Abstract
Increased production of inflammatory cytokines and myeloid-derived suppressor cells occurs in patients with coronavirus disease 2019. These inversely correlated with perforin-expressing natural killer (NK) and CD3+ T cells. We observed a lower number of perforin-expressing NK cells in intensive care unit (ICU) patients compared with non-ICU patients, suggesting an impairment of the immune cytotoxic arm as a pathogenic mechanism.
Keywords: COVID-19, inflammation, NK cells, cytotoxic cells, MDSC
The novel coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), may cause fatal acute respiratory disease [1]. The pathogenesis of coronavirus disease 2019 (COVID-19) remains unknown. A wide range of clinical manifestations occur, ranging from mild disease with a good prognosis (fever, runny nose, malaise, dry cough, and fatigue) to severe pneumonia characterized by acute respiratory distress syndrome and multiorgan failure with a high rate of intensive unit care (ICU) admission [2].
As in other lethal zoonotic infections of humans, such as severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus [3], several data support a role of immune-based pathogenic mechanisms characterized by hyperinflammatory response and lymphopenia. Patients with severe pneumonia requiring ICU admission showed high serum levels of inflammatory mediators and severe lymphopenia, suggesting an unbalanced deleterious immune response [1]. T cells play an important role in the antiviral immune responses and in conferring long-lasting protection. COVID-19 patients with severe disease show a higher level of T-cell activation and high levels of inhibitory molecules associated with a reduction of T-cell functionality, suggesting an ineffective immune response [4–6]. Moreover, cytotoxic cells, such as innate natural killer (NK) cells and adaptive CD8 T lymphocytes, express high levels of the inhibitory receptor NKG2A and reduced perforin and granzyme content, suggesting an impaired cytotoxic immune response during SARS-CoV-2 infection [6]. The mechanisms underlying functional immune impairment deserve further investigation. We evaluated the possible involvement of an inflammatory storm in reducing cytotoxic T-cell profiles.
MATERIALS AND METHODS
Patients
Forty-eight patients with COVID-19 disease admitted to the National Institute for Infectious Diseases “L. Spallanzani” in Rome were enrolled in the study. The local ethical committee approved the study and all patients provided written consent. Blood samples were collected within 5 days from the admission. Clinical features of enrolled patients are summarized in Table 1 (see also Supplementary Table 1). Twenty healthy donors (HDs) were used as controls.
Inflammatory and Immunological Parameters
Plasma samples were obtained after speed centrifugation for 10 minutes at 2000 rpm and immediately stored at −80°C. Interleukin 1β (IL-1β), interleukin 6 (IL-6), interleukin 8 (IL-8), and tumor necrosis factor alpha (TNF-α) were measured in plasma samples by using an automated enzyme-linked immunosorbent assay (ELLA microfluidic analyzer, Protein Simple).
Perforin expression was evaluated on whole blood by flow cytometry. In brief, 50 µL of whole blood was stained with a surface antibody cocktail (anti-Vδ2 FITC, anti- CD3 PB, anti-CD56 PeCy7) for 20 minutes at 4°C, lysed with Lysing Solution (BD Biosciences) for 15 minutes at room temperature in the dark, and washed with phosphate-buffered saline 1X. Finally, cells were intracellularly labeled with anti-perforin PE antibody (BD Biosciences) for 20 minutes at room temperature in the dark. After 1 wash, cells were acquired using a FACSCanto II flow cytometer (BD Biosciences).
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll gradient density and characterized by flow cytometry. Myeloid-derived suppressor cell (MDSC) identification was performed on PBMCs stained by using customized Duraclone Tubes (CD11b FITC, HLA-DR ECD, CD14 PC5.5, CD33 PC7, CD80 APC, DRAQ7, CD56 APC-Alexa750, CD19 APC-Alexa750, CD3 APC-Alexa750, CD15 Pacific Blue, CD45KrO, Beckman Coulter) following the manufacturer’s instruction, and acquired on a Navios flow cytometer (Beckman Coulter). Data were analyzed by Kaluza software (Beckman Coulter).
Statistical Analysis
Principal component analysis (PCA) including 48 COVID-19 subjects was performed to identify the relevant information and visualize major trends inherent to the immunological profile. Data were analyzed using RStudio software from http://www.rstudio.org with the libraries FactoMineR (for the analysis) and factoextra (for ggplot2-based visualization). Clusters of variables were identified using k-means clustering algorithm. To visualize a correlation matrix in R, we used the corrplot function and generated a heatmap object using correlation coefficients (computed using the Spearman correlation test) as input to the heatmap. The heatmap was produced with the R package heatmap3. Quantitative variables were compared with nonparametric Mann-Whitney test. P values < .05 were considered statistically significant. Statistical analyses were performed using GraphPad Prism version 8.0 software.
RESULTS
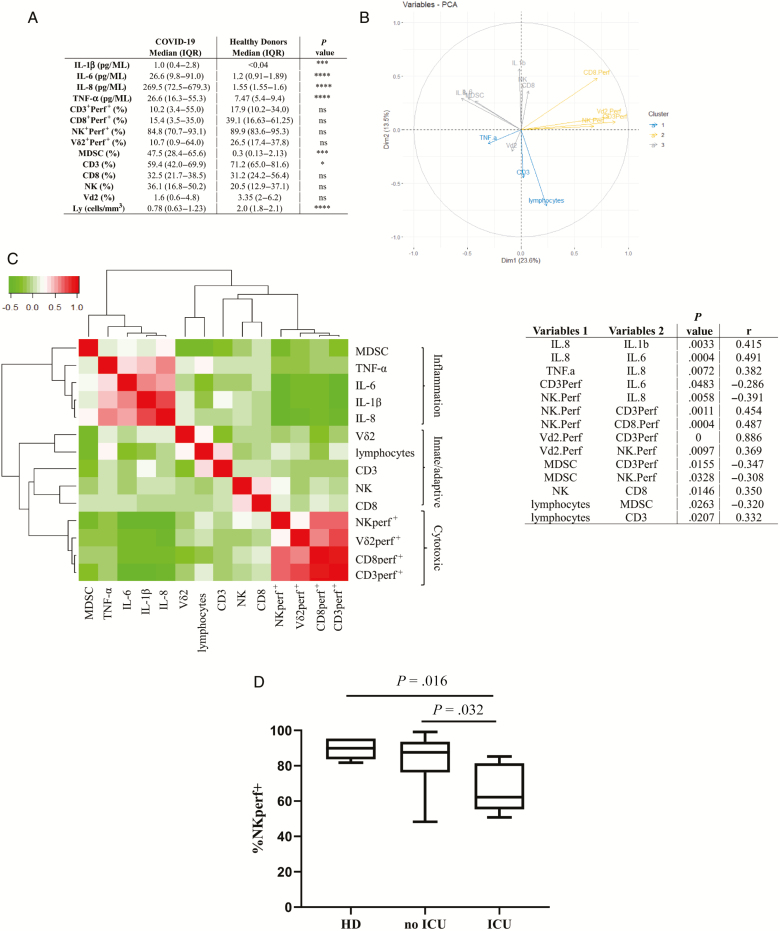
To define a possible involvement of inflammatory environment in shaping the immune response, immune subsets (lymphocyte count, percentage of CD3, CD8, NK, Vδ2, MDSCs), inflammatory cytokines (IL-1β, IL-6, IL-8, TNF-α), and cytotoxic cells (percentage of perforin+ CD3 T cells, perforin+ CD8 T cells, perforin+ NK cells, perforin+ Vδ2 T cells) were assessed (Figure 1A). Higher levels of inflammatory cytokines were observed in COVID-19 patients than in HDs, consistently with an inflammatory profile being a hallmark of SARS-CoV-2 infection [1, 5]. Moreover, we confirmed the occurrence of lymphopenia in COVID-19 patients, mainly affecting CD3+ T cells. No significant differences were observed in other immune subsets and in the frequency of perforin+ cells.
Figure 1.
A, Data are reported as median and interquartile range for immunological subset frequency, inflammatory cytokines, and cytotoxic cell percentage. Statistical differences were assessed by Mann-Whitney test. *P < .05; ***P < .001; ****P < .0001. B, Principal component analysis of the immunological parameters of patients with coronavirus disease 2019 (COVID-19) patients. Graph of variables: directions and magnitude of each vector indicate the contribution of the corresponding mediator levels to principal component 1 and principal component 2. Different colors represent different correlation groups between mediator levels and principal components. C, Immunological correlations among inflammatory and immunological variables (innate, adaptive, and cytotoxic cells). The nonparametric Spearman test was applied to evaluate multiple correlations. A color map matrix (left panel) shows the strength and direction of these correlations (−0.5 [green] to +1 [red]). Statistically significant correlations (P < .05) between variables are shown in the right panel. Variables 1 indicate the parameters shown on the x-axis; variables 2 indicate the parameters shown on the y-axis. D, Frequency of perforin-expressing natural killer cells in non–intensive care unit (ICU) patients, in ICU patients, and in healthy donors. Statistical differences were assessed by Mann-Whitney test. Abbreviations: HD, healthy donors; ICU, intensive care unit; IL, interleukin; IQR, interquartile range; MSDC, myeloid-derived suppressor cells; NK, natural killer; ns, not significant; PCA, principal component analysis; Perf+, perforin-expressing; TNF-α, tumor necrosis factor alpha.
To identify the relevant information and visualize major trends inherent to the immunological profile in COVID-19 patients, PCA analysis was performed (Figure 1B and 1C). The variables graph allows visualizing all parameters (immune cells, inflammatory mediators, and cytotoxic profile) and drawing conclusions about their correlations (Figure 1B). The longer vectors represent the variables that mainly contribute to the variance. Cluster analysis classified the parameters into 3 main groups: group 1 clustered TNF-α, CD3, and lymphocyte count; group 2 included cytotoxic cells (perforin+ CD3, perforin+ CD8, perforin+ Vd2, and perforin+ NK); and group 3 included inflammatory cytokines (IL-1β, IL-6, and IL-8), innate immunity (Vδ2 T and NK cells), and suppressor cells (MDSCs). Among the analyzed variables, the cytotoxic group contributed mainly to the disease profile.
To define possible associations among immunological parameters, we therefore performed a correlation matrix among the same 14 continuous variables (Figure 1C). All results were represented in a color map matrix, with statistically significant associations (P < .05) listed in Figure 1C. Results allowed the identification of 3 main clusters that could be identified as inflammatory markers, immune subsets, and cytotoxic cells. A strong positive correlation was observed among IL-8 and IL1-β, IL-6 and TNF-α, suggesting a coordinated inflammatory response. Interestingly, both IL-6 and IL-8 were negatively correlated with perforin content in both innate (NK) and adaptive (CD3) immune cells. A similar negative effect was also observed between MDSCs and innate and adaptive cytotoxic cells, suggesting that inflammatory environment and suppressor cells may contribute to decrease cytotoxic activity of cells of innate and adaptive immunity. To evaluate whether inflammatory mediator levels and/or cytotoxic activity were associated with severe disease, we grouped ICU patients (n = 7) and non-ICU patients (n = 41) and we analyzed proinflammatory cytokines, MDSCs, and cytotoxic cells. We found that neither inflammatory mediators nor MDSC levels differed between the 2 groups, suggesting that hyperinflammation is a hallmark of symptomatic COVID-19. Interestingly, the percentage of perforin+ NK cells was lower in ICU patients as compared to non-ICU patients and HDs (Figure 1D), suggesting an impairment of NK cell activity in patients with severe illness.
DISCUSSION
In this study, we show for the first time that the increase in inflammatory mediators is correlated with a reduction of innate and adaptive cytotoxic antiviral function. Our data are in line with other studies reporting an inflammatory profile in patients with COVID-19 [1, 5] shown by an increase in IL-1β, IL-6, IL-8, and TNF-α serum levels in comparison to HDs and with no significant differences between ICU and non-ICU patients.
Our results suggest that IL-8 and IL-6 may impact the protective immune response by reducing the number of perforin+ NK and T cells and consequently cytotoxic activity against infected cells. Indeed, a correlation between IL-6 level and cytotoxic impairment has been demonstrated in IL-6–overexpressing mice [7], in several human infectious diseases [8], and in diseases characterized by hyperinflammation, such as hemophagocytic lymphohistiocytosis [9]. Moreover, excessive IL-6 observed during viral infection may also promote the development and differentiation of other T-cell subsets, such as Th17, that could contribute to viral persistence by protecting virus-infected cells from apoptosis and CD8 T cell–mediated destruction [10].
The hyperinflammatory response induced by SARS-CoV-2 infection prompted us to evaluate the frequency of MDSCs in patient peripheral blood. They have been described as a response to inflammatory processes to curb excessive and potentially harmful immune response in several conditions, including cancer, inflammatory diseases, viral infections, and autoimmune disorders [11]. They are able to inhibit several immune cell subsets, in particular, T lymphocytes and NK cell functions [12]. Our data suggest that the expansion of MDSCs paralleled by a high level of inflammatory mediators in COVID-19 patients can both contribute to decrease the cytotoxic arm of the immune system. A similar analysis of patients with severe pneumonia could highlight whether our findings are truly related to SARS-CoV-2 infection.
Overall, we found that during symptomatic SARS-CoV-2 infection, a hyperinflammatory environment and MDSC expansion were induced, in turn correlating to a reduced cytotoxic function mainly in patients with more severe disease. Therapeutic approaches targeting interleukin 6R, interferon gamma, and interleukin 1 are currently under investigation in patients with COVID-19, with the goal of controlling the hyperinflammation potentially responsible for lung damage. A secondary beneficial effect on these cytokine-targeting treatments could be that of improving cytotoxic function of both NK and T cells.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors gratefully acknowledge the Collaborators Members of the National Institute for Infectious Diseases (INMI) COVID-19 study group: Maria Alessandra Abbonizio, Amina Abdeddaim, Chiara Agrati, Fabrizio Albarello, Gioia Amadei, Alessandra Amendola, Mario Antonini, Tommaso Ascoli Bartoli, Francesco Baldini, Raffaella Barbaro, Barbara Bartolini, Rita Bellagamba, Martina Benigni, Nazario Bevilacqua, Gianlugi Biava, Michele Bibas, Licia Bordi, Veronica Bordoni, Evangelo Boumis, Marta Branca, Donatella Busso, Marta Camici, Paolo Campioni, Maria Rosaria Capobianchi, Alessandro Capone, Cinzia Caporale, Emanuela Caraffa, Ilaria Caravella, Fabrizio Carletti, Concetta Castilletti, Adriana Cataldo, Stefano Cerilli, Carlotta Cerva, Roberta Chiappini, Pierangelo Chinello, Carmine Ciaralli, Stefania Cicalini, Francesca Colavita, Angela Corpolongo, Massimo Cristofaro, Salvatore Curiale, Alessandra D’Abramo, Cristina Dantimi, Alessia De Angelis, Giada De Angelis, Maria Grazia De Palo, Federico De Zottis, Virginia Di Bari, Rachele Di Lorenzo, Federica Di Stefano, Gianpiero D’Offizi, Davide Donno, Francesca Faraglia, Federica Ferraro, Lorena Fiorentini, Andrea Frustaci, Matteo Fusetti, Vincenzo Galati, Roberta Gagliardini, Paola Gallì, Gabriele Garotto, Saba Gebremeskel Tekle, Maria Letizia Giancola, Filippo Giansante, Emanuela Giombini, Guido Granata, Maria Cristina Greci, Elisabetta Grilli, Susanna Grisetti, Gina Gualano, Fabio Iacomi, Giuseppina Iannicelli, Giuseppe Ippolito, Eleonora Lalle, Simone Lanini, Daniele Lapa, Luciana Lepore, Raffaella Libertone, Raffaella Lionetti, Giuseppina Liuzzi, Laura Loiacono, Andrea Lucia, Franco Lufrani, Manuela Macchione, Gaetano Maffongelli, Alessandra Marani, Luisa Marchioni, Andrea Mariano, Maria Cristina Marini, Micaela Maritti, Alessandra Mastrobattista, Giulia Matusali, Valentina Mazzotta, Paola Mencarini, Silvia Meschi, Francesco Messina, Annalisa Mondi, Marzia Montalbano, Chiara Montaldo, Silvia Mosti, Silvia Murachelli, Maria Musso, Emanuele Nicastri, Pasquale Noto, Roberto Noto, Alessandra Oliva, Sandrine Ottou, Claudia Palazzolo, Emanuele Pallini, Fabrizio Palmieri, Carlo Pareo, Virgilio Passeri, Federico Pelliccioni, Antonella Petrecchia, Ada Petrone, Nicola Petrosillo, Elisa Pianura, Carmela Pinnetti, Maria Pisciotta, Silvia Pittalis, Agostina Pontarelli, Costanza Proietti, Vincenzo Puro, Paolo Migliorisi Ramazzini, Alessia Rianda, Gabriele Rinonapoli, Silvia Rosati, Martina Rueca, Alessandra Sacchi, Alessandro Sampaolesi, Francesco Sanasi, Carmen Santagata, Alessandra Scarabello, Silvana Scarcia, Vincenzo Schininà, Paola Scognamiglio, Laura Scorzolini, Giulia Stazi, Fabrizio Taglietti, Chiara Taibi, Roberto Tonnarini, Simone Topino, Francesco Vaia, Francesco Vairo, Maria Beatrice Valli, Alessandra Vergori, Laura Vincenzi, Ubaldo Visco-Comandini, Pietro Vittozzi, and Mauro Zaccarelli. The authors thank also Andrea Passavanti Pizzuto for help in bioinformatic analysis.
Financial support. INMI authors are supported by the Italian Ministry of Health (Ricerca Corrente Linea 1). G. I. and A. Z. are co–principal investigators of the Pan-African Network on Emerging and Re-emerging Infections, funded by the European and Developing Countries Clinical Trials Partnership, supported under Horizon 2020. A. Z. is in receipt of a National Institute for Health Research senior investigator award. M. M. is a member of the innate immunity advisory group of the Bill & Melinda Gates Foundation, and is funded by the Champalimaud Foundation.
Potential conflicts of interest. F. D. reports unrestricted research grants from Novartis, Roche, Novimmune, Sanofi, Sobi, and Pfizer, outside the submitted work. N. P. reports speaker fees from Becton & Dickinson, and speaker/scientific board member fees from MSD, Pfizer, Shionogi, and Cepheid, outside the submitted work. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Huan C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China [manuscript published online ahead of print 7 February 2020]. JAMA 2020. doi:10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Memish ZA, Perlman S, Van Kerkhove MD, Zumla A. Middle East respiratory syndrome [manuscript published online ahead of print 4 March 2020]. Lancet. 2020;395:1063–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng HY, Zhang M, Yang CX, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol 2020; 17:541–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China [manuscript published online ahead of print 12 March 2020]. Clin Infect Dis 2020. doi:10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng M, Gao Y, Wang G, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol 2020;17:533–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cifaldi L, Prencipe G, Caiello I, et al. Inhibition of natural killer cell cytotoxicity by interleukin-6: implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheumatol 2015; 67:3037–46. [DOI] [PubMed] [Google Scholar]
- 8. Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, Borca MV. The role of interleukin 6 during viral infections. Front Microbiol 2019; 10:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kogawa K, Lee SM, Villanueva J, Marmer D, Sumegi J, Filipovich AH. Perforin expression in cytotoxic lymphocytes from patients with hemophagocytic lymphohistiocytosis and their family members. Blood 2002; 99:61–6. [DOI] [PubMed] [Google Scholar]
- 10. Hou W, Jin YH, Kang HS, Kim BS. Interleukin-6 (IL-6) and IL-17 synergistically promote viral persistence by inhibiting cellular apoptosis and cytotoxic T cell function. J Virol 2014; 88:8479–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009; 9:162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bronte V, Brandau S, Chen SH, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 2016; 7:12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.