Abstract
In 2014, our research network was involved in the evaluation of favipiravir, an anti‐influenza polymerase inhibitor, against Ebola virus. In this review, we discuss how mathematical modeling was used, first to propose a relevant dosing regimen in humans, and then to optimize its antiviral efficacy in a nonhuman primate (NHP) model. The data collected in NHPs were finally used to develop a model of Ebola pathogenesis integrating the interactions among the virus, the innate and adaptive immune response, and the action of favipiravir. We conclude the review of this work by discussing how these results are of relevance for future human studies in the context of Ebola virus, but also for other emerging viral diseases for which no therapeutics are available.
Specific aspects of modernization, such as rapid air transit, as well as demographic trends, including urbanization, have accelerated both the (re)emergence and the spread of viruses, 1 exemplified in the last years by the large outbreaks of Zika, Chikungunya, Lassa, or Ebola viruses. Besides these iconic examples, about 200 human pathogenic viruses have been discovered and 200–300 species remain to be discovered, 2 representing a threat for public health. Although nearly 100 antiviral drugs have been approved since the approval of idoxuridine in 1963 (an anti‐herpes virus drug), these molecules are licensed in a very narrow range of only nine human viruses (HIV, hepatitis C virus (HCV), herpes simplex virus, influenza, hepatitis B virus, Varicella zoster virus, human cytomegalovirus, respiratory syncytial virus, and human papillomavirus), with anti‐HIV and anti‐HCV drugs representing more than half of these. 3 Although some agents in development recently showed encouraging results in animal models against Ebola virus (EBOV) 4 , 5 or Zika virus, 6 there are no antiviral drugs available against the vast majority of human pathogenic viruses, including the emerging ones. Given the cost of drug development and the epidemiology of these infections that mostly affect low‐income countries, it is unlikely that the drug industry will invest largely in the development of specific antiviral compounds. In this context, it is the role of academic research groups to look for innovative antiviral strategies against emerging viral infections.
In the last years, our group was involved in the evaluation of favipiravir, an anti‐influenza drug approved in Japan, that has shown activity against a variety of other RNA viruses, in particular Ebola, Lassa fever, Marburg, Nipah, and Zika viruses. 7 , 8 , 9 The main advantages of applying an approved agent to treat a new indication, a process called “drug repurposing,” are that the drug is available in large quantities and that safety studies have already been conducted on large populations. 10 Consequently, it can be used more rapidly on a large scale and be particularly relevant as a first line of protection to administer to suspect or contact cases during a pathogen outbreak. However, the data available for a specific pathogen are often limited, and specific methodologies are needed to leverage the information that has been collected with other pathogens or in animal models. This is particularly crucial as the antiviral efficacy of a repurposed drug can be lower than what could be obtained with a specific antiviral agent.
Following what has been done for other chronic or acute viral infections, 11 , 12 , 13 we here show how the techniques of mathematical modeling have been instrumental to better understand the immune‐pathogenesis of Ebola virus disease (EVD) and to optimize the use of favipiravir. In addition, we discuss the implications of this work for other emerging viruses.
EBOLA VIRUS DISEASE
EVD is caused by infection with a virus of the family Filoviridae, genus Ebolavirus. Four highly pathogenic species have been identified in humans: Zaire ebolavirus, Sudan ebolavirus, Taï Forest ebolavirus, and Bundibugyo ebolavirus. 14 Zaire ebolavirus is the causative specie of the 2013–2016 West Africa outbreak and the current Democratic Republic of Congo outbreak, responsible of > 95% of the reported fatality cases since the first description of the virus in 1976, and is, therefore, the specie of choice to develop animal models of the disease. Depending on the viral strain and available medical care, case fatality rate ranges from 30% up to 90% 15 , 16 and it was 40% during the 2013–2016 West Africa outbreak. After an incubation period of 2–21 days, symptomatic patients enter an acute phase of infection during which they are highly contagious. 14 Early symptoms include fever, asthenia, myalgia, which then evolve to severe gastrointestinal syndrome and possibly multi‐organ failure leading to severe renal impairment, hemorrhage syndrome, and shock. 17
The virus has a broad cell tropism; it disseminates preferentially in blood and lymph circulation early in the infection, but then infects monocytes, hepatocytes, adrenocortical cells, fibroblast, and epithelial cells. 18 , 19 In vitro experiments suggest that the eclipse phase ranges between 2 and 15 hours. 20 , 21 Cell apoptosis is caused by direct viral‐induced cytopathic effects but also by the release of multiple pro‐inflammatory mediators, including TNFα, interleukin (IL)‐1, IL‐6, and nitric oxide by activated monocytes/macrophages. 22 Consistent with EVD pathogenesis, high levels of viral load and cytokines (IL6, IL10, IL1β, TNFα, MIP1α, MIP1β, and MCP1) at study inclusion were associated with a poor EVD prognosis. 23 , 24 , 25 , 26 , 27 A global immunosuppression state along with altered adaptive responses, as suggested by the high level of T cells expressing inhibitory molecules CTLA‐4 and PD‐1, was also associated with fatal outcome. 25 In contrast, a strong CD8 T cell response was observed in survivor patients, 28 suggesting that the adaptive immune response is involved in viral clearance.
FAVIPIRAVIR IN HUMANS INFECTED WITH EVD
The 2013–2016 Ebola epidemic has been the largest outbreak of the virus since its discovery in 1976. Spread over Guinea, Liberia, and Sierra Leone, 28,616 cases of patients with EVD have been reported, of which 11,310 were fatal. 29 Although the epidemics started late 2013, it took until the summer 2014 for the main health authorities, including the World Health Organization (WHO), to realize that this outbreak was unprecedented and required a coordinated and global response in terms of public health and therapeutic interventions. In September 2014, the WHO established a list of drug candidates for testing in patients with EVD. 30 Among those candidates was favipiravir (T‐705), an RNA polymerase inhibitor approved in Japan for the treatment of noncomplicated influenza infections and in clinical development in the United States. Favipiravir is an oral drug with a fast absorption (time of maximum plasma concentration (Tmax) ~ 1 hour) and high bioavailability (> 90%), a low distribution volume (~ 15 L), and a short half‐life (2–5 hours). This drug was characterized by dose‐dependent and time‐dependent pharmacokinetics (PKs) due to the auto‐inhibition of the main enzymatic pathway, mediated by aldehyde oxidase, and ethnic discrepancies of exposure, with lower plasma concentration of ~ 50% reported in white and African patients as compared with Japanese patients. 31 , 32 Favipiravir was at that time the only molecule to meet the three following criteria: (i) documented antiviral activity against EBOV in vitro and in a laboratory mouse model, 33 , 34 (ii) favorable safety profile when administrated per os to > 2,000 healthy volunteers or patients with influenza worldwide, 31 and (iii) immediate availability in large quantities.
Mathematical modeling to support the dosing regimen of favipiravir
In line with the efforts of a global response against the spreading Ebola epidemic, a European consortium, Reaction! (horizon 2020 grant 666092), was set up to evaluate favipiravir efficacy in a context of emergency. The first aim of mathematical modeling was to find the optimal dosing regimen of favipiravir in patients with EVD, for which the drug’s half‐maximal effective concentration (EC50) was much larger than against most strains of influenza virus. 8 The dosing regimen of favipiravir approved in Japanese patients for influenza was 1,600 mg twice daily the first day followed by 600 mg twice daily from the second to the fifth day. The target exposure was identified from a preclinical experiment, where mice infected by EBOV were successfully treated with favipiravir 150 mg/kg b.i.d. initiated 6 days after viral challenge. 35 PK data from manufacturer allow to identify PK parameters (C average and C trough) of favipiravir in mice treated with this dosing regimen and to compute corresponding values in patients taking into account the species plasma protein binding. Targeted C average and C trough were, respectively, 113 and 10 µg/mL. Using a PK model developed by the manufacturer, we identified that a loading dose of 6,000 mg the first day (split over three intakes), followed by 1,200 mg b.i.d. afterward for 10 days could achieve similar average and trough concentrations (83.3 and 57 µg/mL, respectively) in African and white patients as that predicted in mice successfully treated with favipiravir, taking into account the species plasma protein binding. 35 A similar approach, using the same target concentrations and the population PK model with weight‐based allometric scaling of model parameters, was then used to find optimal dose in children above 1 year old, 36 based on the fact that maturation profile of aldehyde oxidase is fully achieved at this age.
Results of the JIKI trial
It is this dosing regimen that was chosen in “JIKI,” an open multicenter nonrandomized clinical trial that took place in Guinea from December 2014 to April 2015. 37 JIKI was the largest therapeutic clinical trial performed during the epidemic, with 126 patients included. The first result of the JIKI trial was that mortality was strongly associated with baseline viremia, and accordingly the result analyses should be stratified on the baseline cycle threshold (CT) value, with a cutoff for baseline viremia of 20 CT (corresponding to 7.7 log10 EBOV RNA copies/mL). The mortality rate was equal to 20.0% (95% confidence interval (CI) 11.6–32.4%) in patients with CT ≥ 20 and 90.9% (95% CI 78.8–96.4%) in patients with CT < 20. Both CIs for the mortality rates included the predefined target values (defined on values observed in the same centers before the trial) of 30% and 85%, respectively, providing no evidence that favipiravir monotherapy has a favorable benefit/risk ratio in patients with very high viral load at onset. However, in those having a baseline CT value ≥ 20, there was a small signal toward a better survival rate, albeit not significant. Consistent results were reported in a retrospective analysis of 163 patients admitted in an Ebola treatment center in Coyah, Guinea, where lower but no significant case fatality rate and significant longer survival time were found in patients receiving favipiravir on a compassionate use basis. 38 Several clinical trials evaluating other candidate treatments from different therapeutic classes, including ZMapp, TKM‐Ebola, brincidofovir, and convalescent plasma were implemented in Gunea, Liberia, and Sierra Leone during the 2013–2016 outbreak, yet none of them could demonstrate a significant improvement of survival rate related to the antiviral treatment. 39 , 40
Pharmacokinetic analysis
To better understand the results of the JIKI trial, we analyzed the drug concentrations in 66 patients and we compared them with those predicted by the PK model taking into account patient’s individual characteristics. 41 At day 2 post‐treatment initiation, the observed drug concentrations were slightly lower than the model predictions (median value of 46.1 vs. 54.3 µg/mL for observed and predicted concentrations, respectively; P = 0.012). However, the concentrations dropped at day 4, which was not anticipated by the model (median values of 25.9 and 64.4 µg/mL for observed and predicted concentrations, respectively; P < 10−6). There was no significant relationship between favipiravir concentrations and EBOV viral kinetics or mortality. Overall, these results suggested that favipiravir plasma concentrations in the JIKI trial failed to achieve the target exposure defined before the trial, defined as C trough and C average equal to 10 and 113 µg/mL, respectively. Furthermore, the drug concentration showed an unanticipated drop between day 2 and day 4. The reasons why concentrations were lower than anticipated are not fully understood, but may be due to the fact that the PK model could not capture well the nonlinear kinetic for the dosing regimen used in the JIKI trial, as this model had been built on patients receiving much lower doses of favipiravir than in the JIKI trial (up to 800 mg b.i.d., as compared with 1,200 mg b.i.d. in the JIKI trial). An additional nonexclusive reason could be an effect of the disease on vital functions, altering the PKs of the drug.
In summary, both the clinical and the PK results supported the conclusion that favipiravir, at this dosing regimen, did not achieve a strong antiviral activity against EBOV in the JIKI trial.
MATHEMATICAL MODELING SUPPORTS HIGH DOSES OF FAVIPIRAVIR IN NONHUMAN PRIMATES
In parallel to the JIKI trial, and anticipating the limitations due to the study design, in particular the absence of a control group, nonhuman primate (NHP) studies were planned to optimize the use of favipiravir for subsequent trials. In these experiments, the first objective was to characterize favipiravir PKs and to use modeling techniques to identify relevant dosing regimens and drug concentrations.
Setting up an experimental model of EVD in NHPs
A model of EVD was set up in cynomolgus macaques from a Mauritian colony, which well recapitulates the disease in humans. 42 In brief, animals were infected intramuscularly with 10 to 1,000 focus forming units with Ebola virus Gabon 2001 strain, belonging to Zaire ebolavirus species. Viral load was detectable in most animals at day 3 postinfection (D3), deterioration of the clinical score was visible in most animals at D5 and peak viremia was achieved at D7 with levels around 8 ± 2 log10 EBOV RNA/mL (i.e., close to the levels observed in humans (see above)). The model is fully lethal with death occurring between D8 and D11. This is an important difference with the human infection, for which the pathogenesis is less rapid; however, the time between disease onset, defined as the observation of the first clinical signs, and peak viremia (2–4 days) is close to what has been estimated in humans (5 days). 43
Favipiravir 100 mg/kg b.i.d. slightly reduces viral load peak but does not significantly increase survival
Preliminary PK studies conducted in uninfected cynomolgus macaques from a Chinese colony found that a loading dose of 200 mg/kg b.i.d. the first day followed by 100 mg/kg b.i.d. achieved drug concentrations that were comparable to those predicted in successful mice experiments and expected in the JIKI trial. Accordingly, a first experiment was designed where cynomolgus macaques from Mauritius Island were infected 2 days after the initiation of treatment (pre‐exposure prophylaxis). Although treated animals had a median peak viremia lower than untreated animals, there was no benefit of favipiravir in terms of survival (Table 1 first experiment). The analysis of drug concentrations revealed levels of exposure lower than anticipated from PK studies performed by the manufacturer in Cynomolgus macaque from China, when parallel experiment in noninfected Mauritian cynomolgus also underlined lower C average (10 µg/mL) than the targeted computed from mice experiment (113 µg/mL). Based on these results, detailed PK studies were planned to characterize favipiravir PK and to evaluate the possibility to use higher doses of favipiravir. 8
Table 1.
Viral load at D7 in animals infected with 1,000 PFU and left untreated or receiving 100 mg/kg b.i.d., 47 predictions for the two subsequent experiments with higher dosing regimens obtained using Eq. 1, viral load observations obtained in subsequent experiments
| Median viral load (log10 RNA/mL) | ||
|---|---|---|
| Day 5 | Day 7 | |
| Observations in the first two experiments (min‐max) | ||
| Untreated | 6.5 (5.2–8.5) | 8.9 (7.1–9.2) |
| 100 mg/kg b.i.d. | 6.2 (5.0–6.2) | 6.9 (6.0–8.0) |
| Predictions for the next two subsequent experiments | ||
| 150 mg/kg b.i.d. | 4.4 | 5.8 |
| 180 mg/kg b.i.d. | 3.5 | 4.6 |
| Observations in next two subsequent experiments (min‐max) | ||
| 150 mg/kg b.i.d. | 4.1 (3.9–7.0) | 5.9 (3.5–7.5) |
| 180 mg/kg b.i.d. | 3.6 (1.0–5.0) | 4.4 (3.5–7.0) |
Predicting the effect of higher doses of favipiravir on viremia
In addition to historical preclinical data available from the manufacturer, new studies evaluating high dose levels of i.v. favipiravir in Mauritian cynomolgus macaques were conducted by the Reaction! consortium. Altogether, favipiravir PKs was characterized in 30 uninfected cynomolgus macaques from Chinese (n = 17) or Mauritian (n = 13) colonies treated with i.v. favipiravir for 7–14 days with maintenance doses of 60–180 mg/kg b.i.d. 8 Favipiravir was found to exhibit a complex nonlinear kinetics in the early days of treatment with, in addition, a progressive reduction in drug concentrations over time that could be related to the auto‐inhibition of the aldehyde oxidase, the main enzyme involved in favipiravir metabolism. 31 This PK model included an enzyme inhibition process accounting for concentration‐dependent aldehyde oxidase inhibition, 31 and the enzyme‐dependent elimination rate, denoted k enz, that increases over time to capture the decline in drug levels observed in the days following treatment initiation (Figure 1 ). Interestingly, k enz was significantly larger in NHPs from the Mauritian than from the Chinese colony (P < 10−4), explaining in part the low drug concentrations found in our NHP experiment as compared with preliminary PK studies. 8
Figure 1.
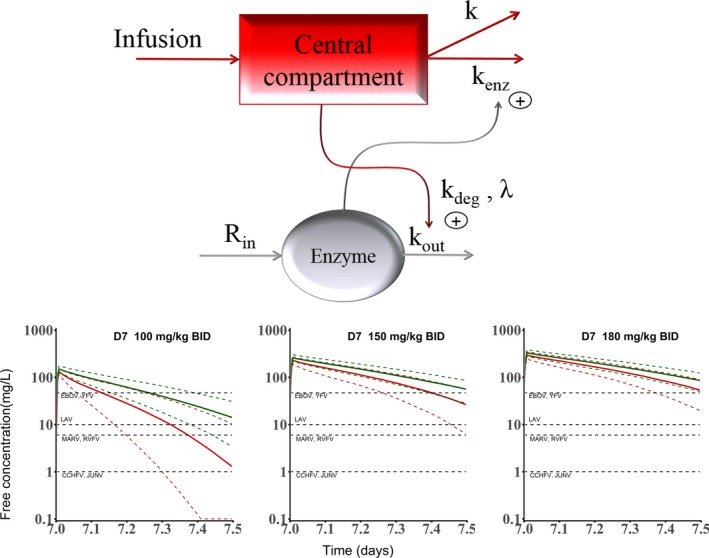
Top: Pharmacokinetic model for favipiravir in non‐human primates; bottom: predicted drug concentrations at D7 for three levels of dosing regimens and comparisons with drug half‐maximal effective concentration for various emerging viruses. CCHFV, Crimea Congo haemorrhagic fever virus; EBOV, Ebola virus; JUNV, Junin virus; LAV, Lassa fever virus; MARV, Marburg virus; RVFV, Rift valley fever virus; YFV, Yellow fever virus. Reproduced from ref. 8.
Next, we used these predictions and the viral load already collected in NHPs to elaborate a simplified mathematical model of Ebola viral dynamics where we assumed no target cell replenishment (such that dT/dt = −βVT in Eq. 2, see below), allowing us to derive an approximate solution for the viral load kinetics to the peak of viremia 44 :
| (1) |
where V 0 is the initial viral load level, R 0 is the basic reproductive number (see below), δ is the loss rate of infected cells, C ave is the average drug concentration (as predicted by the PK model), and EC50 is the drug concentrations leading to 50% efficacy in reducing the basic reproductive number. Using this simple model to fit the viral load obtained in untreated NHPs and in those treated with 100 mg/kg b.i.d. (with δ fixed to 1/day to ensure parameter identifiability), we estimated R 0 to about 3.25 and EC50 to 334 µg/mL. Then, we plugged into the model the predicted concentrations obtained with doses of 150 and 180 mg/kg b.i.d. (administered prophylactically) to predict that median reduction of 3–4 log10 RNA/mL at D7 could be obtained with these dosing regimens compared with untreated animals (Table 1 : Prediction for the two subsequent experiments). Such reduction could significantly extend survival, as observed in rhesus macaques treated with the polymerase inhibitor galidesivir (BCX‐4430), where all treated animals that had a peak viremia below 106 RNA copies/mL survived the infection. 45
High doses of favipiravir used for pre‐exposure prophylaxis significantly reduce viremia and increase survival rate
Based on these predictions, two additional studies at the doses of 150 or 180 mg/kg were conducted in cynomolgus macaques from Mauritius Island using the same design (i.e., with treatment starting 2 days prior to infection and continuing for 14 days; i.e., 12 days postinfection). We reported in the primary analysis the results obtained in animals challenged with 1,000 focus forming units. This included 13 animals left untreated and 13 treated with favipiravir doses of 100 (described above), 150 and 180 mg/kg (N = 3, 5, and 5, respectively). All animals left untreated or treated with 100 mg/kg died within 10 days postinfection, as already discussed above, whereas animals receiving 150 and 180 mg/kg had extended survival (P < 0.001 and 0.001, respectively, compared with untreated), leading to a survival rate of 40% (2/5) and 60% (3/5), respectively, at day 21 postinfection (study termination, Figure 2 ). Further, we found that favipiravir inhibited viral replication (molecular and infectious viral loads) in a drug concentration‐dependent manner. Remarkably, the observed viremia at D7 in the 150 and 180 mg/kg b.i.d. were very close to the predictions made beforehand (Table 1 ). These results allowed us to identify that favipiravir plasma trough concentrations larger than 70–80 µg/mL, which are about twice as much as found in patients of the JIKI trial (see above), were associated with reduced viral loads, lower infectious titers, and extended survival. The mechanism of action by which favipiravir reduces viral replication may be direct inhibition of the viral polymerase via chain termination, but also by the dramatic increase in virus mutagenesis revealed by genomic deep‐sequencing analyses. 46
Figure 2.
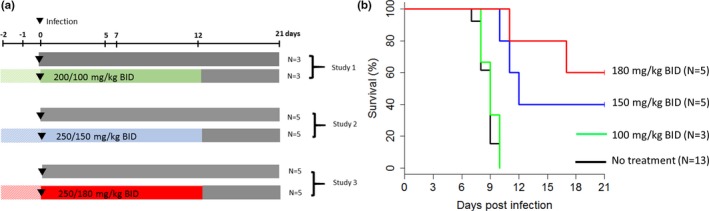
Survival obtained in 26 nonhuman primates infected with 1,000 focus forming units Ebola virus and treated with ascending doses of favipiravir. Left: Study design (note that treatment is administered 2 days prior to infection). Right: Survival curves. Reproduced from ref. 47.
In summary, these results established that favipiravir can be effective against EBOV in NHPs in a prophylactic setting and with drug concentrations about twice larger than in the JIKI trial, 47 and allowed to define new target concentrations for the evaluation in patients. They support the implementation of dose‐ranging studies in healthy volunteers to assess the concentrations and the tolerance that could be achieved with doses higher than those used in the JIKI trial.
GENERAL CONCEPTS ON MODELING VIRAL DYNAMICS AND ANTIVIRAL TREATMENT DURING AN ACUTE INFECTION
The standard model of viral dynamics
The basic model of viral infection in absence of treatment, called in the following the “target cell limited model,” is given by the following set of equations (Eq. 1) 48 , 49
| (2) |
The model considers three populations of cells: target cells, T, infected cells in eclipse phase, I 1, and productively infected cells, I 2 (Figure 3 ). Target cells are produced at rate s, are eliminated with per capita rate d, and become de novo infected by circulating virions, V, with rate constant β. Once infected, these cells are cleared with per capita rate δ. Virions are released from infected cells at a rate of p per cell per day and are cleared from the circulation with a rate c. Based on this model, the basic reproduction number, R 0, as the number of cell infections that occur from a single infected cell at the beginning of the infection and given by .
Figure 3.
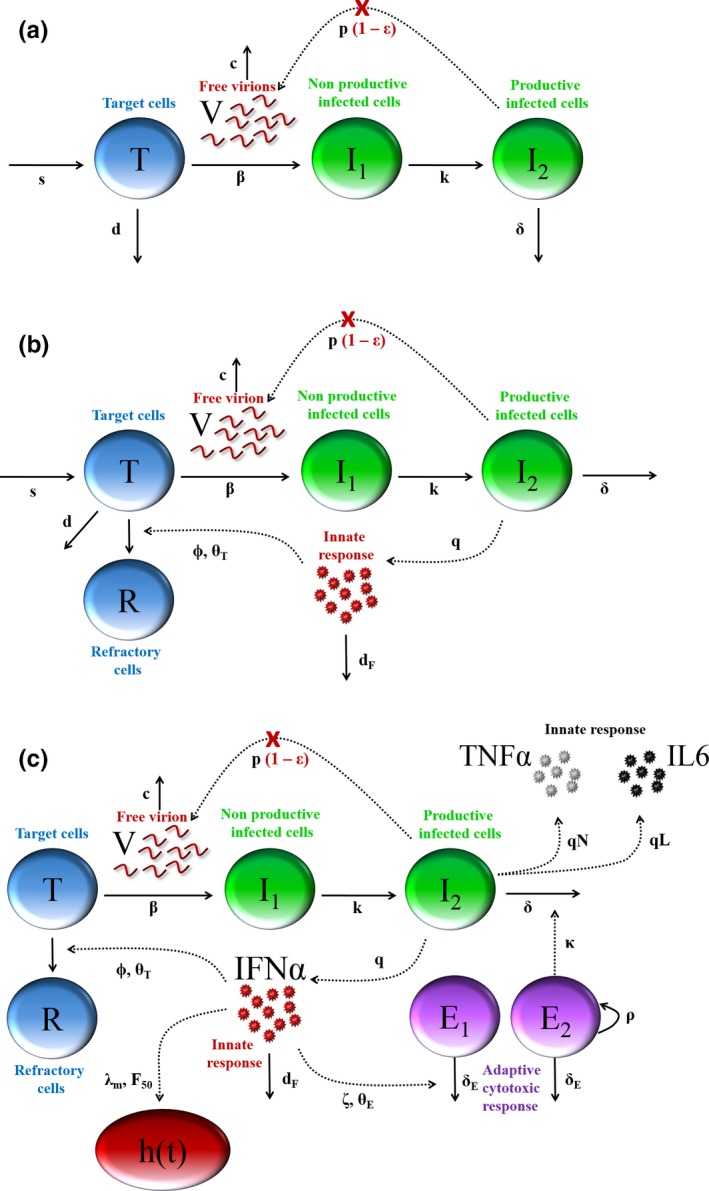
Model structures used to described acute viral infection dynamics (a) “Target cell limited” model of viral dynamics considers the interaction between free virions (V) and three cell populations, namely target cells (T), infected cells in the eclipse phase (I1) and productively infected cells (I2). In this model, polymerase inhibitor, such as favipiravir, act by blocking viral replication from infected cells with efficacy noted ϵ. (b) Extended model to account for the role of cytokines, in particular IFNα, that can decrease the cell susceptibility to infection, modeled by a compartment of refractory cells (R). (c) Integrated model of Ebola virus (EBOV) infection. In addition to compartments presented in Figure 3 , the model also considers IL6 and TNF. In parallel, the cytokine release increases the apoptosis of nonspecific CD8 T‐cells (E1), giving room for EBOV‐specific T‐cells (E2) to grow and increase the elimination rate of actively infected cells. Cytokine levels increase the instantaneous rate of death, h(t). Modified from ref. 69.
Antiviral treatment during chronic infection
During chronic infection, such as HIV or HCV, 49 , 50 where these models have been mostly used, viral load levels, V, are roughly constant in absence of treatment and reflect the equilibrium between the process of viral production and viral elimination. Likewise, the number of infected cells is constant, which reflects the fact that the loss of infected cells, due to virus cytopathicity or elimination by the immune response, is compensated by de novo infection of target cells that constantly replenish the pool of infected cells. In these models, the initiation of an antiviral drug may primarily block new infection with an effectiveness η, or block viral production with an effectiveness ε. These treatment effect parameters vary between 0 (no drug effect) and 1 (full suppression).
In chronic infection, the role of an antiviral treatment is to reduce viral replication, making the few remaining virus particles uncapable to compensate the continuous loss of infected cells and to support infection. Viral clearance can be achieved if treatment effectiveness passes a threshold, called the “critical efficacy,” and equal to . In HCV treatment, the mean R0 estimated in > 2,000 patients was found to be 7 on average, with large interindividual variation 51 ; thus, treatment efficacy needs to be in the order of 90–99% to generate viral clearance. It is remarkable that similar R 0 values were found for HIV or HBV infections, with values of about 5–8. 52 , 53 Obviously, the success of a therapy will also depend on other parameters that are not taken into account in this model, such as the treatment duration, the presence of viral reservoirs, and the barrier to resistance. Nonetheless, this shows that high antiviral activity will be needed to compensate the impaired immune response and to achieve viral clearance in chronic infection.
Antiviral treatment during acute infection and the role of the innate immune response
In an acute infection, such as influenza or EVD, the role of an antiviral treatment is not so much to clear the infection, because the virus does not persist and can be naturally eliminated by the host in a couple of days or weeks. Rather, using an antiviral treatment may be relevant even if it does not reduce R 0 below 1, as long as it can delay the virus growth to such extent that viral load levels and disease symptoms, which are closely related, 54 can be alleviated, giving time for the immune response to be fully effective and clear the pathogen. This can be formalized by using a mathematical model that explicitly takes into account the interaction among the virus, the immune system, and the antiviral treatment. Several models for the innate and the adaptive immune response to acute viral infection have been proposed (see for instance refs. 55, 56 essentially in the context of influenza), but the common characteristic of these models is that the action of the immune response results in limiting cell infection (for instance via an interferon (IFN) response, see ref. 57) or in increasing elimination of infected cells (for instance via a CD8 or an NK cell response, see ref. 58). For instance, the role of the innate immune response can be modeled as follows:
| (3) |
In this model, cytokine levels, F, are proportional to the number of infected cells and their release in the circulation increases the number of cells that are refractory to the infection, noted R. When the cytokine levels exceed a certain threshold, denoted θ, cell receptors are saturated and the rate of cell “conversion” from susceptible to refractory state saturates to a level noted ϕ. Of note, several model extensions can be proposed, such as a reversion of refractory to susceptible cells, or also considering the effect of IFN in reducing viral production. 12 Further extensions may include the cellular or humoral adaptive immune responses. In acute infections, most of the proposed adaptive response models focused on cytotoxic lymphocytes and/or antibodies responses and are structurally close to those proposed for the innate response, but need to incorporate the delay characterizing the development of these responses. 59 , 60 Including adaptive response may help explore the mechanisms leading to the outcome of the infection, viral clearance, host death, or chronic infection.
As shown in Figure 4 (top), the model given by Eq. 3 can reproduce the patterns of viral load observed in clinical data where the peaks of viremia and cytokines coincide. 56 , 61 Consistent with the standard model of viral dynamics, viral decline after peak viremia results from a lack of susceptible cells; however, in this model, the peak viremia does not correspond to an exhaustion of noninfected cells, but rather to the fact that most noninfected cells have been set into an antiviral state. Thus, the model allows one to quantify the “race” between the virus and the innate immune response to “get first in contact” with the susceptible cells. In absence of treatment, this race triggers an inflammatory response to counter the exponential viral replication and this “cytokine storm” is directly related to clinical symptoms, as shown in influenza infection. 62 Thus, in this dynamic interaction between the host and the virus, an antiviral treatment can be beneficial even if it does not abrogate viremia, as long as it can reduce it to such an extent that transmission risk is reduced and an exacerbated immune response is averted. This is illustrated in Figure 4 , where a treatment initiated at D0 with an effect of 50%, which is lower than the critical efficacy (here equal to 83%) is sufficient to reduce viral growth, giving time for the IFNα response to put more cells into a refractory state, leading to both a delay and a reduction in peak viremia by about 4 logs. This prediction is different from the target cell limited model (Eq. 2), where ε = 50% would impair viral growth and, hence, delay peak viremia, but with only little effect on the peak viremia value shown (Figure 4 bottom).
Figure 4.
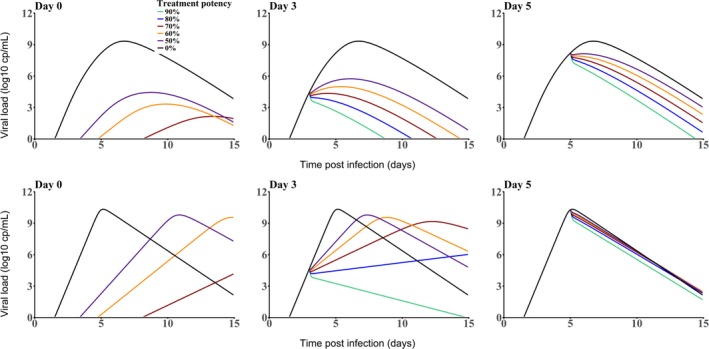
Effect of an antiviral treatment on viral dynamics according to the timing of treatment initiation (left: day 0, middle: day 3; right: day 5) and the level antiviral effectiveness. Top: Model including an innate immune response (Eq. 3); bottom: target cell limited model (Eq. 2) according to the timing of treatment initiation. The following parameter values were used to generate these curves: T 0 = 109 cell/mL; c = 22/day, p = 1,000 virions/cell/day; V 0 = 10−4 virions/mL, δ = 2/day, ϕ = 0.3 mL/pg/day, d = 1/day, q = 100 pg/cell/day, R 0 = 6, and θ = 200 pg/mL. In the target cell limited model, q was set to 0.
The timing of treatment initiation is a key parameter in both models, with delayed treatment intervention requiring higher efficacy to achieve a similar reduction in peak viremia. In our example, if treatment is initiated at D5, the treatment efficacy ε will need to be > 90% to reduce peak viremia by 3 logs or more (Figure 4 ). Treatment initiated after peak viremia will have very little or no effect on viral dynamics. 63
In practice, the use of a model integrating immune response dynamics has been limited by the fact that parameter estimation requires to measure several markers longitudinally (such as IFN and viremia) during the short period of time where these markers are detectable. 56 , 64 This is obviously difficult in humans (due to the difficulty to identify viral infection before symptoms occur) but also in animal models, due to the limitation in the frequency and the volume that can be performed.
Of note, in these models, the viral load does reach 0 (i.e., eradication, only asymptotically, and cannot address the question of optimal treatment duration). A paradox of these models is that, in case of high antiviral efficacy, viral growth is almost entirely blocked, thus the immune system does not step in and all cells remain targets when treatment is stopped. Because viral dynamic models are deterministic, it cannot reproduce viral eradication and even a small quantity of virus can be sufficient to reignite the infection, provided that there are a large number of target cells are present. Thus, if treatment is stopped, virus dynamics will be similar to an acute infection without any treatment. To address the question of treatment duration needed to achieve cure, one would need to incorporate in these models the possibility of viral eradication, using stochastic events 65 or including a “cure boundary,” as was done in the context of HCV to predict cure after end of antiviral treatment. 50 , 66 , 67
TARGET CELL LIMITED MODEL IN EBOV INFECTED MICE TREATED WITH FAVIPIRAVIR
The first use of an EBOV kinetic model in vivo was to characterize the curative efficacy of favipiravir in a mice model. 33 Twenty IFNαR−/− mice infected with Makona strain EBOV received favipiravir 300 mg/day initiated at D6 (N = 5) or D8 (N = 5), or were left untreated (N = 10), leading to survival rates at D25 of 100%, 0%, and 0%, respectively.
A target cell limited model was sufficient to capture the viral kinetic of the acute infection, consistent with the fact that IFN response is impaired in these animals. 68 Viral kinetics in untreated animals was characterized by an exponential growth of virus until D8 followed by rapid death, leading to an estimated R0 of 9, which suggests that a treatment efficacy larger than 90% would be needed to block viral replication. Viral growth was accompanied with high levels of transaminases, 33 suggesting a large cell loss, and accordingly the half‐life of infected cells was short and estimated to about 6 hours (e.g., δ = 2.6/day).
The antiviral activity of favipiravir was modeled using a maximum effect (Emax) model, where , with Ce defining the active drug concentration, and EC50 the drug concentration needed to achieve 50% of the maximal effect. In order to capture the delay observed between the drug administration and the beginning of viral decline, we assumed an effect compartment between drug concentration in plasma, C(t), and the active concentration where k is the rate of transfer between the plasma and the effect concentration. We estimated that it took about 2 days to achieve a high antiviral effectiveness of about 95%. In mice starting treatment at D6 (i.e., 2 days before peak viremia), the treatment period was sufficient to reduce the peak viremia by 1.5 log. However, in mice treated at D8, the treatment initiation was too late and the activity of favipiravir was not sufficient (estimated to 80% at peak viremia) to substantially perturb viral dynamics and increase survival. This suggests that favipiravir treatment, in order to be effective, has to be initiated as early as possible and before peak viremia.
MODELING EVD PROGRESSION IN NHPS TREATED WITH FAVIPIRAVIR USING AN INTEGRATED APPROACH OF THE HOST‐PATHOGEN‐DRUG INTERACTION
Model building
Although the results shown in Section “Mathematical modeling supports high doses of favipiravir in nonhuman primates” demonstrated that favipiravir is effective in EVD in NHP, a number of open questions remained: how and to what extent does favipiravir modify the course of the infection? How does it potentiate the immune response? What can be expected with more potent drugs? What could be the benefit of antivirals outside prophylaxis or post exposure treatment? We aimed to address these questions using a host pathogen modeling approach, described in ref. 69
We modeled the virologic and PK data that had been collected in the 44 animals of different experiments conducted in the BSL4 (see above), and complemented them with additional data on the innate and the adaptive immune responses. 69 A number of cytokines were measured longitudinally in a subset of macaques (n = 20) using Luminex technology or enzyme‐linked immunosorbent assay, in particular IFNα, IL6, and TNFα, and cytometry measures were performed in the last experiment (n = 10, 5 untreated and 5 treated with 180 mg/kg b.i.d.) to characterize lymphocyte populations.
At D7, the levels of several cytokines were highly and negatively associated with survival times, in particular cytokines having pro‐inflammatory or anti‐inflammatory effects. In fact, the levels of IL6, IL10, IL1‐RA, IFNα, IFNγ, and G‐CSF, were more significantly associated with survival than viral load levels, suggesting that those cytokines may be more predictive of outcome than viral load. Furthermore, in the four animals that had an extended survival for which cytometry data were available, most of the CD4 and CD8 lymphocyte population counts rapidly increased after D10. In particular, the CD8 T cells expressing cytotoxic surface markers (granzyme B, perforin, and NKp80) increase showed a trend to be associated to viral load decline after D10.
As described in Section “Mathematical modeling supports high doses of favipiravir in nonhuman primates”, favipiravir delayed and reduced peak viremia in a concentration dependent manner, which, as explained in Section “General concepts on modeling viral dynamics and antiviral treatment during an acute infection”, cannot be satisfactorily explained by a basic target cell model. Therefore, mathematical models of increasing complexity were used to fit EBOV viremia, and we tested several models 55 , 59 , 64 assuming that the various cytokine dynamics could reflect an effect of the innate immune response in either (i) increasing cell refractoriness to infection, 57 , 70 (ii) reducing virion production from infected cells, 70 , 71 (iii) increasing target cell availability, 72 , 73 or (iv) increasing the loss rate of infected cells. 58 , 71 To identify the most relevant cytokine associated with viral dynamics, we successively fitted the model to viral load data and each cytokine found significantly associated with survival at D7. Thus, pro‐inflammatory cytokines IL6, IFNα, and TNFα were successively incorporated in models (i), (ii), and (iii) (leading to a total of 12 possible candidate models), whereas IFNγ was used for model (iv). Models assuming that pro‐inflammatory cytokines increased cell refractoriness to infection consistently provided the best description of the viral load in untreated and treated NHPs, allowing us to better capture the dose effect relationship on viremia compared with the target cell limited model. Given their high level of correlations, a similarly good fit to the viral load data could be obtained when assuming that this effect was driven by either of the three pro‐inflammatory cytokines IFNα, IL6, or TNFα. Because the effects of IFNα is supported by in vitro experiments, we included only the effect of IFNα but we kept IL6 and TNFα in the model as instrumental variables reflecting the overall level of cytokine response.
Then the model was extended to include the adaptive response, assuming a decline of nonspecific cells and an expansion of specific cytotoxic T‐cells. Following the same strategy, models building was performed assuming that this effect was primarily supported by cytotoxic CD8 T lymphocytes subpopulation. CD8 T lymphocytes expressing granzyme B, perforin, or NKp80 were successively fitted to the data and we found that CD8 T lymphocytes expressing perforin provided the best improvement of the data description and could reproduce both the cytokine‐mediated lymphopenia observed in early infection and the rapid viral decline in NHPs after peak viremia. The schematic representation of the final selected viral dynamic model is given in Figure 3c .
Impact of viral dynamics and cytokine storm on disease progression
Finally, we used previous developments in the field of joint modeling 74 , 75 to model disease dynamics (i.e., viral load and cytokine dynamics) and survival. Joint modeling is a statistical approach that aims to estimate the impact of a time‐dependent biomarker on the instantaneous rate of death (also called the hazard function in statistics), noted h(t), and the probability to survive up to time t, noted S(t). The best model was:
| (4) |
In this model, λm is the maximal instantaneous rate of death, X is the viral load or cytokine levels, and Xe(t) is the lag‐value of the viral load or cytokine, and X50 value leading to 50% of the maximal hazard, and γ is a Hill coefficient. As hinted in the transversal analysis at D7, a model assuming the hazard function to be dependent on IFNα rather than viral load provided the best description of the time to death (Figure 5 ), suggesting that cytokine storm, rather than viral replication per se, was the main driver of death.
Figure 5.
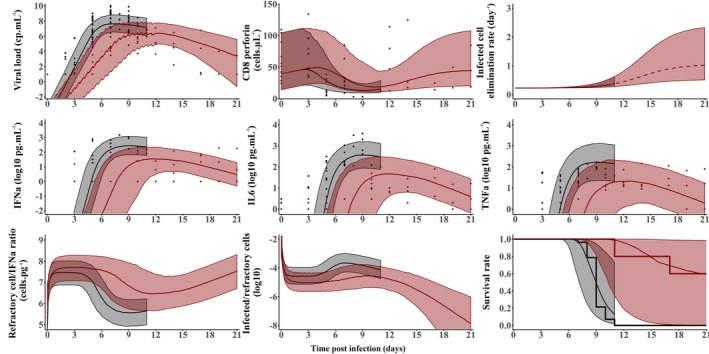
Model predictions (median and 95% prediction interval) for the compartments of the integrated model given in Figure 3c in animals left untreated (black) or treated with favipiravir 180 mg/kg BID. 69 The dots are the individual data in each compartment. Reproduced from ref. 69.
Model analysis
The estimate of parameter θ conferring 50% of the maximal cell protection, was equal to 1.7 pg/mL. This implied that IFNα concentration greater than about 10‐fold this value (17 pg/mL) provides little additional benefit for cell protection. Thus, in untreated animals, the infection affects a significant proportion of the target cells, and consecutively trigger a massive release of IFNα (with median levels of 400 pg/mL at peak viremia; Figure 5 ) and other pro‐inflammatory cytokines that negatively affects survival rate in spite of limiting cell infection. Conversely, in treated animals, favipiravir reduces viral production and slows the infection progression. Hence, the number of infected cells and IFNα concentrations are lower. These levels of IFNα concentrations are sufficient to confer a nearly similar effect on cell protection than in untreated macaques. The impairment of the viral replication in treated macaques gives time to the organism to protect the targets cells and, thus, reduces and delays peak viremia while limiting the deleterious effect of a cytokine storm on survival (Figure 5 ).
In the days that follow infection, the half‐life of infected cells was estimated to about 3 days (δ = 0.22/day), suggesting that in absence of an adaptive response, it would take several weeks to clear the virus in the blood. However, the loss rate of infected cells was reduced continuously with the concomitant increase in specific CD8 T lymphocyte levels, in particular those expressing perforin. This leads to a much shorter half‐life of about 16 hours (δ = 1/day) at D21 in animals that survived, which explains the rapid clearance of viremia observed after peak in surviving animals.
Given the PKs of the drug, we estimated the in vivo drug efficacy, which was close to 50% with maintenance doses 180 mg/kg b.i.d. With this level of efficacy, our integrated model reproduced the 60% survival rate observed at D21, and predicted that this level would remain largely similar even if treatment initiation is delayed up to D3, but not afterward (Figure 6 ). Finally, we used the model to predict the impact of more effective drugs. With a drug effectiveness of 90% (such as GS‐5734 4 ), we predicted that 100% (resp. 70%) survival could be obtained if treatment is initiated up to D4 (resp. D5), consistent with the 100% survival obtained in animals treated with 10 mg/kg 3 days after viral challenge (Figure 6 ). 4 However, treatment given after D5 would not affect survival, regardless of drug efficacy (Figure 6 ).
Figure 6.
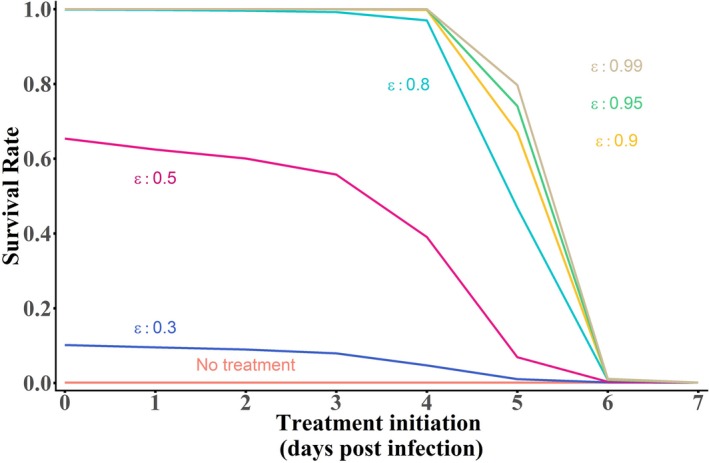
Median survival rate of n = 1,000 in silico macaques simulated with the integrated model given in Figure 3c depending on the day of treatment initiation, for various treatment potency.
LESSONS FOR THE FUTURE EVALUATION AND USE OF FAVIPIRAVIR
Beside EBOV, the knowledge gained on the drug PK/pharmacodynamic (PD) could also be useful in other viral infections for which favipiravir has shown antiviral efficacy in vitro or in vivo.
Combining favipiravir and ribavirin in Lassa fever
Favipiravir was also reported to have antiviral effect on Lassa fever virus, providing 100% protection in the lethal NHP model of Lassa fever. 76 It could be an alternative to ribavirin, the only recommended drug against Lassa, or be used in combination to it. Indeed. several in vitro studies and mice studies showed that ribavirin may potentiate the efficacy of favipiravir against various RNA viruses, 9 , 77 which prompted its use to treat several patients with severe Lassa fever virus infection. 78 In order to better understand how ribavirin may potentiate the efficacy of favipiravir in mice, we re‐analyzed data obtained in mice treated at different doses of favipiravir and/or ribavirin 9 to decipher the mechanism of action of ribavirin.
Similar to the mice model used in EBOV, these animals were chimeric IFNαR−/− C57BL/6 animals and, thus, lacked type‐I IFN receptor function and, therefore, the model used was a simple target cell limited model. In order to gain information on the potential role of ribavirin on loss cell death, we assumed that the level of aspartate amino transferase (AST, noted A) reflected cell loss, following models previously developed in HCV to quantify the levels of hepatocytes injury using alanine aminotransferase dynamics 79 :
where sx and cA are the natural production and loss rate of AST, respectively, and α is the rate of AST release by dying infected cells. We assumed four potential mechanisms of action for ribavirin that could (i) act as a mutagen limiting the infectivity of virions; (ii) reduce viral production; (iii) reduce cell loss by limiting inflammation; and (iv) increase cell loss by enhancing antiviral immunity.
Using these models, we consistently estimated an R 0 of Lassa fever virus of about 5–6, and an efficacy of favipiravir to larger than 90% at all doses considered, consistent with our results obtained for Ebola infection in mice. 80 Our analyses did not find a statistically significant effect of ribavirin in reducing viral production, however, the best description to the data was obtained by assuming an effect of ribavirin in preventing infected cells from dying. This possibly explains why the AST levels remained lower in ribavirin‐treated mice and could also explain the viral plateau observed at peak viremia in ribavirin monotherapy group. 80 Thus, our model suggested that ribavirin did not act much as a direct antiviral against Lassa virus infection, but rather by reducing the elimination of infected cells, which could limit inflammation and extend survival.
Potential applications to Zika and Marburg viruses
Given the broad antiviral activity of favipiravir, we also evaluated in vitro its efficacy against Zika and Marburg viruses and reported antiviral EC50 of 6.5 and 6.8 µg/mL, respectively. 7 , 8 Interestingly these levels are markedly 2–10 times lower than what has been reported for EBOV, 31 suggesting that favipiravir could also be relevant against these pathogens.
In 2016, the group of James Whitney published the first model of Zika viral dynamics in NHPs. 81 By using the target cell limited model (Eq. 2), the parameters of Zika viral dynamics could be estimated, in particular R 0 found equal to 10.7. 7 Then, the effect of favipiravir on blocking viral production was plugged into this model assuming a dosing regimen of 250 mg/kg b.i.d. the first day followed by 150 mg/kg b.i.d., using the PK model developed previously (see Figure 3 ). 8 Interestingly, the model predicted that favipiravir administered at day 0 had the potential to reduce the median peak viremia by about 3 logs, corresponding to a median efficacy of 90%, and, therefore, close to the critical efficacy to abrogate Zika replication (Figure 7 ). Given that peak viremia occurred 2–3 days after subcutaneous infection in this animal model, the window for treatment is nonetheless narrow and the model predicted that treatment needs to be administered no later than day 1 to have a strong effect on systemic levels of viruses, at least in NHPs. In humans, lower viremia levels are often observed 58 , 82 , 83 and the peak of plasma viremia occurs later, potentially allowing for later antiviral treatment initiation. Furthermore, as favipiravir penetrates the sexual compartments and crosses the blood brain barrier, 31 an important benefit of the treatment may be to purge the reservoirs of the virus, another feature that can be tested in the NHP model. 84 If the probable teratogenicity of favipiravir prevent its use in pregnant women, it might be of interest for symptomatic severe central nervous system injury in adults with acute infections. However, the impact of the timing initiation on the kinetics in these compartments is unknown.
Figure 7.
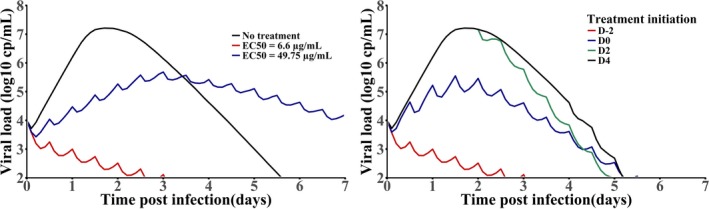
Viral dynamic predictions in Zika infections. Left: Prophylactic treatment assuming two different levels of drug half‐maximal effective concentration (EC50); right: effect of delayed treatment initiations on Zika dynamics.
This dosing regimen was also recently evaluated in an NHP model of Marburg infection treated in prophylaxis. Interestingly, although the 6 untreated animals died between D7 and D9, 5 of 6 treated animals survived until study termination at D28 (P = 0.0013) and only one treated animal died at D14. 85
Implication of these results for clinical use
How to interpret our results in the context of antiviral therapy, in particular in EVD? The first aspect is obviously the question of the target drug concentration. The results found in NHPs, where significant protection was conferred with a drug efficacy of 50% corresponds to free drug concentrations of ~ 100 μg/mL. This is greater than the value determined prior to the trial based on mice experiments, and is, more importantly, greater than the level achieved in patients of the JIKI trial. This strongly suggests that doses larger than those used in the JIKI trial will be needed to achieve a high level of antiviral efficacy against EBOV. Given the complexity of the drug PKs, we now need to find safe and tolerable dosing regimen that could generate such exposure in humans and this will be evaluated in a clinical trial.
Even if drug exposure is satisfactory, the translation of results from NHP experiments to patients needs caution. First, unlike what is obtained in the NHP model, EVD is not uniformly lethal and the mortality rate during the 2013–2016 outbreak was close to 40%. Second, no supportive care was administered in infected animals, which was shown to be critical for patient outcome. Thus, it is possible that the NHP models are more stringent and that treatment initiation of a potent drug, closer to peak viremia, may nonetheless have an impact on the disease and survival. In the 2013–2016 outbreak, the time from symptom onset to admission in the Ebola treatment centers was between 3 and 5 days 37 , 86 when the maximal viral load at admission was 4–5 days after symptom onset. 43 Based on these observations, it is likely that most patients included in EVD clinical trials, such as JIKI (favipiravir) or Prevail II (ZMapp), 86 initiated antiviral treatment close to the peak viremia and several days after virus replicated at high levels. Consistent with our prediction that antiviral drugs should be initiated as early as possible to reach maximal therapeutic benefit, the effect on survival in these studies was modest for favipiravir, 37 and larger but not statistically significant for ZMapp. 86 In the context of the North Kivu EBOV outbreak, the PALM trial, a randomized controlled trial of four investigational agents (ZMapp, remdesivir, mAb114, and REGN‐EB3), was initiated in November 2018. 87 In August 2019, an interim analysis on the first 499 included patients showed favorable results with two of four candidates, REGN‐EB3 or mAb114, which are, respectively, monoclonal antibodies cocktail and single monoclonal antibodies. 88 This result is the first demonstration in patients of the benefit of antiviral strategy on survival rate. Yet, the association of the duration of symptoms before admission and the lower viral load at baseline to survival 88 suggest that their efficacy could remain largely contingent on the timing of treatment initiation, and monoclonal antibodies remain uneasy to use in limited resource settings and expensive to produce. In that respect, and following what has been done in chronic viral infections, mathematical modeling can be used to better understand the efficacy of antiviral drugs. In spite of the difficulty to collect data in treatment centers during epidemics, more modelers should be involved in the design and the analysis of clinical trials to accelerate the development of effective therapies.
In the future, it will thus remain critical to administer drugs not only to confirmed or suspect cases, but also to contact individuals as early as possible, in a context where clinical trials or NHP models demonstrated their efficacy. This may meet the request of field teams during the Western African EVD outbreak to propose an early oral treatment to suspected cases, even before their transfer to care centers. In addition, having a drug active even only when administered early may contribute to empower local communities and improve their collaborations with non‐government organizations and local authorities. 89 An antiviral strategy may also offer an alternative to a vaccine, or be complementary to it, for exposed people. The promising vesicular stomatitis virus‐based vaccine used in Guinea provided high level protection after a 10‐day delay, but its efficacy in postexposure in both humans and NHPs is less clear. 90 , 91 Interestingly, favipiravir has been used alone or with monoclonal antibodies as postexposure prophylaxis in at least five healthcare workers with percutaneous accidents and suspected EBOV exposures during the West Africa outbreak. None of these individuals developed laboratory or clinical evidence of EBOV infection, but whether any infections were prevented by the use of postexposure prophylaxis is not possible to determine from this small number of uncontrolled cases. 92 There remains, more generally, to find drug combinations that could potentiate favipiravir against EBOV. Given the cost of experiments, this will essentially rely on in vitro experiments, that can be optimized using modeling, as was done for instance in HCV. 93 Importantly, and unlike what was found in mice infected with Lassa virus, it is unlikely that the combination of favipiravir and ribavirin is relevant against EBOV, as no gain in terms of survival or viral replication was found in animals receiving the combination as compared with those receiving favipiravir alone (unpublished results).
Beyond EVD, several other hemorrhagic fevers may benefit from the evaluation of favipiravir. For instance, the promising effects on favipiravir in the NHP model of Marburg disease encouraged the implementation of a clinical trial targeting exposed people in a prophylaxis or postexposure setting. During the last Marburg outbreak in Uganda in October 2017, favipiravir was considered for such a trial, and a protocol was developed and submitted to the Inserm ethics committee and the non‐government organization Médecins Sans Frontières. Despite the fact that the rupture of the transmission chain prevented the need of the intervention, the protocol remains usable in case of new epidemic clusters arising. Likewise, the combination of favipiravir and ribavirin in Lassa virus hemorrhagic fever could be of relevance for future clinical use. However, a better understanding of the PKs and the tolerance of favipiravir in humans is now needed to identify dosing regimens that are safe and achieve the target concentrations identified in animal studies.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 666092 and 666100, and from the St. Luke’s International University (Tokyo, Japan) in the framework of Research Program on Emerging and Re‐emerging Infectious Diseases of the Japan Agency for Medical Research and Development (AMED).
Conflict of Interest
As Editor‐in‐Chief for CPT: Pharmacometrics & Systems Pharmacology, France Mentré was not involved in the review or decision process for this paper. The other authors declared no conflict of interest.
Acknowledgment
All data and models developed in NHPs are available in the Supplementary Material s of ref. 69
References
- 1. Marston, H.D. , Folkers, G.K. , Morens, D.M. & Fauci, A.S. Emerging viral diseases: confronting threats with new technologies. Sci. Transl. Med. 6, 253ps10–253ps10 (2014). [DOI] [PubMed] [Google Scholar]
- 2. Woolhouse, M. , Scott, F. , Hudson, Z. , Howey, R. & Chase‐Topping, M. Human viruses: discovery and emergence. Phil. Trans. R. Soc. B 367, 2864–2871 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Clercq, E. & Li, G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 29, 695–747 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Warren, T.K. et al Therapeutic efficacy of the small molecule GS‐5734 against Ebola virus in rhesus monkeys. Nature 531, 381–385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Warren, T. et al Efficacy of galidesivir against Ebola virus disease in rhesus monkeys. Open Forum Infect. Dis. 4, S302 (2017). [Google Scholar]
- 6. Lim, S.‐Y. et al Galidesivir, a direct‐acting antiviral drug, abrogates viremia in rhesus macaques challenged with Zika virus. Open Forum Infect. Dis. 4 (suppl. 1), S55 (2017). [Google Scholar]
- 7. Best, K. et al Zika plasma viral dynamics in nonhuman primates provides insights into early infection and antiviral strategies. Proc. Natl. Acad. Sci. 114, 8847–8852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Madelain, V. et al Favipiravir pharmacokinetics in nonhuman primates and insights for future efficacy studies of hemorrhagic fever viruses. Antimicrob. Agents Chemother. 61, e01305‐ 16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oestereich, L. et al Efficacy of favipiravir alone and in combination with ribavirin in a lethal, immunocompetent mouse model for Lassa fever. J. Infect. Dis. 213, 934–938 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pushpakom, S. et al Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18, 41–58 (2019). [DOI] [PubMed] [Google Scholar]
- 11. Clapham, H.E. , Tricou, V. , Chau, N.V.V. , Simmons, C.P. & Ferguson, N.M. Within‐host viral dynamics of dengue serotype 1 infection. J. R. Soc. Interface 11, 20140094 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baccam, P. , Beauchemin, C. , Macken, C.A. , Hayden, F.G. & Perelson, A.S. Kinetics of influenza A virus infection in humans. J. Virol. 80, 7590–7599 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith, A.M. et al Kinetics of coinfection with influenza A virus and Streptococcus pneumoniae . PLoS Pathog. 9, e1003238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ansari, A.A. Clinical features and pathobiology of Ebolavirus infection. J. Autoimmun. 55, 1–9 (2014). [DOI] [PubMed] [Google Scholar]
- 15. Feldmann, H. & Geisbert, T.W. Ebola haemorrhagic fever. Lancet 377, 849–862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malvy, D. , McElroy, A.K. , de Clerck, H. , Günther, S. & van Griensven, J. Ebola virus disease. Lancet 393, 936–948 (2019). [DOI] [PubMed] [Google Scholar]
- 17. Schieffelin, J.S. et al Clinical illness and outcomes in patients with Ebola in Sierra Leone. N. Engl. J. Med. 371, 2092–2100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Olejnik, J. , Ryabchikova, E. , Corley, R.B. & Mühlberger, E. Intracellular events and cell fate in filovirus infection. Viruses 3, 1501–1531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baseler, L. , Chertow, D.S. , Johnson, K.M. , Feldmann, H. & Morens, D.M. The pathogenesis of Ebola virus disease. Annu. Rev. Pathol. 12, 387–418 (2017). [DOI] [PubMed] [Google Scholar]
- 20. Towner, J.S. et al Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high‐throughput antiviral drug screening. Virology 332, 20–27 (2005). [DOI] [PubMed] [Google Scholar]
- 21. Hoenen, T. , Groseth, A. , Callison, J. , Takada, A. & Feldmann, H. A novel Ebola virus expressing luciferase allows for rapid and quantitative testing of antivirals. Antiviral Res. 99, 207–213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caballero, I.S. et al In vivo Ebola virus infection leads to a strong innate response in circulating immune cells. BMC Genom. 17, 707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baize, S. et al Inflammatory responses in Ebola virus‐infected patients. Clin. Exp. Immunol. 128, 163–168 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wauquier, N. , Becquart, P. , Padilla, C. , Baize, S. & Leroy, E.M. Human fatal Zaire Ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl. Trop. Dis. 4, e837 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruibal, P. et al Unique human immune signature of Ebola virus disease in Guinea. Nature 533, 100–104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vernet, M.‐A. et al Clinical, virological, and biological parameters associated with outcomes of Ebola virus infection in Macenta, Guinea. JCI Insight 2, e88864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reynard, S. et al Immune parameters and outcomes during Ebola virus disease. JCI Insight 4, pii: 125106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McElroy, A.K. et al Human Ebola virus infection results in substantial immune activation. Proc. Natl. Acad. Sci. USA 112, 4719–4724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. World Health Organization . WHO|Ebola situation reports [Internet]. <http://apps.who.int/iris/bitstream/10665/208883/1/ebolasitrep_10Jun2016_eng.pdf?ua=1>.
- 30. World Health Organization . Categorization and prioritization of drugs for consideration for testing or use in patients infected with Ebola. In: Ebola treatments and interventions. <http://www.who.int/medicines/ebola-treatment/2015_0703TablesofEbolaDrugs.pdf?ua=1> (2015). Accessed September 7, 2015.
- 31. Madelain, V. et al Ebola virus infection: review of the pharmacokinetic and pharmacodynamic properties of drugs considered for testing in human efficacy trials. Clin. Pharmacokinet. 55, 907–923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Japanese Pharmaceuticals and Medical Devices Agency (PMDA) Report on the Deliberation Results (English version). <https://www.pmda.go.jp/files/000210319.pdf>.
- 33. Oestereich, L. et al Successful treatment of advanced Ebola virus infection with T‐705 (favipiravir) in a small animal model. Antiviral Res. 105, 17–21 (2014). [DOI] [PubMed] [Google Scholar]
- 34. Smither, S.J. et al Post‐exposure efficacy of oral T‐705 (Favipiravir) against inhalational Ebola virus infection in a mouse model. Antiviral Res. 104, 153–155 (2014). [DOI] [PubMed] [Google Scholar]
- 35. Mentré, F. et al Dose regimen of favipiravir for Ebola virus disease. Lancet Infect Dis. 15, 150–151 (2015). [DOI] [PubMed] [Google Scholar]
- 36. Bouazza, N. et al Favipiravir for children with Ebola. Lancet 385, 603–604 (2015). [DOI] [PubMed] [Google Scholar]
- 37. Sissoko, D. et al Experimental treatment with favipiravir for Ebola virus disease (the JIKI Trial): a historically controlled, single‐arm proof‐of‐concept trial in Guinea. PLoS Med. 13, e1001967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kerber, R. et al Laboratory findings, compassionate use of favipiravir, and outcome in patients with Ebola virus disease, Guinea, 2015‐a retrospective observational study. J. Infect. Dis. 220, 195–202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cardile, A.P. , Warren, T.K. , Martins, K.A. , Reisler, R.B. & Bavari, S. Will there be a cure for Ebola? Annu. Rev. Pharmacol. Toxicol. 57, 329–348 (2017). [DOI] [PubMed] [Google Scholar]
- 40. Cross, R.W. , Mire, C.E. , Feldmann, H. & Geisbert, T.W. Post‐exposure treatments for Ebola and Marburg virus infections. Nat. Rev. Drug Discov. 17, 413–434 (2018). [DOI] [PubMed] [Google Scholar]
- 41. Nguyen, T.H.T. et al Favipiravir pharmacokinetics in Ebola‐Infected patients of the JIKI trial reveals concentrations lower than targeted. PLoS Negl. Trop. Dis. 11, e0005389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Piorkowski, G. et al Implementation of a non‐human primate model of Ebola disease: Infection of Mauritian cynomolgus macaques and analysis of virus populations. Antiviral Res. 140, 95–105 (2017). [DOI] [PubMed] [Google Scholar]
- 43. Faye, O. et al Use of viremia to evaluate the baseline case fatality ratio of Ebola virus disease and inform treatment studies: a retrospective cohort study. PLoS Med. 12, e1001908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bonhoeffer, S. , May, R.M. , Shaw, G.M. & Nowak, M.A. Virus dynamics and drug therapy. Proc. Natl. Acad. Sci. 94, 6971–6976 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Warren, T.K. et al Protection against filovirus diseases by a novel broad‐spectrum nucleoside analogue BCX4430. Nature 508, 402–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Furuta, Y. , Komeno, T. & Nakamura, T. Favipiravir (T‐705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 93, 449–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guedj, J. et al Antiviral efficacy of favipiravir against Ebola virus: a translational study in cynomolgus macaques. PLoS Med. 15, e1002535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Neumann, A.U. et al Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon‐alpha therapy. Science 282, 103–107 (1998). [DOI] [PubMed] [Google Scholar]
- 49. Perelson, A.S. Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2, 28–36 (2002). [DOI] [PubMed] [Google Scholar]
- 50. Perelson, A.S. & Guedj, J. Modelling hepatitis C therapy‐predicting effects of treatment. Nat. Rev. Gastroenterol. Hepatol. 12, 437–445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Snoeck, E. et al A comprehensive hepatitis C viral kinetic model explaining cure. Clin. Pharmacol. Ther. 87, 706–713 (2010). [DOI] [PubMed] [Google Scholar]
- 52. Whalley, S.A. et al Kinetics of acute hepatitis B virus infection in humans. J. Exp. Med. 193, 847–854 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ribeiro, R.M. et al Estimation of the initial viral growth rate and basic reproductive number during acute HIV‐1 infection. J. Virol. 84, 6096–6102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carrat, F. et al Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am. J. Epidemiol. 167, 775–785 (2008). [DOI] [PubMed] [Google Scholar]
- 55. Li, Y. & Handel, A. Modeling inoculum dose dependent patterns of acute virus infections. J. Theor. Biol. 347, 63–73 (2014). [DOI] [PubMed] [Google Scholar]
- 56. Saenz, R.A. et al Dynamics of influenza virus infection and pathology. J. Virol. 84, 3974–3983 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stetson, D.B. & Medzhitov, R. Type I interferons in host defense. Immunity 25, 373–381 (2006). [DOI] [PubMed] [Google Scholar]
- 58. Waggoner, S.N. et al Roles of natural killer cells in antiviral immunity. Curr. Opin. Virol. 16, 15–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith, A.M. & Perelson, A.S. Influenza A virus infection kinetics: quantitative data and models. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 429–445 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Perelson, A.S. & Ribeiro, R.M. Modeling the within‐host dynamics of HIV infection. BMC Biol. 11, 96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hayden, F.G. Experimental human influenza: observations from studies of influenza antivirals. Antivir. Ther. 17, 133–141 (2012). [DOI] [PubMed] [Google Scholar]
- 62. Hayden, F.G. et al Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J. Clin. Invest. 101, 643–649 (1998).9449698 [Google Scholar]
- 63. Vegvari, C. et al How can viral dynamics models inform endpoint measures in clinical trials of therapies for acute viral infections? PLoS One 11, e0158237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pawelek, K.A. et al Modeling within‐host dynamics of influenza virus infection including immune responses. PLoS Comput. Biol. 8, e1002588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pearson, J.E. , Krapivsky, P. & Perelson, A.S. Stochastic theory of early viral infection: continuous versus burst production of virions. PLoS Comput. Biol. 7, e1001058 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dixit, N.M. , Layden‐Almer, J.E. , Layden, T.J. & Perelson, A.S. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature 432, 922–924 (2004). [DOI] [PubMed] [Google Scholar]
- 67. Dahari, H. et al HCV kinetic and modeling analyses indicate similar time to cure among sofosbuvir combination regimens with daclatasvir, simeprevir or ledipasvir. J. Hepatol. 64, 1232–1239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Madelain, V. et al Ebola virus dynamics in mice treated with favipiravir. Antiviral Res. 123, 70–77 (2015). [DOI] [PubMed] [Google Scholar]
- 69. Madelain, V. et al Ebola viral dynamics in nonhuman primates provides insights into virus immuno‐pathogenesis and antiviral strategies. Nat. Commun. 9, 4013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bekisz, J. , Schmeisser, H. , Hernandez, J. , Goldman, N.D. & Zoon, K.C. Human interferons alpha, beta and omega. Growth Factors 22, 243–251 (2004). [DOI] [PubMed] [Google Scholar]
- 71. Koyama, S. , Ishii, K.J. , Coban, C. & Akira, S. Innate immune response to viral infection. Cytokine 43, 336–341 (2008). [DOI] [PubMed] [Google Scholar]
- 72. Helming, L. Inflammation: cell recruitment versus local proliferation. Curr. Biol. 21, R548–R550 (2011). [DOI] [PubMed] [Google Scholar]
- 73. Shi, C. & Pamer, E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11, 762–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Desmée, S. , Mentré, F. , Veyrat‐Follet, C. & Guedj, J. Nonlinear mixed‐effect models for prostate‐specific antigen kinetics and link with survival in the context of metastatic prostate cancer: a comparison by simulation of two‐stage and joint approaches. AAPS J. 17, 691–699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Desmée, S. , Mentré, F. , Veyrat‐Follet, C. , Sébastien, B. & Guedj, J. Using the SAEM algorithm for mechanistic joint models characterizing the relationship between nonlinear PSA kinetics and survival in prostate cancer patients. Biometrics 73, 305–312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rosenke, K. et al Use of favipiravir to treat Lassa virus infection in macaques. Emerg. Infect. Dis. 24, 1696–1699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Westover, J.B. et al Low‐dose ribavirin potentiates the antiviral activity of favipiravir against hemorrhagic fever viruses. Antiviral Res. 126, 62–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Raabe, V.N. et al Favipiravir and ribavirin treatment of epidemiologically linked cases of Lassa fever. Clin. Infect. Dis. 65, 855–859 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ribeiro, R.M. , Layden‐Almer, J. , Powers, K.A. , Layden, T.J. & Perelson, A.S. Dynamics of alanine aminotransferase during hepatitis C virus treatment. Hepatology 38, 509–517 (2003). [DOI] [PubMed] [Google Scholar]
- 80. Carrillo‐Bustamante, P. et al Determining Ribavirin’s mechanism of action against Lassa virus infection. Sci. Rep. 7, 11693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Osuna, C.E. et al Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat. Med. 22, 1448–1455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lanciotti, R.S. et al Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 14, 1232–1239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Barzon, L. et al Infection dynamics in a traveller with persistent shedding of Zika virus RNA in semen for six months after returning from Haiti to Italy, January 2016. Eurosurveillance 21, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Osuna, C.E. & Whitney, J.B. Nonhuman primate models of Zika virus infection, immunity, and therapeutic development. J. Infect. Dis. 216, S928–S934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bixler, S.L. et al Efficacy of favipiravir (T‐705) in nonhuman primates infected with Ebola virus or Marburg virus. Antiviral Res. 151, 97–104 (2018). [DOI] [PubMed] [Google Scholar]
- 86. Prevail, I. ; Multi‐National PREVAIL II Study Team . A randomized, controlled trial of ZMapp for Ebola virus infection. N. Engl. J. Med. 375, 1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Investigational Therapeutics for the Treatment of People with Ebola Virus Disease – Full Text View – ClinicalTrials.gov. <https://clinicaltrials.gov/ct2/show/NCT03719586>.
- 88. Mulangu, S. et al A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 381, 2293–2303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gomez‐Temesio, V. & La Le Marcis, F . La mise en camp de la Guinée. L’Homme 57–90, (2017). [Google Scholar]
- 90. Henao‐Restrepo, A.M. et al Efficacy and effectiveness of an rVSV‐vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster‐randomised trial. Lancet 386, 857–866 (2015). [DOI] [PubMed] [Google Scholar]
- 91. Marzi, A. et al VSV‐EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science 349, 739–742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fischer, W.A. et al Ebola virus disease: an update on post‐exposure prophylaxis. Lancet Infect. Dis. 18, e183–e192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Koizumi, Y. et al Quantifying antiviral activity optimizes drug combinations against hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 114, 1922–1927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


