Abstract
The co-precipitation of calcium phosphate nanoparticles (CaPs) in the presence of nucleotide chains such as polynucleotides (i.e., plasmid DNA and siRNA) and oligonucleotides has been extensively used for pre-clinical gene or drug delivery and immunotherapy studies. However, the exact role of these molecules in mineralization and tuning the physicochemical characteristics of the synthesized CaPs is still not entirely clear. In this study, we evaluated the effects of three different CpG oligodeoxynucleotides (ODN) and two representative nucleic acids (siRNA and DNA), when used as templates for the formation of CaPs. We examined the influence of CpGs with naturally-occurring phosphodiester or modified phosphorothioate backbones on the homogeneous formation of CaPs from a modified simulated body fluid solution. The hydrodynamic size, size polydispersity, morphology and surface charge of the CaPs were used as the most critical checkpoints to unravel the involved mechanisms. Our results show that the characteristics of CaPs are highly dependent on the composition, backbone, sequence and concentrations of the CpGs. The CpG type and concentration control the size distribution of the mineralized CaPs and their immunostimulation performance as verified by the activation of dendritic cells and secretion of the pro-inflammatory interleukin-6 (IL-6) cytokine, type I interferon-α (IFN-α) and co-stimulatory CD80, CD86 and CD40 markers. This study paves the way for better design of more efficient CaPs loaded with different types of CpGs for immunostimulation applications as vaccine adjuvants.
1. Introduction
Biodegradability, biocompatibility and more predictable body clearance mechanisms are the major factors distinguishing calcium phosphate nanoparticles (CaPs) as one of the most favorable nanomedicine candidates for clinical translations. CaPs loaded with different types of nucleotide chains (e.g., nucleic acids and oligonucleotides) have been extensively used as nanoplatforms for gene, drug and vaccine delivery applications.1–4 However, systematic studies are required to better tune the mineralization and physicochemical properties of these nanoparticles for each specific biomedical application.
Nucleic acids are biodegradable macromolecules with a well-defined and broad range of molecular weights and have been studied as crystal modifiers in the synthesis of calcium phosphate and calcium carbonate nanoparticles.5,6 Nucleic acids are building blocks of genetic materials that have high water solubility at room temperature. This hydrophilicity makes them feasible templates for the controlled synthesis of calcium-based minerals from biological solutions (i.e., biomineralization). The anionic phosphate groups of these molecules can efficiently bond and interact with cations such as Ca2+ within the mineralization solution.7 For example, the ability of DNA and RNA to sequester the divalent metals made them effective templates for the synthesis of metal nanoparticles.8,9 Also, long strands of these molecules have a unique ability to fold and alter their configuration and these variations can change the kinetics of nucleation as well as the physicochemical properties of the mineralized nanoparticles. Finally, the use of these molecules does not require stringent synthesis conditions and they can be easily analyzed quantitatively by different spectroscopy methods.10,11
Graham and Van Der Eb12 used DNA molecules as templates for the biomineralization of calcium phosphates and gene delivery applications more than three decades ago. Different strategies for more efficient incorporation of DNA or other polynucleotides and their derivatives such as aptamers and oligonucleotides into CaPs have been reported recently.3,13,14 However, the exact role of these molecules in the mineralization of CaPs is still unknown and a roadmap is needed for a better selection and design of these molecules in order to improve the efficacy of CaPs for gene and vaccine delivery applications. This is mainly because the physicochemical characteristics of CaPs (e.g., hydrodynamic size, surface charge, morphology, percentage of loaded polynucleotides, size stability and degradation rate) change significantly when different molecular structures of nucleotide chains are used as templates for their mineralization. These variations change the biomedical performance of the CaPs in different pre-clinical therapeutic studies. Recent progress in the preparation and more accurate characterization of different polynucleotides, oligonucleotides and synthetic peptides can provide better control on the mechanisms involved in the mineralization of CaPs, and therefore expand the library of different CaP formulations for diverse nanomedicine applications.
In this study, we developed a facile and low-cost method to prepare spherical and monodisperse calcium phosphate nanoparticles from simulated body fluids,15,16 utilizing five different types of single- and double-stranded nucleotide chains (i.e., oligonucleotides, DNA and siRNA) as templates for mineralization. We used three types of CpG oligodeoxynucleotides as representatives of oligonucleotides. These CPG molecules are specific for the activation of dendritic cells, a process generally called immunostimulation. Compared to other types of immunostimulants, the CaP nanoparticles reported in this work have the ability to entrap the CpGs in situ (Scheme 1), which eliminates the need for additional conjugation steps (i.e., synthesis of nanoparticles and conjugation of CpG molecules are accomplished in a single step). The synthesis settings are mild and mimic the physiological conditions, and therefore prevent the denaturation and damage to the CpG molecules. In addition, the electrostatic attractions between positively charged Ca2+ and negatively charged CpGs during synthesis enable the nearly 100% incorporation efficiency of CpG molecules. Finally, unlike most of the inorganic and metallic nanoparticles, CaPs are generally considered biodegradable and dissolve into their ionic constituents and as a result they do not accumulate in the cells and tissues for long periods after in vivo administration.
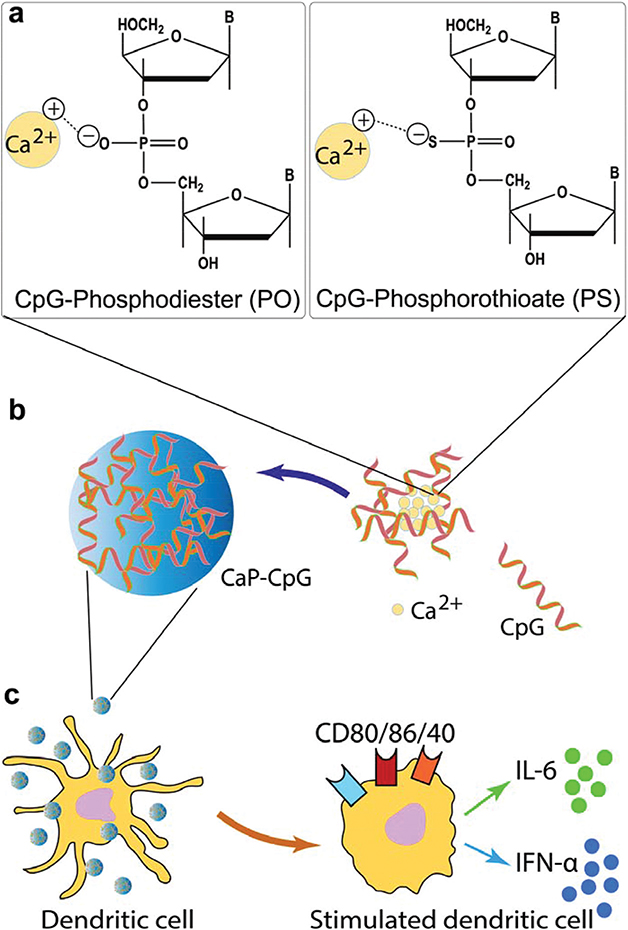
Scheme 1.
(a) Phosphodiester (PO) and phosphorothioate (PS) groups on the CpG oligodeoxynucleotide (ODN) backbone structure and electrostatic interaction between negatively charged PO or PS groups and calcium cations. (b) Controlled mineralization of CaP nanoparticles in the presence of CpGs. (c) Activation of immature dendritic cells by CaPs incorporating CpGs (CaP-CpG), as verified by the secretion of pro-inflammatory IL-6 cytokine and IFN-α interferon or expression of co-stimulatory CD80, CD86 and CD40 markers.
We also used DNA- and siRNA-templated mineralization as references for comparison. Our control on the spherical shape and size monodispersity of these particles enabled quantitative comparisons when different CpGs and nucleic acids were used as templates for CaP mineralization. Previous studies have only been focused on the use of single or double-stranded DNA with a naturally-occurring phosphodiester (PO) backbone and did not investigate the details of mineralization mechanisms.13,14 Here, we chose various types of CpG oligodeoxynucleotides with either PO or phosphorothioate (PS) functional groups to evaluate the effect of the backbone structure on the formation of CaP particles and their effectiveness in the activation of dendritic cells (i.e., immunostimulation). We evaluated the effect of time, concentration, backbone and sequence on the physical properties of the CaPs. By proper selection of mineralizing conditions, CaPs with well-controlled size and morphology were obtained and their immunostimulation characteristics were evaluated in correlation with their physicochemical properties.
While this study is focused to evaluate CaP-CpG nanoparticles as potential adjuvants for enhanced vaccine delivery applications, we envision that our results will also enable better understanding of mechanisms involved in the incorporation of a wider range of single- and double-stranded nucleotide chains (e.g., CpG, DNA, siRNA) into different calcium-based biominerals such as calcium phosphates or carbonates. This will further expand the applications of these biodegradable nanoparticles in nanomedicine.
2. Experimental section
2.1. Materials
All chemical reagents were purchased from Sigma-Aldrich (St Louis, MO) unless otherwise specified.
2.2. Formation of CaP-CpG and CaP-nucleic acid complexes
CaP nanoparticles were prepared in 50 ml Falcon tubes (BD Biosciences; San Jose, CA) by adding certain concentrations of CpG oligodeoxynucleotides (CpG ODN, TriLink BioTechnologies; San Diego, CA), DNA (Aldevron; Fargo, ND) or siRNA (Cell Signaling Technology, Danvers, MA) in Tris-EDTA (TE) buffer to 1 ml of supersaturated calcium phosphate solution (Table 1). The mixtures were placed at 37 °C on a Rotomix Type 50800 speed rotator (Barnstead/Thermolyne; Dubuque, IA) at a speed of 115 rpm for a specified period of times. Supersaturated calcium phosphate solutions were prepared based on previously reported procedures.3,15,17 The following materials: CaCl2, KH2PO4, NaCl, KCl, MgSO4, NaHCO3, and MgCl2 were added together in Milli-Q water, except CaCl2 that was added last in a dropwise manner to prevent spontaneous precipitation. The resulting solution was filtered with a 0.2 μm Millipore membrane and buffered to pH 7.4 with Tris-HCl and stored at 4 °C.
Table 1.
Composition of the modified simulated body fluid (SBF) used for the mineralization of CaPs in the presence of CpGs, siRNA and DNA
| Concentration (mM) | Simulated body fluid (SFB) | Modified SFB |
|---|---|---|
| CaCl2·H2O | 2.5 | 3.7 |
| NaCl | 141 | 141 |
| KH2PO4 | 1 | 1 |
| MgSO4·6H2O | 0.5 | 0.5 |
| KCl | 4 | 4 |
| NaHCO3 | 4.2 | 4.2 |
| MgCl2·6H2O | 1 | 1 |
Three types of CpG ODNs were used in this study including ODN 1826-PO (5′TCC ATG ACG TTC CTG ACG TT3′), ODN 1826-PS (5′tcc atg acg ttc ctg acg tT3′) and ODN 2216 (5′ggG GGA CGA TCG TCg ggg gG3′). The uppercase and lowercase letters denote phosphodiester (PS) and phosphorothioate (PS) backbones, respectively.
2.3. Characterization of CaP nanoparticles
At the end of designated times, the hydrodynamic size and zeta potential (ZP) of CaPs were measured using a Zetasizer Nano ZS (Malvern Instruments; Westborough, MA) equipped with a zeta-potential analyzer. Three autocorrelation functions from the scattered intensities were obtained for each sample and the average diameter of the nanoparticles was determined using the Stokes–Einstein equation.18 Zeta potentials of the nanoparticles were measured after removing the excess mineralizing solution and re-dispersing them in KNO3 solution (10 mM, pH = 7.4). All measurements were conducted at room temperature. The morphology of CaPs was analyzed by scanning electron microscopy (SEM, JEOL 7000, Peabody, MA). The samples were sputter-coated with platinum and imaged at 10 kV. The amount of calcium ions at various time points was determined using a modified colorimetric method based on complex formation with ortho-cresolphthalein as previously described.19
2.4. Evaluation of CpG and DNA/siRNA binding efficiency
At the end of mineralization, the mixtures were centrifuged at 13 000 rpm for 10 min and the supernatants were removed. The OliGreen Assay kit for ssDNA (Invitrogen, Frederick, MD) and Quant-iT™ PicoGreen dsDNA reagent (Invitrogen; Carlsbad, CA) were respectively used to measure the amount of CpGs and DNA/siRNA in supernatants according to the manufacturer’s protocols. The quantity of the CpG or DNA/siRNA incorporated into the CaPs was calculated by subtracting the amount of CpG or DNA/siRNA in supernatants from their initial quantity added to each sample.
2.5. Quantification of nanoparticle uptake by flow cytometry
BC-1 dendritic cells (a gift from Dr Yoshiki Yanagawa) were cultured into 24-well plates at a density of 2 × 105 cells per well and incubated for 24 h. The cell culture medium was then removed and the cells were incubated for 6 h with 500 μl of culture medium containing CaPs that were tagged with fluorescently labeled CpGs (FITC-CpG 2216 and FITC-CpG 1826, TriLink BioTechnologies, San Diego, CA). The culture medium was then removed and the cells were washed three times with Dulbecco’s phosphate buffered saline (DPBS) before harvesting them using trypsin–EDTA (0.1% trypsin, 0.4% EDTA·4Na) at 37 °C for 10 min. Cell suspensions were washed three times with DPBS containing 1% FBS and immediately analyzed using flow cytometry (BD FACScan2, San Jose, CA). Trypan blue (0.4%) was added to quench the fluorescence due to cell surface-associated particles. The amount of CaP uptake was evaluated using FlowJo software (Treestar, Inc., Ashland, OR) and expressed as the geometric mean of fluorescence intensity (MFI).
2.6. Immunostimulation studies
We investigated the activation of BC-1 dendritic cells to evaluate the immunostimulation performance of CaPs mineralized with CpG ODNs. BC-1 cells (2.5 × 105) were cultured at 37 °C under an atmosphere with 5% CO2 in 24-well plates in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 20% supernatant of fibroblast NIH-3T3 cells and 5% supernatant containing a colony-stimulating factor (GM-CSF) obtained from the GM-CSF-secreting J558L mouse cell line grown separately. Other components added to the media were as follows: 2-mercaptoethanol (50 mM), L-glutamine (4 mM), penicillin (100 units per ml), and streptomycin (100 mg ml−1). NIH-3T3 cells were grown at 37 °C under an atmosphere with 5% CO2 in IMDM with fetal bovine serum (FBS, 10%), penicillin (100 units per ml) and streptomycin (100 mg ml−1). Different concentrations of CaPs mineralized with CpG ODNs were added to dendritic cells with 500 μl medium in each well (n = 5). The supernatants were collected after 24 h incubation and the morphology of the dendritic cells was evaluated using an inverted microscope. Enzyme-linked immunosorbent assay (ELISA) was used for the quantification of the pro-inflammatory IL-6 cytokine and type I interferon IFN-α in obtained supernatants using a modified manufacturer’s protocol (eBiosciences, San Diego, CA). For flow cytometric analysis of the CD80, CD86 and CD40 markers, the cells were collected, resuspended and stained with anti-CD80, anti-CD40 (560523 and 553791 respectively, from BD Biosciences) and anti-CD86 (105031 from BioLegend) monoclonal antibodies (mAbs) before and after activation with CpG-incorporated nanoparticles. The cells were washed twice after incubation with these antibodies (~25 minutes at 4 °C) and were then subjected to flow cytometric analysis.
2.7. Statistical analysis
All presented data are the result of a minimum of three independent experiments. The data reported are the average values with error bars showing the standard deviation (SD) of the samples. GraphPad Prism 7 for MacOSX (GraphPad Software, La Jolla California USA) was used to perform the statistical analysis and the significant differences between samples were calculated by one-way analysis of variance (ANOVA) using Dunnett’s multiple comparison test. Statistical significance was assumed for p < 0.05, unless otherwise noted. Error bars in the graphs represent standard deviations.
3. Results and discussion
3.1. Effect of varying CpG 2216 concentration and mineralization time on particle size, polydispersity index and zeta potential
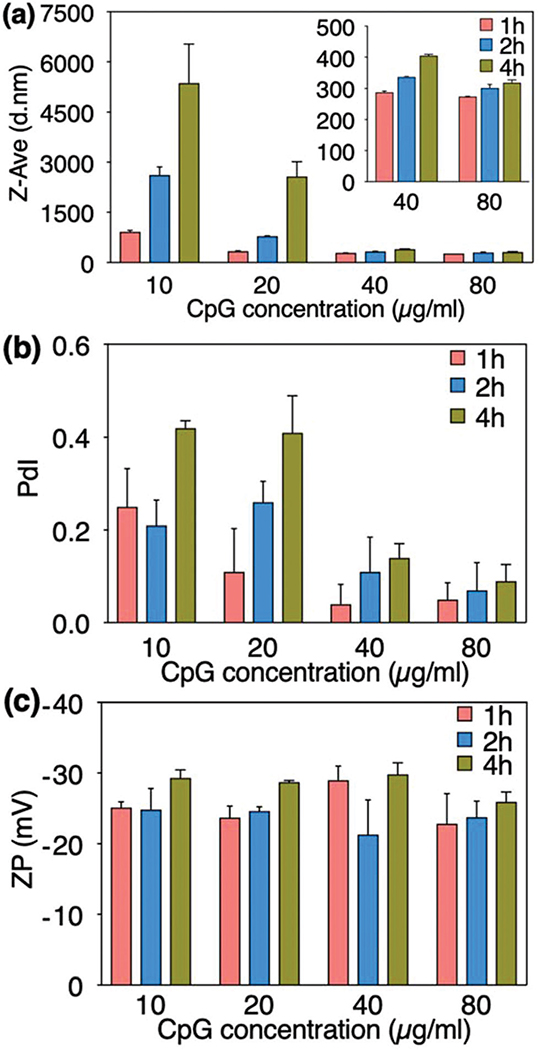
A dynamic light scattering (DLS) technique was used to evaluate the hydrodynamic size (Z-Ave) and size distributions (polydispersity index or PdI) of CaP nanoparticles prepared with varying concentrations of CpG 2216 (Fig. 1). The PdI is a quality control factor to evaluate the synthesized nano-based drugs, and should be carefully considered besides the Z-Ave values in any DLS measurement.20 A recent study by Shlaferman et al.13 showed the effects of DNA aptamer concentration on the mineralization kinetics and morphology of CaPs. Here, we found that the average hydrodynamic size of the CaP precipitates (i.e., the most critical physicochemical property of nanoparticles defining their efficiency in different nanomedicine applications) is directly proportional to the molecular structure and initial concentration of the CpGs. The average particle size was about 5 μm at lower CpG concentration (10 μg ml−1) and it was significantly decreased to 300 nm at a higher CpG range (40–80 μg ml−1). The particle size reduction was more pronounced when the concentration of the CpG was increased beyond 40 μg ml−1 (Fig. 1a-inset). The size distribution analysis (PdI) of CaP nanoparticles revealed the same reduction trend at higher CpG concentrations (Fig. 1b). The PdI value of 0.4 was observed in a lower range and it was noticeably reduced to 0.05 at higher CpG concentrations (40 and 80 μg ml−1). It is worth mentioning that this reduction in particle size and size distributions was less noticeable for the CpG concentrations in the range of 40 and 80 μg ml−1, denoting the possibility of reaching the saturation range of CpG adsorption on the CaPs.
Fig. 1.
Effect of CpG 2216 concentration and mineralization time on hydrodynamic size (a), polydispersity index or PdI (b), and zeta potential (c) of the mineralized CaP nanoparticles. Increasing the CpG concentration decreased the size and PdI steadily, and did not have a significant effect on the surface charge of the CaPs.
The zeta potential of the nanoparticles was determined by the Laser Doppler Velocimetry (LDV) method. The CaP nanoparticles showed highly negative zeta potentials (−21 to −29 mV) indicating the adsorption of CpGs onto their surface (Fig. 1c). There was no significant difference in the zeta potential values of the nanoparticles prepared with different CpG concentrations. Studies have shown that colloidal stability is strongly dependent on the zeta potential values in which higher values, either positive or negative, create considerable electrostatic repulsion forces that prevent the particles from aggregation.21 The remarkably low PdI of the CaP nanoparticles can be attributed to their strong negative zeta potential.
The formation of CaP nuclei is a dynamic and reversible process and requires sufficient reaction time to produce stable particles. To determine the effect of mineralization time on the physicochemical properties of the particles, three different mineralization time points (1, 2 and 4 h) were selected (Fig. 1). We observed a steady increase in the particle size and PdI over time, however, no apparent changes were detected in the particle surface charge (ZP) during the extended mineralization times.
3.2. Effect of varying CpG 2216 concentrations and mineralization times on binding efficiency
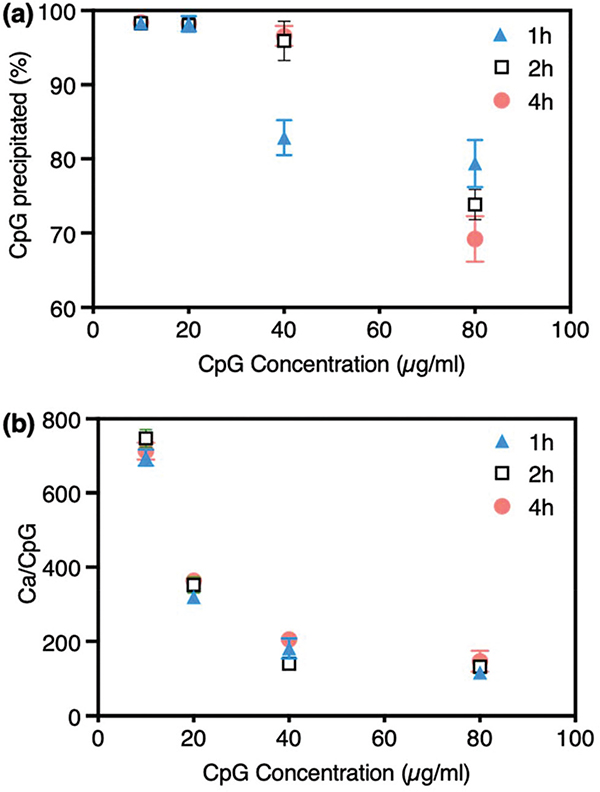
The efficiency of CpG 2216 incorporation into the CaP particles was evaluated by measuring the amount of unbound CpG molecules in the supernatant of the reactions and subtracting them from the initial values added for mineralization. These results (Fig. 2a) show that CpG 2216 molecules were readily adsorbed on nanoparticles and changed the characteristics of the precipitates, providing them with greater colloidal stability, due to the relatively high zeta potential values. The binding efficiency of nearly 100% was observed for nanoparticles prepared in the presence of 10 and 20 μg ml−1 CpG at 1 h mineralization time. The binding efficiency values were ~84% and ~70% for CaP particles precipitated in the presence of higher CpG concentrations (40 and 80 μg ml−1, respectively) at 1 h mineralization time, showing that all CpG molecules were not precipitated with CaP nanoparticles at such a short mineralization time. However, the binding efficiency was ~96%, when the mineralization time was increased to 2 h, for samples prepared in the presence of 40 μg ml−1 CpG. This shows that 2 h mineralization time was sufficient to adsorb a major part of the CpG molecules at this concentration and increasing the mineralization time to 4 h was not necessary in order to achieve a higher binding efficiency. Finally, the CpG adsorption rate did not increase significantly by prolonging the mineralization time to 2 and 4 h, when CpG concentration was 80 μg ml−1.
Fig. 2.
Variation of CaP precipitation efficiency by the concentration of CpG 2216 and mineralization time. (a) The percentage of CpG precipitated on CaPs decreased when the CpG concentration was increased. (b) The Ca/CpG ratio also decreased steadily by increasing the CpG concentration to 40 μg mL−1 and did not change significantly when the CpG concentration was doubled (80 μg mL−1). Some error bars are very small and not visible in the graphs.
In addition, using a modified colorimetric method we quantified the remaining calcium ions in mineralization solutions, and used the values to calculate the Ca/CpG ratios in precipitated nanoparticles (Fig. 2b). Ca/CpG ratios decreased steadily, as we increased the concentration of CpG from 10 to 40 μg ml−1. The magnitude of this decline was directly proportional to the amount of CpG added for mineralization. When we doubled the concentration of CpG for each mineralization reaction, the value of Ca/CpG was also dropped to about half. This steady rate was not observed when the CpG concentration was increased from 40 to 80 μg ml−1, and approximately similar values were obtained for both 40 and 80 μg ml−1 CpG concentrations (plateau shape in the diagram). This was possibly due to the fact that nanoparticles reached their saturation points in adsorbing the CpG molecules, when 40 μg ml−1 CpG was used for their mineralization. Moreover, we found that these trends were independent of mineralization duration and only negligible differences in Ca/ CpG ratios were observed by changing the mineralization times from 1 to 2 and 4 h. On the other hand, as shown in Fig. 1, increasing the mineralization time resulted in larger size CaP nanoparticles, which was not desirable for optimal cell uptake and immunostimulation. Therefore, based on the results shown in Fig. 1 and 2, we chose 2 h as the optimum mineralization time and used it for the follow-on studies.
3.3. Effect of the CpG backbone structure on CaP precipitation
Electrostatic attractions are considered to be the main interaction forces between Ca2+ and negatively charged phosphate groups on the nucleic acid backbones.14,22,23 This provided the motivation to systematically study the molecular factors that can affect the formation of CaP-CpG precipitates. Two types of CpGs from the same ODN family, one containing naturally-occurring phosphodiester (CpG 1826-PO) and the other one with phosphorothioate groups (CpG 1826-PS), were chosen to evaluate the effect of the CpG backbone on the formation of CaP particles (Fig. 3). CpGs with phosphorothioates are a modified ODN backbone (Scheme 1a) in which one of the oxygens in the phosphate group is substituted by a sulfur.
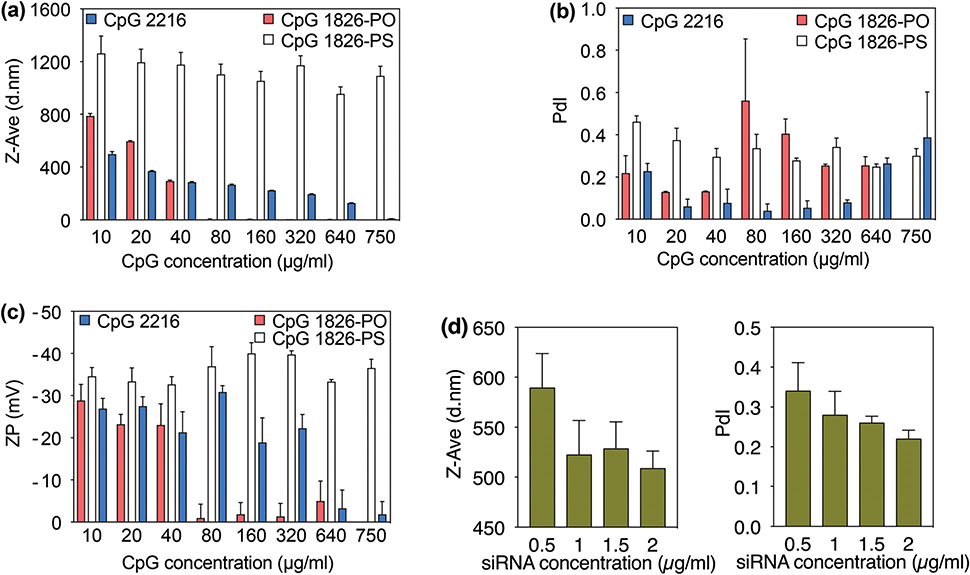
Fig. 3.
Effect of CpG type on the hydrodynamic size (a), PdI (b) and zeta potential (c) of mineralized CaPs. (d) Hydrodynamic size and PdI of the CaPs mineralized in the presence of siRNA.
The particle sizes of the CaP precipitates synthesized using various concentrations of CpG 1826-PO and CpG 1826-PS were determined using the DLS method. The particle sizes for the normal PO and modified PS backbones were in the range of 290–780 nm and 990–1170 nm, respectively (Fig. 3a). The average particle size of the CaPs was found to be directly proportional to the initial concentration of the CpG with the PO backbone (CpG 1826-PO). In particular, the size of particles steadily decreased from 780 nm for a CpG 1826-PO concentration of 10 μg ml−1 to 290 nm when CpG concentration was 40 μg ml−1. It is worth noting that only negligible variations were observed among different measurements as indicated by the small error bars on the graph bars. Also, we observed that further increasing the CpG 1826-PO concentration has inhibitory effects on the formation of CaPs and almost no particles were formed beyond the 40 μg ml−1 concentration point. This observation was also verified by a significant increase in the particles polydispersity index (Fig. 3b) and a substantial decline in the zeta potential (Fig. 3c). Increasing the concentration of this CpG from 10 μg ml−1 to 40 μg ml−1 decreased the PdI from 0.22 to 0.13. However, beyond this point, we observed a significant increase and irregular variations in polydispersity indices measured from final mixtures. These results were further verified by the surface charge measurements. At 10–40 μg ml−1 CpG 1826-PO concentrations, particles showed strong negative charges ranging from −28 to −23 mV, and these values shifted to neutral ranges when higher CpG concentrations were used for mineralization. Negative zeta potential indicates that CpGs were adsorbed on the surface of nanoparticles and this value was significantly decreased with an increase in the CpG concentration. These observations together suggest the inhibitory effect of high CpG-PO concentrations on CaP mineralization, in which the formation of stable CaP precipitates was completely blocked when CpG 1826-PO concentrations were more than 40 μg ml−1.
Previous studies on the co-precipitation of calcium phosphates with plasmid DNA have shown that the amount of DNA can have a major effect on the mineralization reaction, as a result of the physicochemical characteristics of negatively charged DNA,24–26 and the fact that DNA molecules use phosphate groups on their backbone to interact with CaPs.14 Sommerdijk et al.27 also demonstrated the strong inhibitory effect of DNA on the crystallization of calcium carbonate thin films, when the amount of DNA is increased. At high concentrations the DNA molecules can be detrimental to particle formation and at very low concentrations their positive effects cannot achieve their full potential for inducing mineralization.22 Consistent with the previous studies based on plasmid DNA, we observed that CpGs with backbones similar to DNA (i.e., CpG 1826-PO and CpG 2216) exhibited a similar trend. As suggested by Jordan et al.,25 these results indicate that CpGs with a normal PO backbone (CpG-PO) precipitate with the CaP nuclei at the first instances of nucleation. However, the growth of these preliminary nuclei is inhibited at very high CpG-PO concentrations, as shown in Fig. 3a. A significantly higher number of phosphate groups at very high CpG-PO concentrations results in a burst of nucleation (i.e., formation of thermodynamically unstable nuclei). However, the presence of a large number of CpGs around each nuclei limits the accessibility and diffusion of additional Ca2+ ions to these preliminary nuclei, resulting in the suppression of growth of nanoparticles.22 This results in relatively smaller hydrodynamic sizes when a higher concentration of CpG-PO molecules (i.e., CpG 1826-PO and CpG 2216) are present in the mineralization solution (Fig. 3a). This phenomenon completely blocks the mineralization when the concentration of CpGs with PO backbones is more than a certain value (i.e., 40 and 640 μg mL−1 for CpG 1816-PO and CpG 2216, respectively), since the thermodynamically unstable nuclei cannot grow and therefore get dissolved in the mineralization solution. However, as demonstrated in Fig. 3a, we did not observe such a mineralization blocking phenomenon at a higher concentration of the same CpG molecules with PS backbones (i.e., CpG 1826-PS). Also, these CpG-PS molecules did not limit the growth of preliminary nuclei, resulting in the formation of larger particles, as evidenced by the hydrodynamic size and SEM results shown in Fig. 3a and 4b. This can be attributed to the larger anionic radius and lower electronegativity of sulfur ions in the CpG-PS backbone compared with oxygen ions in CpG-PO, which leads to relatively weaker attraction between Ca2+ and CpG molecules with PS groups. Additionally, as a result of this phenomenon, we only observed small differences in size, PdI and zeta potential of the particles prepared in the presence of different CpG 1826-PS concentrations. Overall, CaPs formed in the presence of CpG 1826-PS were larger and their PdI and zeta potential values were higher than those prepared in the presence of CpGs with a PO backbone.
Fig. 4.
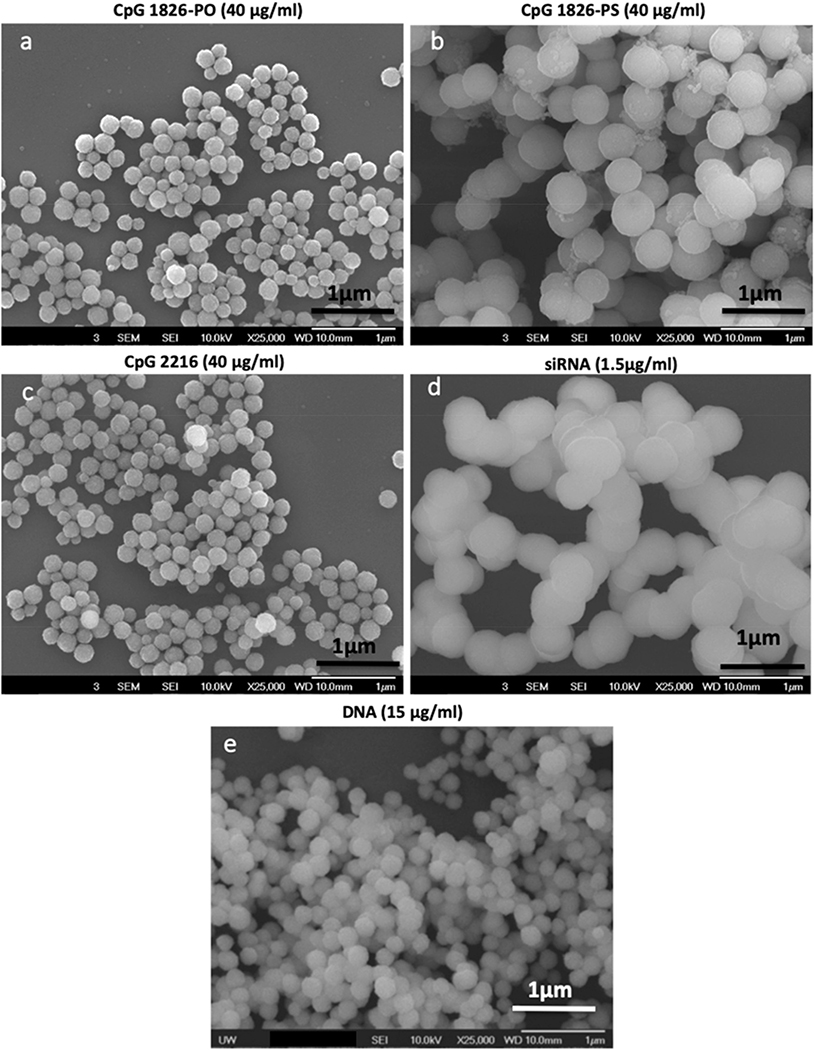
Scanning electron microscopy (SEM) images showing the morphology of CaP nanoparticles formed in the presence of CpG 1826-PO (a) CpG 1826-PO (b), CpG 2216 (c), siRNA (d), and DNA (e).
3.4. Role of CpG sequence on the formation of CaP precipitates
We used two different sequences of CpGs with full or partial phosphodiester backbones (i.e., CpG 1826-PO and CpG 2216, respectively) and evaluated the effects of sequence variation on the formation of CaP particles (Fig. 3). We observed a similar trend of size reduction and small changes on the surface charge of particles, by increasing the concentration of these CpGs. However, as discussed in the previous section, the formation of particles was completely blocked at higher CpG 1826-PO concentrations. This mineralization blocking concentration was significantly higher when ODN 2216 was used for the synthesis of CaPs. The concentration limits for the particles prepared with ODN 1826-PO or ODN 2216 were 40 and 640 μg ml−1, respectively (Fig. 3a). This variation was further confirmed by a sharp rise in the polydispersity index (Fig. 3b) and a steep drop in the associated zeta potential values (Fig. 3c) of the CaPs prepared at different concentrations of these two CpGs. CpG 2216 includes both PO and PS groups on its backbone and therefore its influence on CaP mineralization was a combination of what was observed when CpG 1826-PO and 1826-PS were used separately for CaP mineralization. Overall, the particles synthesized in the presence of CpG 2216 showed a lower PdI compared with those prepared in the presence of the same concentration of CpG 1826-PO, indicating that it can be used as a template when the highest size monodispersity (i.e., extremely low PdI range of (0.04 to 0.22) is required for a specific application.
We further explored the effect of double-stranded siRNA on the mineralization of CaP nanoparticles and compared it with single-stranded CpGs. Both siRNA and CpGs have relatively similar lengths, which allows us to compare the effect of single- or double-stranded biomacromolecules on CaP mineralization. We repeated the same mineralization experiments using siRNA as the template for the formation of CaPs and the size and PdI values are shown in Fig. 3d. We observed a similar size reduction trend when siRNA molecules were used for CaP mineralization. However, the formation of CaPs occurred at much lower siRNA concentration ranges compared with CpGs shown in previous figures (i.e., 0.5–1.5 μg ml−1 for siRNA vs. 10–40 μg ml−1 for CpG 1826-PO or 10–640 μg ml−1 for CpG 2216). The differences between CaP mineralization in the presence of single-stranded CpGs and double-stranded siRNA molecules can be possibly attributed to the spatial configurations of the phosphate groups in their backbones. These distinctive configurations result in different charge density distributions in double-stranded siRNA molecules,23 which increases their binding affinity to CaP particles and helps to mineralize CaPs at much lower siRNA concentrations. These results are consistent with previous studies10,22,23 that demonstrated the binding mechanism of single- and double-stranded DNA with calcium phosphates. It has been shown that double-stranded DNA molecules bind to CaPs only as a result of electrostatic forces. However, single-stranded DNA molecules bind to CaPs based on both hydrophobic and electrostatic interactions, due to the simultaneous presence of hydrophobic bases and negatively charged phosphate groups in their backbone. Consistent with these reports, our results suggest that double-stranded siRNA molecules also have a relatively stronger binding affinity to CaPs compared with single-stranded CpGs. This can be due to the presence of molecularly-aligned phosphate groups, which are exposed on the exterior parts of a more rigid siRNA backbone structure, compared with single-stranded CpG molecules.
SEM was used for the qualitative assessment of the size and morphology of the CaP particles prepared in the presence of various CpG, siRNA and DNA molecules. As shown in Fig. 4, the particles synthesized with these macromolecules were all spherical in nature. We observed a negligible variation in size distribution, indicating that the particles were well defined and highly uniform. The particles synthesized with CpG 1826-PS had larger sizes compared with those prepared with CpG 1826-PO and CpG 2216. The SEM micrographs showed discrete particles for all the macromolecules except for those prepared in the presence of siRNA. These particles were prepared in a relatively lower concentration of siRNA in comparison with our other studied macromolecules and the presence of a lower number of siRNA molecules on their surface resulted in their less stability and higher tendency to aggregation.
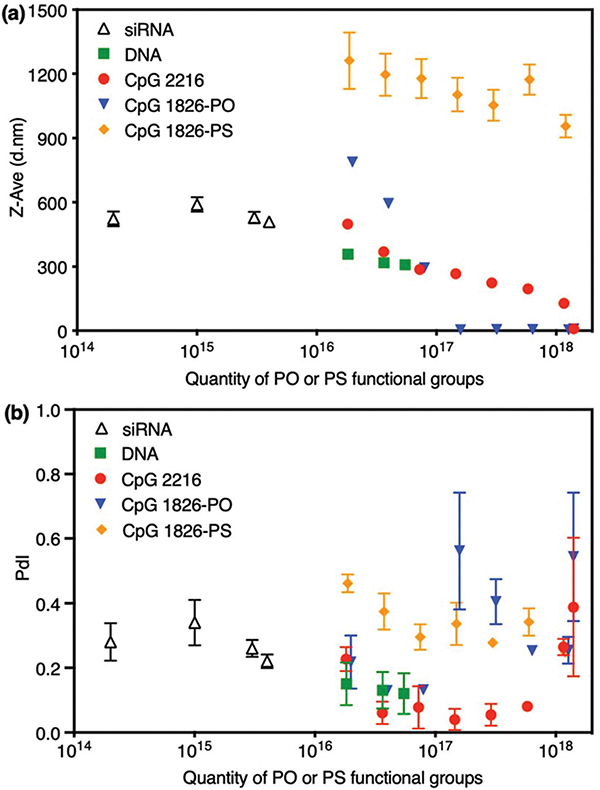
3.5. Comparing various macromolecules based on the quantity of their PS or PO functional groups
The quantity of PS or PO functional groups was calculated using the number of mers for CpGs and quantity of base pairs for siRNA and DNA. The overview of this quantification and the resulting particles sizes and PdI values for various macromolecules are shown in Fig. 5. In each case, we observed that the quantity of PO functional groups plays an important role in the particle size, size distributions and mineralization blocking point. However, the particle size did not change significantly as the number of PS functional groups was increased and a relatively steady decline of the hydrodynamic size was observed with an increase in the total number of PO and PS groups available in the mineralization solution, regardless of the type of molecule used as the synthesis template. Interestingly, we observed that double-stranded DNA (dsDNA) resulted in the smallest particle sizes in the graph, when compared with single-stranded CpG oligonucleotides (i.e., ssODN) with the same amounts of PO functional groups. Previous studies have shown that dsDNA is more rigid than ssDNA and this is known to impact DNA compaction.28,29 We speculate that this molecular rigidity may lead to differences in the accessibility of PO functional groups by Ca2+ ions, limiting the particle growth when dsDNA was used for CaP mineralization instead of ssODNs. Both siRNA and DNA are double-stranded and were able to induce the mineralization at relatively lower concentrations compared with CpGs. In addition, they can effectively suppress the nanoparticle growth and therefore, the selection of the appropriate siRNA or DNA concentration ranges is critical in order to achieve CaPs with mono-disperse size distribution.
Fig. 5.
Variation of CaPs hydrodynamic size (a) and PdI (b) values with the amount of PO or PS functional groups in their mineralization solution, when different molecules (CpG, DNA and siRNA) were used as CaP synthesis templates. The much lower amount of functional groups when siRNA was used for CaP mineralization is noted.
3.6. Immunostimulation using CpG-incorporated calcium phosphate nanoparticles
Toll-like receptors (TLRs) play a major role in the recognition of pathogen associated molecular patterns (i.e., PAMPs). Among different types of TLRs, TLR-9 has been identified to recognize single-stranded DNA molecules such as CpGs discussed in the previous sections of this manuscript, which makes them essential agents for mediating immune responses. When CpG motifs are unmethylated they can be recognized by TLR-9 receptors on dendritic and B cells and activate them, a phenomenon known as immunostimulation.30 This characteristic makes these molecules effective adjuvants for different vaccine delivery applications. However, the incorporation of these molecules on biodegradable nanoplatforms such as CaP nanoparticles enables their targeted delivery into the cells through the endosome–lysosome pathway, if the nanoparticle size, surface charge and stability are optimized.31
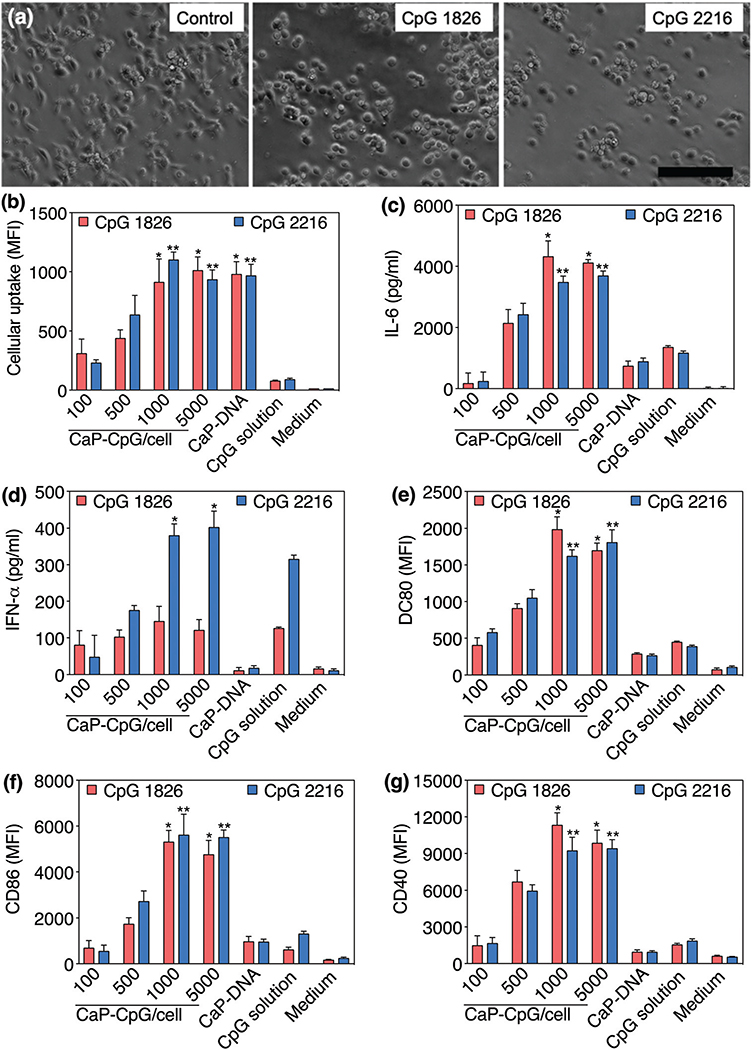
In previous sections, we showed that the molecular structure and concentration of CpGs change the physicochemical properties of the CaP nanoparticles. CaPs prepared in the presence of 40 μg mL−1 of CpG 2216 and CpG 1826-PO had very close hydrodynamic sizes (287 vs. 295 nm), PdI values (0.08 vs. 0.13) and zeta potentials (−21 vs. −23 mV), as shown in Fig. 3. Therefore, we used these two batches for the direct comparison of cellular uptake and immunostimulation efficacy. We incubated these two CaP-CpG formulations with immature BC-1 dendritic cells, to evaluate their stimulation efficacy. The activation of dendritic cells was identified by changes in their morphology, secretion of the proinflammatory IL-6 cytokine and type I interferon IFN-α and the presence of co-stimulatory CD80, CD86 and CD40 immuno-markers. Both CaP-CpG formulations changed the morphology of the cells to a round shape as a typical sign indicating the activation of these immature dendritic cells (Fig. 6a).32 Stimulations of these dendritic cells were further confirmed by an increased amount of IL-6 secretion, when they were incubated with CaP nanoparticles incorporating CpG 2216 or CpG 1826-PO (Fig. 6c). There is a statistically significant higher amount of IL-6 production for the cells treated with 1000 or 5000 CaP-CpG per cells for both types of CpGs, as compared to all other samples. The differences between the levels of IL-6 production in these two sample groups were not statistically significant. On the other hand, only CaPs prepared with CpG 2216 showed a statistically significant higher amount of IFN-α production as compared to all other samples (Fig. 6d). It is know that unlike CpG 2216, CpG 1826 is only effective to activate the secretion of proinflammatory cytokines such as IL-6, without noticeable stimulation of IFN secretion.33–35
Fig. 6.
(a) Activation of immature BC-1 dendritic cells with CaPs incorporating CpG 1826-PO (middle) and CpG 2216 (right). Stimulation of the cells was qualitatively verified by their round-shape morphology after activation vs. relatively elongated morphology before the activation (scale bars = 100 μm). (b) Cellular uptake of CaPs. There was no statistically significant difference in cellular uptake between DCs incubated with 1000 or 5000 nanoparticles per cell (α = 0.05). (c–g) Variation of IL-6, IFN-α, CD80, CD86 and CD40 after activation of dendritic cells with different amounts of CaPs. Single and double asterisks denote statistically significant differences, as compared to all other samples, for CaPs incorporating CpG 1826-PO and CpG 2216, respectively (α = 0.05).
We also evaluated the extent of the cellular uptake of these CaPs using flow cytometry (Fig. 6b). The cellular uptake was increased by increasing the amount of CaP concentration up to 1000 CaP-CpG per cell. There was no statistically significant difference in the amount of cellular uptake between 1000 and 5000 CaP-CpG per cell results (α = 0.05). However these two sample groups showed a statistically significant higher amount of uptake as compared to other sample groups except for CaP-DNA, which was used as our control. The level of IL-6 production for both types of CpG 2216 and CpG 1826-PO showed a strong dependence on the amount of nanoparticle uptake. On the other hand, IFN-α production was dependent on nanoparticle uptake for CpG-2216 sample groups only, in which higher levels of IFN-α were produced by increasing the amount of CaP uptake using 1000 CaP-CpG per cell concentration for incubation.
The immunostimulation efficacy of the CaPs formed in the presence of CpG 1826-PS (40 μg mL−1) is also shown in Fig. S1.† The CaP-related IL-6 and IFN-α values were almost similar to those of free CpG-1826-PS, indicating that these nanoparticles did not increase the immunostimulation efficacy, due to their large hydrodynamic size (i.e., ~1178 nm), which generally decreases the uptake of nanoparticles. In addition, the effect of hydrodynamic size on the immunostimulation efficacy of CaP CpG 1826-PO and CaP CpG-2216 nanoparticles is shown in Fig. S2a–d.† The nanoparticles with larger hydrodynamic sizes had relatively lower immunostimulation rates, due to their decreased uptake. Note that CaP CpG 1816-PS particles did not have significantly different sizes when different concentrations of CpG 1816-PS were used for their mineralization (Fig. 3), and therefore we did not evaluate their size-dependent immunostimulation efficacy.
Additionally, we investigated the presence of co-stimulatory CD80, CD86 and CD40 markers on our stimulated dendritic cells. These molecules bind to specific receptors on T-cells to activate their proliferation and differentiation and their enhanced expression on stimulated BC-1 dendritic cells were shown before.36,37 The stimulation of the dendritic cells with both types of CaPs incorporating CpG 1826-PO and CpG 2216 enhanced the expression of these markers significantly, as evidenced by the flow cytometry of the cells before and after stimulation (Fig. 6e–g). We observed similar expression trends, when these two specific CpG derivatives were used for the stimulation of our dendritic cells. A statistically significant higher amount of immune-related markers (CD40, CD80, CD86) were observed for samples incubated with 1000 or 5000 CaP-CpG per cells for both types of CpGs, as compared to all other sample groups. However, the difference between these two sets of sample groups was not statistically significant.
Finally, Ca2+ ions are known to activate dendritic cells as reported in previous studies.38,39 Therefore, we used DNA-coated CaPs3 as controls, to study the possible role of Ca2+ ions that might be released from these nanoparticles. Note that CaP nanoparticles are not stable and tend to aggregate quickly when they are not coated with macromolecules such as CpG, DNA and siRNA. Also, bare CaP nanoparticles are not feasible to be tagged with fluorescent molecules for any flow cytometric uptake analysis. The analysis of the cells incubated with these DNA-coated CaPs (CaP-DNA) showed that their uptake was in the range of CaP-CpG nanoparticles (Fig. 6b). However, their uptake did not activate the dendritic cells as verified by IL-6, IFN-α, CD80, CD86 and CD40 analysis, shown in Fig. 6c–g. This can be due to much slower CaP degradation kinetics and delayed release of Ca2+ from the core of the nanoparticles compared with the coating CpG layer. This phenomenon generates a faster stimulation response as a result of released CpG molecules only, compared with the bare CaP core of the nanoparticles.
4. Conclusion
We analyzed the synthesis of calcium phosphate nanoparticles (CaPs) using various types of CpG oligodeoxynucleotides and nucleic acids as templates for mineralization. We found that the ability of these macromolecules to control and regulate the precipitation reaction was highly dependent on their backbone structures as well as their concentrations in mineralization solutions. The PO backbone of the CpGs appeared to distinctly influence the mineralization and its concentration played an important role in defining the size and size distributions of the particles. CaP mineralization was blocked at PO concentrations more than a certain range, while this effect was not observed for the increased concentration of PS groups. CaPs synthesized in the presence of CpG 1826-PO and CpG 2216 molecules lead to an enhanced immunostimulatory effect as verified by the activation of the immature dendritic cells and quantification of the proinflammatory IL-6 cytokine, type I interferon INF-α and co-stimulatory CD80, CD86 and CD40 markers. This research shows that oligonucleotide and nucleic acids could mediate and regulate the nucleation and growth of the CaP nanoparticles significantly and careful considerations are required before the selection of a specific nucleotide chain and designing its relevant synthesis parameters. This study provides new insight into the control of calcium phosphate mineralization in the presence of different derivatives of nucleic acids, a commonly neglected parameter which could improve the repeatability and effectiveness of a wide range of CaP-based immunotherapy and gene delivery studies over the last decade. Our results can also be generalized and used as a preliminary guideline for co-precipitation of other inorganic materials in the presence of various natural or synthetic nucleic acids and oligonucleotide molecules.
Supplementary Material
Acknowledgements
The authors acknowledge financial support from the NIH AI088597. This work was in part supported by National Institute of Health (NIH) K99/R00 Pathway to Independence award (grant No. 1K99CA234208-01A1). RK and HA acknowledge supports from NIH T32 CA196585 (Cancer-Translational Nanotechnology Training, Cancer-TNT) and NIH T32 CA009695 (Stanford Cancer Imaging Training, SCIT) Postdoctoral Fellowship Programs at Stanford University, respectively. HA acknowledges support by the Marie Skłodowska-Curie MINDED Project [grant agreement no. 754490]. The authors also acknowledge support from the Department of Chemical Engineering at University of Washington and helpful discussions with Dr H. Shen.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9nr09782a
References
- 1.Zhou Z, Li H, Wang K, Guo Q, Li C, Jiang H, Hu Y, Oupicky D and Sun M, ACS Appl. Mater. Interfaces, 2017, 9, 14576–14589. [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Zhang X, Liao H, Sun Y, Ding L, Teng Y, Zhu W-H, Zhang Z and Duan Y, Adv. Funct. Mater, 2018, 28, 1706124. [Google Scholar]
- 3.Khalifehzadeh R and Arami H, ACS Biomater. Sci. Eng, 2019, 5, 3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hesse C, Kollenda S, Rotan O, Pastille E, Adamczyk A, Wenzek C, Hansen W, Epple M, Buer J, Westendorf AM and Knuschke T, Mol. Cancer Ther, 2019, 18, 1069–1080. [DOI] [PubMed] [Google Scholar]
- 5.Kato T, Suzuki T, Amamiya T, Irie T and Komiyama N, Supramol. Sci, 1998, 5, 411–415. [Google Scholar]
- 6.Cheng B, Cai WQ and Yu JG, J. Colloid Interface Sci, 2010, 352, 43–49. [DOI] [PubMed] [Google Scholar]
- 7.Sokolova V and Epple M, Angew. Chem., Int. Ed, 2008, 47, 1382–1395. [DOI] [PubMed] [Google Scholar]
- 8.Feldheim DL and Eaton BE, ACS Nano, 2007, 1, 154–159. [DOI] [PubMed] [Google Scholar]
- 9.Gugliotti LA, Feldheim DL and Eaton BE, Science, 2004, 304, 850–852. [DOI] [PubMed] [Google Scholar]
- 10.Ngourn SC, Butts HA, Petty AR, Anderson JE and Gerdon AE, Langmuir, 2012, 28, 12151–12158. [DOI] [PubMed] [Google Scholar]
- 11.Meng Y, Liu P, Zhou W, Ding J and Liu J, ACS Nano, 2018, 12, 9070–9080. [DOI] [PubMed] [Google Scholar]
- 12.Graham FL and van der Eb AJ, Virology, 1973, 54, 536–539. [DOI] [PubMed] [Google Scholar]
- 13.Shlaferman J, Paige A, Meserve K, Miech JA and Gerdon AE, ACS Biomater. Sci. Eng, 2019, 5, 3228–3236. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Zhang Z, Liu B, Liu Y, Lopez A, Wu J and Liu J, Langmuir, 2018, 34, 14975–14982. [DOI] [PubMed] [Google Scholar]
- 15.Shen H, Tan J and Saltzman WM, Nat. Mater, 2004, 3, 569–574. [DOI] [PubMed] [Google Scholar]
- 16.Tan J and Saltzman WM, Biomaterials, 2004, 25, 3593–3601. [DOI] [PubMed] [Google Scholar]
- 17.Sun B, Tran KK and Shen H, Biomaterials, 2009, 30, 6386–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stetefeld J, McKenna SA and Patel TR, Biophys. Rev, 2016, 8, 409–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin LG, Am. J. Clin. Pathol, 1974, 61, 114–117. [DOI] [PubMed] [Google Scholar]
- 20.McNeil SE, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol, 2009, 1, 264–271. [DOI] [PubMed] [Google Scholar]
- 21.Berg JC, An introduction to interfaces & colloids : the bridge to nanoscience, World Scientific, Hackensack, N.J., 2010. [Google Scholar]
- 22.Okazaki M, Yoshida Y, Yamaguchi S, Kaneno M and Elliott JC, Biomaterials, 2001, 22, 2459–2464. [DOI] [PubMed] [Google Scholar]
- 23.Chen WY, Lin MS, Lin PH, Tasi PS, Chang Y and Yamamoto S, Colloids Surf., A, 2007, 295, 274–283. [Google Scholar]
- 24.Olton D, Li J, Wilson ME, Rogers T, Close J, Huang L, Kumta PN and Sfeir C, Biomaterials, 2007, 28, 1267–1279. [DOI] [PubMed] [Google Scholar]
- 25.Jordan M, Schallhorn A and Wurm FM, Nucleic Acids Res, 1996, 24, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan M and Wurm F, Methods, 2004, 33, 136–143. [DOI] [PubMed] [Google Scholar]
- 27.Sommerdijk N, van Leeuwen ENM, Vos MRJ and Jansen JA, CrystEngComm, 2007, 9, 1209–1214. [Google Scholar]
- 28.Zhou SQ, Liang DH, Burger C, Yeh FJ and Chu B, Biomacromolecules, 2004, 5, 1256–1261. [DOI] [PubMed] [Google Scholar]
- 29.Sennato S, Bordi F, Cametti C, Diociaiuti M and Malaspina P, Biochim. Biophys. Acta, Biomembr, 2005, 1714, 11–24. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi H, Misato K, Aoshi T, Yamamoto Y, Kubota Y, Wu X, Kuroda E, Ishii KJ, Yamamoto H and Yoshioka Y, Front. Immunol, 2018, 9, 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran TH, Tran TTP, Nguyen HT, Phung CD, Jeong JH, Stenzel MH, Jin SG, Yong CS, Truong DH and Kim JO, Int. J. Pharm, 2018, 542, 253–265. [DOI] [PubMed] [Google Scholar]
- 32.Cougoule C, Lastrucci C, Guiet R, Mascarau R, Meunier E, Lugo-Villarino G, Neyrolles O, Poincloux R and Maridonneau-Parini I, Front. Immunol, 2018, 9, 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C and Taniguchi T, Nature, 2005, 434, 1035–1040. [DOI] [PubMed] [Google Scholar]
- 34.Lipford GB, Sparwasser T, Bauer M, Zimmermann S, Koch ES, Heeg K and Wagner H, Eur. J. Immunol, 1997, 27, 3420–3426. [DOI] [PubMed] [Google Scholar]
- 35.Krug A, Rothenfusser S, Hornung V, Jahrsdorfer B, Blackwell S, Ballas ZK, Endres S, Krieg AM and Hartmann G, Eur. J. Immunol, 2001, 31, 2154–2163. [DOI] [PubMed] [Google Scholar]
- 36.Yanagawa Y, Iijima N, Iwabuchi K and Onoe K, J. Leukocyte Biol, 2002, 71, 125–132. [PubMed] [Google Scholar]
- 37.Iijima N, Yanagawa Y, Iwabuchi K and Onoe K, Immunology, 2003, 110, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saez PJ, Saez JC, Lennon-Dumenil AM and Vargas P, Curr. Opin. Immunol, 2018, 52, 74–80. [DOI] [PubMed] [Google Scholar]
- 39.Shumilina E, Huber SM and Lang F, Am. J. Physiol.: Cell Physiol, 2011, 300, C1205–C1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.