate (green solid line), its TPs omethoate (blue dashed line) and desmethyldimethoate (red dashed line, confirmed level 3) in the March rain event, and B) the herbicide dithiopyr (green solid line) and its predicted TP with CAS #: 128294–56-4 (blue dashed line) in the January rain event. Bottom: annotated plausible MS/MS spectra of the identified transformation products. C) desmethyldimethoate (MSC score 71.4) and D) dithiopyr TP with CAS #: 128294–56-4 (MSC score 92.6). § predicted by MSC; * predicted by CFM-ID.
Re-running the samples in targeted MS/MS mode, evaluating the MS/MS spectra using the MSC software, comparing measured fragments to those predicted by CFM-ID, and manual inspection eliminated two compounds in positive mode and five in negative mode because they had implausible MS/MS spectra (i.e., fragments that could not be explained by the molecular structure).Seven compounds had plausible MS/MS fragments and were initially identified with confidence level 314. Two examples are shown in Fig. 2 (remaining compounds in SI-7). The insecticide dimethoate had two TPs with matching abundance patterns (top left): i) omethoate which was already found in the All-Ions workflow and was later confirmed by a reference standard, and ii) O-desmethyl dimethoate (CAS # 2700–77-8) for which no reference standard was available but which had plausible MS/MS fragments (Agilent MSC score 71.4); three of them were also predicted by CFM-ID (bottom left). Omethoate is the key metabolite of dimethoate and is formed in soil29. The perfectly matching concentration pattern between parent and TP indicates that the transformation happened at the source. O-desmethyl dimethoate is a known plant or water metabolite29 which to the authors’ knowledge has not been found in surface waters previously. The second example, the herbicide dithiopyr, which was frequently found in the All-Ions workflow, had one unknown TP with CAS # 128294–56-4 with matching abundance pattern (Fig. 2, top right), and multiple plausible MS/MS fragments (Agilent MSC score 92.6); six of them were also predicted by CFM-ID (bottom right). In addition, norflurazon-desmethyl, azoxystrobin acid, trifloxystrobin acid, and 2,4-dichlorophenol (TP of 2,4-D) were detected and all were fully confirmed by a reference standard (see SI-7 for MS/MS spectra). In addition to the TPs found by the extended screening, five TPs that were not predicted by EAWAG-PPS were detected by either target analysis or suspect screening. Four fipronil TPs were detected by target analysis on LC-QTOF-MS and GC-QTOF-MS, and the diuron metabolite 3,4-dichlorophenylisocyanate was tentatively confirmed by the GC-QTOF-MS suspect screening.
Significance of Suspect Screening
By applying both target and suspect screening approaches using LC-QTOF-MS and GC-QTOF-MS, 132 unique compounds were detected at least once in the 51 water samples during the two rain events in the Cache Slough Complex (Fig. 1, SI-6). Analysis for the 48 target pesticides (27 LC-QTOF-MS, 16 GC-QTOF-MS, 5 both instruments), identified only 37 compounds; thus 95 compounds that were identified by suspect screening would have been missed.
75 of the 132 detected compounds were uniquely detected by LC-QTOF-MS, 29 uniquely by GC-QTOF-MS and 28 on both instruments. From the uniquely detected compounds by GC-QTOF-MS, five were also on the LC-QTOF-MS suspect list, while 17 of the uniquely detected compounds by LC-QTOF-MS were also on the GC-QTOF-MS suspect list. The reason why these compounds were not detected by the other instruments is most likely that they were above detection limits due to low environmental concentrations. This highlights the importance of measuring samples on both separation platforms (LC & GC) and implementing comprehensive suspect screening approaches in routine monitoring programs to assess chemical contamination in a holistic manner.
The use of an All-Ions approach allowed for collection of MS and MS/MS level data in one injection, while the availability of spectral libraries was critical for positive compound identification. The development of more curated exact mass spectral libraries, especially for GC-EI-MS, is strongly suggested. Despite software advances that perform automated peak picking, compound identification and structure elucidation, manual review of data still allows refinement especially for low abundance features to reduce false positives and negative reporting. The extraction, analysis, data processing and reporting workflow shown here is highly effective for quantification of targeted compounds and identification of suspects and TPs in water samples.
Environmental Relevance
As might be anticipated for a surface water sampling program triggered by impending storms, the majority of detected compounds mainly entered via non-point sources (65 pesticides, 14 TPs), likely released by runoff during the rain events. However, a significant additional number of compounds were identified, including some that were expected to be present in WWTP effluent (22 pharmaceuticals, 5 flame retardants, 5 PFCs, 13 various) and 8 other compounds with unknown sources30–32 (see SI-6). Most compounds (109/132) could be quantified by a reference standard; 81 of these had an EC50 value available allowing calculation of an RQ. The top 10 compounds based on RQ, maximum concentration and number of detections in this study are listed in Table 1 (complete list in SI-6).
Table 1.
Prioritized compounds from this study, including the 10 compounds with the highest risk quotient (RQ), maximum measured environmental concentration (Max MEC, ng/L), and number of detections (# Det.). The table is sorted by RQ, maximum concentration and detection frequency, respectively.
| Compound Name | Compound Class | CASRN | Work-flow | Insturment | Max RQ | Max MEC | # Det. |
|---|---|---|---|---|---|---|---|
| Cypermethrin | Insecticide | 52315–07-8 | T | GC | 16 | 33 | 6 |
| Cyfluthrin | Insecticide | 68359–37-5 | T | GC | 2.5 | 29 | 18 |
| Bifenthrin | Insecticide | 82657–04-3 | T | GC | 0.6 | 5.4 | 20 |
| Cyhalothrin | Insecticide | 91465–08-6 | T | GC | 0.5 | 6.3 | 23 |
| Malathion | Insecticide | 121–75-5 | S | LC+GC | 0.4 | 236 | 4 |
| Dimethoate | Insecticide | 60–51-5 | T+S | LC+GC | 0.2 | 493 | 27 |
| Diazinon | Insecticide | 333–41-5 | S | GC | 0.2 | 60 | 4 |
| Esfenvalerate | Insecticide | 66230–04-4 | T | GC | 0.2 | 1.9 | 6 |
| Deltamethrin | Insecticide | 52918–63-5 | T | GC | 0.2 | 1.0 | 13 |
| Permethrin | Insecticide | 52645–53-1 | T | GC | 0.1 | 5.5 | 2 |
| Sucralose | Food additive | 56038–13-2 | S | LC | - | >5000 | 51 |
| Iohexol | PPCP | 66108–95-0 | S | LC | - | >5000 | 51 |
| Metformin | PPCP | 657–24-9 | S | LC | 9E-05 | >5000 | 39 |
| 2,4-dichlorophenol | Herbicide TP | 120–83-2 | S | LC | - | >1000 | 22 |
| Triclopyr | Herbicide | 55335–06-3 | S | LC | 4E-04 | >1000 | 44 |
| 2,4-Dinitrophenol | different uses | 51–28-5 | S | LC | 0.003 | >1000 | 1 |
| Tolyltriazole | Corrosion inhibitor | 136–85-6 | S | LC | - | >1000 | 45 |
| 9-Octadecenamide | Endogenous | 301–02-0 | S | LC | - | 940 | 26 |
| TCPP2 | Flame Retardant | 13674–84-5 | S | LC | - | 930 | 40 |
| TDCPP1 | Flame Retardant | 13674–87-8 | S | LC | - | 890 | 51 |
| 2,4-D | Herbicide | 94–75-7 | T | LC | 5E-05 | 778 | 51 |
| Metoprolol | PPCP | 37350–58-6 | S | LC | 7E-05 | 487 | 51 |
| Boscalid | Fungicide | 188425–85-6 | T+S | LC+GC | 3E-04 | 368 | 51 |
| Diuron | Herbicide | 330–54-1 | T | LC | 0.08 | 199 | 51 |
| Fluxapyroxad | Fungicide | 907204–31-3 | S | LC | 3E-05 | 76 | 51 |
| DEET | Insect repellent | 134–62-3 | T+S | LC+GC | 7E-07 | 53 | 51 |
| fipronil | Insecticide | 120068–37-3 | T | LC+GC | 0.01 | 14 | 51 |
| Fipronil amide | Insecticide TP | 205650–69-7 | T | GC | - | 13 | 51 |
| Fipronil-sulfone | Insecticide TP | 120068–36-2 | T | LC+GC | 4E-04 | 9.0 | 51 |
| Fipronil-desulfinyl | Insecticide TP | 205650–65-3 | T | LC+GC | 9E-05 | 4.5 | 51 |
| PFHxS3 | PFCs | 355–46-4 | S | LC | - | 4.2 | 51 |
| Chlorthal-dimethyl | Herbicide | 1861–32-1 | S | GC | 5E-07 | 3.1 | 51 |
| Dichlobenil | Herbicide | 1194–65-6 | S | GC | - | - | 51 |
| Dithiopyr TP | Herbicide TP | 128294–56-4 | S | LC | - | - | 51 |
Tris(1,3-dichloroisopropyl)phosphate,
Tris(2-chloroisopropyl)phosphate,
Perfluorohexanesulfonic acid, T: Target Method, S: Suspect Screening, GC: GC-QTOF-MS, LC: LC-QTOF-MS, TP: transformation product, no toxicity data available or not quantified.
Substances with the highest concentrations (maxima >890 ng/L) were mainly waste water derived (e.g., the artificial sweetener sucralose, the X-ray contrast media iohexol, and the pharmaceutical metformin), but included one herbicide (triclopyr) and one herbicide TP (2,4-dichlorophenol). For seven of the ten compounds with the highest concentration, no toxicity data were available, precluding risk assessment. Surprisingly, 17 compounds from different substance classes were detected in all 51 samples and nearly half of the detected compounds were found in more than 50% of the samples.
The results clearly show that the ten most critical compounds for this catchment are insecticides, mainly pyrethroids (7 out of 10), with RQ>0.1, hence, at concentrations close to or above the EC50 concentration for aquatic invertebrates. Another six insecticides (chlorpyrifos, imidacloprid, flubendiamine, novaluron, chlorantraniliprole and fipronil) and the pharmaceutical venlafaxine had RQs between 0.01 and 0.1 based on invertebrate toxicity data. At or below these concentrations, reduced survival was observed in the field4, 33 and in the European Union, the Uniform Principle requires that RQs are below 0.01 for invertebrates and fish34. In addition, synergistic mixture effects resulting from the large number of co-occurring chemicals are expected to negatively affect the ecosystem 3, 4, 26, 35, 36. This study highlighted a potential risk for aquatic organisms in the Cache Slough complex during rain events, mainly caused by multiple insecticides.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the State and Federal Contractors Water Agency (SFCWA Contract 15–16) and by the National Institute of Environmental Health Sciences under Award Number P42ES004699. The content is solely the responsibility of the authors and does not necessarily represent the official views of the SFCWA or the National Institutes of Health. We thank Agilent Technologies, Inc. for technical assistance in instrument setup and applications, especially Phil Wylie, Agilent Technologies, for his support on the suspect screening using GC-QTOF-MS. We also thank Don Weston, UC Berkeley, Richard Connon, UC Davis, and Helen Poynton, University of Boston, MA, for the coordination of the project and the sampling campaign; Chris Alaimo, UC Davis, for significant laboratory assistance; Henry Calanchini, UC Davis, for sampling assistance; Kathrin Fenner for assistance with Eawag-PPS; the US EPA National Pesticide Standard Repository and Michelle Hladik, USGS, for providing reference standards.
Footnotes
ASSOCIATED CONTENT
Supporting Information
(1) Additional sampling information, (2) additional target compound information, (3) optimized analytical parameters for LC- QTOF-MS and GC-QTOF-MS methods, (4) Quality control parameters for target compounds, (5) optimized parameter settings for suspect screening with Agilent MassHunter Find by Formula, (6) detected targets and suspects (LC- QTOF-MS and GC-QTOF-MS), (7) Concentration pattern and MS/MS spectra of identified transformation products
The authors declare no competing financial interest.
References
- 1.Schwarzenbach RP; Escher BI; Fenner K; Hofstetter TB; Johnson CA; von Gunten U; Wehrli B, The Challenge of Micropollutants in Aquatic Systems. Science 2006, 313, (5790), 1072. [DOI] [PubMed] [Google Scholar]
- 2.Richardson SD; Kimura SY, Water Analysis: Emerging Contaminants and Current Issues. Anal Chem 2016, 88, (1), 546–82. [DOI] [PubMed] [Google Scholar]
- 3.Moschet C; Wittmer I; Simovic J; Junghans M; Piazzoli A; Singer H; Stamm C; Leu C; Hollender J, How a complete pesticide screening changes the assessment of surface water quality. Environ Sci Technol 2014, 48, (10), 5423–32. [DOI] [PubMed] [Google Scholar]
- 4.Bundschuh M; Goedkoop W; Kreuger J, Evaluation of pesticide monitoring strategies in agricultural streams based on the toxic-unit concept - Experiences from long-term measurements. Science of the Total Environment 2014, 484, (1), 84–91. [DOI] [PubMed] [Google Scholar]
- 5.Krauss M; Singer H; Hollender J, LC–high resolution MS in environmental analysis: from target screening to the identification of unknowns. Analytical and Bioanalytical Chemistry 2010, 397, (3), 943–951. [DOI] [PubMed] [Google Scholar]
- 6.Schymanski EL; Singer HP; Slobodnik J; Ipolyi IM; Oswald P; Krauss M; Schulze T; Haglund P; Letzel T; Grosse S; Thomaidis NS; Bletsou A; Zwiener C; Ibanez M; Portoles T; de Boer R; Reid MJ; Onghena M; Kunkel U; Schulz W; Guillon A; Noyon N; Leroy G; Bados P; Bogialli S; Stipanicev D; Rostkowski P; Hollender J, Non-target screening with high-resolution mass spectrometry: critical review using a collaborative trial on water analysis. Anal Bioanal Chem 2015, 407, (21), 6237–55. [DOI] [PubMed] [Google Scholar]
- 7.Gago-Ferrero P; Schymanski EL; Hollender J; Thomaidis NS, Chapter 13 - Nontarget Analysis of Environmental Samples Based on Liquid Chromatography Coupled to High Resolution Mass Spectrometry (LC-HRMS) In Comprehensive Analytical Chemistry, Sandra Pérez PE; Damià B, Eds. Elsevier: 2016; Vol. Volume 71, pp 381–403. [Google Scholar]
- 8.Acena J; Stampachiacchiere S; Perez S; Barcelo D, Advances in liquid chromatography-high-resolution mass spectrometry for quantitative and qualitative environmental analysis. Anal Bioanal Chem 2015, 407, (21), 6289–99. [DOI] [PubMed] [Google Scholar]
- 9.Gago-Ferrero P; Schymanski EL; Bletsou AA; Aalizadeh R; Hollender J; Thomaidis NS, Extended Suspect and Non-Target Strategies to Characterize Emerging Polar Organic Contaminants in Raw Wastewater with LC-HRMS/MS. Environ Sci Technol 2015, 49, (20), 12333–41. [DOI] [PubMed] [Google Scholar]
- 10.Hug C; Ulrich N; Schulze T; Brack W; Krauss M, Identification of novel micropollutants in wastewater by a combination of suspect and nontarget screening. Environ Pollut 2014, 184, 25–32. [DOI] [PubMed] [Google Scholar]
- 11.Moschet C; Piazzoli A; Singer H; Hollender J, Alleviating the reference standard dilemma using a systematic exact mass suspect screening approach with liquid chromatography-high resolution mass spectrometry. Anal Chem 2013, 85, (21), 10312–20. [DOI] [PubMed] [Google Scholar]
- 12.Schymanski EL; Singer HP; Longree P; Loos M; Ruff M; Stravs MA; Ripolles Vidal C; Hollender J, Strategies to characterize polar organic contamination in wastewater: exploring the capability of high resolution mass spectrometry. Environ Sci Technol 2014, 48, (3), 1811–8. [DOI] [PubMed] [Google Scholar]
- 13.Hernández F; Ibáñez M; Bade R; Bijlsma L; Sancho JV, Investigation of pharmaceuticals and illicit drugs in waters by liquid chromatography-high-resolution mass spectrometry. TrAC Trends in Analytical Chemistry 2014, 63, 140–157. [Google Scholar]
- 14.Schymanski EL; Jeon J; Gulde R; Fenner K; Ruff M; Singer HP; Hollender J, Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol 2014, 48, (4), 2097–8. [DOI] [PubMed] [Google Scholar]
- 15.NORMAN Association NORMAN MassBank. www.massbank.eu (07/22/16),
- 16.Zedda M; Zwiener C, Is nontarget screening of emerging contaminants by LC-HRMS successful? A plea for compound libraries and computer tools. Anal Bioanal Chem 2012, 403, (9), 2493–502. [DOI] [PubMed] [Google Scholar]
- 17.Chapman JD; Goodlett DR; Masselon CD, Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrometry Reviews 2014, 33, (6), 452–470. [DOI] [PubMed] [Google Scholar]
- 18.NIST NIST Standard Reference Database 1A. http://www.nist.gov/srd/nist1a.cfm (accessed: 04/0½016),
- 19.Kwiecien NW; Bailey DJ; Rush MJP; Cole JS; Ulbrich A; Hebert AS; Westphall MS; Coon JJ, High-Resolution Filtering for Improved Small Molecule Identification via GC/MS. Analytical Chemistry 2015, 87, (16), 8328–8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Alba A; Uclés S; Belmonte-Valles N; Riener J Screening for Hundreds of Pesticide Residues Using a GC/Q-TOF with an Exact Mass Pesticide Database inFood. Agilent Technologies Application Note; 2015. [Google Scholar]
- 21.California Department of Pesticide Regulation Pesticide Use Reporting (PUR). http://www.cdpr.ca.gov/docs/pur/purmain.htm (accessed: 08/24/2016),
- 22.Hladik ML; Kuivila KM, Assessing the occurrence and distribution of pyrethroids in water and suspended sediments. Journal of agricultural and food chemistry 2009, 57, (19), 9079–85. [DOI] [PubMed] [Google Scholar]
- 23.Feo ML; Eljarrat E; Barcelo D, Performance of gas chromatography/tandem mass spectrometry in the analysis of pyrethroid insecticides in environmental and food samples. Rapid Commun Mass Spectrom 2011, 25, (7), 869–76. [DOI] [PubMed] [Google Scholar]
- 24.EAWAG-BBD Pathway Prediction System website. http://eawag-bbd.ethz.ch/predict/ (accessed: 06/10/2016),
- 25.Allen F; Greiner R; Wishart D, Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 2015, 11, (1), 98–110. [Google Scholar]
- 26.Nowell LH; Norman JE; Moran PW; Martin JD; Stone WW, Pesticide Toxicity Index--a tool for assessing potential toxicity of pesticide mixtures to freshwater aquatic organisms. Sci Total Environ 2014, 476–477, 144–57. [DOI] [PubMed] [Google Scholar]
- 27.Weston DP; Lydy MJ, Urban and Agricultural Sources of Pyrethroid Insecticides to the Sacramento-San Joaquin Delta of California. Environmental Science & Technology 2010, 44, (5), 1833–1840. [DOI] [PubMed] [Google Scholar]
- 28.Moschet C; Vermeirssen EL; Seiz R; Pfefferli H; Hollender J, Picogram per liter detections of pyrethroids and organophosphates in surface waters using passive sampling. Water Res 2014, 66, 411–22. [DOI] [PubMed] [Google Scholar]
- 29.University of Hertfordshire The Pesticide Properties DataBase (PPDB) developed by the Agriculture & Environment Research Unit (AERU), University of Hertfordshire, 2006–2013. http://sitem.herts.ac.uk/aeru/footprint/ (accessed: 06/30/2016),
- 30.Kolpin DW; Furlong ET; Meyer MT; Thurman EM; Zaugg SD; Barber LB; Buxton HT, Pharmaceuticals, hormones, and other organic waste contaminants in U.S. streams, 1999–2000: a national reconnaissance. Environmental Science and Technology 2002, 36, (6), 1202–1211. [DOI] [PubMed] [Google Scholar]
- 31.Anumol T; Wu S; Marques dos Santos M; Daniels KD; Snyder SA, Rapid direct injection LC-MS/MS method for analysis of prioritized indicator compounds in wastewater effluent. Environmental Science: Water Research & Technology 2015, 1, (5), 632–643. [Google Scholar]
- 32.Dickenson ERV; Snyder SA; Sedlak DL; Drewes JE, Indicator compounds for assessment of wastewater effluent contributions to flow and water quality. Water Res. 2011, 45, (3), 1199–1212. [DOI] [PubMed] [Google Scholar]
- 33.Schafer RB; von der Ohe PC; Rasmussen J; Kefford BJ; Beketov MA; Schulz R; Liess M, Thresholds for the effects of pesticides on invertebrate communities and leaf breakdown in stream ecosystems. Environ Sci Technol 2012, 46, (9), 50134–42. [DOI] [PubMed] [Google Scholar]
- 34.European Commission, Implementing regulation (EC) No 1107/2009 of the European Parliament and of the Council as regards uniform principles for evaluation and authorisation of plant protection products. Off J Eur 2011, L155:127–75. [Google Scholar]
- 35.Backhaus T; Faust M, Predictive environmental risk assessment of chemical mixtures: a conceptual framework. Environ Sci Technol 2012, 46, (5), 2564–73. [DOI] [PubMed] [Google Scholar]
- 36.Lydy M; Belden J; Wheelock C; Hammock B; Denton D, Challenges in Regulating Pesticide Mixtures. Ecology and Society 2004, 9 (http://www.ecologyandsociety.org/vol9/iss6/art1) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.