Abstract
Internode elongation is one of the key agronomic traits determining a plant’s height and biomass. However, our understanding of the molecular mechanisms controlling internode elongation is still limited in crop plant species. Here, we report the functional identification of an atypical basic helix-loop-helix transcription factor (OsbHLH073) through gain-of-function studies using overexpression (OsbHLH073-OX) and activation tagging (osbhlh073-D) lines of rice. The expression of OsbHLH073 was significantly increased in the osbhlh073-D line. The phenotype of osbhlh073-D showed semi-dwarfism due to deficient elongation of the first internode and poor panicle exsertion. Transgenic lines overexpressing OsbHLH073 confirmed the phenotype of the osbhlh073-D line. Exogenous gibberellic acid (GA3) treatment recovered the semi-dwarf phenotype of osbhlh073-D plants at the seedling stage. In addition, quantitative expression analysis of genes involving in GA biosynthetic and signaling pathway revealed that the transcripts of rice ent-kaurene oxidases 1 and 2 (OsKO1 and OsKO2) encoding the GA biosynthetic enzyme were significantly downregulated in osbhlh073-D and OsbHLH073-OX lines. Yeast two-hybrid and localization assays showed that the OsbHLH073 protein is a nuclear localized-transcriptional activator. We report that OsbHLH073 participates in regulating plant height, internode elongation, and panicle exsertion by regulating GA biosynthesis associated with the OsKO1 and OsKO2 genes.
Keywords: bHLH transcription factor, gibberellin, GA homeostasis, internode elongation, OsbHLH073, plant height, rice
1. Introduction
Gibberellin (GA) plays pivotal roles in many developmental processes including seed germination, root growth, stem and hypocotyl elongation, the promotion of cell division and elongation, flower induction, and internode elongation [1,2,3,4]. The GA biosynthetic pathway has been analyzed in detail in genetic and chemical studies in higher plants [4,5,6]. In higher plants, GA biosynthesis can generally be divided into three subcellular compartmentalization: Plastids, the endoplasmic reticulum (ER) membrane, and the cytoplasm. In plastids, two enzymes, ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS) are converted from geranylgeranyl diphosphate to ent-kaurene through a two-step cyclization [7,8]. Two cytochrome p450 monooxygenases, ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO), convert ent-kaurene into GA12 [8,9,10,11]. GA13 oxidase converts GA12 into GA53 [12]. In the cytoplasm, GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox) convert from GA12 and GA53 to various GA intermediates (GA44, GA19, and GA20) and bioactive GAs (GA1, GA3, and GA4), respectively [13,14]. GA 2-oxidase deactivates active GAs [15,16,17,18,19].
Semi-dwarfism is a very important trait for breeding cereal crops because it is associated with improved lodging resistance and a very good harvest index [14,20,21]. Mutations of GA20ox2 (sd1) and GA3ox2 (d18) display a loss of function, resulting in dwarfism in rice [16]. The overexpression of GA2-oxidase (GA2ox) genes in Arabidopsis, rice, and other plants also results in a dwarf phenotype [18,22,23,24]. The TOMATO INTERNODE ELONGATED1-1(TIE1-1) and ELONGATED INTERNODE (EI) mutants display internode elongation in tomato. TIE1-1/EI encodes a class III GA 2-oxidase, a GA2oxidase 7 [25,26]. The overexpression of the ELONGATED UPPERMOST INTERNODE1 (EUI1) gene causes the dwarf phenotype, and Null mutations of the EUI1 gene result in the accumulation of active GAs and elongated uppermost internodes and increased plant height [2,3,12,27]. A class I homeodomain-leucine zipper, HOX12, positively controls the expression of EUI1 by directly binding to the promoter region [28]. In addition, a C2H2 zinc finger, PREMATURE INTERNODE ELONGATION 1, plays a negative role in internode elongation in rice [29].
Basic helix-loop-helix (bHLH) transcription factors (TFs) play diverse roles in controlling various biological processes in both plants and in animals. In rice, there are 177 bHLH genes, while there are 183 in poplar, 167 in Arabidopsis, and 98 in moss [30]. The bHLH domain consists of 60 amino acids organized in a basic region and an HLH region. The basic region is needed for DNA binding and the HLH domain is needed for protein–protein interaction [31]. bHLH TFs are divided into two groups according to their DNA-binding activity, the atypical non-DNA-binding and the typical DNA-binding bHLH family [32]. Both atypical and typical bHLH TFs regulate various biological processes via protein–protein interaction. For example, in Arabidopsis, PHYTOCHROME INTERACTING FACTORS 4 and 5 (PIF4 and PIF5), encoding typical DNA-binding bHLH TFs, function in light signaling and hypocotyl growth. It has recently been reported that PIF3, PIF4, PIF5, and PIF3-LIKE 5 (PIL5) are involved in the GA biosynthesis and signaling pathway in Arabidopsis [4]. An atypical bHLH TF, LONG HYPOCOTYL IN FAR-RED1, modulates phytochrome signaling via heterodimerization with PIF4 and PIF5 [33]. In rice, typical bHLH TFs are involved in the development of the tapetum, internode elongation, grain size, iron homeostasis, and hormone signaling. Thus, UNDEVELOPED TAPETUM1 is needed for early tapetum development in rice [34], and overexpression of OsPIL1/OsPIL13 increases internode cell size and promotes internode elongation in rice [35]. POSITIVE REGULATOR OF GRAIN LENGTH1, 2 (PGL1, 2) and ANTAGONIST OF PGL1 (APG) are implicated in the processes determining grain length and weight in rice [36,37]. OsbHLH107 and its homolog, OsPIL11, regulates the grain size [38]. OsbHLH057 and OsbHLH058 positively regulated the iron deficiency responses [39]. Atypical bHLH TFs, INCREASED LAMINAR INCLINATION (ILI) and ILI1 BINDING bHLH1 (IBH1), regulate cell elongation and lamina joint bending in rice [40]. A complex consisting of BRASSINOSTEROID (BR) UPREGULATED1-LIKE1 (OsBUL1) and OsBUL1 COMPLEX1 (OsBC1) regulates leaf angle and grain size [41], and LAX PANICLE (LAX) is needed for the initiation and maintenance of axillary meristems in rice panicle [42]. OsbHLH035 involved in the seed germination through ABA-dependent and independent manners, respectively [43].
In rice, although some atypical bHLH TFs are involved in hormone signaling [41,44,45], shoot branching [46], and the determination of grain length and weight [36,37], only a few atypical bHLH genes have been functionally identified. In this study, we identified and characterized an atypical bHLH TF, OsbHLH073, that negatively regulates plant height by suppressing internode elongation. Yeast two-hybrid and protoplast assays suggest that OsbHLH073 has a role as a nuclear localized-transcriptional activator. In addition, expression analysis of the GA biosynthetic genes in the activation and overexpression lines of OsbHLH073 proposes the molecular mechanism on the function of OsbHLH073 associated with GA biosynthesis.
2. Results
2.1. Isolation of a Semi-Dwarf Phenotype Mutant by Simulating OsbHLH073
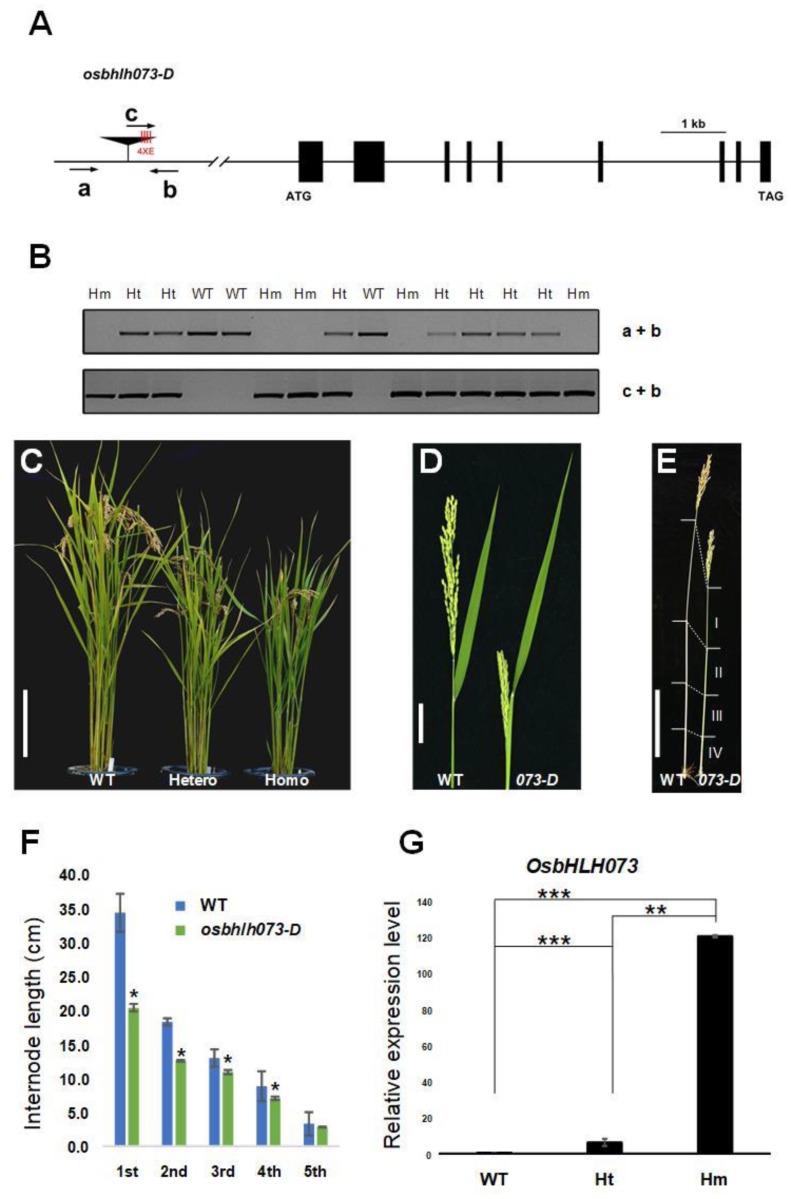
We isolated a semi-dwarf phenotype mutant line, PFG_4A-02508, from our T-DNA tagging population [47,48,49]. Flanking sequence analysis revealed that T-DNA was inserted 8489 bp upstream of the start codon of LOC_Os05g14010 on chromosome 5 (Figure 1A). The gene encodes OsbHLH073, an atypical bHLH TF of the predicted 177 bHLH genes of rice (Figure S1) [30,32]. Segregation and genotyping analysis of the PFG_4A-02508 plants showed dominant semi-dwarf phenotypes observed in heterozygous and homozygous progeny. Collectively, these results indicate that the phenotype is likely caused by gain-of-function mutations (Figure 1B,C). qRT-PCR analysis displayed that the transcript of OsbHLH073 was significantly increased in all heterozygous and homozygous plants (Figure 1G). We named this line osbhlh073-D.
Figure 1.
Phenotype of osbhlh073-D plants and the expression level of OsbHLH073. (A) Schematic diagram of the OsbHLH073 gene structure and relative T-DNA insertion positions. Black boxes denote exons. 4XE denotes four tandem copies of CaMV 35S enhancer. Primers (a, b, and c) used for genotyping. (B) Genotyping analysis of osbhlh073-D plants. WT, Ht, and Hm mean wild-type segregant, heterozygote, and homozygote for T-DNA insertion, respectively. (C) Phenotype of wild-type, heterozygous, and homozygous plants. Bar = 20 cm. (D) Panicle exsertion phenotype of wild-type and osbhlh073-D (073-D) plants. Bar = 5 cm. (E) Photograph showing internode length of wild-type and osbhlh073-D (073-D) plants. Bar = 20 cm. (F) Average internode length of wild-type and osbhlh073-D plants. Data are mean ± standard deviation (SD) from at least 10 plants. (* p < 0.001, Student’s t-test) (G) Relative expression level of OsbHLH073 in wild-type, heterozygous, and homozygous plants. Expression levels were normalized to Actin (LOC_Os03g50885) (** p < 0.01, *** p < 0.001, Student’s t-test).
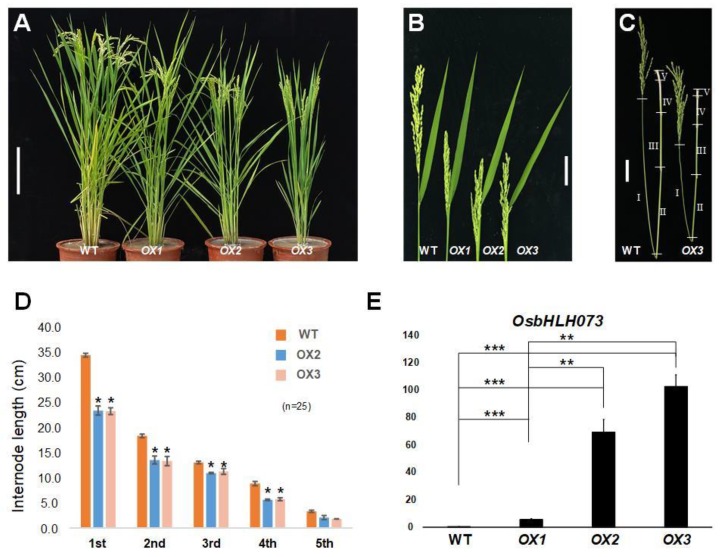
At the reproductive stage, osbhlh073-D plants showed the defective panicle exsertion phenotype besides a semi-dwarf compared to wild-type (Figure 1C,D). The poor panicle exsertion was mostly caused by inhibition of the elongation of the first and second internodes, resulting in the failure of the panicles of osbhlh073-D to fully emerge from the leaf sheath (Figure 1E,F). To confirm mutant phenotypes, we generated OsbHLH073-overexpressing (OX) plants constitutively expressing OsbHLH073 under the control of the maize ubiquitin promoter via Agrobacterium-mediated transformation in a Dongjin rice background [50]. We selected three lines among 6 T0 plants. Those selected lines were used for the generation of T2 homozygous lines and all displayed the similar phenotype to the osbhlh073-D mutant (Figure 2A–D). We also confirmed that the mRNA level of OsbHLH073 in the T2 OX plants (OX1- OX3) was correlated to the semi-dwarf phenotype (Figure 2A,E). This result indicates that OsbHLH073 regulates internode elongation in a negative way.
Figure 2.
Phenotype of OsbHLH073-overexpression (OX) plants and the expression levels of OsbHLH073. (A) Phenotype of wild-type and OsbHLH073-OX plants. Bar = 20 cm. (B) Panicle exsertion phenotype of wild-type and OsbHLH073-OX plants. Bar = 5 cm. (C) Photograph showing internode length of wild-type and OsbHLH073-OX plant. Bar = 5 cm. (D) Average internode length of wild-type and OsbHLH073-OX3 plant. Data are mean ± SD from 25 plants (* p < 0.001, Student’s t-test). (E) Relative expression level of OsbHLH073 in wild-type and OsbHLH073-OX plants. Expression levels were normalized to OsActin (LOC_Os03g50885) (** p < 0.01, *** p < 0.001, Student’s t-test).
2.2. OsbHLH073 and OsbHLH074 Knockout Mutants Have no Visible Phenotype
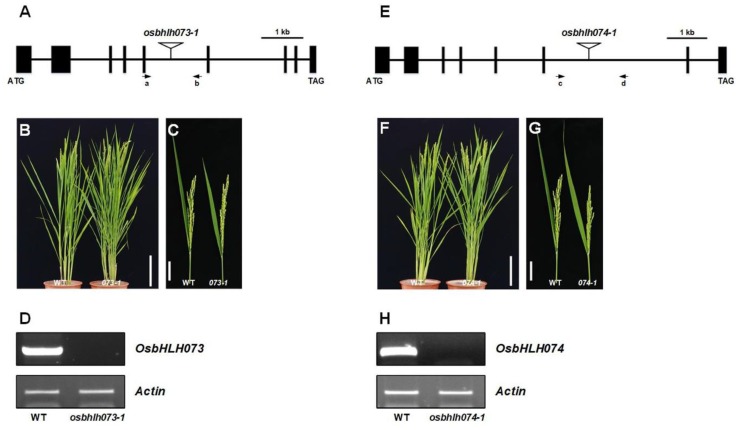
To investigate the function of OsbHLH073, we screened T-DNA insertional mutant pools and selected a mutant in which T-DNA was inserted into the fifth intron of OsbHLH073 (PFG_3A-17056) (Figure 3A). OsbHLH073 transcript was not detected in this line, indicating that osbhlh073-1 is a knockout mutant of this gene (Figure 3D). At the heading stage, we examined the phenotype of the osbHLH073-1 mutant, but did not observe any visible differences in phenotype (Figure 3B,C). Plant height and panicle exsertion were almost the same as in the wild-type. One possible explanation for this result could be that paralogous genes of OsbHLH073 gene might result in functional redundancy. Phylogenetic analysis showed that two homologous genes, OsbHLH072 and OsbHLH074, were present in the rice genome (Figure S2) [51]. The OsbHLH073 and OsbHLH074 protein sequences were 52% identical and had 61% similarity over the full length of the protein sequence, respectively, while the OsbHLH073 and OsbHLH072 protein had 41% identity and 51% similarity, respectively (Figure S3). Since OsbHLH074 was revealed as the closest homologue of OsbHLH073, we isolated a T-DNA insertional mutant of OsbHLH074 (PFG_5A-00405). T-DNA was inserted at the sixth intron of OsbHLH074 (LOC_Os01g13000) on chromosome 1 (Figure 3E). Intact OsbHLH074 transcript was not detectable in this mutant, indicating that osbhlh074-1 is also a null mutant of this gene (Figure 3H). However, osbhlh074-1 plant also did not show any visible phenotype, and the height and panicle exsertion of osbhlh074-1 were quite similar to those of wild-type at the heading stage (Figure 3F,G). Since there were no obvious phenotypic alterations in each osbhlh073 and osbhlh074 mutant, genetic redundancy can be considered for explaining the results based on their sequence similarity.
Figure 3.
Phenotypes of osbhlh073-1 and osbhlh074-1 mutants. (A) Genomic structure of OsbHLH073 and the positions of T-DNA insertion. Black boxes denote exons. Primers (a, b) were used for genotyping. (B) Phenotype of wild-type and osbhlh073-1(073-1) plants. (C) Panicle exsertion phenotype of wild-type and 073-1 mutant. Bar = 5 cm. (D) RT-PCR analyses of OsbHLH073 transcripts in wild type (WT) and 073-1 mutant. (E) Scheme diagram of the OsbHLH074 gene and the T-DNA insertion positions. Black boxes denote exons. Primers (c, d) were used for genotyping. (F) Phenotype of wild-type and osbhlh074-1(074-1) plants. Bar = 20 cm. (G) Panicle exsertion phenotype of wild-type and 074-1 mutant. Bar = 5 cm. (H) RT-PCR analyses of OsbHLH074 transcripts in wild-type (WT) and 074-1 mutant. Actin (LOC_Os03g50885) was used as the control.
2.3. Exogenous GA3 Application Rescues the Dwarf Phenotype of Osbhlh073-D Plants
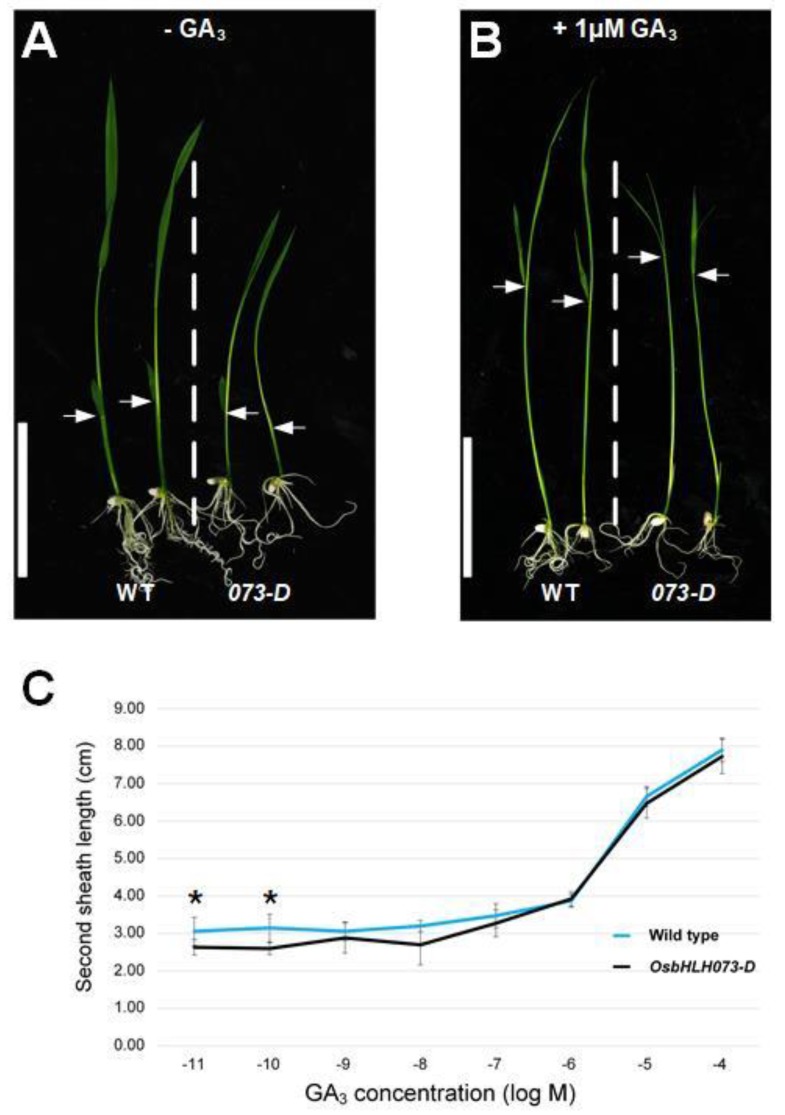
In plants, various hormones are associated with plant height [20,52]. Among these hormones, GAs are very important factors in determining plant height [14,20,53]. At the reproductive stage, osbhlh073-D showed a semi-dwarf phenotype. To test whether the semi-dwarf phenotype of osbhlh073-D was caused by insensitivity to or deficiency of GA, we measured the height (from the shoot base to the top leaf tip) (Figure 4A,B), and analyzed the response to exogenous GA3 application, and measured the second leaf sheaths of both wild-type and mutant growing on 1/2 MS median containing from 10−11 to 10−4 M GA3 in the continuous light condition for seven days (Figure 4C) [54]. Although osbhlh073-D showed a semi-dwarf phenotype compared to wild-type (Figure 4A), the response to application with GA3 in the length of the second leaf sheath of both wild-type and mutant was quite similar (Figure 4B,C). These results indicate that exogenous GA3 rescues the semi-dwarf phenotype of osbhlh073-D, indicating that osbhlh073-D may be a GA-deficient mutant. This observation is consistent with results in other GA-deficient mutants in rice [1,16,28,55].
Figure 4.
Rescue of the osbhlh073-D semi-dwarf phenotype by exogenous gibberellic acid (GA3) application. (A) Phenotype of seven-day-old plants in the light without GA3. Arrow indicates the second leaf sheath. (B) Phenotype of seven-day-old plants in the light with 1 µM of GA3 added to the medium. Bar = 5 cm. Arrow indicates the second leaf sheath. (C) Measurement of second leaf sheath length after GA3 treatment. Data are mean ± SD from at least 10 plants. (* p < 0.05, Student’s t test).
2.4. OsbHLH073 is Preferentially Expressed in Developing Organs at Both the Seedling and Reproductive Stages
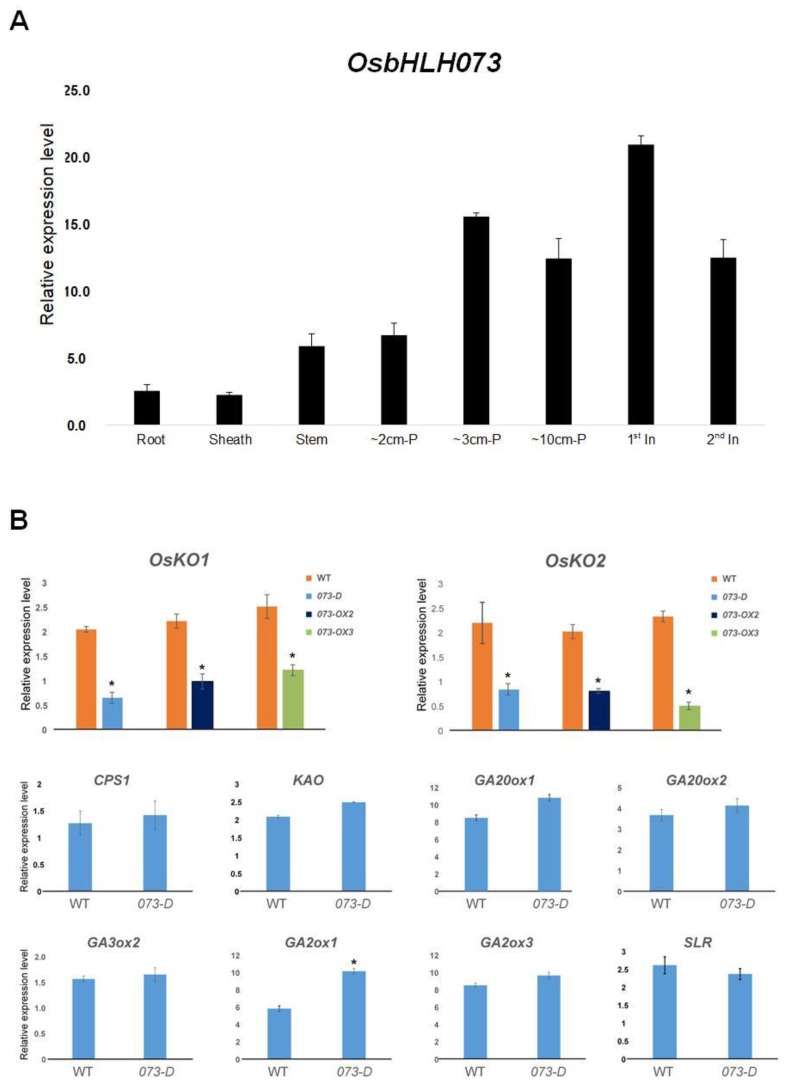
To analyze the biological roles of OsbHLH073, we examined the OsbHLH073 transcripts in various organs using real-time PCR. The various organs included the root, shoot base, and leaf sheath in 14-day-old plants, young panicles, the first four internodes, and mature panicles. At the seedling stage, we found that OsbHLH073 transcripts were more abundantly expressed in the shoot base than in the root or leaf blade. At the reproductive stage, its expression was more abundant in the young panicle and the first internode than in the mature panicle and the second internode (Figure 5A). This observation is consistent with reports of many genes involved in GA biosynthesis in rice. For example, EUI1 transcripts are mainly expressed in the young seedling shoot base and young panicles [2,3], and OsKO1 transcripts are abundant in young panicles and elongated stems [56].
Figure 5.
Expression patterns of OsbHLH073 and GA metabolic genes. (A) Expression patterns of OsbHLH073 in various organs. Fourteen-day-old plants were used for the root, sheath, and stems. Stem: Shoot base (around 2 cm); P: Panicle; In: Internode. (B) Expression pattern of OsKO1, OsKO2, and other GA biosynthetic and signaling pathway genes using mature panicles in osbhlh073-D (073-D) and OsbHLH073-OX (073-OX2, 073-OX3) plants, respectively. Expression levels were normalized to OsActin (LOC_Os03g50885). (* p < 0.05, Student’s t-test).
2.5. osbhlh073-D Alters the Expression of KO1 and KO2
It is well known that genes involved in GA biosynthesis and catabolism are negatively and positively regulated by active GAs [6]. To investigate the functional involvement of OsbHLH073 in the GA biosynthetic or signaling pathways, we monitored the relative expression levels of GA biosynthesis-related genes using mature panicles of osbhlh073-D and OsbHLH073-OX plants. As shown in Figure 5, the OsKO1 and OsKO2 transcripts were decreased in the osbhlh073-D and OsbHLH073-OX lines compared to wild-type (Figure 5B). The expression of gibberellin deactivation enzyme, GA2ox1, was increased in the osbhlh073-D mutant (Figure 5B). However, the relative expression levels of other GA biosynthetic pathway-related genes did not show a significant difference between osbhlh073-D and wild-type plants (Figure 5B). We also monitored the expression patterns of gene in the GA signaling pathway, and found that the expression of a negative regulator of gibberellin signaling, DELLA protein SLR1 was not significantly different from that in wild-type plants (Figure 5B) [56].
Previously, it was shown that an OsKO2-weak allele mutant (d35Tan-Ginbozu) exhibited defective panicle exsertion and the semi-dwarf phenotype, and that knocking out OsKO2 resulted in a more severe dwarf phenotype [1]. The phenotype of d35Tan-Ginbozu is quite similar to that of osbhlh073-D (Figure 1B–E). This result indicates that OsbHLH073 may control the expression of the OsKO1 and OsKO2 genes to regulate internode elongation and GA homeostasis.
2.6. OsbHLH073 is a Nuclear Localized-Transcription Activator
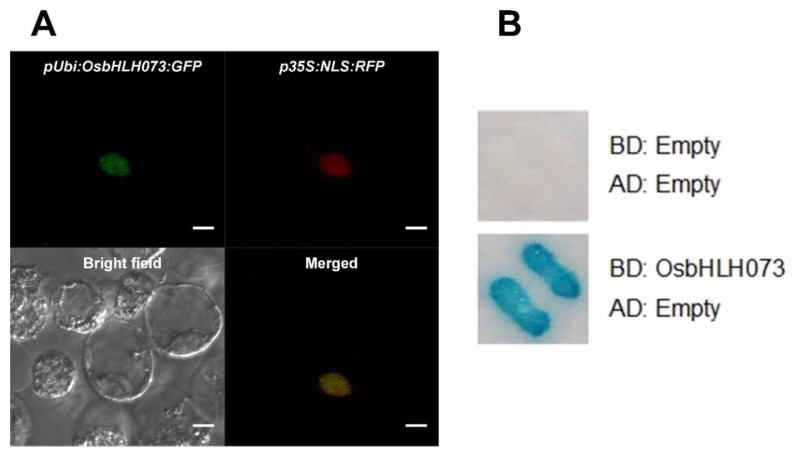
Most of the bHLH proteins are TFs. Some bHLH TFs, such as AtbHLH112 [57] and OsBC1, are activators [41], whereas some others, such as JA-ASSOCIATED MYC2-LIKE2 (JAM2) and JAM3 [58], are repressors. To investigate the function of OsbHLH073, we constructed the OsbHLH073-GFP fusion construct under the maize ubiquitin1 promoter and transfected it into a rice protoplast. To control the localization in the nucleus, 35S::NLS-mRFP vector was transfected into the rice protoplast. As expected, NLS-mRFP was localized in the nucleus and the OsbHLH073-GFP fusion protein was also detected in the nucleus (Figure 6A). We then examined the transcriptional activity of OsbHLH073 using the yeast two-hybrid system [41]. As shown in Figure 6B, OsbHLH073 showed transcriptional activation activity in yeast, even though it was not likely to contain a DNA-binding region.
Figure 6.
Localization of OsbHLH073::GFP protein in the nucleus and transcriptional activation assay. (A) Rice protoplast was co-transfected with 35S::NLS-mRFP (positive control) and pUbi::OsbHLHL073-GFP. Bar = 5 μm. (B) OsbHLH073 displays transcriptional activity in yeast. An empty vector was used as the control. GAL4-DNA binding domain and transcription activation domain for yeast two-hybrid system are presented as BD and AD, respectively. OsbHLH073 was fused to the BD to generate BD:OsbHLH073 fusion protein.
3. Discussion
In this study, the function of OsbHLH073 gene was investigated through gain-of-function approaches by activation tagging and overexpression analysis. We obtained and generated two classes of mutants (osbhlh073-D and OsbHLH073-OX, respectively), both of which displayed a semi-dwarf phenotype at the seedling stage, and poor panicle exsertion due to a defect in the elongation of the first and second internodes at the reproductive stage. Exogenous GA3 application rescued the dwarf phenotype of osbhlh073-D plants at the seedling stage, indicating that OsbHLH073 is involved in the GA biosynthetic pathway. However, there are no reports elucidating the functions of atypical bHLH TFs involved in GA signaling or biosynthesis pathways in rice. In Arabidopsis, PHYTOCHROME-INTERACTING FACTOR 1 (PIF1), a typical bHLH TF, regulates GA responsiveness by directly binding to the promoter regions of GIBBERELLIC-ACID INSENSITIVE (GAI) and REPRESSOR OF THE GAI during seed development or germination [59]. Many bHLH TFs including PIF1 have a conserved DNA-binding site, the E-box (5′-CANNTG-3′), to regulate cell-type-specific and developmental expression. OsbHLH073 belongs to the atypical bHLH TFs without DNA-binding motif, but this bHLH displays transcriptional activity as a nuclear protein. Thus, the function of OsbHLH073 might require the interaction with typical bHLHs like Arabidopsis PIF1.
The expression patterns of genes in the GA biosynthesis or signaling pathways revealed that both OsKO1 and OsKO2 genes were downregulated in the mature panicle of the osbhlh073-D and OsbHLH073-OX lines. From these findings, we propose that, at least in part, the OsbHLH073 gene regulates plant growth and participates in GA homeostasis via regulation of the OsKO1 and OsKO2 genes. However, we did not perform the experiment to test a direct interaction of OsbHLH073 with the OsKO1 and OsKO2 promoter regions because OsbHLH073 belongs to the non-DNA-binding bHLH family, reflecting the existence of potential OsbHLH073-interacting TFs. T-DNA insertion mutants of OsbHLH073 and OsbHLH074, the closest homolog to OsbHLH073, did not show clear visible phenotype, indicating that OsbHLH073 and OsbHLH074 can be functionally redundant. Investigating the phenotype by making a double mutant and observing the variant phenotype may give a better understanding on the function of these genes. Functional redundancy is very general in this gene family [60]. For example, the pif1pif3pif4pif5 quadruple mutant displays a strong pleiotropic phenotype associated with constitutive-photomorphogenesis in the dark [61], and the triple mutant of BR ENHANCED EXPRESSION 1, 2, AND 3 (BEE1, BEE2, and BEE3) shows reduced hypocotyl elongation phenotype in response to simulated shade [44].
Previous studies have shown that the regulation of KO2 expression by directly binding to the KO2 promoter leads to alteration of the GA level and causes the dwarf phenotype. Tobacco transcription factor, REPRESSION OF SHOOT GROWTH, activates the expression of KO2 gene in Arabidopsis [62]. GIBBERELLIN-DEFICIENT DWARF1/BRITTLE CULM 12 directly binds to the promoter of OsKO2 and causes the impaired cell elongation with dwarf phenotype [63]. O. SATIVA LSD-ONE-LIKE 1 (OsLOL1; C2C2-type zinc finger protein) and OsbZIP58 activates OsKO2, thereby inducing seed germination by affecting GA biosynthesis [64]. Rice NAC DOMAIN-CONTAINING PROTEIN 2 (OsNAC2) overexpression reduces plant height and internode length, and OsNAC2 represses the expression of OsKO2 by directly binding to its promoter, as shown in Chip-seq analysis and yeast one-hybrid analysis [65]. OsbZIP48 directly binds to the promoter of OsKO2 and regulates its expression to control internode elongation [66].
To identify how to regulate OsKO1 and OsKO2 gene expression and control the growth and GA homeostasis associated with OsbHLH073, it will be necessary to identify the proteins interacting with OsbHLH073. Previous studies reported that atypical bHLH proteins interact with typical bHLH TFs, which control the downstream genes [33,67,68,69]. For example, the Arabidopsis atypical bHLH TFs PACLOBUTRAZOL RESISTANCE1 and ILI1 BINDING bHLH PROTEIN1 (IBH1) interact with each other and regulate the expression of many genes antagonistically by the interaction of IBH1 with the DNA-binding bHLH factor, HOMOLOG OF BEE2 INTERACTING WITH IBH1 [68]. We manually analyzed the promoter sequences of OsKO1 and OsKO2 and revealed that there were three and nine conserved E-box binding sites within the ~2 kb promoter regions of OsKO1 and OsKO2, respectively (Figures S4 and S5). We hypothesize that typical bHLH TFs, interacting partners of OsbHLH073, may bind to some of these sites to negatively regulate the expression of OsKO1 and OsKO2 in order to modulate GA homeostasis and cell elongation.
4. Materials and Methods
4.1. Plant Growth
T2 seeds of osbhlh073-D, osbhlh073-1, osbhlh074-1 (T-DNA insertional mutant, PFG_4A-02508, PFG_3A-17056, PFG_5A-00405), and wild-type (Oryza sativa cv. Japonica Dongjin) were germinated on half strength (1/2) Murashige and Skoog (MS) medium including 0.4% phytagel and 3% sucrose [70]. Plants were grown in the greenhouse and then transplanted into a field at Kyungpook National University (36o N). PCRs for genotyping were conducted as explained previously [71,72]. The PCR primers are described in Table S1.
4.2. Hormone Treatment Analysis
T2 seeds of osbhlh073-D and wild-type were germinated in 1/2 MS medium in the light condition without or with 1 µM of GA3 added to the medium. For second leaf sheath analysis, seeds of osbhlh073-D and Dongjin wild-type were grown into 1/2 MS containing various concentrations of GA3 under continuous light at 30 °C for seven days.
4.3. Vector Construction and Rice Transformation
The full-length OsbHLH073 cDNA was amplified by PCR using the primer sets listed in Table S1. The OsbHLH073 cDNA was cloned into the binary vector pGA3428 or pGA3438 [73], which contained the maize (Zea mays) ubiquitin (GRMZM2G409726) promoter. To generate the transgenic plants, Agrobacterium tumefaciens LBA4404 harboring the pGA3428 or pGA3438 was used for rice transformation [74].
4.4. RNA Isolation, RT-PCR, and Quantitative RT-PCR Analyses
Total RNA was extracted from various organs (seedling shoot, root, stem, young panicles, and internode) using QIAzol lysis reagent following by the manufacturer’s manual (Qiagen; https://www.qiagen.com). For cDNA synthesis, 2 µg of total RNA, reverse transcriptase (Promega, Madison, WI, USA), 10 ng of the oligo (dT) primers, and 2.5 mM of dNTP were used. Synthesized cDNAs were used as templates for RT-PCR and quantitative RT-PCR (qRT-PCR). SYBR green premix (Enzynomics, Daejeon, KOREA) and the Bio-rad instrument system (Bio-rad, USA) were used for qRT-PCR. Rice actin 1 was used as an internal control. At least three biological replicates used for experiments and data were analyzed by Student’s t-test. The comparative Ct (2−∆∆Ct) method was used to calculate the change of relative gene expression [74]. All primer sets are presented in Table S1 in this study.
4.5. Localization Assay and Transcriptional Activation Assay in Yeast
To investigate the cellular localization, we cloned an OsbHLH073-green fluorescent protein (GFP) fusion vector under the control of the maize ubiquitin promoter using pGA3452 [73]. The nuclear localization signal-tagged monomeric red fluorescent protein (NLS-mRFP) vector under the control of the cauliflower mosaic virus (CaMV) 35S promoter was used as marker to control localization in the nucleus. The OsbHLH073-GFP vector and the nuclear marker were transfected into the rice Oc suspension protoplasts using an electroporation method [75]. Protoplasts from Oc suspension cells were isolated as described previously [76,77]. Expression of the OsbHLH073-GFP fusion protein was monitored using a fluorescence microscope (Zeiss, Germany). To test the transcriptional activation assay, the OsbHLH073 full-length ORF (Open Reading Frame) was cloned in-frame in the pBD-GAL4 Cam vector (Stratagene, La Jolla, CA, USA) to generate a BD:OsBHLH073 construct and the pAD-GAL4 vector (Stratagene, La Jolla, CA, USA) was used for AD:empty. An X-gal filter assay was conducted as explained previously [41].
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/4/547/s1, Figure S1; Multiple sequence alignment of bHLH transcription factors, Figure S2; Phylogenetic tree of atypical and typical bHLH transcription factors, Figure S3; Multiple sequence alignment of the OsbHLH072, OsbHLH073, and OsbHLH074 proteins, Figure S4; Sequence of the OsKO1 promoter region from the start codon, Figure S5; Sequence of the OsKO2 promoter region from the start codon, Table S1: Primers used in this study.
Author Contributions
J.L., S.M., S.J., K.-H.J., and S.K.P. conceived and designed the experiments. J.L., S.M., and S.J. performed the experiments. S.L. and G.A. provided the resources and the discussion. J.L., S.M., and S.J. wrote the original draft. J.L., S.J., K.-H.J. and S.K.P. revised the manuscript. All authors commented on the article. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the Next-Generation BioGreen 21 program (Project No. PJ01369001 to S.K.P.), Rural Development Administration, Republic of Korea.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Itoh H., Tatsumi T., Sakamoto T., Otomo K., Toyomasu T., Kitano H., Ashikari M., Ichihara S., Matsuoka M. A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 2004;54:533–547. doi: 10.1023/B:PLAN.0000038261.21060.47. [DOI] [PubMed] [Google Scholar]
- 2.Luo A., Qian Q., Yin H., Liu X., Yin C., Lan Y., Tang J., Tang Z., Cao S., Wang X., et al. EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 2006;47:181–191. doi: 10.1093/pcp/pci233. [DOI] [PubMed] [Google Scholar]
- 3.Zhu Y., Nomura T., Xu Y., Zhang Y., Peng Y., Mao B., Hanada A., Zhou H., Wang R., Li P., et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell. 2006;18:442–456. doi: 10.1105/tpc.105.038455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao X., Zhang Y., He Z., Fu X. Hormone metabolism and signaling in plants. In: Li J., Li C., Smith S.M., editors. Hormone Metabolism and Signaling in Plants. 1st ed. Academic Press; Cambridge, MA, USA: 2017. pp. 406–429. [DOI] [Google Scholar]
- 5.Hedden P., Phillips A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/S1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 6.Olszewski N., Sun T.P., Gubler F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell. 2002;14(Suppl. 1):S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aach H., Bode H., Robinson D.G., Graebe J.E. ent-kaurene synthase is located in proplastids of meristematic shoot tissues. Planta. 1997;202:211–219. doi: 10.1007/s004250050121. [DOI] [Google Scholar]
- 8.Helliwell C.A., Chandler P.M., Poole A., Dennis E.S., Peacock W.J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA. 2001;98:2065–2070. doi: 10.1073/pnas.98.4.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helliwell C.A., Sheldon C.C., Olive M.R., Walker A.R., Zeevaart J.A., Peacock W.J., Dennis E.S. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA. 1998;95:9019–9024. doi: 10.1073/pnas.95.15.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helliwell C.A., Poole A., Peacock W.J., Dennis E.S. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999;119:507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrone D., Chen X., Coates R.M., Peters R.J. Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem. J. 2010;431:337–344. doi: 10.1042/BJ20100597. [DOI] [PubMed] [Google Scholar]
- 12.Magome H., Nomura T., Hanada A., Takeda-Kamiya N., Ohnishi T., Shinma Y., Katsumata T., Kawaide H., Kamiya Y., Yamaguchi S. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. USA. 2013;110:1947–1952. doi: 10.1073/pnas.1215788110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh H., Ueguchi-Tanaka M., Sentoku N., Kitano H., Matsuoka M., Kobayashi M. Cloning and functional analysis of two gibberellin 3 beta -hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA. 2001;98:8909–8914. doi: 10.1073/pnas.141239398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., Ishiyama K., Saito T., Kobayashi M., Khush G.S., et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto T., Kobayashi M., Itoh H., Tagiri A., Kayano T., Tanaka H., Iwahori S., Matsuoka M. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 2001;125:1508–1516. doi: 10.1104/pp.125.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto T., Miura K., Itoh H., Tatsumi T., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Agrawal G.K., Takeda S., Abe K., et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.H., An G., Kitano H., Ashikari M., et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- 18.Lo S.F., Yang S.Y., Chen K.T., Hsing Y.I., Zeevaart J.A., Chen L.J., Yu S.M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20:2603–2618. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan C., Mei Z., Duan J., Chen H., Feng H., Cai W. OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS ONE. 2014;9:e87110. doi: 10.1371/journal.pone.0087110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng J., Richards D.E., Hartley N.M., Murphy G.P., Devos K.M., Flintham J.E., Beales J., Fish L.J., Worland A.J., Pelica F., et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- 21.Spielmeyer W., Ellis M.H., Chandler P.M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schomburg F.M., Bizzell C.M., Lee D.J., Zeevaart J.A., Amasino R.M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15:151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee D.J., Zeevaart J.A.D. Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 2005;138:243–254. doi: 10.1104/pp.104.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J., Tang D., Shen Y., Qin B., Hong L., You A., Li M., Wang X., Yu H., Gu M., et al. Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.) J. Genet. Genom. 2010;37:23–36. doi: 10.1016/S1673-8527(09)60022-9. [DOI] [PubMed] [Google Scholar]
- 25.Schrager-Lavelle A., Gath N.N., Devisetty U.K., Carrera E., Lopez-Diaz I., Blazquez M.A., Maloof J.N. The role of a class III gibberellin 2-oxidase in tomato internode elongation. Plant J. 2019;97:603–615. doi: 10.1111/tpj.14145. [DOI] [PubMed] [Google Scholar]
- 26.Sun X., Shu J., Ali Mohamed A.M., Deng X., Zhi X., Bai J., Cui Y., Lu X., Du Y., Wang X., et al. Identification and Characterization of EI (Elongated Internode) Gene in Tomato (Solanum lycopersicum) Int. J. Mol. Sci. 2019;20:2204. doi: 10.3390/ijms20092204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Zhu Y., Peng Y., Yan D., Li Q., Wang J., Wang L., He Z. Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice. Cell Res. 2008;18:412–421. doi: 10.1038/cr.2008.28. [DOI] [PubMed] [Google Scholar]
- 28.Gao S., Fang J., Xu F., Wang W., Chu C. Rice HOX12 Regulates Panicle Exsertion by Directly Modulating the Expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell. 2016;28:680–695. doi: 10.1105/tpc.15.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez-Ariza J., Brambilla V., Vicentini G., Landini M., Cerise M., Carrera E., Shrestha R., Chiozzotto R., Galbiati F., Caporali E., et al. A transcription factor coordinating internode elongation and photoperiodic signals in rice. Nat. Plants. 2019;5:358–362. doi: 10.1038/s41477-019-0401-4. [DOI] [PubMed] [Google Scholar]
- 30.Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martinez-Garcia J.F., Bilbao-Castro J.R., Robertson D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atchley W.R., Fitch W.M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Duan X., Jiang H., Sun Y., Tang Y., Yuan Z., Guo J., Liang W., Chen L., Yin J., et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung K.H., Han M.J., Lee Y.S., Kim Y.W., Hwang I., Kim M.J., Kim Y.K., Nahm B.H., An G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 2005;17:2705–2722. doi: 10.1105/tpc.105.034090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todaka D., Nakashima K., Maruyama K., Kidokoro S., Osakabe Y., Ito Y., Matsukura S., Fujita Y., Yoshiwara K., Ohme-Takagi M., et al. Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc. Natl. Acad. Sci. USA. 2012;109:15947–15952. doi: 10.1073/pnas.1207324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heang D., Sassa H. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS ONE. 2012;7:e31325. doi: 10.1371/journal.pone.0031325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heang D., Sassa H. An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed. Sci. 2012;62:133–141. doi: 10.1270/jsbbs.62.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X., Ren Y., Cai Y., Niu M., Feng Z., Jing R., Mou C., Liu X., Xiao L., Zhang X., et al. Overexpression of OsbHLH107, a member of the basic helix-loop-helix transcription factor family, enhances grain size in rice (Oryza sativa L.) Rice. 2018;11:41. doi: 10.1186/s12284-018-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi T., Ozu A., Kobayashi S., An G., Jeon J.S., Nishizawa N.K. OsbHLH058 and OsbHLH059 transcription factors positively regulate iron deficiency responses in rice. Plant Mol. Biol. 2019;101:471–486. doi: 10.1007/s11103-019-00917-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L.Y., Bai M.Y., Wu J., Zhu J.Y., Wang H., Zhang Z., Wang W., Sun Y., Zhao J., Sun X., et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang S., An G., Li H.Y. Rice Leaf Angle and Grain Size Are Affected by the OsBUL1 Transcriptional Activator Complex. Plant Physiol. 2017;173:688–702. doi: 10.1104/pp.16.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., Kyozuka J. LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA. 2003;100:11765–11770. doi: 10.1073/pnas.1932414100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H.C., Cheng W.H., Hong C.Y., Chang Y.S., Chang M.C. The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively. Rice. 2018;11:50. doi: 10.1186/s12284-018-0244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedrichsen D.M., Nemhauser J., Muramitsu T., Maloof J.N., Alonso J., Ecker J.R., Furuya M., Chory J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S., Lee S., Yang K.Y., Kim Y.M., Park S.Y., Kim S.Y., Soh M.S. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- 46.Komatsu M., Maekawa M., Shimamoto K., Kyozuka J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 2001;231:364–373. doi: 10.1006/dbio.2000.9988. [DOI] [PubMed] [Google Scholar]
- 47.Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., Kim C., Jang S., Yang K., Nam J., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- 48.Jeong D.H., An S., Kang H.G., Moon S., Han J.J., Park S., Lee H.S., An K., An G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002;130:1636–1644. doi: 10.1104/pp.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong D.H., An S., Park S., Kang H.G., Park G.G., Kim S.R., Sim J., Kim Y.O., Kim M.K., Kim S.R., et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- 50.Lee S., Jeon J.S., Jung K.H., An G. Binary vectors for efficient transformation of rice. J. Plant Biol. 1999;42:310–316. doi: 10.1007/BF03030346. [DOI] [Google Scholar]
- 51.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tong H., Jin Y., Liu W., Li F., Fang J., Yin Y., Qian Q., Zhu L., Chu C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009;58:803–816. doi: 10.1111/j.1365-313X.2009.03825.x. [DOI] [PubMed] [Google Scholar]
- 53.Ji H., Han C.D., Lee G.S., Jung K.H., Kang D.Y., Oh J., Oh H., Cheon K.S., Kim S.L., Choi I., et al. Mutations in the microRNA172 binding site of SUPERNUMERARY BRACT (SNB) suppress internode elongation in rice. Rice. 2019;12:62. doi: 10.1186/s12284-019-0324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsukura C., Itoh S., Nemoto K., Tanimoto E., Yamaguchi J. Promotion of leaf sheath growth by gibberellic acid in a dwarf mutant of rice. Planta. 1998;205:145–152. doi: 10.1007/s004250050306. [DOI] [Google Scholar]
- 55.Cho S.H., Kang K., Lee S.H., Lee I.J., Paek N.C. OsWOX3A is involved in negative feedback regulation of the gibberellic acid biosynthetic pathway in rice (Oryza sativa) J. Exp. Bot. 2016;67:1677–1687. doi: 10.1093/jxb/erv559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y., Ji X., Nie X., Qu M., Zheng L., Tan Z., Zhao H., Huo L., Liu S., Zhang B., et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 2015;207:692–709. doi: 10.1111/nph.13387. [DOI] [PubMed] [Google Scholar]
- 58.Nakata M., Ohme-Takagi M. Two bHLH-type transcription factors, JA-ASSOCIATED MYC2-LIKE2 and JAM3, are transcriptional repressors and affect male fertility. Plant Signal. Behav. 2013;8:e26473. doi: 10.4161/psb.26473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19:1192–1208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009;5:e1000440. doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fukazawa J., Sakai T., Ishida S., Yamaguchi I., Kamiya Y., Takahashi Y. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell. 2000;12:901–915. doi: 10.1105/tpc.12.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J., Jiang J., Qian Q., Xu Y., Zhang C., Xiao J., Du C., Luo W., Zou G., Chen M., et al. Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell. 2011;23:628–640. doi: 10.1105/tpc.110.081901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu J., Zhu C., Pang J., Zhang X., Yang C., Xia G., Tian Y., He C. OsLOL1, a C2C2-type zinc finger protein, interacts with OsbZIP58 to promote seed germination through the modulation of gibberellin biosynthesis in Oryza sativa. Plant J. 2014;80:1118–1130. doi: 10.1111/tpj.12714. [DOI] [PubMed] [Google Scholar]
- 65.Chen X., Lu S., Wang Y., Zhang X., Lv B., Luo L., Xi D., Shen J., Ma H., Ming F. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J. 2015;82:302–314. doi: 10.1111/tpj.12819. [DOI] [PubMed] [Google Scholar]
- 66.Burman N., Bhatnagar A., Khurana J.P. OsbZIP48, a HY5 Transcription Factor Ortholog, Exerts Pleiotropic Effects in Light-Regulated Development. Plant Physiol. 2018;176:1262–1285. doi: 10.1104/pp.17.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bai M.Y., Fan M., Oh E., Wang Z.Y. A Triple Helix-Loop-Helix/Basic Helix-Loop-Helix Cascade Controls Cell Elongation Downstream of Multiple Hormonal and Environmental Signaling Pathways in Arabidopsis. Plant Cell. 2012;24:4917–4929. doi: 10.1105/tpc.112.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Capella M., Ribone P.A., Arce A.L., Chan R.L. Arabidopsis thaliana HomeoBox 1 (AtHB1), a Homedomain-Leucine Zipper I (HD-Zip I) transcription factor, is regulated by PHYTOCHROME-INTERACTING FACTOR 1 to promote hypocotyl elongation. New Phytol. 2015;207:669–682. doi: 10.1111/nph.13401. [DOI] [PubMed] [Google Scholar]
- 70.Lee Y.S., Yi J., Jung K.H., An G. Comparison of rice flowering-time genes under paddy conditions. J. Plant Biol. 2016;59:238–246. doi: 10.1007/s12374-016-0029-0. [DOI] [Google Scholar]
- 71.Kim S.R., Jeon J.S., An G. Development of an efficient inverse PCR method for isolating gene tags from T-DNA insertional mutants in rice. Methods Mol. Biol. 2011;678:139–146. doi: 10.1007/978-1-60761-682-5_11. [DOI] [PubMed] [Google Scholar]
- 72.Lee J., Jang S., Ryu S., Lee S., Park J., Lee S., An G., Park S.K. Impaired Plastid Ribosomal Protein L3 Causes Albino Seedling Lethal Phenotype in Rice. J. Plant Biol. 2019;62:419–428. doi: 10.1007/s12374-019-0380-z. [DOI] [Google Scholar]
- 73.Kim S.R., Lee D.Y., Yang J.I., Moon S., An G. Cloning Vectors for Rice. J. Plant Biol. 2009;52:73–78. doi: 10.1007/s12374-008-9008-4. [DOI] [Google Scholar]
- 74.Lee J., Jang S., Ryu S., Lee S., Park J., Lee S., An G., Park S.K. Mutation of plastid ribosomal protein L13 results in an albino seedling-lethal phenotype in rice. Plant Breed. Biotechnol. 2019;7:395–404. doi: 10.9787/PBB.2019.7.4.395. [DOI] [Google Scholar]
- 75.Cho L.H., Yoon J., Pasriga R., An G. Homodimerization of Ehd1 Is Required to Induce Flowering in Rice. Plant Physiol. 2016;170:2159–2171. doi: 10.1104/pp.15.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee J., Park J.J., Kim S.L., Yim J., An G. Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 2007;65:487–499. doi: 10.1007/s11103-007-9196-1. [DOI] [PubMed] [Google Scholar]
- 77.Choi S.C., Lee S., Kim S.R., Lee Y.S., Liu C., Cao X., An G. Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol. 2014;164:1326–1337. doi: 10.1104/pp.113.228049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.