Abstract
Mass spectrometry has revolutionized the ability to study posttranslationally modified proteoforms from biologic samples, yet we still lack methods to systematically predict, or even prioritize, which modification sites may perturb protein function. Here we describe a proteomic method to detect the effects of site-specific protein phosphorylation on the thermal stability of thousands of native proteins in live cells. This massively parallel biophysical assay unveiled shifts in overall protein stability in response to site-specific phosphorylation sites, as well as trends related to protein function and structure. This method can detect both intrinsic changes to protein structure as well as extrinsic changes to protein-protein, and protein-metabolite interactions resulting from the diminutive introduction of a phosphate onto large proteins. Finally, we show that functional “hotspot” protein modification sites can be discovered and prioritized for study in a high-throughput and unbiased fashion. This approach is applicable to diverse organisms, cell types and posttranslational modifications.
Introduction
The posttranslational modification (PTM) of protein backbone and side-chains affects the function of target proteins by perturbing the three-dimensional structure of the protein itself, as well as by altering intermolecular interactions with other biomolecules. PTMs regulate protein-protein interactions1, sub-cellular compartmentalization2, substrate and co-factor interactions, protein turnover3, and other functions to collectively impact normal4 and dysregulated5-7 signal transduction in cells. The quantitative interrogation of endogenous PTMs has been greatly accelerated by methods such as liquid chromatography-mass spectrometry (LC-MS)8. Subsequent integration of front-end, PTM-specific enrichment protocols with data-dependent and de-novo PTM discovery algorithms has resulted in the discovery of tens-of-thousands of unique PTM sites on proteins from diverse organisms9-11, as well as quantification of PTM dynamics in diverse biological contexts and responses to stimuli12-14. Central questions around how minute changes to chemical structure alters the biophysical properties of a much larger protein, and how these changes lead to signaling and phenotypic consequences remain only partially explored. This dearth of mechanistic information on PTMs exists because we lack general methods with which to predict, or even prioritize, which modification sites are likely to be functional in the proteome, and therefore the one-by-one study of PTMs is intrinsically inefficient and low-throughput. Here we sought to develop a proteomic method that enables direct interrogation of protein stability in response to endogenous PTMs for thousands of proteins in parallel. This method, Hotspot Thermal Profiling (HTP), couples PTM-specific enrichments and isotopic labeling with global profiling by LC-MS/MS to measure the thermal stability of endogenous modified proteoforms, which we refer to herein as “modiforms,” in live cells (Fig. 1). We apply this approach to a ubiquitous yet diminutive PTM – phosphorylation – and demonstrate that HTP can detect both inter- and intramolecular interactions mediated by site-specific PTMs on a wide range of proteins. Moreover, we demonstrate that perturbation of protein stability can be used as a means to discover functionally important, previously uncharacterized “hotspot” modification sites. The resulting dataset represents a rich and generalizable resource to prioritize existing datasets of phosphorylation sites in diverse biological contexts.
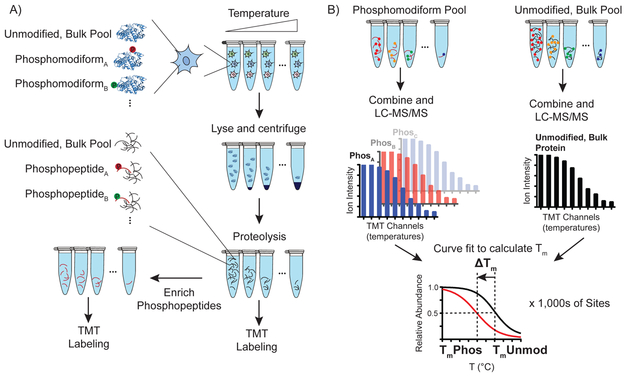
Fig. 1. Hotspot thermal profiling workflow.
A) Aliquots of live cells are exposed to short, varied temperature challenge, lysed, and soluble/insoluble proteins separated by ultracentrifugation. Downstream trypsinization, and separation of aliquots for bulk proteome (5%), and phosphopeptide enrichment (95%) differentiates bulk, unmodified and phosphomodiform interrogation. B) Bulk and enriched peptide aliquots are labeled with unique tandem-mass tag channels, pooled and analyzed by LC-MS/MS. The output of each mass spectrometry run is searched using an integrated platform for peptide identification and quantification, Tm curve fitting, and integration for bulk, unmodified protein- or modification site-level Tm determination. Unique phosphomodiforms and their corresponding phosphopeptides are abbreviated as PhosA, PhosB…Phosx.
RESULTS
Hotspot thermal profiling detects changes in protein stability in response to site-specific modification.
The HTP workflow begins with pulsed exposure of live cell aliquots to temperatures ranging from 37-67 °C for 3 minutes, cooling to room temperature, rapid cell lysis, and centrifugation to remove unfolded proteins, in line with reports mapping ligand-protein15-18 and protein-protein19 interactions by thermal proteome profiling (Fig. 1a). The soluble proteome of each aliquot is then proteolyzed, and 5% of each peptide aliquot is labeled with specific tandem mass tag (TMT) isotope channels and reserved for quantification of the “unmodified, bulk” protein pool (Fig. 1a). The remaining 95% of each aliquot is used for phosphopeptide enrichment by TiO2 resin, followed by TMT isotope labeling (Fig. 1b). All aliquots of the unenriched and phosphorylated TMT tryptic peptides are subsequently combined and used for LC-MS/MS detection and quantitation of relative peptide levels across the temperature range. Peptide-level measurements are analyzed en masse with identification, quantification and automated curve fitting algorithms to generate peptide-specific melting points (Tm) for tens-of-thousands of unique peptides per sample (Fig. 1a). Because quantitative proteomic measurements are performed on individual peptides, measurements made on a unique modified peptide (e.g. phosphorylated peptide) serve as a “bar-code” to report on any protein that contains a modification at that site to the thermal challenge in live cells. Therefore, this method does not report on single “proteoforms” as recently defined20, but instead reports on an inclusive set of the protein pool that are unambiguously phosphorylated at the detected site, which we define as the “phosphomodiform.” This is a key differentiation from measuring the effect of a modification on the bulk protein pool, such that HTP can detect altered thermal stability even if a modiform is only a minor member of all the modified proteoforms present in the cell (Fig. 1b).
We applied the HTP workflow to detect the Tm values of phosphomodiforms and the bulk, unmodified protein pool in HEK293T cells. We detected >250,000 TMT-labeled peptides from thousands of proteins across a handful of LC-MS/MS runs (Supplementary Fig. 1), and statistical filtering through identification, quantification and curve fitting were combined to generate 4,293 “unmodified, bulk” protein Tm values (Fig. 2a, b). Here we define the unmodified, bulk as all unmodified peptides that map to a given protein of interest, such that this Tm reflects the “reference” population of protein present in cells. Protein and peptide Tm measurements were highly stable across replicates, validating the reproducibility of cellular and proteome-level manipulations in the HTP workflow, and indicating that even small shifts in thermal stability (e.g. ~1.5 °C) can be reliably detected with statistical significance (Supplementary Fig. 2a-f).
Fig. 2. HTP identifies thousands of unmodified, bulk and phosphomodiform Tm’s in HEK293T cells.
A) The total number of unique phosphomodiforms detected in our dataset and criteria for identifying high-quality ΔTm values. FLS FDR: localization site false discovery rate. B) Global distribution of unmodified, bulk protein-level, and phosphomodiform Tm values. C) Volcano plot of high-confidence ΔTm values. Phosphomodiforms with significant shifts (P < 0.05, two-sided t-test for within-protein comparisons) are shown in red (negative shift) and blue (positive shift). D-G) Representative Tm curves from phosphomodiforms that do not affect (D), decrease (E), and increase (F), thermal stability. Distinct phosphomodiforms of the same protein exhibit unique ΔTm values, as exemplified in TPI1 (G). Detected phosphorylation sites in TPI1 are shown as spheres; PDB accession: 1WYI. Tm curves and error bars in (D-G) correspond to mean and S.E.M from n = 12, 10, 6, 4, 12, 2, 12, 10, and 4 replicate measurements (in order of Tm curve appearance, top to bottom, left to right). All data were generated from n = 12 (bulk, unmodified) and n = 10 (phospho) total MS technical replicates from n = 6 (bulk, unmodified) and 5 (phospho) independent biological replicates. ΔTm P-values computed with two-sided t-test.
To measure phosphomodiform-specific Tm values, we integrated metal-dependent phosphopeptide enrichment with subsequent TMT-barcoding (Fig. 1). Global phosphorylation was not grossly affected by the short exposure to varied temperature (Supplementary Fig. 3). Duplicate technical replicates from five biological replicates (n = 10 LC-MS/MS runs), as well as grouping phosphopeptides that map to a common site (Supplementary Fig. 4), resulted in the identification of 10,800 phosphomodiforms - again defined as unique proteoforms that contain a defining modification site. Automated Tm curve fitting cutoffs (R2 > 0.8) resulted in a database of 9,695 distinct phosphomodiforms. Finally, filtering of only high-confidence Tm values detected in both phospho- and unmodified, bulk proteomes, as well as stringent modification site localization validation21, yielded 2,883 high-quality ΔTm values that directly compare a specific phosphomodiform to its unmodified, bulk protein (Fig. 2a, b; Supplementary Table 1).
The unmodified and phosphorylated proteomes were globally indistinguishable, with mean Tm values of 51.2 and 51.0°C, respectively (Fig. 2b). By contrast, comparison of a given protein to its specific phosphomodiforms yielded a diverse ΔTm profile that was gaussian in nature, and highlighted the fact that the most phosphorylated modiforms did not exhibit altered thermal stability relative to their parent proteins (Fig. 2c; Supplementary Table 2). There were a sizeable number of phosphosites, however, associated with markedly different thermal stability compared to their unmodified, bulk protein pool in live cells (Fig. 2c). A detailed analysis of specific temperature-response curves verified these trends, with many parent protein-phosphomodiform pairs producing sigmoidal curves that overlay almost exactly (Fig. 2d). In contrast, numerous phosphomodiforms exhibited significantly different temperature-response curves compared to their unmodified, bulk protein counterpart, including pY15 on cyclin-dependent kinase CDK1, and pT926 on kinesin-like protein KIF11 (Fig. 2e-f). Both of these sites have critical roles in regulating the function of their target protein: pY15 has been shown to inactivate CDK1 through structural reorganization and autoinhibition22,23, while pT926 controls intermolecular association of KIF11 with the spindle apparatus during mitosis24. The phosphosite-specific resolution of HTP contrasts with previous reports that have detected shifts in bulk protein Tm values in response to phosphorylation as indirect but related targets of drug action18,19,25. Instead, the direct interrogation of phosphomodiforms with amino-acid resolution validates the ability to distinguish functional effects at different modification sites, as well as minor phosphomodiforms that might otherwise be masked by more abundant proteoforms. For example, the pS58 phosphomodiform of the enzyme TPI1 is associated with significant destabilization, while pT210 has no effect on its stability (Fig. 2g). These data confirmed that modification-dependent changes in protein stability can be detected by thermal profiling of the modified proteome. Most importantly, we hypothesized that “hotspot” modification sites identified in this screen – those significantly shifted relative to the unmodified, bulk and even other phosphomodiforms of the same protein – may represent sites with disproportionate effects on protein structure and function under specific cellular conditions.
Perturbation of protein stability by phosphorylation is site-specific, not systematic
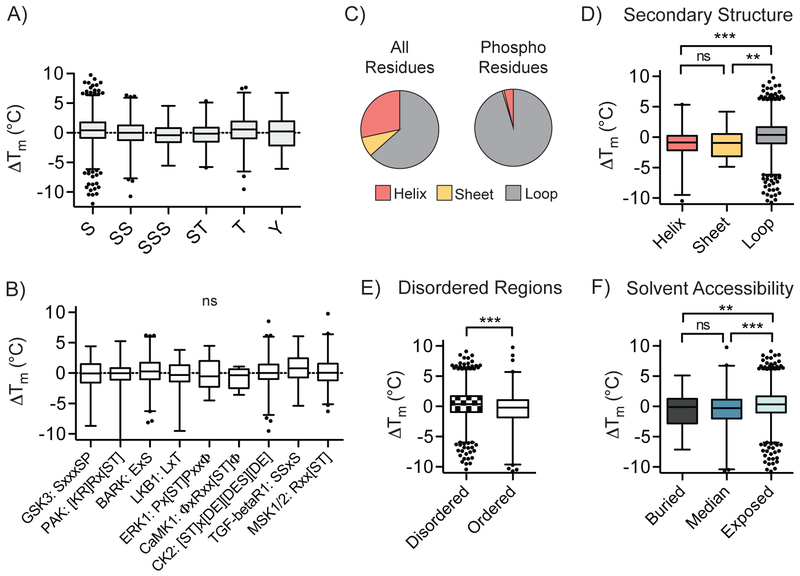
We next asked whether there were any systematic correlations between the types and contexts of protein phosphorylation sites and their effect on protein stability. Comparison of the ΔTm values for phosphorylation on different residues (serine, threonine and tyrosine) across all detected phosphomodiforms did not reveal any general bias for stabilization or destabilization (Fig. 3a). Classes of modiforms harboring multiple phosphorylation sites within the same peptide, and therefore in high proximity on the protein, were also not significantly associated with increased or decreased stability. Global ΔTm values among phosphorylation sites in established kinase motifs also did not reveal significant correlations, indicating that the combination of phosphoamino acid and directly adjacent sequence context does not, at least on a global average in this dataset, impart a systematic stabilizing or destabilizing effect (Fig. 3a, b). As expected, detected phosphosites in our database showed enrichment in predicted loop or coil secondary structural elements (Fig. 3c)26-28. Relative to the majority of sites in coil/loop regions, phosphosites within predicted helical or β-sheet secondary structures demonstrated decreased stability, on average (Fig. 3d). Furthermore, a significant stabilizing trend was observed for phosphosites located in exposed protein regions relative to those located in buried regions, as determined by computational predictions of solvent accessibility and local structure organization (Fig. 3e, f). These global trends imply that the effect of protein phosphorylation – insofar as can be gleaned with this method - is governed by local context within proteins, as opposed to a systemic, biophysical “code” imparted by the phosphoamino acid or linear motif.
Fig. 3. Global relationships between local phosphosite environment and altered phosphomodiform stability.
A) Distribution of ΔTm values for tryptic peptides containing indicated phosphoamino acids or coincidental combinations thereof (P < 0.0001, one-way ANOVA). B) Correlation between detected kinase substrate motifs and ΔTm values for identified sites (P = 0.3011, one-way ANOVA). C) Distribution of all amino acids in the human proteome (left) and detected phosphosites (right) in predicted secondary structural elements. D-F) Comparisons of ΔTm values and predicted secondary structure elements (D; ***, P ≤ 0.0001, two-sided t-test), ordered structural elements surrounding the phosphosite of interest (E; ***, P ≤ 0.0001, two-sided t-test), and solvent accessibility (F; ***, P ≤ 0.0001, two-sided t-test). Box plots (median, 1-99%) are shown in (A-B, D-F), with outliers shown as data points. All data were generated from n = 12 (bulk, unmodified) and n = 10 (phospho) total MS technical replicates from n = 6 (bulk, unmodified) and n = 5 (phospho) independent biological replicates.
Hotspot thermal profiling detects phosphorylation-mediated protein-protein interactions
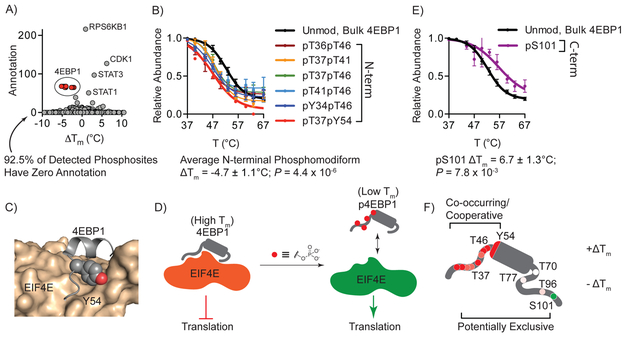
We next asked whether significantly perturbed “hotspot” ΔTm values were correlated with altered intra- or intermolecular interactions in cells, or both. Global correlation of phosphorylation site ΔTm and the number of functional annotations present in the Phosphosite online database highlighted that most sites (92.5%) detected in this dataset had no annotation, as is true for the vast majority of known posttranslational modification sites in the human proteome29. Well-characterized, functional sites in proteins like CDK1, STAT3 and STAT1 stood out on this annotation plot (Fig. 4a). Additionally, a cluster of highly perturbed and well-annotated sites in the 4-enhancer binding protein-1 (4EBP1) were conspicuous. The unmodified, bulk 4EBP1 pool exhibited a Tm of 53.8 ± 0.8°C by HTP (Fig. 4b), whereas the phosphomodiforms containing phosphorylation sites in the N-terminal region were significantly less stable (Fig. 4b). These sites were clustered in a region known to be responsible for direct binding to eukaryotic translation initiation factor 4E (EIF4E), which prevents ribosome binding and translation30,31. A phosphorylation cascade that requires T37, T46 and intervening phosphorylation sites as initiating modifications lead to phosphorylation of Y54, which blocks 4EBP1 binding to EIF4E32. The thermal shifts resulting from phosphorylation around this site are consistent with steric and electronic perturbation of the peptide surrounding Y54 binding the hydrophobic ledge in EIF4E (Fig. 4c), which further suggests that the shift in thermal stability observed for phosphorylated 4EBP1 modiforms may be measuring the bound and unbound forms of this protein on a macromolecular complex, respectively. In this model, 4EBP1 proteins lacking N-terminal phosphorylation demonstrate higher thermal stability due to involvement in the protein-protein interaction with EIF4E and likely other members of a macromolecular complex. This is contrasted by significantly reduced thermal stability of N-terminal phosphorylated modiforms that are not engaged in this interaction, and are either free in solution or engaged in an alternate set of interactions (Fig. 4d). While the data provided by HTP global analysis cannot distinguish these scenarios per se, this method is able to detect these previously annotated functional “hotspot” sites in a global, unbiased manner, and indicate that HTP can detect the consequences of altered intermolecular interactions, which would be impossible to identify in purely in vitro systems. Extending this analysis of 4EBP1, HTP profiling also detected distinct phosphomodiforms, including 4EBP1 phosphorylated at S101, which is associated with increased stability (Fig. 4e). The ability to detect unique phosphomodiforms from the same protein may therefore provide information on the co-occurrence and potential cooperation of modifications, as well as their mutual exclusivity, on the same protein molecules (Fig. 4f).
Fig. 4. HTP detects the known 4EBP1-EIF4E protein-protein interaction mediated by phosphorylation.
A) Plot of phosphosite ΔTm and number of annotations in Phosphosite online database. A cluster of 4EBP1 phosphosites are highlighted in red. B) Representative subset of HTP curves for the unmodified, bulk and N-terminal phosphomodiforms of 4EBP1. C) X-ray structure of the N-terminal helix-turn motif of 4EBP1 (grey) bound to EIF4E (tan); Y54 in 4EBP1is shown as spheres. D) Proposed model correlating observed thermal stability shifts to 4EBP1/EIF4E complexes. E) HTP curve for C-terminal pS101 phosphomodiform. F) Schematic showing the location and relative ΔTm values for all detected phosphomodiforms of 4EBP1, with annotation of potentially co-occurring and exclusive modification patterns based on thermal response. Data points shown in (B, E) represent the mean and S.E.M. from n = 12, 6, 9, 6, 3, 6, 1, 12 and 2 (in order of Tm curve appearance) replicate measurements detected in n = 6 (bulk, unmodified) and n = 5 (phospho) independent biological replicates. ΔTm P-values were calculated with two-sided t-tests.
HTP enables discovery of unannotated, functional phosphorylation sites in the proteome
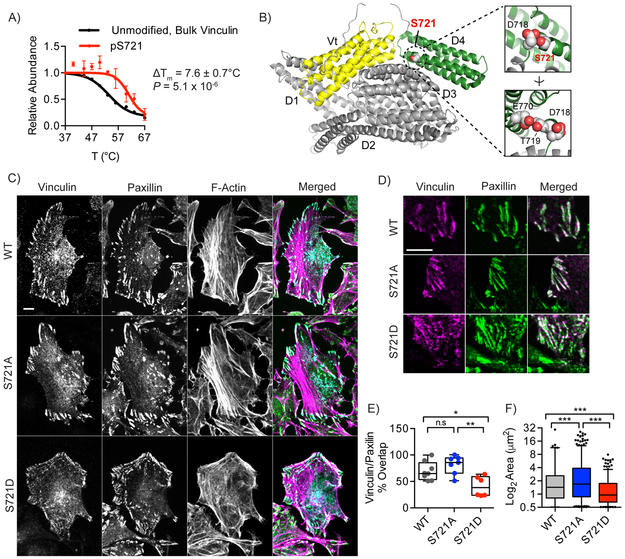
An underlying hypothesis in this study was that the detection of significant modification-site-dependent changes in thermal stability would permit discovery of functional “hotspot” sites that would be otherwise unannotated in the sea of detected sites. We therefore investigated two previously reported yet uncharacterized phosphorylation sites associated with significant ΔTm shifts for functional effects on protein activity. The cytoskeletal protein vinculin exhibited an unmodified, bulk protein Tm that deviated significantly from its pS721 phosphomodiform (Fig. 5a). Published X-ray structures revealed that S721 lies at the N-terminus of an α-helix in the D4 domain between the “head” and “tail” lobes of the protein (Fig. 5b), which regulates conformation switching33 and subsequent vinculin clustering, binding to F-actin stress fibers and recruitment to focal adhesions34.Given the proximity of S721 to contacts within the D4 domain, we hypothesized that the altered stability in response to phosphorylation may reflect altered intramolecular contacts or a functional consequence thereof. We therefore interrogated wild-type, non-phosphorylatable (S721A) and phosphomimetic (S721D) vinculin constructs for the formation of characteristic focal adhesions, the hallmark phenotype for proper function of this protein35. Immunofluorescent visualization of FLAG-tagged, wild-type vinculin in HeLa cells showed high localization to distal membrane regions, which overlapped with F-actin and a marker of focal adhesions, paxillin (Fig. 5c-f). Non-phosphorylatable S721A vinculin exhibited a similar overall distribution, with the majority of this mutant co-localizing to focal adhesions of similar size and number per cell (Fig. 5c, d). The phosphomimetic S721D mutant vinculin displayed a highly significant, two-fold reduction vinculin/paxillin co-localization compared to cells expressing the S721A mutant, as well as a 58% reduction in the average size of vinculin clusters (Fig. 5e, f). These data are consistent with a model where phosphorylation at S721 disrupts contacts in the closed form, and may stabilize an alternative, inactive conformation with diminished recruitment to focal adhesion complexes. Furthermore, we posit that the observed changes in thermal stability of the pS721 vinculin phosphomodiform may be capturing both a conformation change resulting from perturbed intramolecular contacts, as well as the stabilization of an alternate conformational state that is then capable of engaging in new intermolecular interactions. These data provide a proof-of-concept example of the effective prioritization and discovery of uncharacterized, but phenotypically relevant hotspot modification site by HTP.
Fig. 5. HTP discovery of an uncharacterized phosphorylation site in Vinculin that regulates focal adhesion phenotypes.
A) HTP curves for the unmodified, bulk and pS721 phosphomodiform of vinculin. B) X-ray structure of human vinculin with the uncharacterized phosphorylation site S721 shown as spheres packed in between the Vt (yellow) and D4 (green) domains. PDB accession: 1TR2. Inset images show close contacts between S721, D718, T219 and E770, which cap a buried helix in the D4 domain. C) Confocal microscopy of wild-type, S721A and S721D FLAG-tagged vinculin constructs transiently expressed in HeLa cells. Merged, individual, and colocalization channels are shown for vinculin (cyan), paxillin (green), and actin (purple). D) Representative focal adhesions at migrating fronts of the cell membrane are highlighted, and show significant phenotypic differences between the three vinculin constructs. E) Co-localization of vinculin and paxillin at paxillin-positive focal adhesions, with each point represents percent colocalization in a quantified cell (n = 8, 7, and 6 cells, respectively). F) Size of WT, S721A and S721D-vinculin focal adhesions in n = 8, 7, and 6 cells, respectively. Each point represents a distinct focal adhesion. Scale bar in (C) = 10 μm. Data points shown in (A) represent the mean and S.E.M. from n = 6 (bulk, unmodified) and n = 5 (phospho) independent biological replicates. ΔTm P-value in (A) computed with two-sided t-test. Images shown in (C-D) are representative of three independent biological replicates. Box plots (median, 1-99%) are shown in (E-F); *, P ≤ 0.05, **, P ≤ 0.001, ***, P ≤ 0.0001, n.s., P > 0.05, one-way ANOVA.
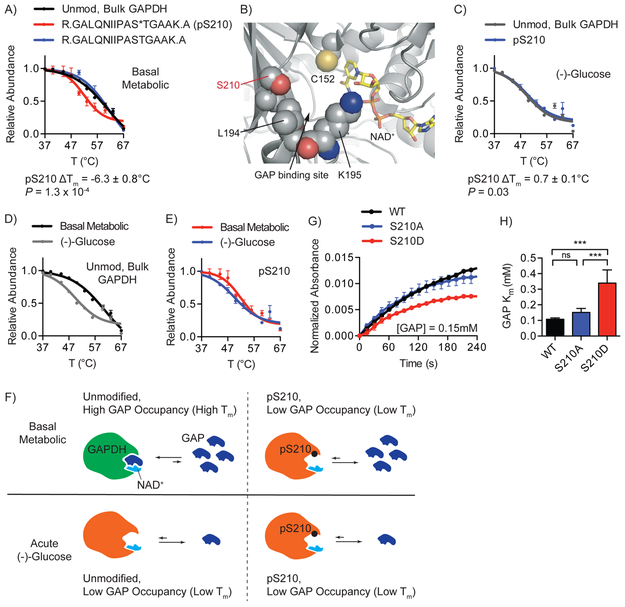
We next interrogated the pS210 phosphomodiform of the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was significantly destabilized relative to the unmodified, bulk GAPDH pool, as well as GAPDH protein that is unambiguously unphosphorylated at S210 (Fig. 6a). Other detected sites, such as pT154, were not associated with a significant ΔTm shift (Supplementary Fig. 5a). Published structures show that S210 packs closely to L194 and by extension K195, which is involved in substrate binding (Fig. 6b). To explore the possibility that phosphorylation may affect GAPDH substrate binding and catalysis, and to extend HTP profiling to kinetic/signaling conditions, we performed HTP on cells under the limiting metabolic environment of acute glucose withdrawal, which significantly depleted cellular glyceraldehyde-3-phosphate (GAP) levels by four-fold (Supplementary Fig. 5b). Under these conditions the thermal shift between the bulk, unmodified GAPDH pool and the pS210 phosphomodiform disappeared (Fig. 6c). Specifically, the stability of the pS210 phosphomodiform was not affected in the presence of lower GAP levels, which contrasted with the significant destabilization of the bulk, unmodified GAPDH protein pool (Fig. 6d, e). These data suggested that GAP binding contributes to the stability of GAPDH protein that is not phosphorylated at S210, and the negative shift in stability associated with the pS210 phosphomodiform may be capturing the reduced substrate occupancy of these proteins in cells (Fig. 6f). To specifically test GAP binding and metabolism we tested the enzyme kinetics of wild-type GAPDH relative to mutants that are incapable of phosphorylation (S210A) or that mimic phosphorylation (S210D). Indeed, the reaction kinetics and KM value for GAP was indistinguishable between the non-phosphorylatable S210A mutant and wild-type enzyme (Fig. 6g, h), whereas the phosphomimetic S210D mutant remained active but exhibited decreased kinetics and a significantly increased KM, consistent with altered ability to bind GAP (Fig. 6f). These effects were unique to S210, as the kinetics of non-phosphorylatable and phosphomimetic mutants for a nearby detected site, T182, did not affect conversion rates or KM value for GAP (Supplementary Fig. 5c-e). These data confirmed that altered modiform-ligand interactions can be discovered and characterized by HTP profiling under basal and altered metabolic conditions.
Fig. 6. Discovery of a previously uncharacterized phosphorylation site affecting GAPDH catalysis.
A) Representative HTP curves for the bulk, unmodified GAPDH and corresponding modiforms in HEK293T cells. B) X-ray structure of human GAPDH active site (grey) bound to NAD+ (tan). The site of phosphorylation, S210, and adjacent residues lining the GAP substrate binding site are shown as spheres. PDB accession: 1ZNQ. C-E) HTP curve comparisons for unmodified (C, D) and pS210 GAPDH (D, E) in cells after acute glucose restriction. F) Proposed model correlating high thermal stability of non-phosphorylated GAPDH bound to GAP in basal metabolic condition, and low thermal stabilities of pS210 in both basal metabolic and acute glucose withdrawal conditions. G-H) Representative in vitro kinetic measurement (G) and Michaelis-Menten kinetic analysis (H) of wild-type, S210A and S210D mutant GAPDH confirmed a significant increase in KM toward GAP for the phosphomimetic- enzyme (**, P ≤ 0.001, two-sided t-test). Data points shown in (A, C-E), represent the mean and S.E.M. from n = 12, 3, 8, 3, and 2 (in order of Tm curve appearance) replicate measurements detected in n = 6 (bulk, unmodified), n = 5 (phospho), and n = 2 (acute (-)-glucose, both bulk, unmodified and phospho) independent biological replicates. Data in (G-H) represent mean and S.E.M. from n = 4 technical replicates from n = 2 independent biological replicates.
DISCUSSION
Here we established a proteomic method enabling the unbiased, global detection of protein stability changes in response to site-specific phosphorylation events on thousands of proteins. In contrast to traditional discovery-mode LC-MS/MS studies, the HTP method provides several novel insights into the functional effects of protein posttranslational modifications. First, it provides site-specific, quantitative relationships between the ‘unmodified, bulk’ population of a protein with its distinct phosphomodiforms in the endogenous context of live cells. We have demonstrated that this format promotes detection of both intrinsic alterations in protein structure, as well as context-specific intermolecular events such as protein-protein, and protein-metabolite interactions15. We posit that the ability to perform HTP profiling under steady state and comparative kinetic conditions in response to small molecules or signaling events can be used to discover and characterize novel PTM-mediated interactions. We demonstrated this attribute of HTP through the identification of previously uncharacterized phosphorylation sites in well-characterized proteins like GAPDH, as well as less studied structural proteins like vinculin. The information gained through HTP profiling of phosphomodiforms adds another functional layer to that measured by quantifying the abundance of modification sites in the proteome, which report on relative “writing” and “erasing” of modifications across the proteome, but provide no information on the dynamic biophysical “reading” of these modifications. HTP profiling alone, or in concert with datasets focused on mapping protein-protein19 and protein-metabolite15,17 interactions will provide novel insights into the functional and interconnected roles of site-specific modification in the proteome.
It is important to consider the limitations of HTP-derived ΔTm values in assuming functional impact on a protein of interest. First, this method specifically interrogates the inclusive pool of proteins that unambiguously contain a defining modification site – what we have described here as a “modiform.” Thus, this method cannot determine the modification status of other sites that do not fall within this peptide on the same protein molecule, a limitation that is intrinsic to massively parallel detection by LC-MS/MS. We do not view this aspect of the approach as a limitation per se, because the primary goal of this approach is to correlate specific modification sites with putative changes to protein structure and function in cells, regardless of the inherent heterogeneity of the protein pool. As such, HTP may under-sample certain combinations of modifications, or miss them entirely, but it can unequivocally connect changes in apparent stability to the presence of site-specific, “hotspot” modifications. Additionally, the response of individual phosphomodiforms to thermal challenge under a defined cellular condition may be used to provide information about the co-occurrence or mutual exclusivity of specific modification sites on the same molecules in the larger protein pool, as we discussed here for TPI1, 4EBP1 and GAPDH. While beyond the scope of the current manuscript, we hope that focused bioinformatic analyses of future datasets could shed light on this aspect of the HTP method. Finally, the major advance with this method is the ability to globally interrogate native modified proteins in relevant biological contexts to identify and prioritize sites for further study and provide mechanistic hypotheses for functional validation. Greater than 90% of modifications detected herein have been previously observed in numerous datasets, but have no functional annotation, underscoring the low-throughput nature of current PTM annotation. We expect the phosphorylation dataset herein, as well as the extension of the HTP method to diverse organisms, cell types, and PTMs will greatly expand our current understanding of the role of protein modifications in regulating protein structure, function and signal transduction.
ONLINE METHODS
Cell Culture
HeLa and HEK293T cell lines were propagated in RPMI 1640 with 2 mM L-glutamine (HyClone) supplemented with 10% fetal bovine serum (Corning) and 1% penicillin-streptomycin (Thermo Fisher Scientific). All cell lines were grown at 37°C in a 5% CO2 humidified incubator.
HTP Proteomic Sample Preparation
HEK293 cells were grown to 90% confluency in 15 cm cell-culture treated plates (Denville). Cells were scraped from plates, briefly washed, and resuspended in PBS with EDTA-free protease inhibitor tablet (Roche) and phosphatase inhibitor cocktail 3 (Sigma-aldrich) before being divided into ten equal aliquots. Each aliquot of live cells was exposed to a steady temperature between 37°C to 67°C for 3 mins in parallel, incubated at 25°C for another 3 mins, and lysed by rapid freeze-thawing. Insoluble proteins and cell debris were removed by centrifugation at 17,000g for 10 mins. Small amount of supernatant from the 37°C aliquot was set aside to use for protein quantification by Bradford’s assay. The remaining fraction of whole cell lysates was denatured in 8 M urea, followed by disulfide reduction with DTT (10 mM, 30 mins, 65°C), alkylation (iodoacetamide, 15 mM, 30 min, room temperature, protected from light) and quenching (DTT, 5 mM, 10 minutes, room temperature). The proteome solution was diluted 4-fold with ammonium bicarbonate solution (50 mM, pH 8.0), CaCl2 added (1 mM) and digested with sequencing grade trypsin (~1:100 enzyme/protein ratio; Thermo Pierce) at 37 ˚C while rotating overnight. Peptide digestion reactions were stopped by acidification to pH 2-3 with 1% formic acid, and peptides were then desalted with Sep-Pak tC18 cartridges (50 mg, Waters), dried under vacuum, resuspended with LC-MS grade water (Sigma Aldrich) and then lyophilized. A small fraction (5%) reserved for bulk unmodified proteome thermal profiling was split from each aliquot and lyophilized separately.
Phosphopeptide Enrichment and TMT Labeling
The major fraction of lyophilized peptides (95%) were re-dissolved in 28.5% lactic acid (Sigma), 57% acetonitrile (LC-MS grade, Sigma), 0.28% TFA (Sigma), and applied to TiO2 spin tips (88303, Thermo Pierce) for phosphopeptide enrichment according to manufacturer’s protocol. Eluted phosphopeptides were acidified with formic acid (pH 2-3), desalted using ZipTip C18 tips (100 μL, Millipore), and lyophilized. The two sets of peptides (phospho-enriched and unenriched) were separately labeled with 10-plex isobaric tandem mass tags (90406, Thermo Scientific) according to manufacturer’s protocol with slight modification. TMT reagents were reconstituted to 8 mg/mL in anhydrous acetonitrile (Sigma) and added to lyophilized peptides dissolved in 100 uL of 200 mM HEPES buffer, pH 8.0 (~8:1 reagent/peptide ratio). Labeling reaction was carried out in room temperature for 1 hr with gentle shaking, and quenched with 5 uL of 5% hydroxylamine (Thermo Scientific). Labeled peptides were combined into a single pool per experiment, acidified with formic acid (pH 2-3), desalted using ZipTip C18 tips (100 μL, Millipore), and lyophilized. The final processed peptides were dissolved in LC-MS/MS Buffer A (H2O with 0.1% formic acid, LC-MS grade, Sigma Aldrich) for LC-MS/MS analysis.
Proteomic LC-MS/MS and Data Analysis.
LC-MS/MS experiments were performed with an Easy-nLC 1000 ultra-high pressure LC system (ThermoFisher) using a PepMap RSLC C18 column (75 μm × 50 cm; 3 μm, 100 Å, Thermo Scientific) heated to 40˚C coupled to a Q Exactive HF orbitrap and Easy-Spray nanosource (ThermoFisher). TMT-labeled digested peptides in MS/MS Buffer A were injected onto the column and separated using the following gradient of buffer B (0.1% Formic acid acetonitrile) at 300 nL/min: 0-10% buffer B in 5 minutes, 10-40% buffer B in 240 minutes, 40-90% buffer B over 6 min, and hold at 90% for 20mins. MS/MS spectra were collected from 0 to 250 minutes using a data-dependent, top-10 ion setting with the following settings: full MS scans were acquired at a resolution of 120,000, scan range of 375-1500 m/z, maximum IT of 60 ms, AGC target of 1e6, and data collection in profile mode. MS2 scans was performed by HCD fragmentation with a resolution of 60,000, AGC target of 1e5, maximum IT of 60 ms, NCE of 30, and data type in centroid mode. Isolation window for precursor ions was set to 1.0 m/z with an underfill ratio of 0.5%. Peptides with charge state 1 and undefined were excluded and dynamic exclusion was set to 20 seconds. Furthermore, S-lens RF level was set to 60 with a spray voltage value of 2.60 kV and ionization chamber temperature of 300˚C.
MS2 files were generated and searched using the ProLuCID algorithm in the Integrated Proteomics Pipeline (IP2) software platform. Human proteome data were searched using a concatenated target/decoy UniProt database (UniProt_Human_reviewed_04-10-2017.fasta). Basic searches were performed with the following search parameters: HCD fragmentation method; monoisotopic precursor ions; high resolution mode (3 isotopic peaks); precursor mass range 600-6,000 and initial fragment tolerance at 600 p.p.m.; enzyme cleavage specificity at C-terminal lysine and arginine residues with 3 missed cleavage sites permitted; static modification of +57.02146 on cysteine (carboxyamidomethylation), +229.1629 on N-terminal and lysine for TMT-10-plex tag; 4 total differential modification sites per peptide, including oxidized methionine (+15.9949), and phosphorylation (+79.9663) on serine, threonine, and tyrosine (only for phospho-enriched samples); primary scoring type by XCorr and secondary by Zscore; minimum peptide length of six residues with a candidate peptide threshold of 500. A minimum of one peptide per protein and half-tryptic peptide specificity were required. Starting statistics were performed with a Δmass cutoff = 10 p.p.m. with modstat, and trypstat settings. False-discovery rates of peptide (sfp) were set to 1%. TMT quantification was peformed using the isobaric labeling 10-plex labeling algorithm, with a mass tolerance of 5.0 p.p.m. or less in cases where co-eluting peptide interfere. Reporter ions 126.127726, 127.124761, 127.131081, 128.128116, 128.134436, 129.131417, 129.13779, 130.134825, 130.141145, and 131.13838 were used for relative quantification. In general all quantified peptides have mass error within 3 p.p.m..
Modification Site Localization Analysis
Phosphorylation site localization analysis was performed using the LuciPHOR algorithm available on the Integrated Proteomics Pipeline (IP2). A global FDR score was calculated for every detected phosphopeptide from each MS run. Phosphosites, and therefore phosphomodiforms, were included in the dataset only if they satisfied a global FDR localization score of < 0.01.
Melting Curve Analyses
All detected phosphopeptides that map to the same phosphorylation sites were grouped together and given a new identifier (Gene_pSite; Supplementary Fig. 4). For example, four different tryptic peptides mapping to the same GAPDH phosphorylation site pS210 were often detected: R.DGRGALQNIIPAS(79.9963)TGAAKAVGK.V, R.DGRGALQNIIPAS(79.9963)TGAAK.A, R.GALQNIIPAS(79.9963)TGAAKAVGK.V, and R.GALQNIIPAS(79.9963)TGAAK.A. These peptides were systematically re-labeled with the new custom identifier “GAPDH_pS210”. TMT reporter ion intensities of phosphopeptides with the same identifier were combined for each temperature fraction from the same MS run (Supplementary Fig. 4). Fold change values were calculated using the lowest temperature fraction as the reference. Similarly, for the bulk unmodified proteome, TMT reporter ion intensities of all unmodified peptides mapped to the same protein were combined and fold change values relative to the lowest temperature condition were calculated. To generate unmodified protein and phosphosite-specific melt curves, relative fold changes as a function of temperature was fitted to the equation derived from the chemical denaturation theory using the R package developed by Savitski et al.18 Tm values were calculated at which the sigmoidal curve crosses the 0.5 fold change level. Only Tm values calculated from melting curves with curve R2 > 0.8 are used in subsequent analyses. Shift in Tm values (e.g. ΔTm) induced by phosphorylation were determined by subtracting the Tm of the unmodified protein from the Tm of the phosphomodiform:
ΔTms could only be calculated for phosphosites belonging to proteins that were also detected in the unmodified proteome. ΔTms were not determined for phosphosites that were detected in fewer than two MS technical replicates. Tm values of all unmodified proteins and phosphorylation sites, and their ΔTm values and P-values are summarized in Supplementary Tables 1 and 2.
Error Analyses
In our workflow, a composite protein-level Tm is calculated by averaging Tm values derived from all detected tryptic peptides of the same protein. To estimate the overall “base error” of our peptide-level Tm measurements, i.e. on average how far a peptide Tm deviates from its composite protein-level Tm, we took 4 representative datasets and calculated the median peptide ΔTm standard deviation (Supplementary Fig. 2). We individually curve fitted every detected tryptic peptide as well as their composite protein to calculate Tm values. Each dataset is comprised of between 10,000 to 16,000 peptide and >2,900 protein melt curves. We then subtracted the composite protein-level Tm from each peptide-level Tm to obtain peptide ΔTms and calculated the standard deviation of each set of peptide ΔTm’s. We plotted the distributions of the peptide ΔTm s.d. of the 4 representative datasets which resulted in median peptide ΔTm s.d. between 1.3 to 1.5°C. The average value of 1.4°C was used as vertical cutoff (aka smallest high confidence ΔTm value) in the ΔTm volcano plot (Fig. 2c).
In situ measurement of polar metabolome by LC-MS/MS
For acute glucose withdrawal studies, confluent 15 cm plates of HEK293T cells were switched from RPMI media containing 10 mM glucose into glucose-free RPMI media and grown for 30 minutes, whereas fresh, glucose-containing media was replaced for control cells. In both conditions, cells were scraped into ice-cold PBS and isolated by centrifugation at 1,400g at 4°C. Cell pellets were resuspended in 300 μl of an 80:20 mixture of MeOH/H2O. Internal deuterated standards, 10 nmol d3-serine, were added to the extraction solution for quantification and sample normalization. LC-MS/MS analysis was then performed as described36.
Gel electrophoresis and western blot
To determine the effect of the 3-min heat pulses on global phosphorylation level, we exposed cells to the thermal profiling heat pulses as detailed above, but directly homogenized whole preoteome by addition of 4X Laemmli buffer containing 50 mM DTT (Supplementary Fig. 3). Samples were prepared for SDS-PAGE by heating to 95˚C for 5 minutes, cooled to room temperature, resolved on 10% SDS-page gel and transferred onto nitrocellulose membranes by standard western blotting methods. Membranes were blocked in 2% BSA in TBS containing 0.1% tween-20 (TBST) and probed with primary and secondary antibodies. Antibodies used in this study include: anti-pS/T/Y (1:1000, ab15556, AbCam). Secondary anti-mouse (Licor) was used at 1:10,000 dilution in 2% BSA-containing TBST and incubated for 1 hr prior to washing and imaging on a Licor infrared scanner. Densitometry measurements were performed with ImageJ software.
Vinculin Expression and Focal Adhesion Confocal Microscopy Assays
Full-length Myc-DDK-tagged-human vinculin (NM_003373, Origene) was used to generate mutants with PCR primers in Supplementary Table 3 according to the QuikChange II Site-Directed Mutagenesis Kit manufacturer protocol (200523, Agilent). HeLa cells were grown to 70% confluency in 6-well plates before transfection of 500 ng plasmid with Lipofectamine 2000 (Invitrogen). After 4 hours, cells were reseeded onto round glass cover slides. After 24 hr cells were washed once with PBS and fixed at 4°C for 10 min in 4% paraformaldehyde-containing PBS. Fixed cells were then washed three times with PBS, and blocked with 2% BSA with 0.05% Triton in PBS for 30 mins in room temperature. Slides were stained with primary antibodies rabbit α-FLAG (1:500, Cell Signaling) and mouse α-Paxillin (1:100, Millipore) overnight at 4°C. Cells were washed three times with 0.05% Tween in TBST for 5 mins and then stained with secondary antibodies Alexafluor488-conjugated goat-α-rabbit (1:200; Sigma Aldrich), Alexa568-conjugated goat α-mouse (1:200; Sigma Aldrich), and Alexa647-conjugated phalloidin (1:25, Invitrogen) for 30 mins in 37°C. Finally, slides were washed three times with TBST and quickly dipped in DI water before mounting in Slow Fade Gold Antifade (ThermoFisher).
Images were taken using Metamorph (Molecular Devices) acquisition software on Nikon Eclipse Ti-PFS inverted microscope with a Yokogawa CSU-X1 confocal scanhead with Spectral Applied Research Laser Merge Module (491 nm, 561 nm, and 643 nm Lasers). Images were obtained using Andor Zyla sCMOS camera with a 60x Plan Apo TIRF 1.49NA objective (Nikon). Image processing and quantification was done in ImageJ. A single focal plane was acquired in which the lamella was in focus, as indicated by basal actin stress fibers stained with phalloidin and Paxillin. Paxillin and Vinculin channels were background subtracted. Paxillin channel was thresholded and made into a binary mask to calculate the area of focal adhesions (FAs) as indicated by Paxillin. The binary mask was then overlaid onto the vinculin channel for segmentation and area calculation (threshold at 0.5 μm2) of Vinculin incorporation into the FAs. For linescans analysis, fluorescence intensity line profiles over vinculin and paxillin channels were merged (raw images that were background subtracted) and done in ImageJ.
GAPDH Expression, Purification and Kinetic Assays
Full-length, human GAPDH (NM_002046, Origene) was used to generate GAPDH mutants using the QuikChange II Site-Directed Mutagenesis Kit manufacturer protocol (200523, Agilent). Mutant GAPDH plasmids were verified by sequencing [CMV (forward), and XL39 (reverse)]. Wild type and GAPDH mutants were transiently expressed from a pCMV6 entry vector with a C-terminal Myc-DDK tag in confluent 10 cm plates of HEK293T cells by transfection of 2 μg plasmid with Lipofectamine 2000 (Invitrogen) according to manufacturer’s protocol. After 24 hours, cells were harvested by scraping, pelleted by centrifugation, washed twice with PBS and lysed in PBS containing EDTA-free complete protease inhibitor (Roche), pH 8.0. After ultracentrifugation, lysates were added to Anti-Flag M2 affinity resin (Sigma Aldrich) and rotated overnight at 4˚C. Resins were washed 5x with PBS before eluting bound protein with 3X-FLAG peptide (100 μg/mL, 150 μL, twice, Sigma Aldrich) in PBS for 1 hours. Relative protein levels were assessed by SDS-PAGE and enzymes were used without further purification.
Michaelis-Menten kinetic assays to assess the catalytic activities of GAPDH proteins were performed on a Tecan Infinite M200 plate reader. GAPDH proteins were supplemented with constant NAD+ (0.278 mM) and serial three-fold dilutions of D/L-glyceraldehyde-3-phosphate (1.36 to 0.0056 mM) and NADH production (340nm absorbance) was monitored. Enzyme velocity at different GAP concentrations was measured in the first 4 minutes and Michaelis constants (KM) were generated by applying a Michaelis-Menten curve fit to the plot of enzyme velocity as a function of [GAP] in Prism 5 software.
Kinase Substrate Motif Analysis
Motifs analysis was performed by comparing peptide sequence containing the target phosphorylation site to commonly described kinase/phosphatase substrate consensus motifs listed in the Human Protein Reference Database (http://www.hprd.org/serine_motifs). Nine common kinase motifs were chosen for this analysis.
Secondary Structure, Solvent Accessibility, and Protein Disorder Prediction
Secondary structure, solvent accessibility, and protein disorder were predicted using RaptorX (http://raptorx.uchicago.edu)28 Protein FASTA sequences were uploaded to RaptorX Structure Prediction portal as described. To calculate the secondary structure percentage, the number of residues predicted to be alpha-helix, beta-sheet, or loop was divided by the total number of residues of all detected proteins. The secondary structure percentage of the phosphosites was calculated by dividing the number of sites predicted to be alpha-helix, beta-sheet, or loop over the total number of detected phosphosites. Secondary structure elements, solvent accessibility and degree of disorder of detected phosphosites were plotted against their ΔTm values.
Phosphosite Annotation Analysis
The number of annotations with site-specific biochemical studies was extracted from the PhosphoSite database (https://www.phosphosite.org) and plotted against ΔTm values.
Data Availability.
Primary data for proteomic analyses are available for download at: ftp://massive.ucsd.edu/MSV000083786; DOI: 10.25345/C5CS7H. All other data and materials are available upon request.
Supplementary Material
Acknowledgements.
We thank M. Rust for discussions surrounding the manuscript, and G. Li for discussions regarding microscopy. We are grateful for financial support of this work from the following: Kwanjeong Educational Fellowship (G.L.); NIH training grant GM007183, and National Academy of Sciences Ford Foundation Fellowship to (K.E.C); 1R01GM104032-01A1 (M.L.G.); NCI R00CA175399 and DP2GM128199-01 (R.E.M.); The Dale F. Frey Award for Breakthrough Scientists from the Damon Runyon Cancer Research Foundation (R.E.M.); and The University of Chicago.
Footnotes
Competing Interests. The authors declare no competing interests.
References
- 1.Agapakis CM, Boyle PM & Silver PA Natural strategies for the spatial optimization of metabolism in synthetic biology. Nature chemical biology 8, 527–535 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Yu CS, Chen YC, Lu CH & Hwang JK Prediction of protein subcellular localization. Proteins 64, 643–651 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Gawron D, Ndah E, Gevaert K & Van Damme P Positional proteomics reveals differences in N-terminal proteoform stability. Molecular systems biology 12, 858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh C Posttranslational modification of proteins : expanding nature’s inventory, (Roberts and Co. Publishers, Englewood, Colo, 2006). [Google Scholar]
- 5.Hunter T Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol 21, 140–146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White FM & Wolf-Yadlin A Methods for the Analysis of Protein Phosphorylation-Mediated Cellular Signaling Networks. Annu Rev Anal Chem (Palo Alto Calif) 9, 295–315 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Martin L, Latypova X & Terro F Post-translational modifications of tau protein: implications for Alzheimer’s disease. Neurochem Int 58, 458–471 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Aebersold R & Mann M Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen JV & Mann M Status of large-scale analysis of post-translational modifications by mass spectrometry. Molecular & cellular proteomics : MCP 12, 3444–3452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walther TC & Mann M Mass spectrometry-based proteomics in cell biology. The Journal of cell biology 190, 491–500 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humphrey SJ, Azimifar SB & Mann M High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nature biotechnology 33, 990–995 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3, ra3 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Tsai CF, et al. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun 6, 6622 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huber KV, et al. Proteome-wide drug and metabolite interaction mapping by thermal-stability profiling. Nature methods 12, 1055–1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez Molina D, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Piazza I, et al. A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication. Cell 172, 358–372 e323 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Savitski MM, et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Tan CSH, et al. Thermal proximity coaggregation for system-wide profiling of protein complex dynamics in cells. Science (2018). [DOI] [PubMed] [Google Scholar]
- 20.Smith LM, Kelleher NL & Consortium for Top Down, P. Proteoform: a single term describing protein complexity. Nature methods 10, 186–187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fermin D, Walmsley SJ, Gingras AC, Choi H & Nesvizhskii AI LuciPHOr: algorithm for phosphorylation site localization with false localization rate estimation using modified target-decoy approach. Molecular & cellular proteomics : MCP 12, 3409–3419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGowan CH & Russell P Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. The EMBO journal 12, 75–85 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe N, Broome M & Hunter T Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. The EMBO journal 14, 1878–1891 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blangy A, et al. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Azimi A, et al. Targeting CDK2 overcomes melanoma resistance against BRAF and Hsp90 inhibitors. Molecular systems biology 14, e7858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gnad F, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol 8, R250 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jimenez JL, Hegemann B, Hutchins JR, Peters JM & Durbin R A systematic comparative and structural analysis of protein phosphorylation sites based on the mtcPTM database. Genome Biol 8, R90 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallberg M, et al. Template-based protein structure modeling using the RaptorX web server. Nature protocols 7, 1511–1522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TY, et al. dbPTM: an information repository of protein post-translational modification. Nucleic acids research 34, D622–627 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beretta L, Gingras AC, Svitkin YV, Hall MN & Sonenberg N Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. The EMBO journal 15, 658–664 (1996). [PMC free article] [PubMed] [Google Scholar]
- 31.Sekiyama N, et al. Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. Proceedings of the National Academy of Sciences of the United States of America 112, E4036–4045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gingras AC, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & development 13, 1422–1437 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garakani K, Shams H & Mofrad MRK Mechanosensitive Conformation of Vinculin Regulates Its Binding to MAPK1. Biophys J 112, 1885–1893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humphries JD, et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. The Journal of cell biology 179, 1043–1057 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chorev DS, et al. Conformational states during vinculin unlocking differentially regulate focal adhesion properties. Sci Rep 8, 2693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only Reference
- 36.Chang JW, Lee G, Coukos JS & Moellering RE Profiling Reactive Metabolites via Chemical Trapping and Targeted Mass Spectrometry. Analytical chemistry (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary data for proteomic analyses are available for download at: ftp://massive.ucsd.edu/MSV000083786; DOI: 10.25345/C5CS7H. All other data and materials are available upon request.