Abstract
Alterations in normal regulation of gene expression is one of the key features of hematopoietic malignancies. In order to gain insight into the mechanisms that regulate gene expression in these diseases, we dissected the role of the Ikaros protein in leukemia. Ikaros is a DNA-binding, zinc finger protein that functions as a transcriptional regulator and a tumor suppressor in leukemia. The use of ChIP-seq, RNA-seq, and ATAC-seq—coupled with functional experiments—revealed that Ikaros regulates both the global epigenomic landscape and epigenetic signature at promoter regions of its target genes. Casein kinase II (CK2), an oncogenic kinase that is overexpressed in leukemia, directly phosphorylates Ikaros at multiple, evolutionarily-conserved residues. Phosphorylation of Ikaros impairs the protein's ability to regulate both the transcription of its target genes and global epigenetic landscape in leukemia. Treatment of leukemia cells with a specific inhibitor of CK2 restores Ikaros function, resulting in cytotoxicity of leukemia cells. Here, we review the mechanisms through which the CK2-Ikaros signaling axis regulates the global epigenomic landscape and expression of genes that control cellular proliferation in leukemia.
Keywords: Ikaros, Tumor suppressor, Casein kinase II (CK2) inhibitor CX-4945, Leukemia, Epigenetic regulation, Gene transcription
1. Introduction
Complex regulatory mechanisms orchestrate development, lineage commitment, and differentiation of hematopoietic cells. Transcription factors play a very important role in regulating expression of genes involved in hematopoiesis. Dysregulation of gene expression is a hallmark of cancer, including leukemia. Recent advances have identified the role of chromatin remodeling and epigenetics as the key mechanisms that regulate gene expression in leukemia. Here, we discuss the role of the Ikaros transcription factor (Georgopoulos et al., 1992, 1994; Lo et al., 1991) in regulating gene expression and as a tumor suppressor in acute lymphoblastic leukemia. Ikaros exerts its regulatory effect on gene expression via chromatin remodeling (Brown et al., 1997; Cobb et al., 2000; Ge et al., 2015, 2016a, 2016b, 2016c, 2017, 2018a, 2018b; Kim et al., 1999, 2009; Sridharan and Smale, 2007; Su et al., 2005). Moreover, global influence on the epigenomic landscape in T-cell leukemia underscores the role of Ikaros as a tumor suppressor (Ding et al., 2019). Recently published studies have uncovered several novel functions of Ikaros as an epigenetic regulator using dynamic, global, epigenomic, and gene expression analyses (Ding et al., 2019). Ikaros activity as a regulator of chromatin remodeling and gene expression is controlled via direct phosphorylation at multiple serine/threonine residues (Dovat et al., 2002, 2011; Ge et al., 2015, 2016a, 2016c, 2017, 2018a; Gowda et al., 2016, 2017a, 2017b, 2017c; Gurel et al., 2008; Payne and Dovat, 2011; Song et al., 2011; Uckun et al., 2012; Wang et al., 2014a). Functional inactivation of Ikaros by pro-oncogenic protein Casein Kinase II (CK2) via phosphorylation, has been demonstrated in B-cell acute lymphoblastic leukemia (Han et al., 2019; Popescu et al., 2009; Song et al., 2011; Wang et al., 2014b, 2016). Inhibition of CK2 using specific inhibitors results in an anti-leukemic effect via restoration of Ikaros' functions as a tumor suppressor and regulator of gene expression (Song et al., 2015). In this article, we will focus on the various roles of Ikaros as an epigenetic regulator, specifically in acute lymphoblastic leukemia. We will demonstrate that the Ikaros transcription factor can be indirectly targeted, in order to restore its function, by inhibition of CK2. We will also discuss several important oncogenic signaling pathways regulated by Ikaros. Finally, we will demonstrate Ikaros’ influence in regulating several genes known as global epigenetic regulators in leukemia, such as KBM5B.
2. Ikaros structure and function
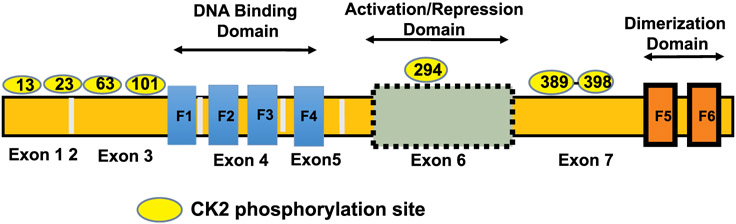
Ikaros is a DNA-binding, zinc finger protein encoded by the IKZF1 gene that is located on chromosome 7 (Georgopoulos et al., 1992, 1994; Lo et al., 1991). Alternate splicing of IKZF1 produces a large number of Ikaros isoforms that are functionally diverse (Hahm et al., 1994; Molnár and Georgopoulos, 1994). Isoform Ik-H or Ik1 is the largest with six zinc finger domains, two in the C-terminal end and four in the N-terminal end (Kim et al., 2009; Ronni et al., 2007). Several combinations of zinc fingers in the N-terminus are seen in the various Ikaros isoforms (Li et al., 2011; Molnár et al., 1996; Sun et al., 1996). Dimerization among several isoforms either potentiates or inhibits Ikaros’ DNA binding affinity, which affects the overall transcriptional activity of Ikaros (Fig. 1) (McCarty et al., 2003; Molnár et al., 1996; Sun et al., 1996).
Fig. 1.
Structure of Ikaros and CK2 phosphorylation sites.
Ikaros is a transcription factor with a critical role in hematopoiesis and development of the immune system (Cobb and Smale, 2005; Georgopoulos, 1997; Georgopoulos et al., 1994, 1997; Papathanasiou et al., 2003; Wang et al., 1996; Winandy et al., 1995). Since its discovery in the early 90's, Ikaros' role as a master regulator of lymphoid development and tumor suppression in leukemia has been extensively studied. The biological function of Ikaros was studied in several Ikaros knockout mice (Nichogiannopoulou et al., 1999; Wang et al., 1996; Wu et al., 1997). Important regulatory functions of Ikaros in hematopoiesis are as follows: First, a pivotal function in early hematopoietic cells called lymphoid-primed multipotent progenitors (LMPPs) where Ikaros regulation of the FLT3-tyrosine kinase receptor and upregulation of the interleukin-7 alpha subunit (IL-7α) promote lymphoid differentiation signals (Busslinger, 2004; Yoshida et al., 2006). Second, Ikaros plays a very important role in B-cell differentiation by regulating pre-B cell receptor component Lambda 5—encoded by gene IGLL1—as well as recombinase activating genes (RAGs) (Reynaud et al., 2008). Third, Ikaros regulates expression of several important genes involved in T-cell differentiation, including terminal deoxynucleotide transferase (TdT), CD4, CD8, and IL2 (Avitahl et al., 1999; Ernst et al., 1996; Georgopoulos et al., 1997; Hahm et al., 1994; Harker et al., 2002; Kirstetter et al., 2002; Trinh et al., 2001; Urban and Winandy, 2004). Mice heterozygous for an inactivating Ikaros mutation develop T-cells that are hyper-proliferative and develop T-cell leukemia with 100% penetrance (Winandy et al., 1995).
3. Clinical significance of IKZF1 alteration
Genome-wide analysis of genetic alterations in acute lymphoblastic leukemia (ALL) has shown that deletion and mutation of IKZF1 is seen in nearly 15% of pediatric B-cell acute lymphoblastic leukemia (B-ALL) and in more than 80% of BCR-ABL positive (Ph +) ALL (Mullighan and Downing, 2008; Mullighan et al., 2007, 2008). ALL with IKZF1 alteration is associated with high risk features, high relapse rate, and poor outcome (Mullighan et al., 2009a). In T-cell acute lymphoblastic leukemia (T-ALL), IKZF1 alteration is seen in nearly 20% of patients and a specific subgroup—early T-cell precursor (ETP) leukemia—has worse outcome and harbors Ikaros mutations in 11% of patients (Zhang et al., 2012). IKZF1 deletions are also common in cases with CRLF2 genomic alterations and in Ph-like (BCR-ABL1-like) ALL (Harvey et al., 2010; Mullighan et al., 2009b). Several clinical trials are currently looking at risk stratifying B-ALL patients with IKZF1 alteration as high-risk and intensifying therapy (Chen et al., 2012; Hunger et al., 2011; Mullighan, 2011a, b; Roberts and Mullighan, 2011). Ikaros deletion and mutation is seen in 5–7% of acute myeloid leukemia (AML) and myeloid dysplastic syndrome (MDS) (Crescenzi et al., 2004; de Rooij et al., 2015; Lavallee et al., 2015; Yagi et al., 2002). Germline deletion and/or mutation of the IKZF1 gene has been associated with development of leukemia (Churchman et al., 2018; Yoshida et al., 2017).
Ikaros' role in regulating the immune response in humans has been demonstrated in several studies. Point mutations at Ikaros’ DNA-binding domain results in severe combined immunodeficiency (SCID) (Goldman et al., 2012). Subsequently, different mutations in the IKZF1 gene were associated with various immunodeficiencies in human (Abdulhay et al., 2016; Berron-Ruiz, 2017; Bigley et al., 2019; Bogaert et al., 2016; Boutboul et al., 2018; Chen et al., 2018; Churchman et al., 2018; Cytlak et al., 2018; Davis et al., 2019; Eskandarian et al., 2019; Hoshino et al., 2017; Kanegane and Hoshino, 2019; Kuehn et al., 2016; Maffucci et al., 2016; Sriaroon et al., 2019). Thus, it has been recognized that germline mutations of Ikaros can cause primary immunodeficiency in humans.
4. The CK2-Ikaros axis
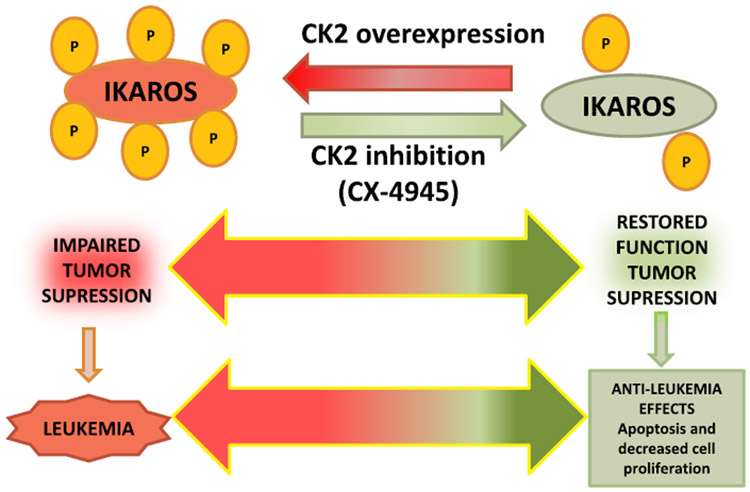
Multiple signaling pathways involving protein phosphorylation by various kinases regulate cellular proliferation and drug sensitivity in human malignancies (Candido et al., 2018; Choi et al., 2018; Drulis-Fajdasz et al., 2018; Geffken and Spiegel, 2018; Jang et al., 2018; Kaushansky and Zhan, 2018; Lee et al., 2018; Pyne et al., 2018; Ramos et al., 2018; Rebello et al., 2018; Saiardi et al., 2018; Sakane et al., 2018; Scarlata et al., 2018). Casein Kinase II (CK2) is a pro-oncogenic kinase that is overexpressed in several cancers, including leukemia. CK2 is a ubiquitous, serine/threonine kinase that is involved in multiple signaling pathways and has over 300 substrates (Ahmad et al., 2005; Gowda et al., 2017a, 2017b; Pinna, 1997; Pinna and Meggio, 1997). CK2 phosphorylates Ikaros at multiple evolutionarily-conserved serine/threonine amino acids (Fig. 1), which impairs Ikaros' ability to bind DNA and localize to peri-centromeric heterochromatin (Gurel et al., 2008; Popescu et al., 2009; Wang et al., 2014b). A fine balance of CK2- and protein phosphatase 1 (PP1)-mediated post-translational modification of Ikaros determines Ikaros' stability and activity (Dovat et al., 2011; Gowda et al., 2017a, 2017c; Popescu et al., 2009; Song et al., 2011; Wang et al., 2014b). In leukemia, as well as other malignancies, the expression and activity of CK2 is increased, which results in functional inactivation of Ikaros and loss of its tumor suppressor function (Barata, 2011; Buontempo et al., 2014; Gomes et al., 2014; Martins et al., 2010, 2011; Silva et al., 2008, 2010). Molecular and pharmacological inhibition of CK2 enhances Ikaros’ tumor suppressive function as seen by restored transcriptional regulation of Ikaros target genes (Ge et al., 2015, 2016a, 2016c, 2017, 2018a; Han et al., 2019; Song et al., 2015; Wang et al., 2016). In high risk B-ALL patient derived xenograft (PDX) models, inhibition of CK2 using CX-4945—a small-molecule, ATP-competitive specific inhibitor—results in a strong in vivo anti-leukemic effect that is a result of restored Ikaros function (Song et al., 2015) (Fig. 2). CK2 inhibition was able to restore Ikaros function in vivo even in cases of high-risk B-ALL with a single copy of Ikaros (IKZF1 deletion) (Song et al., 2015). These results led to discovery of the CK2-Ikaros signaling axis where direct phosphorylation of Ikaros by CK2, and a strong relation between CK2 expression and/or activity and Ikaros function, regulates gene expression and tumor suppression in leukemia and hematopoietic cells (Gowda et al., 2016, 2017a, 2017b, 2017c).
Fig. 2.
Mechanism of action of CK2 inhibitor via Ikaros.
5. Epigenetic regulation of gene expression by Ikaros, HDAC1, and Casein Kinase II in leukemia
The mechanism of Ikaros-mediated transcriptional regulation in leukemia is still not well understood. Ikaros directly associates with components of the histone deacetylase complex (NuRD), HDAC1, HDAC2, and Mi-2 in order to bring about chromatin remodeling (Kim et al., 1999; Koipally et al., 2002; O'Neill et al., 2000; Sridharan and Smale, 2007). In order to study the role of Ikaros and Ikaros-HDAC1 complexes in ALL, we used ChiP-seq and analyzed genome-wide occupancy of Ikaros and HDAC1 using B-ALL cells (Nalm-6 cell line). 12,464 distinct binding sites for Ikaros and 9971 for HDAC1 were identified. 6722 and 6182 target genes were associated with Ikaros and HDAC1, respectively. Only 12% overlap noted with both Ikaros and HDAC1 binding. Quantitative chromatin immunoprecipitation (qChIP) analysis of the high- and low-rank ChIP-seq peak values was used to validate the ChIP-seq data. The effect of Ikaros and HDAC1 DNA binding on the surrounding chromatin was determined by analyzing the genome-wide distribution of histone markers including H3 trimethylation at lysine 4 (H3K4me3), lysine 27 (H3K27me3), lysine 36 (H3K36me3), or lysine 9 (H3K9me3); or acetylation at lysine 9 (H3K9ac) using ChiP-seq (Song et al., 2016).
Ikaros loss-of-function or gain-of-function experiments were used to study the transcriptional regulation and epigenetic signature of Ikaros target genes in primary high-risk B-ALL cells. The epigenetic signature at the promoters of Ikaros and Ikaros-HDAC1 target genes in primary high-risk B-ALL (with loss of Ikaros function), and in primary high-risk B-ALL cells following treatment with CK2 inhibitors (TBB, CX-4945) was analyzed.
DNA binding analysis using the qChIP assay showed that Ikaros DNA binding to the promoters of its target genes is impaired in B-ALL. Treatment with CK2 inhibitor, CX-4945, restored Ikaros DNA binding to promoters and induced a distinct epigenetic signature at Ikaros-only and Ikaros-HDAC1 target genes (Song et al., 2016). High-level H3K9me3, reduced H3K9ac, and the absence of H3K27me3 was noted at the Ikaros-only target genes (e.g. cell cycle progression gene, CDC7). However, for the Ikaros-HDAC1 target genes (e.g. CDC2), restoration of Ikaros binding following CK2 inhibition results in a high level of H3K27me3, the loss of H3K9ac, and largely unchanged H3K9me3. Subsequent studies identified a large number of Ikaros target genes whose expression is regulated at the transcriptional level by Ikaros and/or the CK2-Ikaros signaling axis.
These data led to the conclusion that in B-ALL, chromatin remodeling and target gene expression are regulated by Ikaros alone and in complex with HDAC1 (Song et al., 2016). Ikaros induces the formation of repressive chromatin via direct Ikaros binding resulting in the formation of heterochromatin due to increased H3K9me3 and reduced H3K9ac. Repressive chromatin formation by Ikaros also occurs following Ikaros recruitment of HDAC1, where the most prominent change is a strong increase in H3K27me3 along with reduced H3K9ac. Both mechanisms lead to negative regulation of Ikaros target gene expression. These results demonstrate the strong interplay between Ikaros, HDAC1, and CK2 and underscore the importance of the CK2-Ikaros axis in controlling epigenetic regulation of gene expression in leukemia. Thus, both protein-protein interaction with HDAC1 and DNA binding are essential components of Ikaros’ function as a regulator of gene expression via chromatin remodeling in leukemia (Song et al., 2016). It is important to emphasize that analyses of gene expression by the CK2-Ikaros axis were limited to epigenetic regulation of transcription of the individual Ikaros target genes, and not the global epigenomic effect of Ikaros.
5.1. Regulation of JARID1B/KDM5B by Ikaros and CK2
The above-described studies identified a large number of Ikaros target genes. Among them, several genes are directly involved in global regulation of the epigenetic signature by encoding proteins that function as epigenetic modifiers. One of them is Lysine-specific histone demethylase 5B (KDM5B) also known as JARID1B, a member of the JmjC domain-containing histone demethylases (Kristensen et al., 2012). This protein functions as an epigenetic eraser, as it can demethylate tri-, di-, and mono-methylated lysine 4 of histone H3. Since H3K4me3 is the mark of open chromatin and is associated with positive regulation of gene expression (Santos-Rosa et al., 2002), KDM5B is involved in transcriptional repression of its target genes. KDM5B is overexpressed in several types of malignancies, and it has an important role in regulating genome stability and the DNA double-stranded break response (Albert et al., 2013; Bueno and Richard, 2013; Hayami et al., 2010; Kristensen et al., 2012; Roesch et al., 2010; Santos-Rosa et al., 2002; Wong et al., 2012; Xiang et al., 2007). KDM5B is upregulated in leukemia and inhibition of KDM5B causes cellular growth arrest (Haferlach et al., 2010). Transcriptional repression of JARID1B is associated with increased global levels of H3K4 trimethylation (Wang et al., 2016). Functional experiments showed that Ikaros represses transcription by directly binding to the promoter of KDM5B (Wang et al., 2016). Ikaros-mediated repression of JARID1B is dependent on the activity of the histone deacetylase, HDAC1, which is recruited to the upstream regulatory element of KDM5B in complex with Ikaros. Repression of KDM5B by the Ikaros-HDAC1 protein complex results in an increased level of H3K4me3 in the nucleus. Inhibition of CK2 results in increased DNA-binding affinity of the Ikaros-HDAC1 complex to the promoter of JARID1B. This is associated with increased formation of H3K27me3 and decreased H3K9ac. In high-risk B-ALL that carry deletion of one Ikaros (IKZF1) allele, targeted inhibition of CK2 enhances Ikaros binding and recruitment of HDAC1 to the KDM5B promoter, resulting in repression of KDM5B and increase of global H3K4me3 in cells (Wang et al., 2016). These results demonstrate that the effect of the CK2-Ikaros axis is not limited only to the regulation of epigenetic signature of Ikaros target genes (such as KDM5B), but that it can affect the global epigenomic signature in leukemia via transcriptional regulation of KDM5B and subsequent increase in H3K4me3 in cell.
5.2. Regulation of PHF2 by Ikaros and CK2
Analysis of global genomic occupancy showed that Ikaros binds to the promoter of PHD finger protein 2 (PHF2) (Ge et al., 2018a). PHF2 can also function as a demethylase and an eraser of H3K9me3 epigenetic marks. Since H3K9me3 is a repressive epigenetic mark, PHF2 can positively regulate transcription of its target genes. PHF2 expression is significantly reduced in subsets of ALL patients, and correlates with leukemia cell proliferation. Ikaros positively regulates PHF2 expression directly (Ge et al., 2018a, 2018b). Molecular CK2 inhibition significantly promotes PHF2 expression in an Ikaros-dependent manner. Pharmacological CK2 inhibition by CX-4945 treatment also results in an increase of PHF2 expression and enrichment of the Ikaros protein at PHF2 promoter in ALL. These results demonstrate that the CK2-Ikaros axis can also act as a positive regulator of gene expression of Ikaros target genes.
6. Ikaros regulates the global epigenetic landscape in T-cell leukemia
Next, we studied the direct role of Ikaros in global regulation of the epigenomic landscape. We used T-cell leukemia cells from Ikaros-deficient mice to determine the role of Ikaros in tumor suppression and in global genome-wide regulation of epigenetic signature. Ikaros haplo-knockout mice develop T-cell leukemia, with arrest in T-cell differentiation at the early (DN3) thymocyte stage (Winandy et al., 1995). During the process of malignant transformation, haplo-knockout Ikaros thymocytes lose the remaining wild-type Ikaros allele; the resulting T-ALL has an Ikaros-null genotype. Re-introduction of Ikaros into Ikaros-null T-ALL induces partial T-cell differentiation, along with cessation of leukemia cell growth and tumor suppression (Kathrein et al., 2005). We used this system to study the dynamics of Ikaros' role in regulating global chromatin rearrangement, epigenetic regulation of gene expression, and tumor suppression. Briefly, T-ALL Ikaros-null cells obtained from the spontaneous T-cell leukemia developed in Ikaros knockout mice, were retrovirally transduced with Ikaros. ChIP-seq was used to assess Ikaros’ genome-wide DNA occupancy and global epigenomic signature, ATAC-seq was used to determine chromatin accessibility, while microarrays were used to analyze alterations of gene expression following Ikaros reintroduction. In order to study the dynamic changes in the global epigenomic landscape, Ikaros DNA-binding, and gene expression; the above analyses were done on Ikaros-null T-ALL (day #0), and on days #1, #2, and #3 following Ikaros transduction.
Ikaros’ interaction with DNA elements were studied in four distinct genomic regions as follows:
Gene body region with GENCODE annotation
Promoter region defined as more than or equal to 3 kb upstream or downstream from the transcription start site (TSS) for each annotated gene
Enhancer defined as 3 kb away from TSS with overlapping H3K4me1 peak signal regions
Reminder of the genome without promoter or enhancers, 100 kb apart from the nearest gene, called gene desert.
The total number of Ikaros DNA-binding peaks were noted in all regions, but it was by far the highest in the promoter/enhancer regions on day #1 following Ikaros re-introduction to Ikaros-null T-ALL (Ding et al., 2019). Despite the unchanged Ikaros expression, Ikaros DNA occupancy at promoters, enhancers, and the gene body was severely reduced during subsequent days following Ikaros re-introduction. However, Ikaros occupancy of gene desert regions did not dramatically change. Epigenomic analysis of T-ALL cells following Ikaros re-introduction uncovered several novel Ikaros functions in globally regulating the epigenetic landscape.
6.1. Pioneer activity
Most of the transcription factors are not able to bind DNA if the chromatin is in a compacted, “condensed” structure. These transcriptional factors require chromatin to be in a relaxed, “accessible” state in order to bind to the upstream regulatory elements of their target genes and regulate their transcription (Choukrallah and Matthias, 2014; Mayran and Drouin, 2018). “Pioneer factors” are the small number of transcriptional factors that are capable of binding condensed chromatin and making it accessible for other transcriptional factors and, thus, permissible for transcriptional regulation (Iwafuchi-Doi and Zaret, 2014; Zaret and Carroll, 2011). ATAC-seq analysis showed that Ikaros was able to bind to the chromatin that was condensed in T-ALL in Ikaros-null cells, as evidenced by negative ATAC-seq. Following Ikaros binding, previously condensed chromatin was remodeled into accessible chromatin, as evidenced by positive ATAC-seq. Chromatin remodeling from a condensed into an accessible state was observed at over 3,400 sites. This showed that Ikaros pioneering activity was not restricted to just a few sites, but that it is a global function of Ikaros. Epigenetic analysis showed that a majority of the de novo Ikaros-induced accessible chromatin sites have either promoter (with H3K4me3 marker), or enhancer (with H3K4me1) functions (Ding et al., 2019). Enrichment of H3K27ac in some of the de novo accessible chromatin sites indicated active transcription or active enhancers respectively. Dynamic analysis of Ikaros' pioneering function showed that Ikaros binds transiently to the condensed chromatin; Ikaros binding that was present at Day #1 was not detected on Day #2. However, the accessibility of many of the de novo open chromatin sites remained unchanged for at least 2 days from the initial Ikaros binding. These results confirmed Ikaros’ pioneering activity in chromatin remodeling by demonstrating that Ikaros binding to the condensed chromatin produces accessible chromatin, which remains unchanged for a period of time allowing other transcription factors to bind to these sites and regulate transcription of their target genes. This suggests that Ikaros binding to specific silent chromatin sites facilitates initiation of regulatory activities by promoting chromatin accessibility (Ding et al., 2019). This sets up the T-cell differentiation program, as well as growth cessation of leukemia cells, that is further maintained by other transcription factors. A typical pioneer factor often functions as a master regulator of tissue development, or during stem cell differentiation (Boller et al., 2016; Mayran and Drouin, 2018; Mayran et al., 2018; van Oevelen et al., 2015). Since Ikaros is known to act as a master regulator of T- and B-cell differentiation, the pioneering function of Ikaros explains its biological role in hematopoiesis and controlling of gene expression. The pioneering activity as a part of the tumor suppression process in T-cell leukemia is a relatively novel observation, and the role of chromatin accessibility in leukemogenesis and regulation of T-ALL progression should be further studied.
6.2. Regulation of enhancer formation
Ikaros binding to DNA following its re-introduction into Ikaros-null T-ALL cells is associated with the de novo formation of enhancers, as indicated by de novo enrichment of the H3K4me1 histone modification mark. Despite transient Ikaros DNA-binding, many of the de novo-formed enhancers retained their epigenetic signature during the following days, which indicates that Ikaros binding has a long-lasting effect on determining the enhancer landscape in leukemia and during induction of T-cell differentiation. Ikaros DNA-binding also had opposing effects on the enhancer landscape; it resulted in depletion of the existing enhancers, which was indicated by a loss of the H3K4me1 histone modification mark. Ikaros-induced enhancer depletion persisted following the loss of Ikaros occupancy in subsequent days, which was consistent with the long-lasting effect of Ikaros on the genome-wide enhancer signature. Overall, these results demonstrate the profound effect of Ikaros in regulating the global enhancer landscape in T-ALL and during induction of T-cell differentiation.
6.3. De novo formation of active enhancers
Enhancer regions that have enrichment in H3K4me1, but not in H3K27ac histone modification are in a “poised” state (Choukrallah and Matthias, 2014; Cico et al., 2016; Huang et al., 2016). Such enhancers are “primed” for activation, but do not affect the transcription of their target genes. Comparison of the epigenetic signature of Ikaros-null T-ALL and the same cells following Ikaros re-introduction revealed that Ikaros binding induces the de novo formation of active enhancers. This is characterized by the transition of DNA regions with an absence of H3K4me1 and H3K27ac in Ikaros-null T-ALL, into regions that are enriched with both H3K4me1 and H3K27ac marks. Thus, Ikaros binding to these regions induces the formation of active enhancers at DNA regions that did not have any marks of enhancers in Ikaros-null T-ALL. Overall, 22% of all newly-formed, active enhancers have been occupied by Ikaros, which demonstrates the direct role of Ikaros in chromatin remodeling that results in the formation of active enhancers. Many of the de novo formed activated enhancers remained in active state after the loss of Ikaros occupancy. Formation of de novo activated enhancers resulted in increased expression of nearby genes. These data demonstrated, for the first time, that Ikaros can regulate gene expression not only by binding to their promoters, but also by inducing formation of active enhancers that regulate transcription of their target genes.
6.4. Activation of poised enhancers
During the process of differentiation, a large number of enhancers are in a “poised” state and become activated in order to induce expression of genes that are essential for progression of differentiation (Cico et al., 2016; Heinz et al., 2015; Huang et al., 2016). Since Ikaros-null T-ALL cells are arrested at the DN3 stage of differentiation, many enhancers are in a poised state (with an H3K4me1+/H3K27ac–signature). Re-introduction of Ikaros induces T-cell differentiation, which is associated with activation of a large number of enhancers. Ikaros binding directly induces activation of over 900 poised enhancers. Importantly, over 40% of Ikaros-induced active enhancers remained activated over at least the next 2 days. This indicates that Ikaros has a critical role in activating poised enhancers and that Ikaros binding sets the stage for long-lasting regulation of gene expression that is associated with T-cell differentiation. Overall, the ability of Ikaros to regulate de novo enhancer formation, enhancer depletion, and activation of poised enhancers shows that Ikaros directly regulates the expression of a much larger number of genes than previously hypothesized. These data revealed an Ikaros-controlled regulatory network that regulates expression of genes involved in cellular proliferation and T-cell differentiation.
6.5. Formation of super-enhancers
DNA domains that contain clusters of enhancers are called super-enhancers (Loven et al., 2013; Whyte et al., 2013). These regulatory regions are often occupied by several transcription factors, span large DNA domains (over 10 kB), and often regulate expression of cell-specific genes. The function of super-enhancers was mostly studied during stem cell differentiation, although they are likely involved in all stages of tissue-specific differentiation (Shin, 2018; Wong et al., 2017, 2018). The epigenomic analysis of Ikaros-null T-ALL cells following re-introduction of Ikaros, revealed that Ikaros binding induces the formation of a very large number of super-enhancers (609 following Ikaros re-introduction vs. 24 in Ikaros-null T-ALL cells). Genes that are regulated by super-enhancers have much higher expression, as compare to the genes regulated by “regular” activated enhancers. Dynamic analysis showed that Ikaros binding to the newly-formed super-enhancers is less transient than its binding to a typical enhancers. Since about 90% of the newly-formed super-enhancers retain Ikaros occupancy 24 h following initial Ikaros binding, while less than 5% of regular enhancers retain Ikaros occupancy over the same time period. Ikaros-induced formation of super-enhancers also has a much longer-lasting effect than the formation of regular enhancers, since over 55% of super-enhancers retain their epigenetic signature 2 days after initial Ikaros binding. These data demonstrate that strong effect of Ikaros on the global epigenomic landscape is accomplished by remodeling large DNA domains and inducing the formation of large, potent regulatory elements that control expression of a large number of genes involved in differentiation and regulating cellular proliferation (Ding et al., 2019).
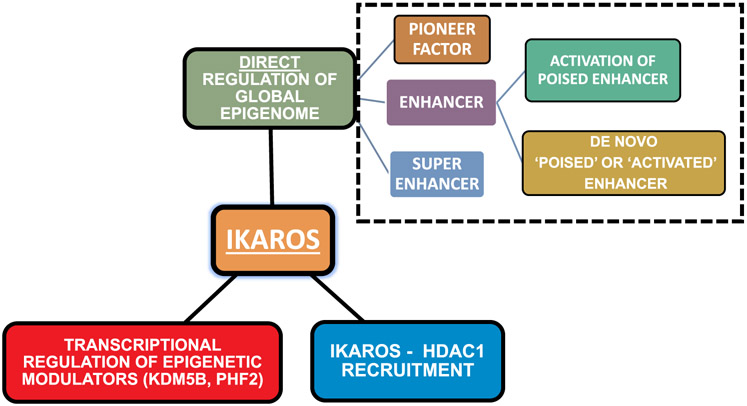
Overall, the studies that analyzed the role of Ikaros re-introduction into Ikaros-null T-ALL using dynamic temporal analysis of the epigenetic landscape following Ikaros re-introduction demonstrated that Ikaros tumor suppressor activity involves complex regulation of the epigenetic signature during T-cell differentiation and in T-ALL (Fig. 3). The complexity of the analysis (dynamic approach involving 4 different time points over 4 days), and specificity of the experimental system (Ikaros-null T-ALL) allowed identification of specific Ikaros functions in epigenomic regulation with minimal background noise. Results of the analysis demonstrated that the major part of Ikaros' tumor suppressor function in leukemia involves global epigenomic control of gene expression. However, whether phosphorylation by CK2 affects Ikaros’ role in regulating the global epigenomic landscape remains unknown.
Fig. 3.
Multifaceted functions of Ikaros in leukemia epigenetic regulation.
7. Conclusion
Functional analysis of the CK2-Ikaros signaling axis in acute lymphoblastic leukemia revealed the interplay between this signaling pathway and epigenetic regulation of gene expression in leukemia. The CK2-Ikaros axis regulates both the epigenetic signature of Ikaros target genes, as well as the global epigenomic signature by regulating expression of epigenetic modulators, such as KDM5B. Dynamic functional studies into Ikaros’ function in tumor suppression in T-ALL and induction of T-cell differentiation showed that Ikaros directly and dramatically regulates the global epigenomic landscape. This is achieved by directly inducing de novo formation and depletion of enhancers, activation of poised enhancers, formation of super-enhancers, and regulation of chromatin accessibility. Future studies will be directed toward further dissecting the role of CK2 and the CK2-Ikaros axis in regulating the epigenomic landscape, cellular differentiation, and leukemia.
Acknowledgements
This work was supported by National Institutes of Health R01CA209829 (KJP and SD); R01CA213912 (SD and CS); F30CA221109 (JLP); Penn State Clinical and Translational Sciences KL2 award (KL2 TR002015) to CG; Hyundai Hope on Wheels Scholar Grant (SD) and Young investigator award (CG), Four Diamonds Fund of the Pennsylvania State University College of Medicine (to SD, CG and CS); Bear Necessities Pediatric Cancer Foundation, Alex’s Lemonade Stand Foundation, and the John Wawrynovic Leukemia Research Scholar Endowment (to SD); and the St. Baldrick's Foundation (SD) career development award (CG) and summer fellowship award (SK) and Bear Necessities (CG); Rally foundation YI award (CG).
Footnotes
Declaration of competing interest
Authors declare no financial or research conflict of interest related to this work.
References
- Abdulhay N, Fiorini G, Kumanovics A, Sun AA, Hansen-Rejali J, Voelkerding KV, Rosenzweig SD, Hill HR, Sankaran VG, 2016. Normal hematologic parameters and fetal hemoglobin silencing with heterozygous IKZF1 mutations. Blood 128 (16), 2100–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad KA, Wang G, Slaton J, Unger G, Ahmed K, 2005. Targeting CK2 for cancer therapy. Anti Canccer Drugs 16 (10), 1037–1043. [DOI] [PubMed] [Google Scholar]
- Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC, Johansen JV, Abarrategui I, Helin K, 2013. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet. 9 (4), e1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, Georgopoulos K, 1999. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity 10 (3), 333–343. [DOI] [PubMed] [Google Scholar]
- Barata JT, 2011. The impact of PTEN regulation by CK2 on PI3K-dependent signaling and leukemia cell survival. Adv. Enzym. Regul 51 (1), 37–49. [DOI] [PubMed] [Google Scholar]
- Berron-Ruiz L, 2017. [Immunological alterations in common variable immunodeficiency]. Rev. Alerg. Mex 64 (1), 87–108. [DOI] [PubMed] [Google Scholar]
- Bigley V, Cytlak U, Collin M, 2019. Human dendritic cell immunodeficiencies. Semin. Cell Dev. Biol 86, 50–61. [DOI] [PubMed] [Google Scholar]
- Bogaert DJ, Dullaers M, Lambrecht BN, Vermaelen KY, De Baere E, Haerynck F, 2016. Genes associated with common variable immunodeficiency: one diagnosis to rule them all? J. Med. Genet 53 (9), 575–590. [DOI] [PubMed] [Google Scholar]
- Boller S, Ramamoorthy S, Akbas D, Nechanitzky R, Burger L, Murr R, Schubeler D, Grosschedl R, 2016. Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity 44 (3), 527–541. [DOI] [PubMed] [Google Scholar]
- Boutboul D, Kuehn HS, Van de Wyngaert Z, Niemela JE, Callebaut I, Stoddard J, Lenoir C, Barlogis V, Farnarier C, Vely F, Yoshida N, Kojima S, Kanegane H, Hoshino A, Hauck F, Lhermitte L, Asnafi V, Roehrs P, Chen S, Verbsky JW, Calvo KR, Husami A, Zhang K, Roberts J, Amrol D, Sleaseman J, Hsu AP, Holland SM, Marsh R, Fischer A, Fleisher TA, Picard C, Latour S, Rosenzweig SD, 2018. Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency. J. Clin. Investig 128 (7), 3071–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG, 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91 (6), 845–854. [DOI] [PubMed] [Google Scholar]
- Bueno MT, Richard S, 2013. SUMOylation negatively modulates target gene occupancy of the KDM5B, a histone lysine demethylase. Epigenetics 8 (11). [DOI] [PubMed] [Google Scholar]
- Buontempo F, Orsini E, Martins LR, Antunes I, Lonetti A, Chiarini F, Tabellini G, Evangelisti C, Evangelisti C, Melchionda F, Pession A, Bertaina A, Locatelli F, McCubrey JA, Cappellini A, Barata JT, Martelli AM, 2014. Cytotoxic activity of the casein kinase 2 inhibitor CX-4945 against T-cell acute lymphoblastic leukemia: targeting the unfolded protein response signaling. Leukemia 28 (3), 543–553. [DOI] [PubMed] [Google Scholar]
- Busslinger M, 2004. Transcriptional control of early B cell development. Annu. Rev. Immunol 22, 55–79. [DOI] [PubMed] [Google Scholar]
- Candido S, Abrams SL, Steelman L, Lertpiriyapong K, Martelli AM, Cocco L, Ratti S, Follo MY, Murata RM, Rosalen PL, Lombardi P, Montalto G, Cervello M, Gizak A, Rakus D, Suh PG, Libra M, McCubrey JA, 2018. Metformin influences drug sensitivity in pancreatic cancer cells. Adv. Biol. Regul 68, 13–30. [DOI] [PubMed] [Google Scholar]
- Chen IM, Harvey RC, Mullighan CG, Gastier-Foster J, Wharton W, Kang H, Borowitz MJ, Camitta BM, Carroll AJ, Devidas M, Pullen DJ, Payne-Turner D, Tasian SK, Reshmi S, Cottrell CE, Reaman GH, Bowman WP, Carroll WL, Loh ML, Winick NJ, Hunger SP, Willman CL, 2012. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children's Oncology Group study. Blood 119 (15), 3512–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QY, Wang XC, Wang WJ, Zhou QH, Liu DR, Wang Y, 2018. B-cell deficiency: a de novo IKZF1 patient and review of the literature. J Investig. Allergol. Clin. Immunol 28 (1), 53–56. [DOI] [PubMed] [Google Scholar]
- Choi S, Houdek X, Anderson RA, 2018. Phosphoinositide 3-kinase pathways and autophagy require phosphatidylinositol phosphate kinases. Adv. Biol. Regul 68, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choukrallah MA, Matthias P, 2014. The interplay between chromatin and transcription factor networks during B cell development: who pulls the trigger first? Front. Immunol 5, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Qian M, Te Kronnie G, Zhang R, Yang W, Zhang H, Lana T, Tedrick P, Baskin R, Verbist K, Peters JL, Devidas M, Larsen E, Moore IM, Gu Z, Qu C, Yoshihara H, Porter SN, Pruett-Miller SM, Wu G, Raetz E, Martin PL, Bowman WP, Winick N, Mardis E, Fulton R, Stanulla M, Evans WE, Relling MV, Pui CH, Hunger SP, Loh ML, Handgretinger R, Nichols KE, Yang JJ, Mullighan CG, 2018. Germline genetic IKZF1 variation and predisposition to childhood acute lymphoblastic leukemia. Cancer Cell 33 (5), 937–948 e938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cico A, Andrieu-Soler C, Soler E, 2016. Enhancers and their dynamics during hematopoietic differentiation and emerging strategies for therapeutic action. FEBS Lett. 590 (22), 4084–4104. [DOI] [PubMed] [Google Scholar]
- Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST, 2000. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 14, 2146–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Smale ST, 2005. Ikaros-family proteins: in search of molecular functions during lymphocyte development. Curr. Top. Microbiol. Immunol 290, 29–47. [DOI] [PubMed] [Google Scholar]
- Crescenzi B, La Starza R, Romoli S, Beacci D, Matteucci C, Barba G, Aventin A, Marynen P, Ciolli S, Nozzoli C, Martelli MF, Mecucci C, 2004. Submicroscopic deletions in 5q- associated malignancies. Haematologica 89 (3), 281–285. [PubMed] [Google Scholar]
- Cytlak U, Resteu A, Bogaert D, Kuehn HS, Altmann T, Gennery A, Jackson G, Kumanovics A, Voelkerding KV, Prader S, Dullaers M, Reichenbach J, Hill H, Haerynck F, Rosenzweig SD, Collin M, Bigley V, 2018. Ikaros family zinc finger 1 regulates dendritic cell development and function in humans. Nat. Commun 9 (1), 1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DA, Shrestha P, Aisabor AI, Stream A, Galli V, Pise-Masison CA, Tagawa T, Ziegelbauer JM, Franchini G, Yarchoan R, 2019. Pomalidomide increases immune surface marker expression and immune recognition of oncovirus-infected cells. OncoImmunology 8 (2), e1546544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij JD, Beuling E, van den Heuvel-Eibrink MM, Obulkasim A, Baruchel A, Trka J, Reinhardt D, Sonneveld E, Gibson BE, Pieters R, Zimmermann M, Zwaan CM, Fornerod M, 2015. Recurrent deletions of IKZF1 in pediatric acute myeloid leukemia. Haematologica 100 (9), 1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Zhang B, Payne JL, Song C, Ge Z, Gowda C, Iyer S, Dhanyamraju PK, Dorsam G, Reeves ME, Desai D, Huang S, Payne KJ, Yue F, Dovat S, 2019. Ikaros Tumor Suppressor Function Includes Induction of Active Enhancers and Super-enhancers along with Pioneering Activity. Leukemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovat S, Ronni T, Russell D, Ferrini R, Cobb BS, Smale ST, 2002. A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 16 (23), 2985–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovat S, Song C, Payne KJ, Li Z, 2011. Ikaros, CK2 kinase, and the road to leukemia. Mol. Cell. Biochem 356 (1–2), 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drulis-Fajdasz D, Rakus D, Wisniewski JR, McCubrey JA, Gizak A, 2018. Systematic analysis of GSK-3 signaling pathways in aging of cerebral tissue. Adv. Biol. Regul 69, 35–42. [DOI] [PubMed] [Google Scholar]
- Ernst P, Hahm K, Trinh L, Davis JN, Roussel MF, Turck CW, Smale ST, 1996. A potential role for Elf-1 in terminal transferase gene regulation. Mol. Cell. Biol 16 (11), 6121–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandarian Z, Fliegauf M, Bulashevska A, Proietti M, Hague R, Smulski CR, Schubert D, Warnatz K, Grimbacher B, 2019. Assessing the functional relevance of variants in the IKAROS family zinc finger protein 1 (IKZF1) in a cohort of patients with primary immunodeficiency. Front. Immunol 10, 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Gu Y, Han Q, Sloane J, Ge Q, Gao G, Ma J, Song H, Hu J, Chen B, Dovat S, Song C, 2018a. Plant homeodomain finger protein 2 as a novel IKAROS target in acute lymphoblastic leukemia. Epigenomics 10 (1), 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Gu Y, Han Q, Zhao G, Li M, Li J, Chen B, Sun T, Dovat S, Gale RP, Song C, 2016a. Targeting high dynamin-2 (DNM2) expression by restoring ikaros function in acute lymphoblastic leukemia. Sci. Rep 6, 38004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Gu Y, Xiao L, Han Q, Li J, Chen B, Yu J, Kawasawa YI, Payne KJ, Dovat S, Song C, 2016b. Co-existence of IL7R high and SH2B3 low expression distinguishes a novel high-risk acute lymphoblastic leukemia with Ikaros dysfunction. Oncotarget 7 (29), 46014–46027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Gu Y, Zhao G, Li J, Chen B, Han Q, Guo X, Liu J, Li H, Yu MD, Olson J, Steffens S, Payne KJ, Song C, Dovat S, 2016c. High CRLF2 expression associates with IKZF1 dysfunction in adult acute lymphoblastic leukemia without CRLF2 rearrangement. Oncotarget 7 (31), 49722–49732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Guo X, Li J, Hartman M, Kawasawa YI, Dovat S, Song C, 2015. Clinical significance of high c-MYC and low MYCBP2 expression and their association with Ikaros dysfunction in adult acute lymphoblastic leukemia. Oncotarget 6 (39), 42300–42311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Han Q, Gu Y, Ge Q, Ma J, Sloane J, Gao G, Payne KJ, Szekely L, Song C, Dovat S, 2018b. Aberrant ARID5B expression and its association with Ikaros dysfunction in acute lymphoblastic leukemia. Oncogenesis 7 (11), 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Zhou X, Gu Y, Han Q, Li J, Chen B, Ge Q, Dovat E, Payne JL, Sun T, Song C, Dovat S, 2017. Ikaros regulation of the BCL6/BACH2 axis and its clinical relevance in acute lymphoblastic leukemia. Oncotarget 8 (5), 8022–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffken K, Spiegel S, 2018. Sphingosine kinase 1 in breast cancer. Adv. Biol. Regul 67, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K, 1997. Transcription factors required for lymphoid lineage commitment. Curr. Opin. Immunol 9 (2), 222–227. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A, 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79 (1), 143–156. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Moore DD, Derfler B, 1992. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 258 (5083), 808–812. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Winandy S, Avitahl N, 1997. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu. Rev. Immunol 15, 155–176. [DOI] [PubMed] [Google Scholar]
- Goldman FD, Gurel Z, Al-Zubeidi D, Fried AJ, Icardi M, Song C, Dovat S, 2012. Congenital pancytopenia and absence of B lymphocytes In a neonate with a mutation in the Ikaros gene. Pediatr. Blood Cancer 58 (4), 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AM, Soares MV, Ribeiro P, Caldas J, Povoa V, Martins LR, Melao A, Serra-Caetano A, de Sousa AB, Lacerda JF, Barata JT, 2014. Adult B-cell acute lymphoblastic leukemia cells display decreased PTEN activity and constitutive hyperactivation of PI3K/Akt pathway despite high PTEN protein levels. Haematologica 99 (6), 1062–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda C, Sachdev M, Muthusami S, Kapadia M, Petrovic-Dovat L, Hartman M, Ding Y, Song C, Payne JL, Tan BH, Dovat S, 2017a. Casein kinase II (CK2) as a therapeutic target for hematological malignancies. Curr. Pharmaceut. Des 23 (1), 95–107. [DOI] [PubMed] [Google Scholar]
- Gowda C, Soliman M, Kapadia M, Ding Y, Payne K, Dovat S, 2017b. Casein kinase II (CK2), glycogen synthase kinase-3 (GSK-3) and ikaros mediated regulation of leukemia. Adv. Biol. Regul 65, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda C, Song C, Kapadia M, Payne JL, Hu T, Ding Y, Dovat S, 2017c. Regulation of cellular proliferation in acute lymphoblastic leukemia by Casein Kinase II (CK2) and Ikaros. Adv. Biol. Regul 63, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda CS, Song C, Ding Y, Kapadia M, Dovat S, 2016. Protein signaling and regulation of gene transcription in leukemia: role of the Casein Kinase II-Ikaros axis. J. Investig. Med 64 (3), 735–739. [DOI] [PubMed] [Google Scholar]
- Gurel Z, Ronni T, Ho S, Kuchar J, Payne KJ, Turk CW, Dovat S, 2008. Recruitment of ikaros to pericentromeric heterochromatin is regulated by phosphorylation. J. Biol. Chem 283 (13), 8291–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, De Vos J, Hernandez JM, Hofmann WK, Mills KI, Gilkes A, Chiaretti S, Shurtleff SA, Kipps TJ, Rassenti LZ, Yeoh AE, Papenhausen PR, Liu WM, Williams PM, Foa R, 2010. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the nnovations in Leukemiaroup. J. Clin. Oncol 28 (15), 2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST, 1994. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol. Cell. Biol 14 (11), 7111–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Ma J, Gu Y, Song H, Kapadia M, Kawasawa YI, Dovat S, Song C, Ge Z, 2019. RAG1 high expression associated with IKZF1 dysfunction in adult B-cell acute lymphoblastic leukemia. J. Cancer 10 (16), 3842–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, Roderick K, Georgopoulos K, Kioussis D, 2002. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol. Cell 10 (6), 1403–1415. [DOI] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ, Kang H, Liu W, Dobbin KK, Smith MA, Carroll WL, Devidas M, Bowman WP, Camitta BM, Reaman GH, Hunger SP, Downing JR, Willman CL, 2010. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood 115 (26), 5312–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayami S, Yoshimatsu M, Veerakumarasivam A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue H, Ponder BA, Nakamura Y, Hamamoto R, 2010. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol. Cancer 9, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK, 2015. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol 16 (3), 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Okada S, Yoshida K, Nishida N, Okuno Y, Ueno H, Yamashita M, Okano T, Tsumura M, Nishimura S, Sakata S, Kobayashi M, Nakamura H, Kamizono J, Mitsui-Sekinaka K, Ichimura T, Ohga S, Nakazawa Y, Takagi M, Imai K, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ogawa S, Kojima S, Nonoyama S, Morio T, Kanegane H, 2017. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J. Allergy Clin. Immunol 140 (1), 223–231. [DOI] [PubMed] [Google Scholar]
- Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan GC, Orkin SH, Xu J, 2016. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell 36 (1), 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger SP, Raetz EA, Loh ML, Mullighan CG, 2011. Improving outcomes for high-risk ALL: translating new discoveries into clinical care. Pediatr. Blood Cancer 56 (6), 984–993. [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS, 2014. Pioneer transcription factors in cell reprogramming. Genes Dev. 28 (24), 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HJ, Suh PG, Lee YJ, Shin KJ, Cocco L, Chae YC, 2018. PLCgamma1: potential arbitrator of cancer progression. Adv. Biol. Regul 67, 179–189. [DOI] [PubMed] [Google Scholar]
- Kanegane H, Hoshino A, 2019. [Inherited lymphoproliferative disorders]. Rinsho Ketsueki 60 (6), 708–715. [DOI] [PubMed] [Google Scholar]
- Kathrein KL, Lorenz R, Innes AM, Griffiths E, Winandy S, 2005. Ikaros induces quiescence and T-cell differentiation in a leukemia cell line. Mol. Cell. Biol 25 (5), 1645–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky K, Zhan H, 2018. The regulation of normal and neoplastic hematopoiesis is dependent on microenvironmental cells. Adv. Biol. Regul 69, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K, 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10 (3), 345–355. [DOI] [PubMed] [Google Scholar]
- Kim JH, Ebersole T, Kouprina N, Noskov VN, Ohzeki J, Masumoto H, Mravinac B, Sullivan BA, Pavlicek A, Dovat S, Pack SD, Kwon YW, Flanagan PT, Loukinov D, Lobanenkov V, Larionov V, 2009. Human gamma-satellite DNA maintains open chromatin structure and protects a transgene from epigenetic silencing. Genome Res. 19 (4), 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S, 2002. Ikaros is critical for B cell differentiation and function. Eur. J. Immunol 32 (3), 720–730. [DOI] [PubMed] [Google Scholar]
- Koipally J, Heller EJ, Seavitt JR, Georgopoulos K, 2002. Unconventional potentiation of gene expression by ikaros. J. Biol. Chem 277 (15), 13007–13015. [DOI] [PubMed] [Google Scholar]
- Kristensen LH, Nielsen AL, Helgstrand C, Lees M, Cloos P, Kastrup JS, Helin K, Olsen L, Gajhede M, 2012. Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals strong substrate recognition in vitro and identifies 2,4-pyridine-dicarboxylic acid as an in vitro and in cell inhibitor. FEBS J. 279 (11), 1905–1914. [DOI] [PubMed] [Google Scholar]
- Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand EW, Maffucci P, Pierce KR, Abbott JK, Voelkerding KV, South ST, Augustine NH, Bush JS, Dolen WK, Wray BB, Itan Y, Cobat A, Sorte HS, Ganesan S, Prader S, Martins TB, Lawrence MG, Orange JS, Calvo KR, Niemela JE, Casanova JL, Fleisher TA, Hill HR, Kumanovics A, Conley ME, Rosenzweig SD, 2016. Loss of B Cells in patients with heterozygous mutations in IKAROS. N. Engl. J. Med 374 (11), 1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavallee VP, Gendron P, Lemieux S, D'Angelo G, Hebert J, Sauvageau G, 2015. EVI1-rearranged acute myeloid leukemias are characterized by distinct molecular alterations. Blood 125 (1), 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Hwang JH, Choi KY, 2018. Interaction of the Wnt/beta-catenin and RAS-ERK pathways involving co-stabilization of both beta-catenin and RAS plays important roles in the colorectal tumorigenesis. Adv. Biol. Regul 68, 46–54. [DOI] [PubMed] [Google Scholar]
- Li Z, Perez Casellas LA, Savic A, Song C, Dovat S, 2011. Ikaros isoforms: the saga continues. World J. Biol. Chem 2 (6), 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K, Landau NR, Smale ST, 1991. LyF-1, a transcriptional regulator that interactswith a novel class of promoters for lymphocyte-specific genes. Mol. Cell. Biol 11, 5229–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA, 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153 (2), 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffucci P, Filion CA, Boisson B, Itan Y, Shang L, Casanova JL, Cunningham-Rundles C, 2016. Genetic diagnosis using whole exome sequencing in common variable immunodeficiency. Front. Immunol 7, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins LR, Lucio P, Silva MC, Anderes KL, Gameiro P, Silva MG, Barata JT, 2010. Targeting CK2 overexpression and hyperactivation as a novel therapeutic tool in chronic lymphocytic leukemia. Blood 116 (15), 2724–2731. [DOI] [PubMed] [Google Scholar]
- Martins LR, Lucio P, Silva MC, Gameiro P, Silva MG, Barata JT, 2011. On CK2 regulation of chronic lymphocytic leukemia cell viability. Mol. Cell. Biochem 356 (1–2), 51–55. [DOI] [PubMed] [Google Scholar]
- Mayran A, Drouin J, 2018. Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem 293 (36), 13795–13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayran A, Khetchoumian K, Hariri F, Pastinen T, Gauthier Y, Balsalobre A, Drouin J, 2018. Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat. Genet 50 (2), 259–269. [DOI] [PubMed] [Google Scholar]
- McCarty AS, Kleiger G, Eisenberg D, Smale ST, 2003. Selective dimerization of a C2H2 zinc finger subfamily. Mol. Cell 11 (2), 459–470. [DOI] [PubMed] [Google Scholar]
- Molnár A, Georgopoulos K, 1994. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol 14 (12), 8292–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár A, Wu P, Largespada DA, Vortkamp A, Scherer S, Copeland NG, Jenkins NA, Bruns G, Georgopoulos K, 1996. The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. J. Immunol 156 (2), 585–592. [PubMed] [Google Scholar]
- Mullighan C, Downing J, 2008. Ikaros and acute leukemia. Leuk. Lymphoma 49 (5), 847–849. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, 2011a. Genomic profiling of B-progenitor acute lymphoblastic leukemia. Best Pract. Res. Clin. Haematol 24 (4), 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, 2011b. New strategies in acute lymphoblastic leukemia: translating advances in genomics into clinical practice. Clin. Cancer Res 17 (3), 396–400. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, Relling MV, Evans WE, Shurtleff SA, Downing JR, 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446 (7137), 758–764. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, Relling MV, Shurtleff SA, Downing JR, 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453 (7191), 110–114. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, Harvey RC, Chen IM, Clifford RJ, Carroll WL, Reaman G, Bowman WP, Devidas M, Gerhard DS, Yang W, Relling MV, Shurtleff SA, Campana D, Borowitz MJ, Pui CH, Smith M, Hunger SP, Willman CL, Downing JR, Children's Oncology G, 2009a. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med 360 (5), 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, Devidas M, Atlas SR, Chen IM, Clifford RJ, Gerhard DS, Carroll WL, Reaman GH, Smith M, Downing JR, Hunger SP, Willman CL, 2009b. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U. S. A 106 (23), 9414–9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K, 1999. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J. Exp. Med 190 (9), 1201–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, Han S, Seong RH, Park SD, Agalioti T , Munshi N, Thanos D, Erdjument-Bromage H, Tempst P, Bank A, 2000. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell Biol 20 (20), 7572–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathanasiou P, Perkins AC, Cobb BS, Ferrini R, Sridharan R, Hoyne GF, Nelms KA, Smale ST, Goodnow CC, 2003. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity 19 (1), 131–144. [DOI] [PubMed] [Google Scholar]
- Payne KJ, Dovat S, 2011. Ikaros and tumor suppression in acute lymphoblastic leukemia. Crit. Rev. Oncog 16 (1–2), 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna LA, 1997. Protein kinase CK2. Int. J. Biochem. Cell Biol 29 (4), 551–554. [DOI] [PubMed] [Google Scholar]
- Pinna LA, Meggio F, 1997. Protein kinase CK2 ("casein kinase-2") and its implication in cell division and proliferation. Prog. Cell Cycle Res 3, 77–97. [DOI] [PubMed] [Google Scholar]
- Popescu M, Gurel Z, Ronni T, Song C, Hung KY, Payne KJ, Dovat S, 2009. Ikaros stability and pericentromeric localization are regulated by protein phosphatase 1. J. Biol. Chem 284 (20), 13869–13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne NJ, El Buri A, Adams DR, Pyne S, 2018. Sphingosine 1-phosphate and cancer. Adv. Biol. Regul 68, 97–106. [DOI] [PubMed] [Google Scholar]
- Ramos AR, Elong Edimo W, Erneux C, 2018. Phosphoinositide 5-phosphatase activities control cell motility in glioblastoma: two phosphoinositides PI(4,5)P2 and PI(3,4)P2 are involved. Adv. Biol. Regul 67, 40–48. [DOI] [PubMed] [Google Scholar]
- Rebello RJ, Huglo AV, Furic L, 2018. PIM activity in tumours: a key node of therapy resistance. Adv. Biol. Regul 67, 163–169. [DOI] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H, 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol 9 (8), 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Mullighan CG, 2011. How new advances in genetic analysis are influencing the understanding and treatment of childhood acute leukemia. Curr. Opin. Pediatr 23 (1), 34–40. [DOI] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M, 2010. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141 (4), 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronni T, Payne KJ, Ho S, Bradley MN, Dorsam G, Dovat S, 2007. Human Ikaros function in activated T cells is regulated by coordinated expression of its largest isoforms. J. Biol. Chem 282 (4), 2538–2547. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Azevedo C, Desfougeres Y, Portela-Torres P, Wilson MSC, 2018. Microbial inositol polyphosphate metabolic pathway as drug development target. Adv. Biol. Regul 67, 74–83. [DOI] [PubMed] [Google Scholar]
- Sakane F, Mizuno S, Takahashi D, Sakai H, 2018. Where do substrates of diacylglycerol kinases come from? Diacylglycerol kinases utilize diacylglycerol species supplied from phosphatidylinositol turnover-independent pathways. Adv. Biol. Regul 67, 101–108. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T, 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419 (6905), 407–411. [DOI] [PubMed] [Google Scholar]
- Scarlata S, Singla A, Garwain O, 2018. Phospholipase Cbeta interacts with cytosolic partners to regulate cell proliferation. Adv. Biol. Regul 67, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HY, 2018. Targeting super-enhancers for disease treatment and diagnosis. Mol. Cells 41 (6), 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A, Jotta PY, Silveira AB, Ribeiro D, Brandalise SR, Yunes JA, Barata JT, 2010. Regulation of PTEN by CK2 and Notch1 in primary T-cell acute lymphoblastic leukemia: rationale for combined use of CK2- and gamma-secretase inhibitors. Haematologica 95 (4), 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, Nowill AE, Leslie NR, Cardoso AA, Barata JT, 2008. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J. Clin. Investig 118 (11), 3762–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Gowda C, Pan X, Ding Y, Tong Y, Tan BH, Wang H, Muthusami S, Ge Z, Sachdev M, Amin SG, Desai D, Gowda K, Gowda R, Robertson GP, Schjerven H, Muschen M, Payne KJ, Dovat S, 2015. Targeting casein kinase II restores Ikaros tumor suppressor activity and demonstrates therapeutic efficacy in high-risk leukemia. Blood 126 (15), 1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Li Z, Erbe AK, Savic A, Dovat S, 2011. Regulation of Ikaros function by casein kinase 2 and protein phosphatase 1. World J. Biol. Chem 2 (6), 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Pan X, Ge Z, Gowda C, Ding Y, Li H, Li Z, Yochum G, Muschen M, Li Q, Payne KJ, Dovat S, 2016. Epigenetic regulation of gene expression by Ikaros, HDAC1 and Casein Kinase II in leukemia. Leukemia 30 (6), 1436–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriaroon P, Chang Y, Ujhazi B, Csomos K, Joshi HR, Zhou Q, Close DW, Walter JE, Kumanovics A, 2019. Familial immune thrombocytopenia associated with a novel variant in IKZF1. Front. Pediatr 7, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Smale ST, 2007. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J. Biol. Chem 282 (41), 30227–30238. [DOI] [PubMed] [Google Scholar]
- Su RC, Sridharan R, Smale ST, 2005. Assembly of silent chromatin during thymocyte development. Semin. Immunol 17 (2), 129–140. [DOI] [PubMed] [Google Scholar]
- Sun L, Liu A, Georgopoulos K, 1996. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 15 (19), 5358–5369. [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, Ferrini R, Cobb BS, Weinmann AS, Hahm K, Ernst P, Garraway IP, Merkenschlager M, Smale ST, 2001. Down-regulation of TdT transcription in CD4+ CD8+ thymocytes by Ikaros proteins in direct competition with an Ets activator. Genes Dev. 15, 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckun FM, Ma H, Zhang J, Ozer Z, Dovat S, Mao C, Ishkhanian R, Goodman P, Qazi S, 2012. Serine phosphorylation by SYK is critical for nuclear localization and transcription factor function of Ikaros. Proc. Natl. Acad. Sci. U. S. A 109 (44), 18072–18077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JA, Winandy S, 2004. Ikaros null mice display defects in T cell selection and CD4 versus CD8 lineage decisions. J. Immunol 173 (7), 4470–4478. [DOI] [PubMed] [Google Scholar]
- van Oevelen C, Collombet S, Vicent G, Hoogenkamp M, Lepoivre C, Badeaux A, Bussmann L, Sardina JL, Thieffry D, Beato M, Shi Y, Bonifer C, Graf T, 2015. C/EBPalpha activates pre-existing and de novo macrophage enhancers during induced pre-B cell transdifferentiation and myelopoiesis. Stem Cell Rep. 5 (2), 232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ouyang H, Lai L, Petrovic-Dovat L, Stankov K, Bogdanovic G, Dovat S, 2014a. Pathogenesis and regulation of cellular proliferation in acute lymphoblastic leukemia - the role of Ikaros. J BUON 19 (1), 22–28. [PubMed] [Google Scholar]
- Wang H, Song C, Ding Y, Pan X, Ge Z, Tan BH, Gowda C, Sachdev M, Muthusami S, Ouyang H, Lai L, Francis OL, Morris CL, Abdel-Azim H, Dorsam G, Xiang M, Payne KJ, Dovat S, 2016. Transcriptional regulation of JARID1B/KDM5B histone demethylase by ikaros, histone deacetylase 1 (HDAC1), and casein kinase 2 (CK2) in B-cell acute lymphoblastic leukemia. J. Biol. Chem 291 (8), 4004–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Song C, Gurel Z, Song N, Ma J, Ouyang H, Lai L, Payne KJ, Dovat S, 2014b. Protein phosphatase 1 (PP1) and casein kinase II (CK2) regulate ikaros-mediated repression of TdT in thymocytes and T-cell leukemia. Pediatr. Blood Cancer 61 (12), 2230–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K, 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5 (6), 537–549. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA, 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153 (2), 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winandy S, Wu P, Georgopoulos K, 1995. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83 (2), 289–299. [DOI] [PubMed] [Google Scholar]
- Wong PP, Miranda F, Chan KV, Berlato C, Hurst HC, Scibetta AG, 2012. Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21(cip) (CDKN1A). Mol. Cell. Biol 32 (9), 1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RWJ, Ishida T, Sanda T, 2018. Targeting general transcriptional machinery as a therapeutic strategy for adult T-cell leukemia. Molecules 23 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RWJ, Ngoc PCT, Leong WZ, Yam AWY, Zhang T, Asamitsu K, Iida S, Okamoto T, Ueda R, Gray NS, Ishida T, Sanda T, 2017. Enhancer profiling identifies critical cancer genes and characterizes cell identity in adult T-cell leukemia. Blood 130 (21), 2326–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Nichogiannopoulou A, Shortman K, Georgopoulos K, 1997. Cell-autonomous defects in dendritic cell populations of Ikaros mutant mice point to a developmental relationship with the lymphoid lineage. Immunity 7 (4), 483–492. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD, 2007. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. U. S. A 104 (49), 19226–19231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Hibi S, Takanashi M, Kano G, Tabata Y, Imamura T, Inaba T, Morimoto A, Todo S, Imashuku S, 2002. High frequency of Ikaros isoform 6 expression in acute myelomonocytic and monocytic leukemias: implications for up-regulation of the antiapoptotic protein Bcl-XL in leukemogenesis. Blood 99 (4), 1350–1355. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Sakaguchi H, Muramatsu H, Okuno Y, Song C, Dovat S, Shimada A, Ozeki M, Ohnishi H, Teramoto T, Fukao T, Kondo N, Takahashi Y, Matsumoto K, Kato K, Kojima S, 2017. Germline IKAROS mutation associated with primary immunodeficiency that progressed to T-cell acute lymphoblastic leukemia. Leukemia 31 (5), 1221–1223. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K, 2006. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol 7 (4), 382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS, 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25 (21), 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG, 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481 (7380), 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]