Abstract
Despite recent advances in targeted therapies, the molecular mechanisms driving breast cancer initiation, progression, and metastasis are minimally understood. Growing evidence indicate that tRNA-derived small RNAs (tsRNA) contribute to biological control and aberrations associated with cancer development and progression. The RUNX1 transcription factor is a tumor suppressor in the mammary epithelium whereas RUNX1 downregulation is functionally associated with breast cancer initiation and progression. We identified four tsRNA (ts-19, ts-29, ts-46, and ts-112) that are selectively responsive to expression of the RUNX1 tumor suppressor. Our finding that ts-112 and RUNX1 anti-correlate in normal-like mammary epithelial and breast cancer lines is consistent with tumor-related activity of ts-112 and tumor suppressor activity of RUNX1. Inhibition of ts-112 in MCF10CA1a aggressive breast cancer cells significantly reduced proliferation. Ectopic expression of a ts-112 mimic in normal-like mammary epithelial MCF10A cells significantly increased proliferation. These findings support an oncogenic potential for ts-112. Moreover, RUNX1 may repress ts-112 to prevent overactive proliferation in breast epithelial cells to augment its established roles in maintaining the mammary epithelium.
Keywords: breast cancer, RUNX1, tRNA-derived small RNA, tsRNA, tRNA-derived fragments, tRF, ncRNA
Graphical Abstract:
We identified four tsRNA (ts-19, ts-29, ts-46, and ts-112) that are selectively responsive to expression of the RUNX1 tumor suppressor. Our findings support an oncogenic potential for ts-112. RUNX1 may repress ts-112 to prevent overactive proliferation in breast epithelial cells to augment the established roles of RUNX1 in maintaining the mammary epithelium.
1. INTRODUCTION
Ongoing advancements in next-generation sequencing sensitivity and corresponding development of optimized computational algorithms for robust ‘big-data’ analyses in the post-genomic era have highlighted the abundance, conservation, and diversity of non-coding RNA (ncRNA) species. It is now understood that the majority of RNA transcription generates ncRNA, including ribosomal RNA (rRNA) and transfer RNA (tRNA), essential for protein-coding messenger RNA (mRNA) translation, as well as a plethora of short and long non-coding RNA (e.g., miRNA, lincRNA), many with unknown or poorly described functions. Examination of ncRNA in deep-sequencing databases (e.g., The Cancer Genome Atlas) reveals disease-specific expression profiles, suggesting both biomarker potential and biologically relevant roles. Further, the activities of RNA polymerases I and III (Pol I and Pol III), responsible for most ncRNA transcription, can be dysregulated by transcription factors associated with RNA polymerase II (Pol II) and mRNA expression (reviewed in (White, 2008)). Of note, high levels of both Pol III and tRNA, solely transcribed by Pol III, have been reported in cancer (reviewed in (Marshall & White, 2008; White, 2008)). Moreover, we and others have shown that short ncRNA molecules derived from cleavage of pre-tRNA transcripts distinguish human tumor samples and cancer cell lines from normal tissue (Balatti et al., 2017; Lee, Shibata, Malhotra, & Dutta, 2009).
ChIP-seq experiment have classified 3 types of Pol III-transcribed genes based on binding of TFIIIA, TFIIIB, and TFIIIC complexes; type 1 — 5S rRNA -- TFIIIA, TFIIIB with BRF1, and TFIIIC; type 2 — tRNA and others - TFIIIB with BRF1 and TFIIIC ; type 3 — U6 snRNA and others - TFIIIB with BRF2 and other factors (Oler et al., 2010). The authors identified nearly 2000 putative transfer DNA (tDNA) loci (hg18) as consensus tRNA, tRNA pseudogene, or tRNA-related, that recruit only TFIIIB with BRF1, TFIIIC, and RNA Pol III (Oler et al., 2010). The resultant pre-tRNA transcripts undergo successive cleavage of the 5-prime and 3-prime ends and intron splicing, if necessary, followed by addition of CCA to the 3-prime tail for amino acid attachment to generate mature tRNA. While once thought to be short-lived by-products, Balatti et al., 2017 recently discovered that the cleaved 3-prime pre-tRNA tails have dynamic levels in breast cancer patient samples with unique expression profiles stratifying normal-like breast epithelial cells from 7 breast cancer cell lines that model pre-malignant through aggressive, triple negative disease (Balatti et al., 2017). No differences were observed in levels of mature tRNA.
Designated tRNA-derived small RNA (tsRNA), these novel ncRNA molecules are a product of normal tRNA biogenesis. RNA Pol III transcription terminates at a sequence of at least four thymidines (TTTT) 10-60 nucleotides downstream of the mature tRNA 3-prime end. Following RNase P cleavage of the 5-prime leader, often only a single nucleotide, this longer 3-prime tail is cleaved by RNase Z to generate tsRNA of 16-48 nucleotides (Haussecker et al., 2010; Pekarsky et al., 2016). Of note, pre-tRNA processing occurs in both the nucleus and mitochondria (Brzezniak, Bijata, Szczesny, & Stepien, 2011; Siira et al., 2018), prior to export into the cytoplasm for animo acid loading. Importantly, tsRNA are only one type of tRNA-derived RNA fragments, tRFs, an abundant class of small ncRNA (Lee et al., 2009). Functional studies from our group and others have reported a role for several tsRNA in cancer following loss-of-function experiments (Balatti et al., 2017; H. K. Kim et al., 2017; Pekarsky et al., 2016). However, compelling questions relevant to mechanisms that regulate tsRNA levels remain unanswered.
Interrogation of ELAC2, the human RNase Z homolog solely responsible for tsRNA cleavage from the pre-tRNA 3-prime tail, in public databases such as The Human Protein Atlas (www.proteinatlas.org) and cBioPortal for Cancer Genomics (www.cbioportal.org) indicate ubiquitous expression with low specificity across normal tissue, cell lines, and cancer specimens, suggesting minimal involvement in regulating tsRNA expression. However, RNA Pol III transcription is reported to be regulated by the local genome context, including histone modifications, chromatin accessibility, and occupancy of proximal RNA Pol II DNA elements (Oler et al., 2010). Further, many individual subunits of TFIIIB and TFIIIC (e.g., BRF1, TBP) are regulated by transcription factors implicated in promoting cancer development and progression (reviewed in (Marshall & White, 2008; White, 2008)). The frequently mutated tumor suppressor TP53 targets TATA-binding protein (TBP), a TFIIIB subunit, inhibiting promoter occupancy and repressing Pol III (Crighton et al., 2003). Studies have shown that another tumor suppressor, the retinoblastoma protein (RB), has homology to TFIIIB subunits TBP and BRF and competes for DNA binding sites (Larminie et al., 1997). Moreover, the proto-oncogene, ETS1, occupies genomic DNA near the transcriptional start site of Pol III tRNA promoters (Oler et al., 2010). Combined, the current body of literature provides evidence for numerous transcription factors that may regulate tsRNA expression.
Over the past decade, numerous studies have identified RUNX1 as a key transcription factor in breast cancer initiation and metastatic progression (reviewed in (Hong et al., 2019; Hong, Fritz, Zaidi, et al., 2018)) with low expression correlating to poor prognosis and aggressive disease (Ferrari et al., 2014; Hong et al., 2017). Expression of RUNX1 has been shown to maintain the mammary epithelium, as loss of RUNX1 in normal-like mammary epithelial cells promoted mesenchymal gene expression and repressed epithelial genes. Thus, the epithelial-to-mesenchymal transition was suppressed by RUNX1 (Hong, Fritz, Zaidi, et al., 2018; Hong et al., 2017). Further, ectopic overexpression of RUNX1 in aggressive breast cancer cells reduced migration, invasion, and stemness, as well as inhibited tumor growth in mice as determined by reduced xenograft size in the mammary fat pad (Hong D. et al., Mol Cancer Res 2018). Moreover, published reports indicate that RUNX1 interacts with TP53 (Wu, Ozaki, Yoshihara, Kubo, & Nakagawara, 2013), ETS1 (Shrivastava et al., 2014), and regulates chromatin and global gene expression profiles in breast cancer (Barutcu et al., 2016). Yet the tumor suppressor-like molecular mechanisms of RUNX1 in breast cancer remain elusive.
Here, we addressed the hypothesis that RUNX1 suppresses breast cancer phenotypes by directly and/or indirectly regulating tsRNA. We interrogated expression of the 113 known tsRNAs in RUNX1-expressing mammary epilithelial cells infected with two different shRNAs against RUNX1 or a non-specific negative control, as well as in RUNX1-negative aggressive breast cancer cells with or without ectopic RUNX1 overexpression. Through differential expression analysis, we identified 4 candidate RUNX1-responsive tsRNA; ts-19, ts-29, ts-46, and ts-112. Of note, ts-112 was the only tsRNA significantly downregulated following RUNX1 overexpression. To determine if reduced ts-112 levels were sufficient to repress breast cancer-related phenotypes, we performed ts-112 gain and loss of function experiments and discovered a role in control of cellular proliferation. These compelling data suggest a novel interaction network in which RUNX1 maintains the normal mammary epithelium by regulating tsRNA expression levels.
2. MATERIALS AND METHODS
2.1. Cell culture
MCF10A cells were maintained in DMEM/F12 (Hyclone, Thermo Fisher Scientific) supplemented with 5% (v/v) horse serum (Gibco, Thermo Fisher Scientific), 10 mg/mL human insulin (Sigma-Aldrich), 20 ng/mL recombinant hEGF (PeproTech), 100 ng/mL cholera toxin (Sigma-Aldrich), 0.5 mg/mL hydrocortisone (Sigma-Aldrich), 50 IU/mL penicillin/50 mg/mL streptomycin (Gibco, Thermo Fisher Scientific), and 2 mmol/L glutamine (Gibco, Thermo Fisher Scientific). MCF10CA1a cells were maintained in DMEM/F12 (Hyclone, Thermo Fisher Scientific), 5% (v/v) horse serum with 50 IU/mL penicillin/ 50 mg/mL streptomycin (Gibco, Thermo Fisher Scientific) and 2 mmol/L glutamine (Gibco, Thermo Fisher Scientific). MCF7 and MDA-MB-231 cells were maintained in DMEM high glucose (Gibco, Thermo Fisher Scientific) supplemented with 10% (v/v) FBS (Atlanta Biologicals) and 50 IU/mL penicillin/50 mg/mL streptomycin (Gibco, Thermo Fisher Scientific). Cell line identities were validated by short tandem repeat analysis using the Promega GenePrint® 10 system in the Vermont Integrated Genomics Resource at the UVM Cancer Center.
2.2. RUNX1 inhibition and overexpression
RUNX1 was inhibited in MCF10A cells or overexpressed in MCF10CA1a cells as previously described (Hong, Fritz, Finstad, et al., 2018; Hong et al., 2017).
2.3. ts-112 inhibition and overexpression
Custom miRIDIAN hairpin inhibitors (Dharmacon, Horizon Discovery) and mirVana inhibitors and mimics (Ambion, Thermo Fisher Scientific) were designed against ts-112. Sequences:
miRIDIAN hairpin inhibitor: proprietary molecule based on ts-112 target sequence
5’ - CUCGGCUUUCCCUGCUAACUGGGCUUU - 3’
mirVana inhibitor: 5’ - AGCCCAGUUAGCAGGGAAAGCCGAGTT - 3’
mirVana mimic: 5’ - AAAGCCCAGUUAGCAGGGAAAGCCGAg - 3’
The appropriate predesigned non-specific controls designed against non-mammalian miRNA, Transfection Control with Dy547 (Dharmacon, Horizon Discovery) or Negative Control #1 (Ambion, Thermo Fisher Scientific), were used as a negative control. MCF10CA1a cells were transfected with 50nM miRIDIAN hairpin inhibitor or 25nM mirVana inhibitor against ts-112, associated negative control, and mock treated (no nucleic acid) with Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific) following the recommended manufacturer protocol. MCF10A cells were transfected with 25nM mirVana ts-112 mimic, Negative Control #1, or mock treated (no nucleic acid) with Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific) following the recommended manufacturer protocol. Transfection efficiency was monitored in negative control samples by fluorescence microscopy (Eclipse TS100 inverted microscope, Nikon).
2.4. RNA isolation
Cells were harvested into TRIzol® Reagent (Invitrogen, Thermo Fisher Scientific) and RNA isolated according to the recommended manufacturer protocol followed by either a sodium acetate RNA precipitation for tsRNA samples or a DNase digestion (Zymo Research) for RNA > 200 nucleotides in length.
2.5. tsRNA microarray profiling
Total RNA was hybridized to custom tsRNA microarrays, spot intensity data extracted, and analyzed as previously described (Balatti et al., 2017). Each tsRNA probe was spotted in triplicate and expression values averaged to generate a single probeset expression value for each interrogated tsRNA. Microarray data have been deposited in GEO datasets (GSE142382)
2.6. Differential expression and clustering analysis
Normalized log2 microarray data were imported into Spotfire® DecisionSite v2 (TIBCO Software), biological replicates averaged, pairwise comparisons performed, and ANOVA p-values determined. Principal component analysis (PCA) was performed using the expression of all 113 probed tsRNA. Hierarchical clustering (UPGMA, Euclidean distance) was performed for 30 tsRNA with significant changes (> 1.5-fold change, ANOVA p < 0.05). Nodes smaller than 3 (i.e., ≤ 2 tsRNA) were combined for cluster identification.
2.7. cDNA synthesis and quantitative RT-PCR
For RNA > 200 nucleotides, cDNA was generated using SuperScript II with random hexamers (Invitrogen, Thermo Fisher Scientific) and then subjected to qPCR using iTaq Universal SYBR® Green Supermix (Bio-Rad) with human specific primers.
RUNX1 Forward: 5’ - TCAGGTTTGTCGGTCGAAG - 3’
RUNX1 Reverse: 5’ - GCCCATCCACTGTGATTTTG - 3’
GAPDH Forward: 5’ - AGGGCTGCTTTTAACTCTGGT - 3’
GAPDH Reverse: 5’ - CCCCACTTGATTTTGGAGGGA - 3’
HPRT Forward: 5’ - TGCTGACCTGCTGGATTACA - 3’
HPRT Reverse: 5’ - TCCCCTGTTGACTGGTCATT - 3’
For RNA < 200 nucleotides, cDNA was generated using the miScript RT II kit (QIAGEN) and miScript SYBR® Green PCR kit (QIAGEN) with target-specific forward primers and the included Universal Reverse Primer (QIAGEN).
ts-112 Forward: 5’ - AAAGCCCAGTTAGCAGGGAAAGCCGAG - 3’
RNU6 Forward: 5’ - ACGCAAATTCGTGAAGCGTTCCATATT - 3’
All qPCR reactions were performed using a ViiA7 thermocycler (Applied Biosystems, Thermo Fisher Scientific). Melt curve data were collected and assessed for primer specificity.
2.8. Proliferation assays
Cell Counting Kit 8 (WST-8/CCK8, Abcam) was used to monitor cell growth following transfection. Briefly, 4 hours after transfection, cells were washed, trypsinized (0.25% trypsin-EDTA, Gibco, Thermo Fisher Scientific), and plated at 5000 cells per well in triplicate in 96-well plates (Corning) with one plate for each time point (0, 24, 48, and 72 hours). For 0hr time point, the WST-8 solution was immediately added, incubated at 37°C for 3 hours (empirically determined), and absorbance read at OD = 450nm on a Victor X4 (PerkinElmer) plate reader; this procedure was repeated for each subsequent time point (24, 48, and 72hr). For growth curves, 50,000 cells per well were plated in triplicate in 6-well plates (Corning) 24 hours after transfection. At each time point (24, 48, and 72hr), cells were trypsinized (0.25% trypsin-EDTA, Gibco, Thermo Fisher Scientific) and counted on a Countess II (Invitrogen, Thermo Fisher Scientific) to determine viable cell number by Trypan Blue (0.4% solution, Invitrogen, Thermo Fisher Scientific) exclusion.
2.9. Statistics and data analysis
All statistics and data visualization were performed in Excel (Microsoft), Prism 8 (GraphPad Software), RStudio (RStudio), or Spotfire DecisionSite v2 (TIBCO Software). Final figures were generated in Illustrator (Adobe). P-values were calculated as per figure legends with p < 0.05 considered significant. R scripts, packages, and version numbers are included as an R Markdown file in the supplemental online material.
3. RESULTS AND DISCUSSION
3.1. Identification of candidate RUNX1-responsive tsRNA
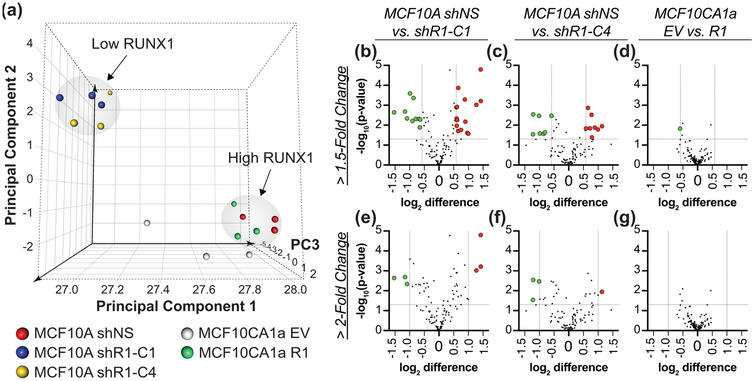
To determine whether tsRNA expression may be regulated by RUNX1 in breast cancer, we interrogated global tsRNA levels using a validated custom microarray (Balatti et al., 2017) in RUNX1-expressing normal-like MCF10A mammary epithelial cells or RUNX1-low MCF10CA1a malignant breast cancer cell lines. Both RUNX1 protein and mRNA were effectively silenced in MCF10A cells with two different shRNA (shR1-C1 and shR1-C4) and exogenously overexpressed in MCF10CA1a cells as previously shown (Hong, Fritz, Finstad, et al., 2018; Hong et al., 2017). After normalization and probeset summarization, relative log2 expression levels for all 113 established tsRNA (Balatti et al., 2017) were subjected to principal component analysis. The six MCF10A samples infected with RUNX1 shRNA cluster together (blue and yellow spheres, top left), disparate from the cluster of MCF10A non-silencing control (red spheres) and MCF10CA1a cells with ectopic RUNX1 expression (green spheres, Figure 1a). One MCF10A shR1-C4 biological replicate (small yellow sphere nearest the arrowhead in top left, Figure 1a, 2a) had reduced RNA quality that likely explains the third principal component (PC3) separation (Z-axis) from the five other tightly clustered shR1 samples (blue and yellow spheres, top left). We observe that overexpression of RUNX1 in MCF10CA1a aggressive breast cancer cells modulates the tsRNA profile to be consistent with tsRNA expression in normal-like MCF10A mammary epithelial cells (Figure 1a, green and red spheres, bottom right). This finding suggests that RUNX1 may directly and/or indirectly regulate tsRNA.
Figure 1: RUNX1 gain and loss of function affects global tsRNA expression profile.
The expression of 113 known human tsRNA were interrogated by microarray in RUNX1-expressing MCF10A normal-like mammary epithelial cells and MCF10A cells with silenced RUNX1 using two different shRNA (shR1-C1, shR1-C4) or non-silencing control (shNS) as well as in RUNX1-negative MCF10CA1a breast cancer cells (EV — empty vector) and MCF10CA1a cells with ectopic RUNX1 expression (R1). (a) Principal component analysis (PCA) based on the expression profiles of all 113 tsRNA generates two distinct clusters distinguished by RUNX1 expression; low RUNX1 (MCF10A shR1-C1 and shR1-C4) and high RUNX1 (MCF10A shNS and MCF10CA1a R1). PC3: Principal Component 3. Volcano plots with 1.5-fold (b-d) and 2-fold (e-g) change, ANOVA p-value < 0.05 cutoffs identify (b) 25, (c) 15, (d) 1, (e) 6, (g) 4, or(g) 0 tsRNA for pairwise comparisons between (b,e) MCF10A shNS and shR1-C1, (c,f) MCF10A shNS and shR1-C4, or (d,g) MCF10CA1a EV and R1. (b-g) Green dots indicate downregulated tsRNA and red dots, upregulated tsRNA as compared to non-silencing (shNS) or empty vector (EV) controls. Mean and ANOVA significance from three biological replicates used to generate plots.
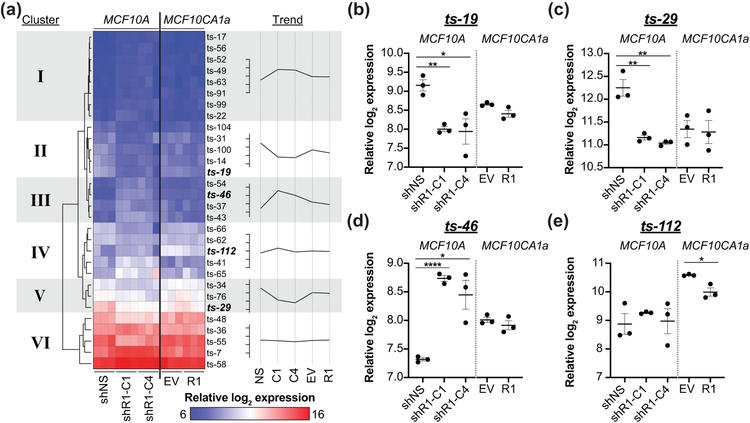
Figure 2: Identification of RUNX1-responsive candidate tsRNA.
(a) Six discernible groups with at least 3 tsRNA are seen following hierarchical clustering of the 30 differentially expressed (ANOVA p < 0.05) identified in Figure 1 Blue represents low/no tsRNA expression and Red represents high tsRNA expression. Three tsRNA have shared differential expression (ANOVA p < 0.05, > 2-fold change) between both RUNX1 shRNAs and non-silencing control in MCF10A normal-like mammary epithelial cells (b-d) with one tsRNA being significantly different (ANOVA p < 0.05, > 1.5-fold change) following RUNX1 overexpression in MCF10CA1a breast cancer cells (bolded). Trend line is the relative log2 tsRNA expression of biological replicate mean with ticks increments of 0.5. (b-e) Expression profiles of these four candidate RUNX1-responsive tsRNA. shR1-C1, shR1-C4 — different shRNA against RUNX1. shNS — non-silencing control. EV — empty vector. R1 — RUNX1 overexpression. Graphs are mean and SEM of three biological replicates with ANOVA significance. * p < 0.05; ** p < 0.01; **** p < 0.0001.
Differential expression was performed for pairwise comparisons of control (MCF10A shNS; MCF10CA1a EV) versus experimental cell lines (MCF10A shR1-C1, shR1-C4; MCF10CA1a R1) to discover potential candidate RUNX1-responsive tsRNA. Inhibition of RUNX1 in MCF10A cells significantly (> 1.5-fold change, ANOVA p < 0.05) changed expression of 22% and 13% of 113 tsRNA (Figure 1b-c, Table 1), shR1-C1 and shR1-C4 respectively, with 5 upregulated and 6 downregulated in both (Table 1). In MCF10CA1a cells following RUNX1 overexpression, only one tsRNA, ts-112, was significantly (> 1.5-fold change, ANOVA p < 0.05) downregulated (Figure 1d, Table 2). Using a more restrictive significance cutoff (> 2-fold change, ANOVA p < 0.05) reduced the number of regulated tsRNA in MCF10A shR1 samples, with 3 tsRNA deregulated in both shR1-C1 and shR1-C4 infected cells (Figure 1e-f, Table 1). No tsRNA was deregulated greater than 2-fold following induced RUNX1 expression in MCF10CA1a cells (Figure 1g, Table 2).
Table 1:
Differentially expressed of tsRNA following RUNX1 inhibition in MCF10A cells
| Comparison | p < 0.05 | ≥ 2-fold | ≥ 2-fold p < 0.05 |
≥ 1.5-fold | ≥ 1.5-fold p < 0.05 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | |||
| shNS (Control) -> shRUNX1-C1 | 43 | 3 | 3 | 3 | 3 | 17 | 12 | 15 | 10 | |
| shNS (Control) -> shRUNX1-C4 | 24 | 2 | 5 | 1 | 3 | 15 | 13 | 8 | 7 | |
| shNS -> both shRUNX1-C1 & -C4 | 21 | 2 | 2 | 1 | 2 | 9 | 8 | 5 | 6 | |
| Identity of tsRNA differentially expressed in both shRUNX1-C1 and shRUNX1-C4 as compared to shNS control | ts-107 ts-52 | ts-46 | ts-19 | ts-46 | ts-19 | ts-41 | ts-100 | ts-43 | ts-14 | |
| ts-14 | ts-54 | ts-65 | ts-29 | ts-29 | ts-43 | ts-104 | ts-46 | ts-19 | ||
| ts-17 | ts-55 | ts-46 | ts-14 | ts-54 | ts-29 | |||||
| ts-18 | ts-56 | ts-54 | ts-19 | ts-56 | ts-48 | |||||
| ts-19 | ts-58 | ts-56 | ts-29 | ts-58 | ts-55 | |||||
| ts-29 | ts-62 | ts-58 | ts-48 | ts-99 | ||||||
| ts-34 | ts-72 | ts-65 | ts-55 | |||||||
| ts-4 | ts-76 | ts-7 | ts-66 | |||||||
| ts-43 | ts-8 | ts-99 | ||||||||
| ts-46 | ts-99 | |||||||||
| ts-48 | ||||||||||
Note: Comparison is fold change and one-way ANOVA between non-silencing (shNS) control and RUNX1 inhibition with two different shRNA (-C1, -C4) in MCF10A normal-like mammary epithelial cells.
Table 2:
Differentially expressed of tsRNA following RUNX1 overexpression in MCF10CA1a cells
| Comparison | p < 0.05 | ≥ 2-fold | ≥ 2-fold p < 0.05 |
≥ 1.5-fold | ≥ 1.5-fold p < 0.05 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | |||
| EV (Control) -> RUNX1 OE | 4 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 1 | |
| Identity of differentially expressed tsRNA | ts-6 | ts-6 | ts-112 | |||||||
| ts-50 | ts-50 | |||||||||
| ts-101 | ts-71 | |||||||||
| ts-112 | ts-101 | |||||||||
| ts-112 | ||||||||||
Note: Comparison is fold change and one-way ANOVA between empty vector (EV) control and RUNX1 overexpression (OE) in MCF10CA1a aggressive breast cancer cells
Hierarchical clustering of the 30 tsRNA differentially expressed by greater than 1.5-fold (ANOVA p < 0.05) in at least one pairwise comparison between control and experimental samples identified 6 tsRNA clusters (Figure 2a). Cluster I and III tsRNA were increased while cluster II and V tsRNA were reduced when RUNX1 was silenced. The relatively flat trends of cluster IV and VI tsRNA may reflect changes occurring in one cell line following RUNX1 deregulation. Of note, the three tsRNA, ts-19, ts-46, and ts-29, with greater than 2-fold change (ANOVA p < 0.05) following RUNX1 silencing, and ts-112 with greater than 1.5-fold change (ANOVA p < 0.05) following RUNX1 overexpression, group to four different clusters (II-V) with moderate expression levels. Decreased ts-19 (cluster II) and ts-29 (cluster V) in MCF10A cells after RUNX1 silencing suggest positive regulation by RUNX1 (Figure 2b-c). In contrast, ts-46 (cluster III) and ts-112 (cluster IV) may be repressed by RUNX1 as they show increased or decreased expression following RUNX1 silencing or overexpression, respectively.
In sum, these experiments demonstrate that loss of RUNX1 coincides with changes in the expression levels of four tsRNA; ts-19 and ts-29 show positive correlation with reduced expression after RUNX1 knock-down and ts-46 and ts-112 show negative correlation with reciprocal expression to RUNX1 knock-down or overexpression, respectively. These findings suggest a functional relationship between RUNX1 and candidate RUNX1-responsive tsRNA.
3.2. RUNX1 and ts-112 have inverse expression trends between pre-malignant and breast cancer cell lines
Our previous studies present compelling evidence that RUNX1 expression in mammary cells and inhibits tumorigenic potential supporting maintenance of an epithelial-like phenotype;RUNX1 protein and mRNA expression are reduced in breast cancer cell lines (Hong, Fritz, Finstad, et al., 2018; Hong et al., 2017). The expression of candidate RUNX1-responsive tsRNA, ts-19, ts-29, ts-46, and ts-112, was curated from our initial microarray study (Balatti et al., 2017) and plotted to discover dynamic regulation in breast cancer cell models (Figure 3a-d). All four tsRNA exhibit significantly different expression (ANOVA p < 0.05) in the triple-negative MDA-MB-231 breast cancer cell line compared to MCF10A cells. Furthermore, ts-29 was reduced and ts-112 elevated more than 2-fold in the four tested breast cancer versus normal-like mammary epithelial cell lines (Figure 3b,d). However, the levels of ts-19 and ts-46 were not changed more than 2-fold among the five tested cell lines (Figure 3a, c). We selected ts-112 for further analysis as it is elevated in breast cancer cells with low/no RUNX1 (Figure 3d) and is the only significant differentially expressed tsRNA, being downregulated in malignant breast cancer cells following ectopic expression of RUNX1 (Figure 1d, 2a,e, Table 2).
Figure 3: RUNX1 and ts-112 have reciprocal expression trends in breast cancer cell lines.
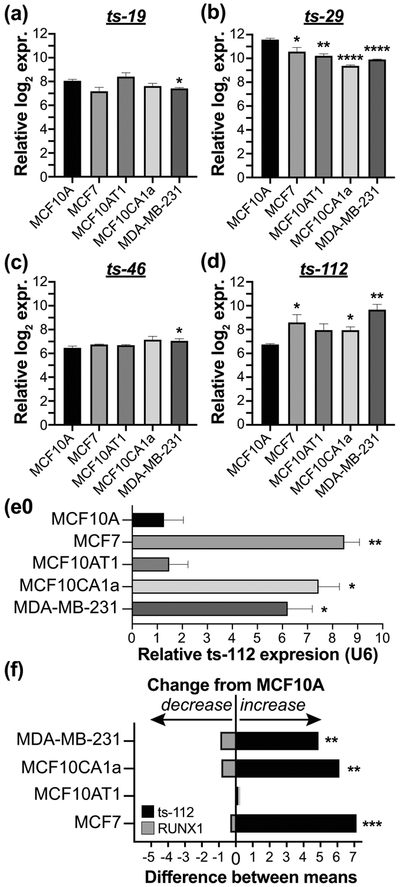
Expression of RUNX1-responsive candidates (a) ts-19, (b) ts-29, (c) ts-46, and (d) ts-112 in MCF10A normal-like mammary epithelial cells and four breast cancer cell lines (MCF7, MCF10AT1, MCF10CA1a, MDA-MB-231). Biological replicate mean (n = 3) and SEM graphed; ANOVA significance. Microarray tsRNA expression data from (Balatti et al., 2017). (e) qPCR validation of ts-112 expression normalized to U6 and average expression in MCF10A biological replicates (n = 3). Mean expression and SEM plotted with unpaired two-tailed t-test significance. (f) qPCR expression difference of RUNX1 (gray - downregulated) and ts-112 (black - upregulated) in four breast cancer cell lines as compared to MCF10A normal-like mammary epithelial cells. Biological replicate mean (n = 3) with Dunnett’s multiple comparisons test significance. RUNX1 qPCR normalized to GAPDH and HPRT. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
To validate and quantify ts-112 expression, we reasoned that quantitative PCR methodology designed and optimized for miRNA expression studies could be used to interrogate tsRNA levels. U6 snRNA was used as a housekeeping gene for data normalization and hsa-miR-93-5p, an oncogenic miRNA (K. Kim et al., 2012), was used as a positive control with pre-validated primers (data not shown). The increased levels of ts-112 were qPCR validated in 3 of 4 breast cancer lines as significantly upregulated, only in malignant breast cancer cell lines (MCF7, MCF10CA1a, MDA-MB-231) but not in premalignant MCF10AT1 cells (Figure 3e). It is notable that qPCR analysis using sequence-specific primers for ts-112 exhibited reciprocal expression to RUNX1 (Figure 3f). Our finding that ts-112 and RUNX1 anti-correlate in multiple breast cancer cell lines is consistent with tumor-related activity of ts-112 and the decreased activity of RUNX1, a tumor suppressor in mammary epithelial cells (Hong, Fritz, Finstad, et al., 2018; Hong et al., 2017).
3.3. ts-112 regulates cell proliferation in normal-like and malignant breast cell line models
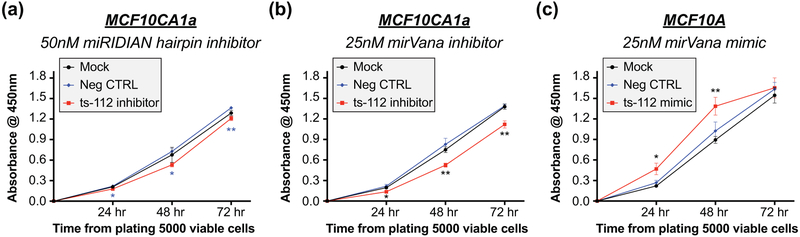
As ts-112 levels are elevated in ER-positive/HER-negative MCF7, ER-low/HER2-positive MCF10CA1a, and triple-negative MDA-MB-231 breast cancer cell lines, we hypothesized a potential role for ts-112 in regulating a hallmark of cancer (Hanahan & Weinberg, 2011). The ability of cancer cells to rapidly grow and evade death can be assayed by monitoring proliferation. Changes in growth rate after treatment are indicative of a potential mechanism. Thus, we designed custom tsRNA inhibitor and mimic molecules using the 28-nucleotide ts-112 sequence to perform gain and loss of function studies. The Dharmacon miRIDIAN hairpin inhibitor was a 27-nucleotide sequence of single-stranded RNA complementary to ts-112 with both 5-prime and 3-prime RNA hairpin structures. The Ambion mirVana inhibitor was a single-stranded RNA molecule complementary to ts-112 with modified RNA bases for added stability. This strategy added rigor to our studies since the molecular composition and structure of the inhibitors are different.
Inhibition of ts-112 in MCF10CA1a cells with 50nM miRIDIAN hairpin inhibitor or 25nM mirVana inhibitor significantly reduced proliferation compared to both mock treated cells and cells with a negative control targeting a non-mammalian small non-coding RNA, over a 72-hour time course assayed with Cell Count Kit-8 (Figure 4a,b). Of note, 25nM mirVana ts-112 inhibitor reduced MCF10CA1a proliferation ~3-fold more than 50nM miRIDIAN ts-112 inhibitor. Supporting viable cell count data show a suggestive, but non-significant, decrease in cell number after ts-112 inhibition (Figure S1a,b). We further observed an increase in population doubling time from 12.5 hours (mock) and 12.9 hours (negative control) to 14.3 hours (ts-112 inhibitor) (Figure S1c).
Figure 4: ts-112 promotes cellular proliferation.
Proliferation of MCF10CA1a cells transfected with (a) 50nM Dharmacon miRIDIAN hairpin inhibitor, (b) 25nM Ambion mirVana inhibitor, or (c) MCF10A cells transfected with 25nM Ambion mirVana mimic assessed by CCK-8 assay. Absorbance values (OD = 450nm) normalized to average replicate expression of cells 0 hour from platting 5000 viable cells, 4 hour from start of transfection. Significance determined by homoscedastic 2-tailed t-test of ts-112 inhibitor (a,b) or mimic (c) transfected cells as compared to (a) negative control or (b,c) mock transfected cells. Mean and SEM from 6 replicate wells across 2 independent experiments. * p < 0.05; ** p < 0.01.
In contrast to loss of ts-112 function in malignant cancer cells, gain of ts-112 in normal-like mammary epithelial MCF10A cells transfected with 25nM mirVana mimic significantly increased viable cell number 24 and 48 hours after plating, corresponding to 48 and 72 hours from-transfection, respectively, relative to control transfected and mock treated cells (Figure 4c). The absence of a significant difference at the 72-hour time point, 96 hours post transfection, may be a result of dilution or loss of the ts-112 mimic as proliferating cells approach confluence.
Taken together, our results show that ts-112 is necessary for enhanced proliferation of a breast cancer cell and promotes proliferation of the normal mammary epithelium. Ongoing and future studies into the global transcriptional changes induced by ts-112 inhibition or overexpression will expand our understanding of the molecular mechanisms and regulatory pathways by which ts-112 mediates biological control of breast cancer.
4. CONCLUSIONS
For the first time, we identify four tRNA-derived small RNAs (tsRNA), ts-19, ts-29, ts-46, and ts-112, that exhibit significant expression changes following deregulation of RUNX1 in cell models of the normal mammary epithelium and malignant breast tumors. Being downregulated, ts-112 was further investigated as the only tsRNA affected by RUNX1 overexpression in cancer cells. Functional assays demonstrated that ts-112 inhibition reduced the proliferative capacity of aggressive breast cancer cells while ts-112 overexpression in normal-like mammary epithelial cells promoted population growth. Combined, these data provide evidence for the oncogenic potential of ts-112. Moreover, RUNX1 expressed by mammary epithelial cells may act as a tumor suppressor by repressing ts-112 to prevent overactive cell proliferation, and thus augmenting the established roles of RUNX1 in maintaining the mammary epithelium (Hong, Fritz, Finstad, et al., 2018; Hong et al., 2017).
Supplementary Material
Figure S1: ts-112 inhibition reduces cellular growth and increases doubling time. MCF10CA1a cells transfected with 50nM Dharmacon miRIDIAN hairpin inhibitor against ts-112 or negative control. Cells seeded at equal density 24 hours post-transfection and viable cell number counted 24, 48, and 72 hours after plating. (a) Number of cells counted at each time point with SEM error shaded in. (b) Graph of modeled exponential growth and associated (c) doubling time. n = 6-12 wells across 2-4 independent experiments. Differences are suggestive (24hr and 48hr p < 0.1) but not statistically significant.
Acknowledgments:
The research reported here was supported by a Pilot Project award (J.L.S., N.H.F.) under grant U54 GM115516 (G.S.S.) from the National Institutes of Health for the Northern New England Clinical and Translational Research network, U01 CA196383 (J.L.S.), the Charlotte E. Perelman Cancer Research Fund (G.S.S.), and the Arthur Jason Perelman Professorship (G.S.S.).
Footnotes
Conflict of Interest Statement:
The authors declare no conflict of interests.
Data Availability Statement: The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request. GEO Datasets accession number for microarray data is GSE142382.
References:
- Balatti V, Nigita G, Veneziano D, Drusco A, Stein GS, Messier TL, … Croce CM (2017). tsRNA signatures in cancer. Proc Natl Acad Sci U S A, 114(30), 8071–8076. doi: 10.1073/pnas.1706908114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu AR, Hong D, Lajoie BR, McCord RP, van Wijnen AJ, Lian JB, … Stein GS (2016). RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochim Biophys Acta, 1859(11), 1389–1397. doi: 10.1016/j.bbagrm.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezniak LK, Bijata M, Szczesny RJ, & Stepien PP (2011). Involvement of human ELAC2 gene product in 3' end processing of mitochondrial tRNAs. RNA Biol, 8(4), 616–626. doi: 10.4161/rna.8.4.15393 [DOI] [PubMed] [Google Scholar]
- Crighton D, Woiwode A, Zhang C, Mandavia N, Morton JP, Warnock LJ, … Johnson DL (2003). p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J, 22(11), 2810–2820. doi: 10.1093/emboj/cdg265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari N, Mohammed ZM, Nixon C, Mason SM, Mallon E, McMillan DC, … Blyth K (2014). Expression of RUNX1 correlates with poor patient prognosis in triple negative breast cancer. PLoS One, 9(6), e100759. doi: 10.1371/journal.pone.0100759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, & Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell, 144(5), 646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, & Kay MA (2010). Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA, 16(4), 673–695. doi: 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Fritz AJ, Finstad KH, Fitzgerald MP, Weinheimer A, Viens AL, … Stein GS (2018). Suppression of Breast Cancer Stem Cells and Tumor Growth by the RUNX1 Transcription Factor. Mol Cancer Res. doi: 10.1158/1541-7786.MCR-18-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Fritz AJ, Gordon JA, Tye CE, Boyd JR, Tracy KM, … Stein GS (2019). RUNX1-dependent mechanisms in biological control and dysregulation in cancer. J Cell Physiol, 234(6), 8597–8609. doi: 10.1002/jcp.27841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Fritz AJ, Zaidi SK, van Wijnen AJ, Nickerson JA, Imbalzano AN, … Stein GS (2018). Epithelial-to-mesenchymal transition and cancer stem cells contribute to breast cancer heterogeneity. J Cell Physiol. doi: 10.1002/jcp.26847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Messier TL, Tye CE, Dobson JR, Fritz AJ, Sikora KR, … Stein GS (2017). Runx1 stabilizes the mammary epithelial cell phenotype and prevents epithelial to mesenchymal transition. Oncotarget, 8(11), 17610–17627. doi: 10.18632/oncotarget.15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, … Kay MA (2017). A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. doi: 10.1038/nature25005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Chadalapaka G, Lee SO, Yamada D, Sastre-Garau X, Defossez PA, … Safe S (2012). Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene, 31(8), 1034–1044. doi: 10.1038/onc.2011.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larminie CG, Cairns CA, Mital R, Martin K, Kouzarides T, Jackson SP, & White RJ (1997). Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J, 16(8), 2061–2071. doi: 10.1093/emboj/16.8.2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, & Dutta A (2009). A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev, 23(22), 2639–2649. doi: 10.1101/gad.1837609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, & White RJ (2008). Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer, 8(12), 911–914. doi: 10.1038/nrc2539 [DOI] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, … Cairns BR (2010). Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol, 17(5), 620–628. doi: 10.1038/nsmb.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Nigita G, … Croce CM (2016). Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A, 113(18), 5071–5076. doi: 10.1073/pnas.1604266113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava T, Mino K, Babayeva ND, Baranovskaya OI, Rizzino A, & Tahirov TH (2014). Structural basis of Ets1 activation by Runx1. Leukemia, 28(10), 2040–2048. doi: 10.1038/leu.2014.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siira SJ, Rossetti G, Richman TR, Perks K, Ermer JA, Kuznetsova I, … Filipovska A (2018). Concerted regulation of mitochondrial and nuclear non-coding RNAs by a dual-targeted RNase Z. EMBO Rep, 19(10). doi: 10.15252/embr.201846198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ (2008). RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet, 24(12), 622–629. doi: 10.1016/j.tig.2008.10.003 [DOI] [PubMed] [Google Scholar]
- Wu D, Ozaki T, Yoshihara Y, Kubo N, & Nakagawara A (2013). Runt-related transcription factor 1 (RUNX1) stimulates tumor suppressor p53 protein in response to DNA damage through complex formation and acetylation. J Biol Chem, 288(2), 1353–1364. doi: 10.1074/jbc.M112.402594 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: ts-112 inhibition reduces cellular growth and increases doubling time. MCF10CA1a cells transfected with 50nM Dharmacon miRIDIAN hairpin inhibitor against ts-112 or negative control. Cells seeded at equal density 24 hours post-transfection and viable cell number counted 24, 48, and 72 hours after plating. (a) Number of cells counted at each time point with SEM error shaded in. (b) Graph of modeled exponential growth and associated (c) doubling time. n = 6-12 wells across 2-4 independent experiments. Differences are suggestive (24hr and 48hr p < 0.1) but not statistically significant.