Abstract
Lack of responsiveness to checkpoint inhibitors is a central problem in the modern era of cancer immunotherapy. Tumor neoantigens are critical targets of the host antitumor immune response and their presence correlates with the efficacy of immunotherapy treatment. Many studies involving assessment of tumor neoantigens principally focus on total neoantigen load, which simplistically treats all neoantigens equally. Neoantigen load has been linked with treatment response and prognosis in some studies, but not others. We developed a Cauchy-Schwarz index of Neoantigens (CSiN) score to better account for the degree of concentration of immunogenic neoantigens in truncal mutations. Unlike total neoantigen load determinations, CSiN incorporates the effect of both clonality and MHC-binding affinity of neoantigens when characterizing tumor neoantigen profiles. By analyzing the clinical responses in 501 treated cancer patients (with most receiving checkpoint inhibitors) and the overall survival of 1,978 cancer patients at baseline, we showed that CSiN scores predict treatment response to checkpoint inhibitors and prognosis in melanoma, lung cancer, and kidney cancer patients. CSiN substantially outperformed prior genetics-based prediction methods of responsiveness and fills an important gap in research involving assessment of tumor neoantigen burden.
One Sentence Summary:
CSiN score is an improved tool to assess the overall quality of tumor neoantigens that predicts response to immunotherapy.
Introduction
Most immunotherapies, including checkpoint inhibitors, benefit only a small subset of patients. For example, anti-PD-1 and anti-PD-L1 agents, which have demonstrated marked clinical benefit in various diseases, have overall response rates ranging from 10%−50% in melanoma and non-small lung cancer (1–5) and higher response rates in some other select tumor types, such as classic Hodgkin lymphoma (65–80%) (6, 7). Similarly, the efficacy of anti-CTLA-4 agents ranges from 10–15% for objective response rates and <3% for complete response (8). Unfortunately, the field is still far from clearly understanding how to distinguish responders and non-responders to immunotherapy. All forms of immunotherapy, such as checkpoint inhibitors and neoantigen vaccines, seek to activate the host immune system to attack the tumor cells. These forms of immunotherapy have different modes of actions, but most are intended to mobilize the cytotoxicity of T cells in the patient. Neoantigens are the primary targets of T cell responses (9–13) and the profiles of tumor neoantigens in each patient are central to determining the responsiveness to immunotherapy treatment.
A major impediment of current research that seeks to correlate neoantigens with immunotherapy treatment response is that most studies have only considered whether a higher neoantigen/mutation load (namely, the total number of neoantigens or mutations) is correlated with better immunotherapy response. This overly simplistic approach fails to take full advantage of the wealth of information contained in the entire repertoire of neoantigens and has been successful in only some studies (2, 4, 14–18), but not others (19–24). Neoantigens are associated with mutations that can be either truncal or subclonal (25). Some neoantigens are also more immunogenic than others. These details are not fully captured by a basic neoantigen/mutation load approach, but could be critical for understanding the responsiveness of cancer patients to immunotherapy treatment. For example, Miao et al showed that cancer patients with a high proportion of clonal mutations have better rates of checkpoint inhibitor treatment response (26). The only other study that we are currently aware of that defined a more sophisticated neoantigen-based predictive metric is the neoantigen fitness model developed based on evolutionary modeling of patient neoantigen profiles (16, 27). This work considered the neoantigen-class I MHC binding affinity and only retained the top neoantigens resulting from missense mutations with the highest binding affinity within each tumor clone. This metric demonstrated excellent predictive power for survival of patients after immunotherapy treatment in a few cohorts; however, its predictive values and prognostic values have not been widely evaluated.
In this study, we developed the Cauchy-Schwarz index of neoantigens (CSiN) to enable a quantitative characterization of the delicate internal structure of patient tumor neoantigen profiles. We assembled 10 immunotherapy-treated patient cohorts and 6 patient cohorts with baseline survival information, both from public sources and our own cohorts. In our unbiased analyses, CSiN achieved almost uniformly significant results on the checkpoint inhibitor cohorts and the baseline cohorts of immunogenic cancers, which is significantly better than the performance of previously described neoantigen-based biomarkers. Taken together, our work fills an important “void” in immuno-oncology research that resulted from prior methods that have not sufficiently integrated data on neoantigen clonal structures.
Results
Constructing the Cauchy-Schwarz index of Neoantigen (CSiN) to characterize the fine structure of patient neoantigen repertoire
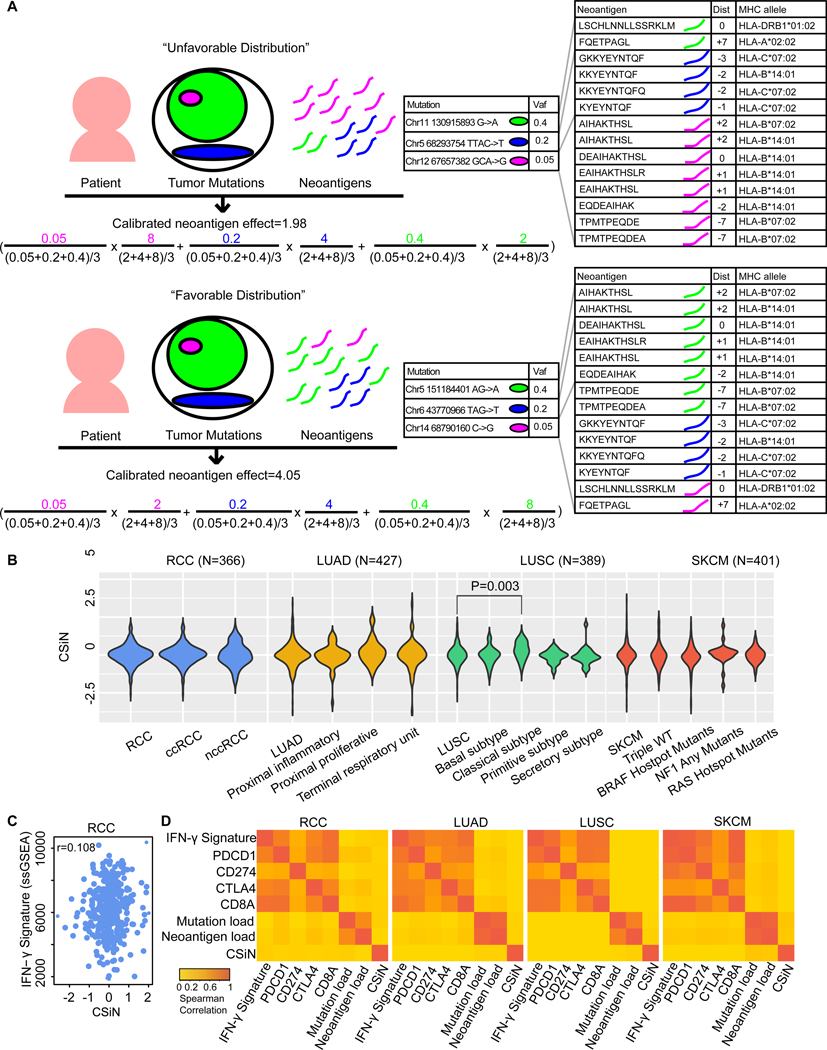
Some somatic mutations are truncal and others are subclonal. Truncal mutations are shared among more tumor clones (i.e. founding and derivative clones) and, if targeted by T cells through neoantigens, will likely result in a stronger cytotoxic effect. Subclonal mutations are unique to different clones, and if a subclonal mutation is in a clone with a larger clonal fraction, the neoantigens associated with this mutation are likely to have a stronger effect on the survival of the tumor cells than subclonal mutations associated with minor clones. Besides, each somatic mutation could generate a different number of neoantigens of different peptide lengths (8 to 11 amino acids for class I MHC-binding peptides, 15 amino acids for class II-binding peptides), with different registers (a sliding window of protein segments around the mutated position) and presented by different HLA alleles (class I and class II). Insertions and deletions usually generate a higher number of neoantigens per mutation than missense mutations as they lead to the translation of completely new segments of protein sequences (shown in Supplementary Information). We hypothesize that a favorable distribution of neoantigens and tumor mutations occurs when immunogenic neoantigens are concentrated on truncal mutations (Fig. 1A), and we developed CSiN to test this prediction.
Fig. 1.
Motivation for CSiN score. (A) Illustration showing the motivation of examining pairings of neoantigens and the tumor mutations with which they are associated. We demonstrated two hypothetical patients, one with an unfavorable distribution of neoantigens and the other with a favorable distribution. The actual mutations and neoantigens shown are based on real data. The outermost circle indicates the whole tumor. Each circle indicates a population of tumor cells with certain mutations. Each different color indicates a distinct mutation, and the area of each circle indicates the proportion of cells having the mutation. For the formula, on the left of each multiplication sign “x”, is the normalized VAF, and on the right of each “x” is the normalized per-mutation neoantigen load. The colorings in the formula correspond to the tumor mutations shown above with the same colorings. The two bigger tables on the right show the neoantigen sequences, registers (“Dist”), and the HLA alleles for each neoantigen. For neoantigens of missense mutations, “Dist” refers to the distance between the altered amino acid and the left end of neoantigen; for neoantigens of insertions/deletions and stoploss mutations, “Dist” refers to the distance between the left end of the mutation and the left end of neoantigen. The “+” sign indicates the left end of neoantigen is on the right of the altered position and vice versa. (B) The distribution of the CSiN scores in the RCC, LUAD, LUSC, and SKCM cohorts. T-tests were used for comparison of CSiN scores between different subtypes of the same tumor cohort. (C) A scatterplot showing the relationship between CSiN and the expression level of the IFN-γ signature in the RCC cohort. Spearman correlation between them is shown. (D) Heatmaps of the pairwise Spearman correlations of the CSiN, mutation load, neoantigen load, and the transcriptomics-based features are shown for the RCC, LUAD, LUSC and SKCM cohorts, which are calculated as in (C).
The core of CSiN is based on the mean of the product of the variant allele frequency (VAF) of each somatic mutation and the number of neoantigens associated with that mutation. When the mutations with higher VAFs are also the mutations that generate more neoantigens (favorable distribution), the product value will be larger (a higher CSiN score). Therefore, a higher CSiN conforms to a favorable neoantigen clonal structure. CSiN is so named because it bears analogy to the Cauchy-Schwarz inequality, which states the inner product of two vectors is maximal when they are in parallel (in the same ranked order). We also considered the immunogenicity of neoantigens. As there are currently no well accepted tools for predicting the potential of both class I and class II neoantigens to induce T cell responses (28), we used the binding affinity of peptides to MHCs, which has been shown to be one of the most important predictors of neoantigen immunogenicity (29). We set a series of cutoffs on the peptide-MHC binding affinity strengths predicted by the IEDB tools (30, 31) to generate several subsets of neoantigens with increasing stringency. We then calculated the means of product of VAF and per-mutation neoantigen load (neoantigens of both class I and class II, all HLA alleles, and all registers from one mutation) for each subset of neoantigens. The final CSiN score is the arithmetic mean of these sub-indices, in which neoantigens with better MHC binding affinity have higher weights (for details, please refer to Supplementary Information). The distributions of the CSiN scores in renal cell carcinoma (RCC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and skin cutaneous melanoma (SKCM), which are all immunogenic tumors (32), are shown in Fig. 1B.
We assessed whether CSiN characterizes unique information that is not captured by the commonly used mutation load and neoantigen load approaches (both classes, all possible lengths and all possible registers) or by candidate transcriptomics-based predictive biomarkers. For transcriptomic features, we examined the expression of PDCD1 (encoding PD-1), CD274 (encoding PD-L1), CTLA4 (33), CD8A, and an IFN-γ gene signature (34). Fig. 1C shows a scatter plot of the CSiN scores with the activation (ssGSEA) of the IFN-γ signature in the RCC cohort, which yielded a Spearman correlation of 0.067. Fig. 1D visualizes these correlations in heatmaps for pairwise comparisons of these biomarkers in each cohort, which shows that CSiN is an independent biomarker (Spearman correlation<0.1 for all comparisons). Further analyses, using the Pearson correlation, threshold comparisons, and mutual information, again demonstrated the independence of the CSiN score (Supplementary Information).
Better response to checkpoint inhibitors in immunogenic cancers is associated with higher CSiN scores
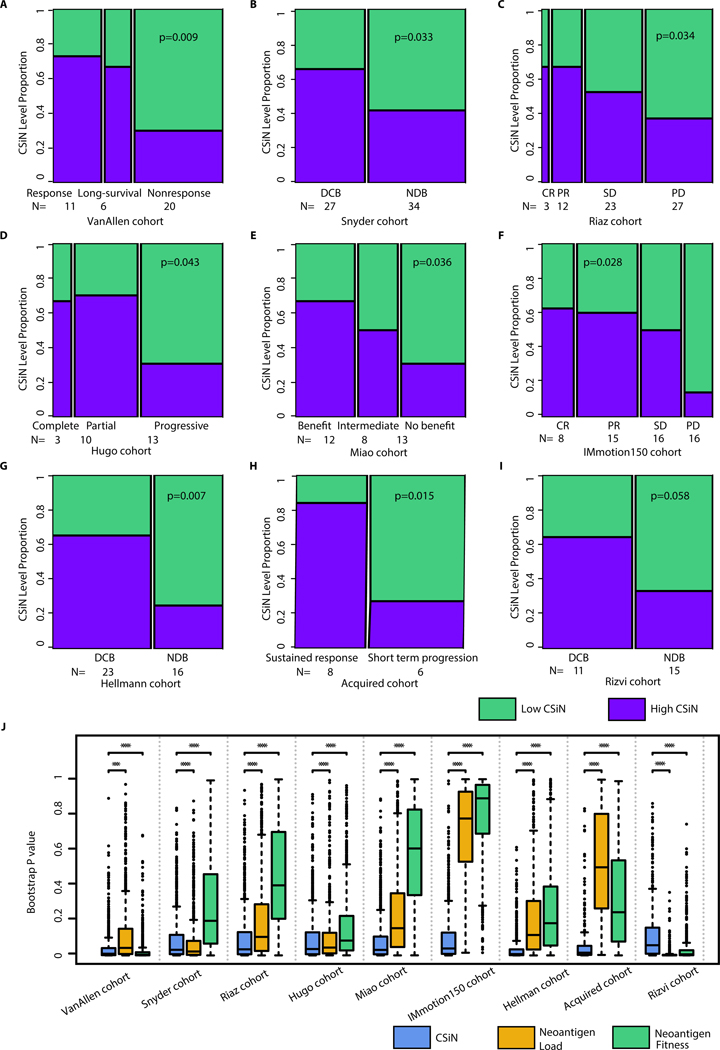
Next we investigated the implications of CSiN for checkpoint inhibitor treatment response. We analyzed the neoantigen profiles of melanoma patients on anti-CTLA-4 therapy from Van Allen et al (35). We observed patients with better responses were more likely to have high CSiN (i.e. higher than median) than patients with worse responses (P=0.009, Fig. 2A). We analyzed another cohort of melanoma patients on anti-CTLA-4 therapy from Snyder et al (36). We observed that patients who received a Durable Clinical Benefit (abbreviated DCB and defined as complete response or partial response or stable disease >6 months) had higher CSiN scores than patients with No Durable Benefit (abbreviated NDB and defined as stable disease <6 months or progressive disease) (P=0.033, Fig. 2B). We analyzed a third cohort of melanoma patients (Riaz cohort) on anti-PD-1 therapy (15). We examined whether patients with better treatment responses had higher CSiN scores. There was a significant positive association in this cohort (P=0.037, Fig. 2C). We analyzed one more cohort (Hugo) of melanoma patients from Hugo et al (14). Fig. 2D shows that there is an overall trend of patients with a better response associated with higher CSiN scores (P=0.043). In clear cell RCC (ccRCC), we examined anti-PD-1/anti-PD-L1-treated ccRCC patients from Miao et al (17). The same significantly positive association of higher CSiN scores with better response was observed (Fig. 2E) (P=0.036). We analyzed metastatic ccRCC patients treated with atezolizumab, an anti-PD-L1 agent (Immotion150 cohort) (37). We found that there was a significant association of higher CSiN with better treatment response for Teff-high patients treated with atezolizumab (Fig. 2F, P=0.028 and Supplementary Information). In contrast, we did not observe this association for Teff-high patients treated with sunitinib (P=0.890). For patients with lower Teff signature expression, there was no significant association for either atezolizumab or sunitinib. Non-small-cell lung cancer (NSCLC) patients (the Hellmann cohort) treated with PD-1 and CTLA-4 inhibitors were available from Hellmann et al (38). Our analyses showed that PD-L1+ patients with Durable Clinical Benefit had higher CSiN scores than patients with No Durable Benefit (Fig. 2G, P=0.007), while this association is insignificant for patients of low PD-L1 expression. We examined another NSCLC cohort (the Acquired cohort) from Anagnostou et al (2) and Gettinger et al (22). All of these patients achieved partial response, except for one patient who exhibited stable disease after initial checkpoint inhibitor treatment. All patients developed acquired resistance after 4 to 40 months. Interestingly, patients with sustained response were more likely to have higher CSiN scores than patients with short term progression (Fig. 2H) (P=0.015). Lastly, another cohort of NSCLC patients on anti-PD-1 therapies from Rizvi et al (4) was analyzed. In Fig. 2I, we showed that more DCB patients had higher CSiN scores than NDB patients (P=0.058). The False Discovery Rates for all the above mentioned cohorts fall under 10% (Table S3).
Fig. 2.
Association of CSiN score with checkpoint inhibitor treatment response. (A) The VanAllen cohort. 11 patients with clinical benefit (response group), 6 patients with long-term survival with no clinical benefit (long-survival) group, and 20 patients with minimal or no clinical benefit (nonresponse) group. (B) The Snyder cohort. 27 patients with DCB, and 34 patients with NDB. (C) The Riaz cohort. 3 patients with complete response (CR), 12 patients with partial response (PR), 23 patients with stable disease (SD), and 27 patients with progressive disease (PD). (D) The Hugo cohort. 3 patients with complete response, 10 patients with partial response, and 13 patients with progressive disease. (E) The Miao cohort. 12 patients with clinical benefit, 8 patients with intermediate benefit, and 13 without clinical benefit. (F) The IMmotion150 cohort. There were 8 patients with CR, 15 patients with PR, 16 patients with SD, and 16 patients with PD. These patients were treated with atezolizumab and possess high Teff signature expression. (G) The Hellmann cohort. There were 23 PD-L1+ (IHC>=3) patients with DCB, and 16 PD-L1+ patients with NDB. (H) The Acquired cohort. There were 8 patients with short term progression (progression<12 month) and 6 patients with sustained response (progression>12 month). (I) The Rizvi cohort. 11 patients with DCB and 15 patients with NCB. Biopsy and genomics data were obtained close to time of progression for all patients, while baseline biopsies were lacking for many patients. For (A)-(I), we tested the association of the dichotomized CSiN scores with the ordered response categories using an ordinal Chi-Square test. (J) Boxplots of bootstrap P values evaluating the robustness of the predictive performance of CSiN, neoantigen load and the neoantigen fitness score, with each P value generated from a bootstrap resample of each cohort. Two-sided Wilcoxon signed-rank test was used to compare the bootstrap P values. NS: P>0.01, *: P=0.01–0.05, **: P=0.001–0.01, ***: P=0.0001–0.001, ****:P<0.0001.
To compare the performance of CSiN score with other widely used metrics for neoantigenicity, we also examined the predictive power of neoantigen load (Fig. S1) and the neoantigen fitness model (Fig. S2) in the same cohorts and by the same statistical tests. For consistency we used the median scores to split the cohorts. For neoantigen load, we also adopted another cutoff (median + 2 x interquartile range), developed by Zehir et al (39) (results shown in Supplementary Information). For this analysis, the neoantigen fitness model was calculated for both class I and class II neoantigens, as was done for CSiN score and neoantigen load. We provided the results of the neoantigen fitness scores calculated using class I 9-mer neoantigens from missense mutations in Supplementary Information, as was done in its original report. Overall, the neoantigen load and neoantigen fitness models were not as strongly predictive of treatment response as CSiN. We used bootstrap analysis to evaluate the statistical significance of the improvement of CSiN compared to the other two approaches, which is an accepted methodology for model comparisons (40, 41). In Fig. 2J, we show that CSiN significantly outperformed neoantigen load in 7 out of the 9 cohorts evaluated, and also outperformed neoantigen fitness in 7 out of the 9 cohorts. Overall, our results show that CSiN is capable of predicting clinical response to checkpoint inhibitors in immunogenic cancers, and demonstrated a significant improvement over existing predictive tools. We also compared the predictive power of CSiN, neoantigen load, and neoantigen fitness models using a survival analysis criterion and observed the same trends. Detailed results are shown in Supplementary Information (Fig. S13).
Finally, we also explored the predictive power of CSiN in other forms of immunotherapies. We generated genomics data for an in-house cohort (the IL-2 cohort) of ccRCC patients treated with concurrent IL-2 and Stereotactic Ablative Body Radiation (SAbR) treatment. IL-2 has pleiotropic activating effects on cytotoxic T cells, Treg cells and NK cells (42). It has been shown that SAbR has multiple immunogenic properties and could enhance the response to IL-2 (43). The neoantigen-mediated cytotoxicity probably partially explains the effects of this regimen. In this cohort, CSiN scores of patients with DCB are higher than CSiN scores of patients with NCB with marginal significance (Fig. S3A, P=0.053), and out-perform the neoantigen load and the neoantigen fitness models (Fig. S3B–C).
Higher CSiN predicts more favorable prognosis in immunogenic cancers
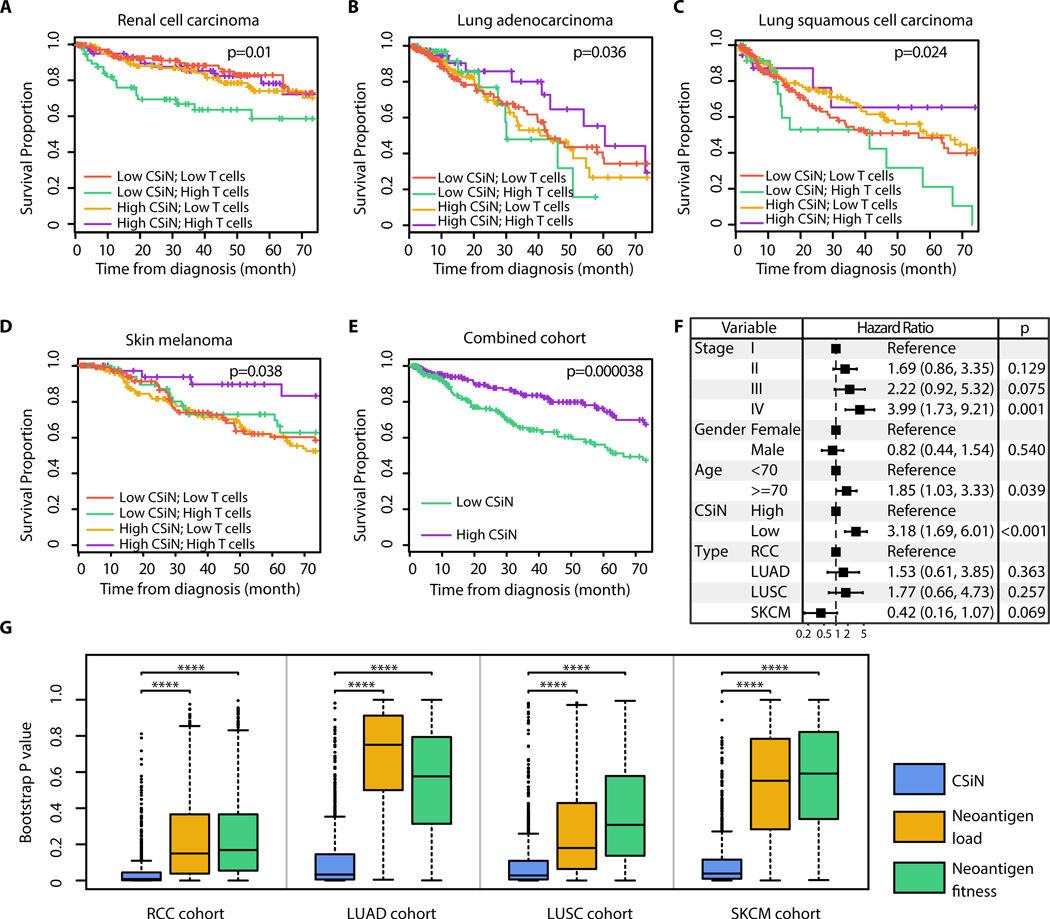
To understand the implications of neoantigen heterogeneity for long term survival of patients, we examined the association between CSiN and prognosis in the RCC, LUAD, LUSC and SKCM cohorts. We first focused on patients with high levels of T cell infiltration, profiled by our recently published empirically defined tumor microenvironment (eTME) gene expression signatures (32). We speculated that the neoantigen-T cell axis is more likely to be functionally active when T cell infiltration is present in the tumor. Interestingly, in these patients, we indeed observed that higher CSiN scores had a significantly positive association with better survival for RCC (P=0.01, Fig. 3A), LUAD (P=0.036, Fig. 3B), LUSC (P=0.024, Fig. 3C), and SKCM (P=0.038, Fig. 3D). The False Discovery Rates for the 4 cohorts fall under 10% (Table S3). However, the overall survival of patients with lower T cell infiltration was indifferent to the levels of CSiN scores, which fits our speculation. We extracted and combined the high T cell infiltration patients from all four cohorts, and carried out survival analyses, which again showed that patients with higher CSiN scores had a significantly better overall prognosis (P=3.8×10−5, Fig. 3E). To further exclude the effect of clinical confounders, we performed multivariate survival analysis adjusted by disease type, stage, gender and age in this combined cohort. The significant association between survival and CSiN was retained (P<0.001, Fig. 3F).
Fig. 3.
Association of CSiN score with overall survival of patients. (A-E) Kaplan-Meier estimator was used to visualize patient overall survival. P values for logrank tests are shown. (A) The RCC cohort. (B) The LUAD cohort. (C) The LUSC cohort. (D) The SKCM cohort. (E) The patients identified as having “High T cells” are extracted from each cohort, combined, and tested together. The high and low CSiN score designations follow those in (A-D). The top 140 RCC patients, 100 LUAD patients, 100 SKCM patients, and 40 LUSC patients with the highest T cell infiltration were designated as having “High T cells”, so the more immunogenic tumor types have more patients selected (83). (F) Forest plot for the coefficients of the multivariate CoxPH analysis of the combined cohort in (D). Disease type, pathological stage, gender, age and the binarized CSiN were included as covariates. The dotted line shows the no effect point. 95% Confidence intervals were shown as bars. (G) Boxplots of bootstrap P values evaluating the robustness of the prognostic performance of CSiN, neoantigen load and the neoantigen fitness score, with each P value generated from a bootstrap resample of each cohort. Two-sided Wilcoxon signed-rank test was used to compare the bootstrap P values. *: P=0.01–0.05, **: P=0.001–0.01, ***: P=0.0001–0.001, ****:P<0.0001.
In contrast, the same analysis for the neoantigen load and the neoantigen fitness models yielded insignificant associations (Fig. S4 and Fig. S5). We also employed bootstrap analysis to evaluate the statistical significance of this comparison. In Fig. 3G, we showed that CSiN significantly outperformed both methods in all 4 cohorts evaluated. Overall, in concordance with several previous studies that reported a lack of association of higher neoantigen load with better prognosis in several cancer types (19–21), our results suggest that the clonal distribution of neoantigens could be more prognostically important.
We also assessed non-immunogenic cancer types. Pediatric acute lymphocytic leukemia (ALL) is an aggressive childhood tumor type with low neoantigen load. We evaluated a cohort of pediatric ALL patients (pALL cohort) and observed that CSiN was not predictive of prognosis (Fig. S6A, P=0.584), with the results for neoantigen load shown in Fig. S6B and neoantigen fitness shown in Fig. S6C. We also studied liver hepatocellular carcinoma patients from The Cancer Genome Atlas (TCGA) (LIHC cohort), although whether liver cancer is an immunogenic cancer type is still under debate (44, 45). We observed patients with higher CSiN scores had a non-significant trend of better survival than patients with low CSiN scores in the high T cell infiltration subset of patients (Fig. S6D, P=0.165), with the results for neoantigen load shown in Fig. S6E and neoantigen fitness shown in Fig. S6F. These observations further confirmed that the predictive value of CSiN score is more apparent in tumor types with higher immunogenicity.
Discussion
The major biological insight from this study is that the neoantigen clonal structure in each tumor specimen and the immunogenicity of the neoantigens (represented by the MHC-binding strength in our study) are predictive of response to checkpoint inhibitors and prognosis. These factors may significantly outweigh the impact of the raw neoantigen count. Our comprehensive analyses show that the CSiN score, which describes these properties of the neoantigen profile quantitatively, possesses substantially better predictive and prognostic performance than other neoantigen-based biomarkers in the majority of evaluated cohorts. Our implementations of the CSiN, neoantigen load, and neoantigen fitness indices have considered both MHC class I and class II neoantigens, and also neoantigens generated from insertions/deletions and stop-loss mutations. This is different from the original publication of the neoantigen fitness model (16, 27) that only considered 9-mer class I neoantigens generated from missense mutations. We believe inclusion of all these potential sources of neoantigens is important for a complete characterization of the neoantigen profiles in each patient (analyses for each class of neoantigens only are shown in Supplementary Information). In alignment with our findings, McGranahan et al. made a qualitative observation that CTLA-4-resistant tumors could be enriched for subclonal mutations, which may enhance total neoantigen burden but not elicit an effective antitumor response due to the subclonal nature of these neoantigens (13). Miao et al also made a similar observation (26). Our study is distinguished from these earlier reports in that we provided a robust quantitative measurement that was subjected to systematic evaluations, and we also evaluated prognosis in addition to treatment response. Overall, CSiN may serve as a valuable predictive tool for medical oncologists treating patients with checkpoint blockade, and has addressed some of the limitations of prior neoantigen-based predictive biomarkers.
CSiN incorporates genetic information that is not captured by the neoantigen load and neoantigen fitness models or by expression-based biomarkers. A number of expression-based biomarkers for immunotherapy have been proposed and validated on different levels including, PD-L1 expression in the tumor microenvironment (46, 47), Th1-type chemokine expression (48), and T cell infiltration (49), But CSiN augments and complements, rather than replaces, these biomarkers. We show that CSiN is associated with longer survival only in patients with sufficient T cell infiltration. Similarly in the treatment cohorts, the P values for testing the correlation between CSiN and treatment response in the high T cell/high PD-L1 patient subsets are generally smaller compared with the P values of the full cohorts (Fig. 2 and Supplementary Information), even though the subset cohorts are much smaller in sample size. These results suggest that it is crucial for all components of the neoantigen-host immune axis to be functionally active, in order to enable efficient immune elimination of tumor cells. The insignificant association of CSiN with prognosis in the cohorts with cases of less immunogenic tumors including liver cancers and pediatric ALL also supports this notion. Our observations may inspire potential future studies to construct more sophisticated predictive and prognostic models that incorporate CSiN, neoantigen load, the neoantigen fitness model and other biomarkers together for improved performance.
Our results reporting the positive correlation between neoantigens and treatment response in RCCs are interesting. Currently, the field is still debating the role of neoantigens in the immune response to RCCs. While RCCs have low neoantigen/mutation loads, Turajlic et al discovered that RCCs have the highest number of insertion/deletion mutations on a pan-cancer basis (50), which tend to encode high quality neoantigens. In terms of predicting survival after immunotherapy treatment, Samstein et al reported a significant correlation between tumor mutation burden and progression-free survival (51), while this observation was not made in the phase 3 JAVELIN Renal 101 trial (52). Cherkasova et al discovered the re-activation of a HERV-E retrovirus in RCCs, which can encode an immunogenic peptide recognizable by cytotoxic T cells (53). It is highly likely both neoantigens and self antigens contribute to the immunogenicity in RCCs. In the future, it will be of interest to investigate in more and larger cohorts whether CSiN is more predictive of patients’ response to immunotherapies when such antigens have been incorporated in the calculation,.
One limitation of our study and, to some extent, this field, is that we used neoantigens predicted from genomics data for correlation with patient phenotypes. Despite our efforts to validate the neoantigen predictions, it is likely there are still false positive and false negative predicted neoantigens that convoluted our analyses. In future studies, incorporating the genomics-based approach with other methods, such as mass spectrometry(54), may improve the sensitivity and specificity of neoantigen detection, and thus further enhance the predictive power of CSiN.
Class I and class II neoantigens represent very different aspects of immune response. But several recent studies have implicated neoantigen-specific CD4+ T-cells in direct tumor clearance (55–57). Our results also suggest that the inclusion of class II neoantigens is important for treatment response prediction. In the future, it will be interesting to carry out comprehensive research into the roles of class II neoantigens in the tumor microenvironment.
The neoantigen repertoire is in constant dynamic evolution (2, 58), with immunoediting and immunotherapy treatments actively modifying its landscape. CSiN offers a new tool to monitor the neoantigen profiles, where different tumor clones could have different growth advantages subject to the pressure of T cell cytotoxicity determined by each clone’s neoantigen composition. Overall, our work offers a rigorous methodology of predicting response to immunotherapy and prognosis from routine patient samples, and should be useful for personalizing medicine in the modern era of immunotherapy.
Materials and Methods
Study Design
The objective of this research project is to study the implication of the clonal structure of neoantigens for predicting treatment response and prognosis. The research subjects were individual cancer patients, some of whom received immunotherapy. As this is a retrospective analysis study, the researchers were not blinded to the allocation labels. We included all available samples and data from either public or private sources in our study. We stopped the data collection on a lock date of September 1, 2019. The endpoints considered were response categories and survival of cancer patients following baseline assessment and in some cases immunotherapy. Usually just one sample per patient was available. In the uncommon cases of more than one sample collected for an individual patient, we averaged the scores (e.g. CSiN) calculated from each sample.
The Cauchy–Schwarz index of Neoantigens and the other neoantigen-based metrics used for comparison
The CSiN score considers the pairing between the repertoire of neoantigens and the tumor mutations to which they belong. One way to characterize this property is to average the product of the variant allele frequencies (VAF) of somatic mutations and the number of neoantigens generated by each mutation, normalized by the average VAF and average mutation-specific neoantigen load in each patient. The CSiN score is >1 under favorable pairing, and vice versa. This forms the backbone of the final CSiN score. The name CSiN was selected because the pairing of tumor mutations and neoantigens and its effect on the overall score bear analogy to the Cauchy-Schwarz inequality, which describes the upper bound of the product sum of two vectors of real numbers and the condition for the equality to be achieved. Refer to Supplementary Information for the implementation details of CSiN and additional analyses involving CSiN which are not shown in the main text. A cartoon showing the workflow of CSiN is shown in Fig. S7.
As a comparison for CSiN scores, we also calculated the neoantigen load and the neoantigen fitness scores. Published studies have calculated neoantigen load in slightly different manners, due to various reasons such as usage of different software for mutation calling (17, 18, 22, 35, 68, 69). Our neoantigen load calculations were based on our own mutation calling and neoantigen calling pipelines when raw genomics data were available, and follow the general standards for neoantigen load calculation. Specifically, we counted the total number of neoantigens of both class I and class II (if class II neoantigens were available from our pipelines or from the original reports when raw genomics data were not available), all HLA alleles, and all registers from all missense, indel, and stop-loss mutations. The neoantigen fitness calculation was done as described previously (16, 27), but included both class I and class II neoantigens (when class II neoantigens are available). In Supplementary Information, we have also shown the results for the neoantigen fitness scores calculated with only class I 9-mer neoantigens from missense mutations.
The QBRC mutation calling pipeline
We used the QBRC mutation calling pipeline for somatic mutation calling, developed in the Quantitative Biomedical Research Center (QBRC) of UT Southwestern Medical Center. In short, exome-seq reads were aligned to the human reference genome by BWA-MEM (59). Picard was used to add read group information and sambamba was used to mark PCR duplicates. The GATK toolkit (60–62) was used to perform base quality score recalibration and local realignment around Indels. MuTect (63), VarScan (64), Shimmer, SpeedSeq (65), Manta, and Strelka2 (66) were used to call SNPs and Indels. A mutation that was repeatedly called by any two of these software programs was retained. Annovar was used to annotate SNPs and Indels, and protein sequence changes (67). All SNPs and Indels were combined and only kept if there were at least 7 total (wild type and variant) reads in the normal sample and at least 3 variant reads in the tumor sample. Somatic mutations and germline mutations were annotated according to the variant allele frequencies in the normal and tumor samples.
The QBRC neoantigen calling pipeline
We used the QBRC neoantigen calling pipeline for neoantigen calling, which follows similar standards as those pipelines used in previous publications (17, 18, 22, 35, 68, 69). The validity of our neoantigen predictions is demonstrated in Supplementary Information. We kept only frameshift, non-frameshift, missense and stop-loss mutations that would lead to protein sequence changes. We kept only somatic mutations whose variant allele frequencies (VAFs) were <0.02 in the normal sample and VAFs>0.05 in the tumor samples. For class I HLA proteins (A, B, C), we predicted putative neoantigens of 8–11 amino acids in length, and for class II HLA proteins (DRB1 and DQB1/DQA1), we predicted putative neoantigens of 15 amino acids in length. Class I and II HLA subtypes were predicted by the ATHLATES tool (70), which has been shown to be accurate for these HLA alleles (71). Samples whose total successfully typed HLA alleles (counting both chromosomes) were <8 were regarded as poor quality data, and were left out from downstream analyses. Putative neoantigens with amino acid sequences exactly matching known human protein sequences were filtered out. For class I bindings, the IEDB-recommended mode (http://tools.iedb.org/main/) was used for prediction of binding affinities, while for class II binding, NetMHCIIpan embedded in the IEDB toolkit was. Neoantigens were kept only if the predicted ranks of binding affinities were ≤2%. Tumor RNA-seq data, if available, were aligned to the reference genome using the STAR aligner (72). FeatureCounts was used to summarize gene expression levels (73). Neoantigens whose corresponding mutations were in genes with expression level <1 RPKM in either the specific exon or the whole transcript were filtered out. For the samples analyzed by our pipeline, we applied the filtering by RNA-seq data on the neoantigen list when RNA-Seq data are available. We showed the results for calculating CSiN with only exome-Seq data in Supplementary Information, which indicated that filtering the neoantigen list by RNA-Seq data is important for the predictive performance of CSiN. Finally, in accordance with our pipeline, Ott et al (74) have shown that neoantigens of class I and class II, and also different registers can all possibly be immunogenic, and IEDB (https://www.iedb.org/) has documented many immunogenic antigens of different peptide lengths.
Patient cohorts
For all patient cohorts, if approval of access to raw exome-seq and RNA-Seq data was obtained, we predicted the somatic mutations and neoantigens using our in-house pipelines. In cases where raw genomics data were not accessible, we analyzed the processed neoantigen and mutation data included in the supplemental files of the original publications. For exploratory analysis and association with overall survival, we collected data from 110 RCC patients from the Kidney Cancer Program at UT Southwestern Medical Center (32), 94 ccRCC patients from Sato et al (23), and 162 ccRCC patients from TCGA (75) (the RCC cohort); 427 lung adenocarcinoma patients from TCGA (76) (the LUAD cohort); 389 lung squamous cell carcinoma patients from TCGA (77) (the LUSC cohort); 401 melanoma patients from TCGA (78) (the SKCM cohort); 103 pediatric and young adult T-lineage acute lymphoblastic leukemia patients (the pALL cohort) from Liu et al (24); and 292 liver cancer patients from TCGA (79) (the LIHC cohort) (Table S1).
For association of CSiN scores with immunotherapy response, 10 patient cohorts were collected that are summarized in Table S1. Table S1 shows the total number of patients within each cohort used in this study for correlation of treatment response and neoantigen-based predictive markers. In the VanAllen cohort, there are 40 patients with matched RNA-Seq and exome-seq data and three of these patients were removed due to lack of response information. For the Riaz cohort, three patients were removed as two of them have lack of response information (“not evaluated” reported by the original study) and the third patient’s HLA alleles cannot be typed accurately from the sequencing data. In the Miao cohort, two patients were removed due to the HLA typing issue. In the Rizvi cohort, three patients were removed due to lack of response information (not reaching six-month follow up) and five patients were removed due an HLA typing issue. In the Snyder cohort, three patients were removed as the neoantigen data were not available from the original publication (we used neoantigens called by the original study for this cohort as we don’t have access to the raw genomics data). In the IMmotion150 cohort, 99 patients on atezolizumab and 50 patients on sunitinib had both exome-seq and RNA-Seq. Three patients treated by atezolizumab and four patients treated by sunitinib were further removed because response information was not available. For the Hellmann cohort, the neoantigen lists were generated by the original authors and used by us in this work. There are 74 patients with neoantigen data generated. Only whole exome-seq was done and only about half of these patients consented to genomics data sharing, so we decided to use these neoantigens called by the original report. For the Hugo cohort, 28 patients had both exome-seq and RNA-Seq data, and two were removed due to an HLA typing issue. Stratified analyses (Fig. 2 and Supplementary Information) were performed for the IMmotion150, VanAllen, Hugo, Riaz and Miao cohorts (with RNA-Seq data available) based on ssGSEA analyses (80) of the Teff gene signature (81) to focus on the top 60% patients, and also performed for the Hellmann cohort based on PD-L1 IHC level with a cutoff on IHC readings (which are given in integers by the original report for 69 patients) chosen such that the top subset is closest to 60% of the whole cohort. For cases in which multiple types of treatment outcomes were recorded in a cohort, we used the primary criterion employed by the original publications. Patient samples that were analyzed but were not shown in Fig. 2 (e.g. IMmotion150 patients on the sunitinib arm) were shown in Supplementary Information.
Statistical analyses
All computations and statistical analyses were carried out in the R computing environment. For all boxplots appearing in this study, box boundaries represent interquartile ranges, whiskers extend to the most extreme data point which is no more than 1.5 times the interquartile range, and the line in the middle of the box represents the median. For association of CSiN score, neoantigen load and the neoantigen fitness model with binary/categorical response variables, we tested the association of the dichotomized CSiN scores with the ordered response categories (e.g. complete response→partial response→stable disease→progressive disease) using an ordinal chi-square test. The three types of predictive scores are each split on the median within each cohort, to ensure comparability and avoid the scaling problem. The alternative hypothesis is that patients with better response and survival outcome are more likely to have higher CSiN scores. We employed the chisq_test function in the R coin package for this purpose. For survival-type analysis, we adopted the log-rank test for evaluating whether patients with higher CSiN scores had better prognosis. T cell infiltrations and activation of the IFN-γ signature were predicted using the single sample gene set enrichment analysis (ssGSEA) method (80). ssGSEA analysis was performed using the R GSVA package by calling the gsva function with parameter method=“ssgsea” and rnaseq=T (82). The forest plot was performed by the forest_model function in the R forestmodel package. For model comparison, 5,000 bootstrap resamples of each cohort were generated, and each resample was used to evaluate the predictive or prognostic performance of CSiN, neoantigen load and the neoantigen fitness score. The P values of 5,000 bootstraps of each approach were compared using two-sided Wilcoxon signed-rank test.
Supplementary Material
Table S2 Processed mutation, expression and neoantigen data of the IL-2 cohort (in Excel spreadsheet).
Fig. S1 Predictive power of neoantigen load.
Fig. S2 Predictive power of the neoantigen fitness model.
Fig. S3 Association of CSiN (a), neoantigen loads (b) and neoantigen fitness (c) with IL-2/SAbR treatment response in ccRCC patients.
Fig. S4 Prognostic power of neoantigen load.
Fig. S5 Prognostic power of the neoantigen fitness model.
Fig. S6 Association of CSiN, neoantigen loads and neoantigen fitness with prognosis of pediatric ALL patients and LIHC patients.
Fig. S7 Cartoon showing the workflow of calculation of CSiN scores.
Table S1 The patient cohorts used in this study.
Table S3 P values and False Discovery Rates of the tested cohorts shown in Fig. 2 and Fig. 3.
Acknowledgements:
We acknowledge Jessie Norris for her helpful comments on the writing of the paper. We acknowledge Dr. Ying Wang for her helpful comments on the scientific contents. We acknowledge the Wakeland sequencing core of UT Southwestern for generating genomics data for the IL-2 cohort. We acknowledge the authors of the phs000452.v2.p1, the phs001493.v1.p1, the phs001464.v1.p1, the phs000218.v1.p1, and the phs000464.v15.p7 datasets, as well as the funding agencies that supported these studies and dbGaP that supported the archival of these datasets.
Funding: This study was supported by grants from the National Institutes of Health (R03 ES026397–01 to T.W.; SPORE 1P50CA19651601 to J.B. and T.W., and CCSG 5P30CA142543 to T.W.), the Cancer Prevention Research Institute of Texas (CPRIT RP180319 to LX and CPRIT RP190208 to T.W.), and American Cancer Society (RSG-16–004-01-CCE to R.H.).
Footnotes
Supplementary Information. A comprehensive explanation of the CSiN score. Includes embedded figures S8–S23.
Competing interests: The authors declare they have no competing interests.
Data and Materials Availability: The QBRC mutation calling and neoantigen calling pipelines are available on GitHub: https://github.com/tianshilu/QBRC-Somatic-Pipeline and https://github.com/tianshilu/QBRC-Neoantigen-Pipeline. The CSiN calculations were embedded in the neoantigen pipeline. Table S1 lists the data source and/or accession methods of these studies. The raw RNA-Seq and exome-seq data of the IL-2 cohort patients can be downloaded from the European Genome-phenome Archive with accession number EGAS00001003605 through controlled access. The processed mutation and neoantigen data of the IL-2 cohort can be found in Table S2.
References and Notes:
- 1.Ascierto PA, Marincola FM, 2015: The Year of Anti-PD-1/PD-L1s Against Melanoma and Beyond., EBioMedicine 2, 92–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, Hruban C, Guthrie VB, Rodgers K, Naidoo J, Kang H, Sharfman W, Georgiades C, Verde F, Illei P, Li QK, Velculescu VE, Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer., Cancer Discov. 7, 264–276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR, KEYNOTE-024 Investigators, Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer., N. Engl. J. Med. 375, 1823–1833 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Chan TA, Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer., Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assi HI, Kamphorst AO, Moukalled NM, Ramalingam SS, Immune checkpoint inhibitors in advanced non-small cell lung cancer., Cancer 124, 248–261 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, Rodig SJ, Chapuy B, Ligon AH, Zhu L, Grosso JF, Kim SY, Timmerman JM, Shipp MA, Armand P, PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma., N. Engl. J. Med. 372, 311–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasamon YL, de Claro RA, Wang Y, Shen YL, Farrell AT, Pazdur R, FDA approval summary: nivolumab for the treatment of relapsed or progressive classical hodgkin lymphoma., Oncologist 22, 585–591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seidel JA, Otsuka A, Kabashima K, Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations., Front. Oncol. 8, 86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R-F, Wang HY, Immune targets and neoantigens for cancer immunotherapy and precision medicine., Cell Res. 27, 11–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yarchoan M, Johnson BA, Lutz ER, Laheru DA, Jaffee EM, Targeting neoantigens to augment antitumour immunity., Nat. Rev. Cancer 17, 209–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirth TC, Kühnel F, Neoantigen Targeting-Dawn of a New Era in Cancer Immunotherapy?, Front. Immunol 8, 1848 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strønen E et al. , Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 352, 1337–1341 (2016). [DOI] [PubMed] [Google Scholar]
- 13.McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TBK, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, Van Allen EM, Miao D, Swanton C, Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade., Science 351, 1463–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS, Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma., Cell 165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH, Bhatia S, Hwu W-J, Gajewski TF, Slingluff CL, Chowell D, Kendall SM, Chang H, Shah R, Kuo F, Morris LGT, Chan TA, Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab., Cell 171, 934–949.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, Rizvi NA, Merghoub T, Levine AJ, Chan TA, Wolchok JD, Greenbaum BD, A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy., Nature 551, 517–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, Norton C, Bossé D, Wankowicz SM, Cullen D, Horak C, Wind-Rotolo M, Tracy A, Giannakis M, Hodi FS, Drake CG, Ball MW, Allaf ME, Snyder A, Hellmann MD, Van Allen EM, Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma., Science 359, 801–806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB, Abu-Akeel M, Liu C, Sauter JL, Rekhtman N, Chang E, Callahan MK, Chaft JE, Voss MH, Tenet M, Li X-M, Covello K, Wolchok JD, Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer., Cancer Cell 33, 843–852.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsushita H, Sato Y, Karasaki T, Nakagawa T, Kume H, Ogawa S, Homma Y, Kakimi K, Neoantigen load, antigen presentation machinery, and immune signatures determine prognosis in clear cell renal cell carcinoma., Cancer Immunol. Res. 4, 463–471 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Miller A, Asmann Y, Cattaneo L, Braggio E, Keats J, Auclair D, Lonial S, MMRF CoMMpass Network SJ Russell, A. K. Stewart, High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma., Blood Cancer J. 7, e612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsushita H, Hasegawa K, Oda K, Yamamoto S, Nishijima A, Imai Y, Asada K, Ikeda Y, Karasaki T, Fujiwara K, Aburatani H, Kakimi K, The frequency of neoantigens per somatic mutation rather than overall mutational load or number of predicted neoantigens per se is a prognostic factor in ovarian clear cell carcinoma., Oncoimmunology 6, e1338996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gettinger S, Choi J, Hastings K, Truini A, Datar I, Sowell R, Wurtz A, Dong W, Cai G, Melnick MA, Du VY, Schlessinger J, Goldberg SB, Chiang A, Sanmamed MF, Melero I, Agorreta J, Montuenga LM, Lifton R, Ferrone S, Politi K, Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer., Cancer Discov. 7, 1420–1435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Ogawa S, Integrated molecular analysis of clear-cell renal cell carcinoma., Nat. Genet. 45, 860–867 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, McCastlain K, Edmonson M, Pounds SB, Shi L, Zhou X, Ma X, Sioson E, Li Y, Rusch M, Gupta P, Pei D, Cheng C, Smith MA, Auvil JG, Mullighan CG, The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia., Nat. Genet. 49, 1211–1218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C, Clonal status of actionable driver events and the timing of mutational processes in cancer evolution., Sci. Transl. Med. 7, 283ra54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao D, Margolis CA, Vokes NI, Liu D, Taylor-Weiner A, Wankowicz SM, Adeegbe D, Keliher D, Schilling B, Tracy A, Manos M, Chau NG, Hanna GJ, Polak P, Rodig SJ, Signoretti S, Sholl LM, Engelman JA, Getz G, Jänne PA, Van Allen EM, Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors., Nat. Genet. 50, 1271–1281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, Wells DK, Cary CIO, Grbovic-Huezo O, Attiyeh M, Medina B, Zhang J, Loo J, Saglimbeni J, Abu-Akeel M, Zappasodi R, Leach SD, Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer., Nature 551, 512–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The problem with neoantigen prediction., Nat. Biotechnol. 35, 97 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, Melief CJ, Oseroff C, Yuan L, Ruppert J, Sidney J, del Guercio MF, Southwood S, Kubo RT, Chesnut RW, Grey HM, Chisari FV, The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes., J. Immunol. 153, 5586–5592 (1994). [PubMed] [Google Scholar]
- 30.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui H-H, Grey H, Sette A, A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus., Nat. Biotechnol. 24, 817–819 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Andreatta M, Karosiene E, Rasmussen M, Stryhn A, Buus S, Nielsen M, Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification., Immunogenetics 67, 641–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Lu R, Kapur P, Jaiswal BS, Hannan R, Zhang Z, Pedrosa I, Luke JJ, Zhang H, Goldstein LD, Yousuf Q, Gu Y-F, McKenzie T, Joyce A, Kim MS, Wang X, Luo D, Onabolu O, Stevens C, Xie Z, Brugarolas J, An empirical approach leveraging tumorgrafts to dissect the tumor microenvironment in renal cell carcinoma identifies missing link to prognostic inflammatory factors., Cancer Discov. 8, 1142–1155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott PA, Hodi FS, Robert C, CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients., Clin. Cancer Res. 19, 5300–5309 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, Yearley J, Seiwert TY, Ribas A, McClanahan TK, IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade., J. Clin. Invest. 127, 2930–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Garraway LA, Genomic correlates of response to CTLA-4 blockade in metastatic melanoma., Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA, Genetic basis for clinical response to CTLA-4 blockade in melanoma., N. Engl. J. Med. 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, Fong L, Joseph RW, Pal SK, Reeves JA, Sznol M, Hainsworth J, Rathmell WK, Stadler WM, Hutson T, Gore ME, Ravaud A, Bracarda S, Suárez C, Danielli R, Powles T, Publisher Correction: Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma., Nat. Med. 24, 1941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB, Abu-Akeel M, Liu C, Sauter JL, Rekhtman N, Chang E, Callahan MK, Chaft JE, Voss MH, Tenet M, Li X-M, Covello K, Wolchok JD, Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer., Cancer Cell 33, 843–852.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, Hellmann MD, Barron DA, Schram AM, Hameed M, Dogan S, Ross DS, Hechtman JF, DeLair DF, Yao J, Mandelker DL, Berger MF, Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients., Nat. Med. 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sieberts SK, Zhu F, García-García J, Stahl E, Pratap A, Pandey G, Pappas D, Aguilar D, Anton B, Bonet J, Eksi R, Fornés O, Guney E, Li H, Marín MA, Panwar B, Planas-Iglesias J, Poglayen D, Cui J, Falcao AO, Mangravite LM, Crowdsourced assessment of common genetic contribution to predicting anti-TNF treatment response in rheumatoid arthritis., Nat. Commun. 7, 12460 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costello JC, Heiser LM, Georgii E, Gönen M, Menden MP, Wang NJ, Bansal M, Ammad-ud-din M, Hintsanen P, Khan SA, Mpindi J-P, Kallioniemi O, Honkela A, Aittokallio T, Wennerberg K, NCI DREAM Community JJ Collins, D. Gallahan, D. Singer, J. Saez-Rodriguez, G. Stolovitzky, A community effort to assess and improve drug sensitivity prediction algorithms., Nat. Biotechnol. 32, 1202–1212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito S, Bollard CM, Carlsten M, Melenhorst JJ, Biancotto A, Wang E, Chen J, Kotliarov Y, Cheung F, Xie Z, Marincola F, Tanimoto K, Battiwalla M, Olnes MJ, Perl S, Schum P, Hughes TE, Keyvanfar K, Hensel N, Muranski P, Barrett AJ, Ultra-low dose interleukin-2 promotes immune-modulating function of regulatory T cells and natural killer cells in healthy volunteers., Mol. Ther. 22, 1388–1395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finkelstein SE, Timmerman R, McBride WH, Schaue D, Hoffe SE, Mantz CA, Wilson GD, The confluence of stereotactic ablative radiotherapy and tumor immunology., Clin. Dev. Immunol. 2011, 439752 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardee AD, Butterfield LH, Immunotherapy of hepatocellular carcinoma: Unique challenges and clinical opportunities., Oncoimmunology 1, 48–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harding JJ, El Dika I, Abou-Alfa GK, Immunotherapy in hepatocellular carcinoma: Primed to make a difference?, Cancer 122, 367–377 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Curiel TJ, Wei S, Dong H, Alvarez X, Cheng P, Mottram P, Krzysiek R, Knutson KL, Daniel B, Zimmermann MC, David O, Burow M, Gordon A, Dhurandhar N, Myers L, Berggren R, Hemminki A, Alvarez RD, Emilie D, Curiel DT, Zou W, Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity., Nat. Med. 9, 562–567 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Lin H, Wei S, Hurt EM, Green MD, Zhao L, Vatan L, Szeliga W, Herbst R, Harms PW, Fecher LA, Vats P, Chinnaiyan AM, Lao CD, Lawrence TS, Wicha M, Hamanishi J, Mandai M, Kryczek I, Zou W, Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression., J. Clin. Invest. 128, 805–815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, Sun Y, Zhao E, Vatan L, Szeliga W, Kotarski J, Tarkowski R, Dou Y, Cho K, Hensley-Alford S, Munkarah A, Liu R, Zou W, Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy., Nature 527, 249–253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN, Rubin SC, Coukos G, Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer., N. Engl. J. Med. 348, 203–213 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, Wong YNS, Rowan A, Kanu N, Al Bakir M, Chambers T, Salgado R, Savas P, Loi S, Birkbak NJ, Sansregret L, Gore M, Larkin J, Quezada SA, Swanton C, Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis., Lancet Oncol. 18, 1009–1021 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Samstein RM, Lee C-H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, Motzer RJ, Hakimi AA, Voss MH, Russo P, Rosenberg J, Iyer G, Bochner BH, Bajorin DF, Morris LGT, Tumor mutational load predicts survival after immunotherapy across multiple cancer types., Nat. Genet. 51, 202–206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choueiri TK, Albiges L, Haanen JBAG, Larkin JMG, Uemura M, Pal SK, Gravis G, Campbell MT, Penkov K, Lee J-L, Ching KA, Mu XJ, Wang X, Zhang W, Wang J, Chudnovsky A, di Pietro A, Robbins PB, Motzer RJ, Biomarker analyses from JAVELIN Renal 101: Avelumab + axitinib (A+Ax) versus sunitinib (S) in advanced renal cell carcinoma (aRCC)., J. Clin. Oncol. 37, 101–101 (2019). [Google Scholar]
- 53.Cherkasova E, Scrivani C, Doh S, Weisman Q, Takahashi Y, Harashima N, Yokoyama H, Srinivasan R, Linehan WM, Lerman MI, Childs RW, Detection of an Immunogenic HERV-E Envelope with Selective Expression in Clear Cell Kidney Cancer., Cancer Res. 76, 2177–2185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bassani-Sternberg M, Bräunlein E, Klar R, Engleitner T, Sinitcyn P, Audehm S, Straub M, Weber J, Slotta-Huspenina J, Specht K, Martignoni ME, Werner A, Hein R, H Busch D, Peschel C, Rad R, Cox J, Mann M, Krackhardt AM, Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry., Nat. Commun. 7, 13404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, Parkhurst MR, Yang JC, Rosenberg SA, Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer., Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schumacher T, Bunse L, Pusch S, Sahm F, Wiestler B, Quandt J, Menn O, Osswald M, Oezen I, Ott M, Keil M, Balß J, Rauschenbach K, Grabowska AK, Vogler I, Diekmann J, Trautwein N, Eichmüller SB, Okun J, Stevanović S, Platten M, A vaccine targeting mutant IDH1 induces antitumour immunity., Nature 512, 324–327 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, Boegel S, Schrörs B, Vascotto F, Castle JC, Tadmor AD, Schoenberger SP, Huber C, Türeci Ö, Sahin U, Mutant MHC class II epitopes drive therapeutic immune responses to cancer., Nature 520, 692–696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verdegaal EME, de Miranda NFCC, Visser M, Harryvan T, van Buuren MM, Andersen RS, Hadrup SR, van der Minne CE, Schotte R, Spits H, Haanen JBAG, Kapiteijn EHW, Schumacher TN, van der Burg SH, Neoantigen landscape dynamics during human melanoma-T cell interactions., Nature 536, 91–95 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform., Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ, A framework for variation discovery and genotyping using next-generation DNA sequencing data., Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data., Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA, From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline., Curr. Protoc. Bioinformatics 11, 1110.1–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G, Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples., Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK, VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing., Genome Res. 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiang C, Layer RM, Faust GG, Lindberg MR, Rose DB, Garrison EP, Marth GT, Quinlan AR, Hall IM, SpeedSeq: ultra-fast personal genome analysis and interpretation., Nat. Methods 12, 966–968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK, Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs., Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Wang K, Li M, Hakonarson H, ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data., Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turajlic S, Xu H, Litchfield K, Rowan A, Chambers T, Lopez JI, Nicol D, O’Brien T, Larkin J, Horswell S, Stares M, Au L, Jamal-Hanjani M, Challacombe B, Chandra A, Hazell S, Eichler-Jonsson C, Soultati A, Chowdhury S, Rudman S, TRACERx Renal Consortium, Tracking cancer evolution reveals constrained routes to metastases: tracerx renal., Cell 173, 581–594.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS, Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma., Cell 168, 542 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Liu C, Yang X, Duffy B, Mohanakumar T, Mitra RD, Zody MC, Pfeifer JD, ATHLATES: accurate typing of human leukocyte antigen through exome sequencing., Nucleic Acids Res. 41, e142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larjo A, Eveleigh R, Kilpeläinen E, Kwan T, Pastinen T, Koskela S, Partanen J, Accuracy of Programs for the Determination of Human Leukocyte Antigen Alleles from Next-Generation Sequencing Data., Front. Immunol. 8, 1815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner., Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features., Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, Chen C, Olive O, Carter TA, Li S, Lieb DJ, Eisenhaure T, Gjini E, Stevens J, Lane WJ, Javeri I, Wu CJ, An immunogenic personal neoantigen vaccine for patients with melanoma., Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cancer Genome Atlas Research Network, Comprehensive molecular characterization of clear cell renal cell carcinoma., Nature 499, 43–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cancer Genome Atlas Research Network, Comprehensive molecular profiling of lung adenocarcinoma., Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cancer Genome Atlas Research Network, Comprehensive genomic characterization of squamous cell lung cancers., Nature 489, 519–525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cancer Genome Atlas Network, Genomic classification of cutaneous melanoma., Cell 161, 1681–1696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu, Cancer Genome Atlas Research Network, Comprehensive and integrative genomic characterization of hepatocellular carcinoma., Cell 169, 1327–1341.e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Fröhling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Hahn WC, Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1., Nature 462, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, Fong L, Joseph RW, Pal SK, Reeves JA, Sznol M, Hainsworth J, Rathmell WK, Stadler WM, Hutson T, Gore ME, Ravaud A, Bracarda S, Suárez C, Danielli R, Powles T, Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma., Nat. Med. 24, 749–757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hänzelmann S, Castelo R, Guinney J, GSVA: gene set variation analysis for microarray and RNA-seq data., BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Şenbabaoğlu Y, Gejman RS, Winer AG, Liu M, Van Allen EM, de Velasco G, Miao D, Ostrovnaya I, Drill E, Luna A, Weinhold N, Lee W, Manley BJ, Khalil DN, Kaffenberger SD, Chen Y, Danilova L, Voss MH, Coleman JA, Russo P, Hakimi AA, Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures., Genome Biol. 17, 231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2 Processed mutation, expression and neoantigen data of the IL-2 cohort (in Excel spreadsheet).
Fig. S1 Predictive power of neoantigen load.
Fig. S2 Predictive power of the neoantigen fitness model.
Fig. S3 Association of CSiN (a), neoantigen loads (b) and neoantigen fitness (c) with IL-2/SAbR treatment response in ccRCC patients.
Fig. S4 Prognostic power of neoantigen load.
Fig. S5 Prognostic power of the neoantigen fitness model.
Fig. S6 Association of CSiN, neoantigen loads and neoantigen fitness with prognosis of pediatric ALL patients and LIHC patients.
Fig. S7 Cartoon showing the workflow of calculation of CSiN scores.
Table S1 The patient cohorts used in this study.
Table S3 P values and False Discovery Rates of the tested cohorts shown in Fig. 2 and Fig. 3.