Summary
In December, 2019, an outbreak of COVID-19 emerged in Wuhan, China and quickly spread globally. As of May 7, 2020, there were 3 672 238 confirmed infections and 254 045 deaths attributed to COVID-19. Evidence has shown that there are asymptomatic carriers of COVID-19 who can transmit the disease to others. The virus incubation time shows a wide range (0–24 days) and the virus displays a high infectivity. It is therefore urgent to develop an effective therapy to treat patients with COVID-19 and to control the spread of the causative agent, severe respiratory syndrome coronavirus 2. Repurposing of approved drugs is widely adopted to fight newly emerged diseases such as COVID-19, as these drugs have known pharmacokinetic and safety profiles. As pathological examination has confirmed the involvement of immune hyperactivation and acute respiratory distress syndrome in fatal cases of COVID-19, several disease-modifying anti-rheumatic drugs (DMARDS), such as hydroxychloroquine and tocilizumab, have been proposed as potential therapies for the treatment of COVID-19. In this Review, we discuss the immunological aspects of COVID-19 and the potential implication of DMARDs in treating this disease.
Introduction
In December, 2019, hospitals in Wuhan, China began to report cases of pneumonia of unknown cause. Most of the initially identified patients were geographically linked to a local wet seafood wholesale market, where living or slaughtered wild animals are sold. The virus then rapidly spread to over 200 countries and territories, resulting in 3 672 238 confirmed cases and 254 045 deaths globally according to a report released by WHO on May 7, 2020. Subsequent deep sequencing of lower respiratory tract samples identified a novel coronavirus distinct from the other strains of coronavirus known to infect humans, subsequently named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—a highly contagious virus that can be transmitted from person to person.1 WHO designated the disease caused by SARS-CoV-2 infection as COVID-19. Similar to other diseases caused by coronaviruses, the main transmission route of SARS-CoV-2 is via aerosolised droplets. Other possible transmission routes such as direct contact, oral–faecal route, and mother-to-child transmission have been proposed, but further proof is needed with regard to these.2 A retrospective study done at the beginning of the pandemic reported an incubation period of SARS-CoV-2 of approximately 5–14 days;3 however, a more recent report indicates that the incubation period could be as long as 24 days.4
There is no effective cure for SARS-CoV-2 infection and the most common treatment for patients with COVID-19 is supportive care. Although multiple anti-viral drugs, including remdesivir and lopinavir plus ritonavir, have been used in clinical practice,5, 6 the safety and efficacy of these are still unclear and are under clinical evaluation. Immune-mediated lung injury and acute respiratory distress syndrome (ARDS) are associated with adverse outcomes in patients with COVID-19.7 Histological examination of lung biopsy tissue from a patient who died of COVID-19 showed bilateral diffuse alveolar damage and fibroblastic proliferation in airspaces, and laboratory tests indicated a hyperactivated status of circulating CD4 and CD8 lymphocytes.7, 8 Due to the hyperactive nature of the immune system in some patients with severe COVID-19, several disease-modifying anti-rheumatic drugs (DMARDs), such as tocilizumab (interleukin [IL]-6 receptor inhibitor), baricitinib (Janus kinase [JAK] inhibitor), anakinra (IL-1 receptor antagonist), and the antimalarial drug hydroxychloroquine (or chloroquine), have been proposed as potential treatments for COVID-19. In this Review, we discuss the immunological aspects of the SARS-CoV-2 virus infection and the potential implication of DMARDs in the treatment of patients with COVID-19.
Overview of coronavirus
Coronaviruses are a group of highly diverse, enveloped, positive-sense, single-stranded RNA viruses that belong to two subfamilies, Coronavirinae and Torovirinae, in the family of Coronaviridae. These viruses were first discovered in the 1960s and can be further classified into four main genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus, on the basis of their phylogenetic relationships and genomic structures.9 Among these four genera, alphacoronaviruses and betacoronaviruses primarily cause respiratory and intestinal infection in mammals, whereas gammacoronaviruses and deltacoronaviruses mainly infect birds. Currently, there are seven strains of coronaviruses that are known to infect humans, including the recently identified SARS-CoV-2, human coronavirus 229E (HCoV-229E), OC43 (HCoV-OC43), NL63 (HCoV-NL63), HKU1 (HCoV-HKU1), severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV).10, 11, 12 Domestic or wild animals could have important roles as zoonotic reservoirs that enable virus transmission to humans. On the basis of current sequence databases, the origins of SARS-CoV, MERS-CoV, HCoV-NL63, HCoV-229E, and SARS-CoV-2 are thought to be bats, whereas HCoV-OC43 and HKU1 probably originated from rodents.13, 14, 15, 16, 17 Although most coronavirus infections cause only mild respiratory symptoms, infection with SARS-CoV, MERS-CoV, and SARS-CoV-2 can be lethal.
SARS-CoV first appeared in southern China and quickly spread around the world between 2002 and 2003. This virus was identified as the causative agent of the global pandemic SARS,18 which led to substantial morbidity and mortality. A decade after SARS, an outbreak of MERS-CoV emerged in 2012.19 Most people with MERS had no previous contact with bats, leading to the identification of camels as an intermediate host.20 Patients with SARS or MERS present with a variety of clinical features, ranging from asymptomatic or mild respiratory illness to fulminant severe ARDS with extra-pulmonary complications.21, 22
SARS-CoV-2 belongs to the genus of Betacoronavirus, and on the basis of evolutionary analysis, is most similar to the SARS-like coronavirus from the Chinese horseshoe bat, with a nucleic acid homology of 84%.3, 10 SARS-CoV-2 also has 78% similarity with SARS-CoV and 50% with MERS-CoV, at the nucleic acid level.3, 10 Only 10 days after the release of the SARS-CoV-2 genome, researchers found a similar coronavirus from fruit bats, BatCoVHKU9–1, based on evolutionary characteristics.23 Several later reports suggested that snakes, mink, and pangolins could be intermediate hosts, based on codon preference and viral infection patterns.24, 25, 26 At the onset of the COVID-19 pandemic, the main symptoms were fever (98%), cough (76%), and myalgia or fatigue (44%).27 About half of the patients developed breathing difficulty in one week and the severely ill patients soon developed ARDS, acute cardiac injury, secondary infections, or a combination thereof.27 The diagnosis of the disease mainly depends on SARS-CoV-2 RNA detection in nasopharyngeal swab by real-time polymerase chain reaction, epidemiological history, clinical manifestations, and lung imaging.
Immune response against SARS-CoV-2
The invasion and pathogenesis of SARS-CoV-2 are associated with the host immune response. The spike glycoprotein (S protein) on the viral envelop binds to its receptor, angiotensin-converting enzyme 2 (ACE2), on the surface of human cells.15, 23 An analysis of the structure of the SARS-CoV-2 S protein and its binding affinity for ACE2 using cryogenic electron microscopy and surface plasmon resonance showed that the structure of SARS-CoV-2 S protein is very similar to that of SARS, although with minor differences.28 The affinity of SARS-CoV-2 S protein binding to ACE2 is 10 to 20 times higher than that of the SARS S protein, suggesting that SARS-CoV-2 might transmit more readily from person to person.28
Innate immunity is the first line of defence against virus invasion. Viral infection of mammals activates intracellular pattern recognition receptors that sense pathogen-associated molecular patterns, such as double-stranded RNA or uncapped mRNA. The recognition of pathogen-associated molecular patterns results in subsequent cytolytic immune responses, mainly through the type I interferons (IFN) and natural killer cells. Adaptive immunity also plays an important part in viral clearance via activated cytotoxic T cells that destroy virus-infected cells and antibody-producing B cells that target virus-specific antigens. Patients with COVID-19, especially those with severe pneumonia, are reported to have substantially lower lymphocyte counts and higher plasma concentrations of a number of inflammatory cytokines such as IL-6 and tumor necrosis factor (TNF).27, 29, 30 Another study29 reported that CD4+ T cells, CD8+ T cells, and natural killer cells were reduced in severely ill patients compared with those with mild disease symptoms. Moreover, a substantial reduction of CD4+ T cell and CD8+ T cell counts in the peripheral blood was also observed in a patient who died.7 Notably, the proinflammatory subsets of T cells, including IL-17-producing CCR4+ CCR6+ CD4+ (T-helper 17 or Th17) cells and perforin and granulysin-expressing cytotoxic T cells were increased, which could be partly responsible for the severe immune injury in the lungs of this patient.7
The anti-viral immune response is crucial to eliminate the invading virus, but a robust and persistent anti-viral immune response might also cause massive production of inflammatory cytokines and damage to host tissues.31 The overproduction of cytokines caused by aberrant immune activation is known as a cytokine storm. In fact, in the late stages of coronavirus disease, including SARS, MERS, and COVID-19, cytokine storms are a major cause of disease progression and eventual death.7, 32, 33 Huang and colleagues27 found increased plasma concentrations of both Th1 (eg, IL-1β and IFNγ) and Th2 (eg, IL-10) cytokines. Notably, patients admitted to the intensive care unit (ICU) had higher plasma concentrations of IL-2, IL-7, IL-10, granulocyte-colony stimulating factor, IFNγ-induced protein-10 (IP-10), macrophage chemoattractant protein-1, macrophage inflammatory protein 1α, and TNF compared to those not admitted to the ICU. Two other studies29, 34 also showed that plasma IL-6 concentrations were above the normal range in patients with severe symptoms of COVID-19 compared with healthy individuals and those with milder symptoms. Mehta and colleagues35 suggest that secondary haemophagocytic lymphohistiocytosis (sHLH) could be associated with severe COVID-19 cases. HLH is a disease entity characterised by an uncontrolled cytokine storm and expansion of tissue macrophages or histiocytes that exhibit haemophagocytic activity.36 HLH can result from genetic defects in cytolytic pathways (familial or primary HLH) or other diseases such as infection, malignancy, and rheumatic disease (sHLH).37 In 1952, Farquhar and Claireaux first described cytokine storm in patients with HLH.38 The characteristics of HLH, including hypercytokinaemia, unremitting fever, cytopenias, hyperferritinaemia, and multi-organ damage, are commonly seen in seriously ill patients with COVID-19.27, 35 It is suggested that alveolar macrophages expressing ACE2 are the primary target cells for SARS-CoV-2 infection. These activated macrophages may play an important part in HLH-like cytokine storm during COVID-19.39 Thus, early identification and appropriate treatment of this hyperinflammatory status is important for reducing the mortality of patients with COVID-19. 35
Potential immunotherapy in COVID-19
Evidence has shown that asymptomatic COVID-19 carriers can transmit the disease to others and that the virus has a wider range of incubation time than initially thought (0–24 days).4 In addition, the virus displays a high infectivity. If the virus continues to mutate to lower its pathogenicity, there is a high possibility that it might coexist with humans. Therefore, there is an urgent need to develop therapies to treat SARS-CoV-2. Repurposing of approved drugs is commonly employed to fight against newly emerged diseases, such as COVID-19, as these drugs have known pharmacokinetic and safety profiles. Due to the importance of immune imbalance in the pathogenesis of SARS-CoV-2 infection, several immune-modulating drugs that regulate different aspects of inflammation (table ) are being tested for their efficacy in the treatment of severe COVID-19. Hyperinflammation is an important determinant of disease outcome in COVID-19, and immunosuppression might be beneficial to reduce the mortality in patients with severe symptoms.7, 35 Therefore, early identification of such patients is crucial. It has been proposed that laboratory tests of ferritin, lymphocyte or leukocyte counts, platelet counts, erythrocyte counts, and sedimentation rate could be used to screen patients at high risk of hyperinflammation. Application of the HScore, used for the evaluation of patients with sHLH, was recommended by Mehta and colleagues35 to identify patients with COVID-19 at high risk of hyperinflammation. The HScore combines both laboratory and clinical parameters, including serum aspartate aminotransferase, triglycerides, fibrinogen, ferritin, cytopaenias, body temperature, organomegaly, haemophagocytosis on bone marrow aspirate, and signs of immunosuppression.35 In addition, evaluation of cytokine profiles and immune cell subsets has important implications for selecting appropriate immunosuppressants (eg, tocilizumab could be considered in patients with high concentrations of serum IL-6). Given the fact that anti-viral immunity is required to recover from COVID-19, the pros and cons of using an immunosuppressant on these patients should be carefully considered. The severity of the hyperinflammation and viral load or replication status needs to be taken into consideration. One way to avoid the suppression of anti-viral immunity is to choose selective instead of broad immunosuppressive drugs. The timing of treatment is also crucial to reduce the side-effects of immunosuppression; unfortunately there is not yet any definitive evidence with regard to the appropriate timing of administration of these agents. Further studies are required to determine the appropriate timing and routes of drug administration.
Table.
Repurposing of immune-modulating therapies for COVID-19
| Mechanism of action | |
|---|---|
| csDMARDs | |
| Chloroquine or hydroxychloroquine | Interference with ACE2 to block virus invasion; increase of endosomal pH required for virus fusion; mild immune suppression |
| Glucocorticoids | Suppression of immune and inflammatory responses |
| Leflunomide | Inhibition of virus replication |
| Thalidomide | Reduction of inflammatory cell infiltration; reduction of cytokine storm; reduction of lung damage and pulmonary interstitial fibrosis |
| bDMARDs | |
| Tocilizumab | Blockade of IL-6 receptor and its downstream signalling pathways |
| Anakinra | Blockade of IL-1 receptor and its downstream signalling pathways |
| tsDMARDs | |
| Baricitinib | JAK inhibitor; blockade of viral invasion through the inhibition of AAK1; immune suppression |
| Ruxolitinib | JAK inhibitor; immune suppression |
| Cell therapy | |
| Stem cells | Suppression of inflammation; proviral silencing |
| Plasma therapy | |
| Convalescent plasma | Promotion of virus elimination via virus-specific antibodies |
AAK1=AP2-associated protein kinase 1. ACE2=angiotensin-converting enzyme 2. bDMARDs=biologic disease-modifying anti-rheumatic drugs. csDMARDs=conventional synthetic disease-modifying anti-rheumatic drugs. IL=interleukin. JAK=Janus kinase. tsDMARDs=targeted synthetic disease-modifying anti-rheumatic drugs.
Biological immuno-modulating drugs
IL-6 is a key inflammatory cytokine that has a critical part in inflammatory cytokine storm and is elevated in patients with COVID-19.29, 34 Tocilizumab, a recombinant humanised monoclonal antibody against the IL-6 receptor, is widely used in treatment for autoimmune diseases, such as rheumatoid arthritis.40 In patients with COVID-19, IL-6-producing CD14+ CD16+ inflammatory monocytes were significantly increased, and numbers of these cells were further increased in patients with COVID-19 admitted to the ICU.41 The authors of this study proposed that hyperactivated Th1 cells producing granulocyte-macrophage colony stimulating factor (GM-CSF) and IFNγ in the lung promote IL-6-producing monocytes through release of GM-CSF, suggesting that both IL-6 and GM-CSF might be potential therapeutic targets in patients with COVID-19.41 Tocilizumab is a first-line drug for the treatment of cytokine release syndrome (a rapid and massive release of cytokines into the blood from immune cells, usually caused by immunotherapy), especially in patients with comorbidities. In terms of mechanism, tocilizumab binds to both the membrane and soluble forms of IL-6 receptor, thereby suppressing the JAK-signal transducer and activator of transcription (STAT) signalling pathway and production of downstream inflammatory molecules.42, 43 There are many ongoing trials assessing the efficacy of tocilizumab in COVID-19 (appendix p 1). However, animal studies have shown that IL-6 is required for the clearance of viruses and control of pulmonary inflammation.44 Therefore, clinicians should pay close attention to the possibility that blocking IL-6 could interfere with viral clearance or exacerbate lung inflammation. A recent observational study45 from China reported that tocilizumab treatment in severe COVID-19 cases resulted in improvement in COVID-19 symptoms, peripheral oxygen saturation, and lymphopenia within a few days. A substantial remission of lung lesion opacity in chest CT scan was observed in 95% of patients (19 of 20) after 5 days of treatment, and all patients were discharged after an average of 15.1 days of hospital stay.45
Blockade of the IL-1 pathway is used for the treatment of some hyperinflammation conditions. The IL-1 receptor antagonist anakinra is approved for rheumatoid arthritis, Still's disease, and cryopyrin-associated periodic syndrome. A phase 3 randomised controlled trial (RCT) for severe sepsis reported that treatment with anakinra was associated with a significantly lower 28-day mortality in patients who were septic with hyperinflammation, without increased adverse events.46 A retrospective analysis47 of 44 patients with sHLH who were treated with anakinra indicated that treatment with anakinra resulted in a 57% decrease of ferritin concentrations, and early initiation of anakinra was associated with reduced mortality. Since IL-1 was reported to be increased in some patients with COVID-19,27 blockade of IL-1 seems a reasonable approach for the treatment of hyperinflammation in these patients.35 Several trials of anakinra are currently underway, including a phase 2/3 clinical trial evaluating the efficacy and safety of anakinra and emapalumab (IFNγ inhibitor) in reducing hyperinflammation and respiratory distress in patients with COVID-19 (NCT04324021; appendix p 1).
Targeted synthetic immunosuppressants
Baricitinib is a small molecule compound that selectively inhibits the kinase activity of JAK1 and JAK2. Baricitinib can be used in combination with one or more TNF inhibitors and is approved for the treatment of rheumatoid arthritis48 and psoriatic arthritis.49 Through searching the BenevolentAI database, Richardson and colleagues50 predicted that baricitinib might effectively reduce the ability of SARS-CoV-2 virus to infect lung cells.51 As noted, SARS-CoV-2 binds to the ACE2 receptor on host cells and enters lung cells through receptor-mediated endocytosis. ACE2 is widely expressed in several tissues, including renal, vascular, heart, and lung. High concentrations of ACE2 expression on pulmonary AT2 alveolar epithelial cells makes these cells particularly susceptible to SARS-CoV-2 infection.52 AP2-associated protein kinase 1 (AAK1) regulates endocytosis via phosphorylation of the clathrin adaptor protein AP2. Richardson and colleagues identified six high-affinity AAK1 inhibitors from 47 clinical candidates in the BenevolentAI database. Baricitinib was then further selected based on its relatively mild side-effects and the feasibility to achieve effective concentrations in the blood. In addition, baricitinib can also bind to cyclin G-related kinases, which also regulate receptor-mediated endocytosis. The immunosuppressive function of baricitinib might also be of benefit to the hyperactive immune status in severe cases of COVID-19 where immune-mediated lung injury and ARDS might occur.
Ruxolitinib, another oral JAK1 and JAK2 inhibitor approved specifically for the treatment of myelofibrosis, has been used for the treatment of sHLH. Ruxolitinib was shown to rapidly improve respiratory, liver, and haemodynamic function in an 11-year-old boy with refractory HLH,53 and to substantially improve serum ferritin, lactate dehydrogenase, fibrinogen, and liver function in a 38-year-old female patient with refractory Epstein-Barr virus-related sHLH.54 An open-label clinical trial55 showed that ruxolitinib was well tolerated and manageable for treating sHLH, with symptoms and cytopenias improved in all (n=5) patients within the first week of ruxolitinib treatment. Concentrations of ferritin, soluble IL-2 receptor, and STAT1 phosphorylation were also reduced after the administration of ruxolitinib.55 Animal studies showed that inhibition of JAK1 and JAK2 using ruxolitinib improved weight loss, organomegaly, anaemia, thrombocytopenia, hypercytokinaemia, and tissue inflammation in animal models of both primary HLH and sHLH by reducing STAT1-dependent CD8+ T-cell expansion.56
Considering the similar hyperinflammatory nature of sHLH and severe COVID-19, JAK1 and JAK2 inhibitors such as baricitinib and ruxolitinib could be potential treatments for the hyperinflammation seen in COVID-19.50 Several registered RCTs are evaluating the efficacy of ruxolitinib and baricitinib in the treatment of COVID-19 (appendix p 2).
Chloroquine and hydroxychloroquine
Chloroquine and hydroxychloroquine, initially used as antimalarial drugs, have been widely used in several infectious (HIV, Q fever, and fungal infections), rheumatological (systemic lupus erythematosus, antiphospholipid antibody syndrome, rheumatoid arthritis, and Sjogren's syndrome), and other immunological diseases.57 The mechanism of action of hydroxychloroquine is diverse and includes anti-inflammatory action, immune regulation, anti-infection, anti-tumour, metabolic regulation, and anti-thrombosis. Chloroquine has been shown to have anti-coronavirus effects in vitro. Based on this, and the immunoregulatory actions of these drugs, chloroquine and hydroxychloroquine were proposed in the treatment of COVID-19. These drugs increase the endosomal pH required for SARS-CoV-2 endocytosis and cell fusion (figure ). Chloroquine also interferes with the glycosylation of ACE2, which is required for virus attachment to host cells.58 Chloroquine was first reported in 2020 to be a potent inhibitor of COVID-19 using an in vitro SARS-CoV-2-infected Vero-E6 cell culture model.59
Figure.
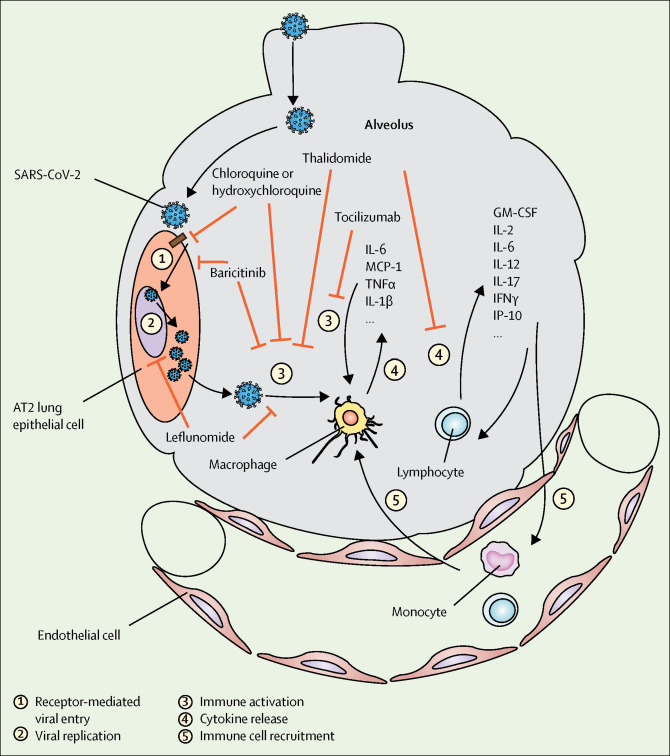
Potential mechanisms of action for DMARDs in COVID-19
DMARDs could target different stages of the virus infection and immune activation. AT2=alveolar type 2. DMARDs=disease modifying anti-rheumatic drugs. GM-CSF= granulocyte-macrophage colony stimulating factor. IFNγ= interferon gamma. IL=interleukin. IP-10=interferon gamma-induced protein 10. MCP-1=monocyte chemoattractant protein-1. SARS-CoV-2=severe respiratory syndrome coronavirus 2. TNFα=tumor necrosis factor alpha.
Hydroxychloroquine is a derivative of chloroquine that has similar pharmacokinetics and mechanism of action as chloroquine, but substantially fewer side-effects.60 Compared with other immunosuppressant drugs such as methotrexate, the use of hydroxychloroquine and chloroquine is associated with a reduced risk of infection, even with chronic use.61, 62 Therefore, hydroxychloroquine is more commonly used in patients with rheumatic diseases and other conditions. Hydroxychloroquine has been used to treat HIV-1 in humans as early as the 1990s. In a randomised, double-blinded, placebo-controlled clinical trial of 40 asymptomatic patients with HIV-1, 800 mg/d hydroxychloroquine treatment for 8 weeks reduced the plasma concentration of HIV-1 RNA, preserved CD4+ T-cell counts and proliferative responses, and lowered serum IL-6 concentrations, compared with the placebo group.63 Although it takes 1–3 months for hydroxychloroquine and chloroquine to fully take effect in patients with rheumatic disease, the drugs' anti-viral effect is relatively rapid. Hydroxychloroquine treatment as short as 3 days was shown to accelerate virus clearance in patients with COVID-19, and azithromycin reinforced the anti-viral effect.64 There are a number of ongoing clinical trials testing the efficacy of hydroxychloroquine and chloroquine in COVID-19 (appendix pp 2–5). Although a recent randomised trial has shown that chloroquine and hydroxychloroquine might improve pneumonia symptoms, laboratory tests, and decrease the progression to severe or critical conditions,65 other studies reported either no benefits66, 67, 68 or hazardous effects after chloroquine or hydroxychloroquine treatment.69Notably, treatment in patients with COVID-19 might cause cardiotoxicity, especially when used at a high dose.70, 71 Therefore, results from ongoing trials are required to assess the efficacy and safety of hydroxychloroquine in COVID-19.
Glucocorticoids
Glucocorticoids and their synthetic analogues have been widely used in rheumatic disease to control autoimmune response.72 Due to their rapid immunosuppressive effect, glucocorticoids are frequently used in hyperinflammatory syndromes, such as ARDS. In patients with ARDS, glucocorticoid treatment improves oxygen saturation, inflammatory markers, ICU length of stay, and ventilator-free days, although its effect on mortality was not consistent between trials.73, 74, 75 In coronavirus disease, inflammation-induced lung injury and ARDS are associated with adverse outcomes.26, 76 Histological investigations showed severe lung inflammation and diffuse alveolar damage in patients with coronavirus disease.7, 77 Therefore, corticosteroids are commonly used in severe cases of coronavirus disease including SARS, MERS, and COVID-19 to control immune-mediated damage of lung tissue.27, 30, 78, 79 However, clinical evidence has not supported a beneficial effect of glucocorticoids in coronavirus illness. In a retrospective study79 of 309 critically ill patients with MERS, after statistical adjustment for time-varying confounders, corticosteroid therapy was not significantly associated with improved 90-day mortality, and resulted in delayed clearance of the MERS coronavirus RNA. In a systematic review of SARS treatments that included 29 studies with cortiocosteroid use, 25 studies were inconclusive with regards to the effect of corticosteroid in SARS and four reported corticosteroids as causing possible harm. Therefore, high-quality RCTs are needed to provide conclusive evidence.
Leflunomide
Leflunomide is a low-molecular weight, synthetic, oral anti-rheumatic drug. The mechanism of its action includes inhibition of pyrimidine synthesis, inhibition of protein tyrosine stimulation, inhibition of nuclear factor kappa beta, and anti-tumour effects.80, 81, 82, 83 Leflunomide has been widely used for the treatment of rheumatoid arthritis,84 and due to its immunosuppressive function, the drug is also used in organ transplantation.85 Another important function of leflunomide is that it inhibits virus replication. In vitro studies have shown that the active metabolite of leflunomide (A77 1726) protects umbilical cord epithelial cells and fibroblasts from infection with human cytomegalovirus.80, 86 Electron microscopy revealed that the morphology of virions in the cytoplasm was abnormal and the assembly of virus particles could not be completed in cytomegalovirus-infected cells treated with A77 1726, indicating that leflunomide interferes with the assembly of virus capsid.80 In a study87 of 17 patients with cytomegalovirus disease, 88% (15) of the patients clinically responded to leflunomide therapy (3 doses of 100 mg/day, followed by 20 mg/day—a dose that maintains blood concentrations of 25–50 ng/mL), with viral clearance from blood and healing of involved organs. Therefore, leflunomide is effective for treatment of cytomegalovirus disease.88
The anti-viral efficacy of leflunomide has also been observed in transplant recipients infected with BK virus89 and human papillomavirus (HPV).90 In patients with polyomavirus type BK nephropathy, those that had blood A77 1726 concentrations higher than 40 μg/mL had either cleared the virus or showed progressive reductions of viral load in blood and urine.89 In another study,90 leflunomide successfully cleared verrucae vulgaris and molluscum lesions in four renal transplant patients with cutaneous warts, usually caused by HPV. However, it is worth noting that leflunomide has been associated with interstitial lung disease.91, 92 Leflunomide-associated interstitial lung disease usually occurs within the first 20 weeks of leflunomide initiation. There was a high mortality in patients with pre-existing interstitial lung disease, diffuse alveolar damage, or ground glass shadowing on high resolution CT.91 The exact mechanism remains unclear, but the presence of pre-existing interstitial lung disease, cigarette smoking, low bodyweight, and the use of methotrexate are risk factors for leflunomide-induced interstitial lung disease.92 It remains unclear if leflunomide would improve or exacerbate the pulmonary lesions in patients with COVID-19. There is currently one clinical trial aiming to evaluate the efficacy and safety of oral leflunomide tablets against pneumonia caused by SARS-CoV-2 (appendix p 2).
Thalidomide
Another DMARD, thalidomide, which has both anti-inflammatory and anti-proliferative activity, has also been used in viral infections. Animal studies have shown that thalidomide inhibits lung injury in mouse models of H1N1 influenza virus infection, with improved survival, reduced inflammatory cell infiltration, reduced concentrations of cytokines (IL-6 and TNF) and chemokines (RANTES and IP-10), and reduced nuclear factor kappa beta activity.93 It was concluded that thalidomide could be an alternative treatment when new influenza viruses emerge, especially before new vaccines are developed. A UK study94 indicated that thalidomide has immunomodulatory and immune remodelling effects by inhibiting TNF, another critical cytokine in COVID-19-associated lung injury. In addition, some studies have shown that thalidomide can treat pulmonary interstitial fibrosis and combat cytokine storm.95, 96 These studies indicate a potential therapeutic value of thalidomide in viral infection. In a case report,97 a 45-year-old female patient with severe COVID-19 and elevated concentrations of circulating cytokines, including IL-6, IL-10, and IFNγ, on admission was treated with oral thalidomide (100 mg once a day) and low-dose methylprednisolone (40 mg intravenously, every 12 h for 3 days; and then 40 mg intravenously once a day for 5 days) due to the severity of clinical manifestations and lack of response to other treatments. The patient's clinical condition, including oxygen index, fever, nausea, and vomiting resolved within 1 week after thalidomide treatment. Concentrations of IL-6, IL-10, and IFNγ all returned to normal range after 6 days of treatment, SARS-CoV-2 tests in swab specimens were negative after 1 week of treatment, and lung lesions disappeared 12 days after treatment.97 Although this is a single case, it could provide some useful insight for further clinical investigation. There are two clinical trials evaluating the therapeutic potential of thalidomide in patients with moderate or severe COVID-19 (appendix p 2).
Other immune-modulating therapies
There are also many other immune-modulating strategies under clinical investigation for the treatment of COVID-19, such as stem-cell therapy and convalescent plasma treatment. Mesenchymal stem cells (MSCs) are of increased importance in inflammatory disease due to their anti-inflammatory properties. Animal experiments showed that MSC treatment was able to reduce influenza A H5N1-induced acute lung injury in vivo.98 Stem cells are able to suppress the activities of viruses via Chaf1a-mediated and Sumo2-mediated epigenetic regulation (termed proviral silencing).99 Several phase 1 and 2 clinical trials have confirmed the safety of MSC therapy in patients with ARDS, and have shown beneficial effects.100, 101 However, several issues have limited MSC use in clinic, such as the lack of clarity with regard to optimal dose and route of MSC delivery, difficulties in large-scale production and cryopreservation, and the potential for substantial variability. There are several ongoing clinical trials testing the efficacy of MSC in COVID-19 (appendix pp 5–6).
Convalescent plasma from patients who have recovered from SARS-CoV-2 infection has also been proposed as a potential treatment for COVID-19. Convalescent plasma has been used in many severe infections such as SARS, MERS, and Ebola, as one of the few therapeutic strategies in the absence of vaccines or other specific treatments.102 The efficacy of such therapy, especially in COVID-19, is being evaluated in ongoing trials (appendix p 6).
Conclusions and outlook
SARS-CoV-2 has spread rapidly since it first emerged in December, 2019, and COVID-19 is characterised as a pandemic by WHO. As a new emerging virus, there is no approved effective drug or vaccine. As of April 16, 2020, several existing drugs are being repurposed for the treatment of patients with COVID-19, with dozens of ongoing clinical trials assessing their potential efficacy. DMARDs, due to their immune-modulating nature, could be a potential treatment option for severe COVID-19. However, there are several issues that need to be taken into consideration. First, the issue of hyperinflammation versus viral replication. Although effective anti-viral immunity is required for the clearance of pathogens, hyperactivation of immune response causes tissue damage and organ failure. Similarly, there are two sides of immunomodulation therapy in COVID-19, and clinicians should determine in which circumstance to use such medications. Second, there are questions about the timing for immunomodulation therapy. As noted, immunosuppressants could affect anti-viral immune response and the timing should be carefully considered. Although early intervention is considered as a key factor for the success of immunomodulation therapy in infection-associated hyperinflammation,103 direct evidence from RCTs are required to determine the appropriate timing for patients with COVID-19. Finally, the pharmacokinetics of oral medications in crucially ill patients merit consideration, as physiological alterations in these patients can substantially affect the pharmacokinetics. Some drugs will need to be given parenterally due to gastrointestinal failure (eg, chloroquine has been used parenterally to treat severely ill patients with malaria, although it is absorbed reliably in these patients).104 Compared with the oral route, parenteral administration of chloroquine results in rapid absorption and transient high plasma concentrations, which is associated with increased risk of acute toxicity.105 In addition, impaired clearance of the drugs might be problematic in patients with hepatic dysfunction and renal failure. Therefore, smaller and more frequent doses, continuous intravenous infusion, or choosing a less toxic drug (eg, using hydroxychloroquine instead of chloroquine) should be considered in patients who are severely ill.104, 105 The many ongoing trials will hopefully provide a better understanding of the potential effects of immunomodulation therapy on COVID-19-associated hyperinflammation.
Search strategy and selection criteria
Two reviewers independently did a computerised literature search of PubMed, Ovid, and Web of Science, using the terms “COVID-19 OR SARS-CoV-2 OR 2019-nCoV”, “DMARDs OR immunosuppressant OR immunomodulation OR anti-rheumatic drugs OR immunotherapy”, and a combination thereof, in English language. We incorporated published and unpublished articles (including preprint articles) into this Review from December 30, 2019, to April 16, 2020. We included publications cited in the papers when relevant. We also referred to related scientific reports, such as the official website of WHO and the Chinese health organisation.
Acknowledgments
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81974254, 81670431, and 81771754) and the Tongji Hospital Clinical Research Flagship Program (2019CR206).
Contributors
JZ and JT wrote the manuscript with input from CY and LD. All authors revised the manuscript and approved the final report.
Declaration of interests
We declare no competing interests.
Supplementary Material
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holshue ML, DeBolt C, Lindquist S. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan WJ, Ni ZY, Hu Y. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen N, Zhou M, Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillaiyar T, Meenakshisundaram S, Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov Today. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Z, Shi L, Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian S, Xiong Y, Liu H. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod Pathol. 2020 doi: 10.1038/s41379-020-0536-x. published online April 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo PC, Lau SK, Lam CS. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan JF-W, Yuan S, Kok K-H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng LFP, Hiscox JA. Coronaviruses in animals and humans. BMJ. 2020;368:m634. doi: 10.1136/bmj.m634. [DOI] [PubMed] [Google Scholar]
- 13.Forni D, Cagliani R, Clerici M, Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su S, Wong G, Shi W. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P, Yang X-L, Wang X-G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Shi Z, Yu M. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 17.Wang LF, Shi Z, Zhang S, Field H, Daszak P, Eaton BT. Review of bats and SARS. Emerg Infect Dis. 2006;12:1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee N, Hui D, Wu A. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 19.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 20.Mohd HA, Al-Tawfiq JA, Memish ZA. Middle East respiratory syndrome coronavirus (MERS-CoV) origin and animal reservoir. Virol J. 2016;13:87. doi: 10.1186/s12985-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Chen P, Wang J. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Q, Li M, Wang C. Host and infectivity prediction of Wuhan 2019 novel coronavirus using deep learning algorithm. bioRxiv. 2020 doi: 10.1101/2020.01.21.914044. published online February 2. (preprint). [DOI] [Google Scholar]
- 25.Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam TT, Shum MH, Zhu HC. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. published online March 26. (preprint). [DOI] [PubMed] [Google Scholar]
- 27.Huang C, Wang Y, Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrapp D, Wang N, Corbett KS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan S, Yi Q, Fan S. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP) medRxiv. 2020 doi: 10.1101/2020.02.10.20021832. published online February 12. (preprint). [DOI] [Google Scholar]
- 30.Wang D, Hu B, Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perlman S, Dandekar AA. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong CK, Lam CW, Wu AK. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang Y, Zhang H, Xu Y, Xie J, Pang P, Ji W. CT manifestations of two cases of 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295:208–209. doi: 10.1148/radiol.2020200280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutchinson M, Tattersall RS, Manson JJ. Haemophagocytic lymphohisticytosis-an underrecognized hyperinflammatory syndrome. Rheumatology (Oxford) 2019;58(suppl 6):vi23–vi30. doi: 10.1093/rheumatology/kez379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halyabar O, Chang MH, Schoettler ML. Calm in the midst of cytokine storm: a collaborative approach to the diagnosis and treatment of hemophagocytic lymphohistiocytosis and macrophage activation syndrome. Pediatr Rheumatol Online J. 2019;17:7. doi: 10.1186/s12969-019-0309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farquhar JW, Claireaux AE. Familial haemophagocytic reticulosis. Arch Dis Child. 1952;27:519–525. doi: 10.1136/adc.27.136.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaofu Wang, Xie J, Lei Zhao. Aveolar macrophage activation and cytokine storm in the pathogenesis of severe COVID-19. Nature Research. 2020 doi: 10.21203/rs.3.rs-19346/v1. published online March 25. (preprint). [DOI] [Google Scholar]
- 40.Choi IA, Lee SJ, Park W. Effects of tocilizumab therapy on serum interleukin-33 and interleukin-6 levels in patients with rheumatoid arthritis. Arch Rheumatol. 2018;33:389–394. doi: 10.5606/ArchRheumatol.2018.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Fu B, Zheng X. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. bioRxiv. 2020 doi: 10.1101/2020.02.12.945576. published online February 20. (preprint). [DOI] [Google Scholar]
- 42.Riegler LL, Jones GP, Lee DW. Current approaches in the grading and management of cytokine release syndrome after chimeric antigen receptor T-cell therapy. Ther Clin Risk Manag. 2019;15:323–335. doi: 10.2147/TCRM.S150524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimabukuro-Vornhagen A, Gödel P, Subklewe M. Cytokine release syndrome. J Immunother Cancer. 2018;6:56. doi: 10.1186/s40425-018-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka T, Narazaki M, Kishimoto T. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy. 2016;8:959–970. doi: 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- 45.Xu X, Han M, Li T. Effective treatment of severe COVID-19 patients with tocilizumab. PNAS. 2020 doi: 10.1073/pnas.2005615117. published online April 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shakoory B, Carcillo JA, Chatham WW. Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of macrophage activation syndrome: reanalysis of a prior phase III trial. Crit Care Med. 2016;44:275–281. doi: 10.1097/CCM.0000000000001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eloseily EM, Weiser P, Crayne CB. Benefit of anakinra in treating pediatric secondary hemophagocytic lymphohistiocytosis. Arthritis Rheumatol. 2020;72:326–334. doi: 10.1002/art.41103. [DOI] [PubMed] [Google Scholar]
- 48.Taylor PC. Clinical efficacy of launched JAK inhibitors in rheumatoid arthritis. Rheumatology (Oxford) 2019;58(suppl 1):i17–i26. doi: 10.1093/rheumatology/key225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Witte T. JAK inhibitors in rheumatology. Dtsch Med Wochenschr. 2019;144:748–752. doi: 10.1055/a-0652-2731. [DOI] [PubMed] [Google Scholar]
- 50.Richardson P, Griffin I, Tucker C. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segler MHS, Preuss M, Waller MP. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555:604–610. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- 52.Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv. 2020 doi: 10.1101/2020.01.26.919985. published online Jan 26. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Broglie L, Pommert L, Rao S. Ruxolitinib for treatment of refractory hemophagocytic lymphohistiocytosis. Blood Adv. 2017;1:1533–1536. doi: 10.1182/bloodadvances.2017007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sin JH, Zangardi ML. Ruxolitinib for secondary hemophagocytic lymphohistiocytosis: first case report. Hematol Oncol Stem Cell Ther. 2019;12:166–170. doi: 10.1016/j.hemonc.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Ahmed A, Merrill SA, Alsawah F. Ruxolitinib in adult patients with secondary haemophagocytic lymphohistiocytosis: an open-label, single-centre, pilot trial. Lancet Haematol. 2019;6:e630–e637. doi: 10.1016/S2352-3026(19)30156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das R, Guan P, Sprague L. Janus kinase inhibition lessens inflammation and ameliorates disease in murine models of hemophagocytic lymphohistiocytosis. Blood. 2016;127:1666–1675. doi: 10.1182/blood-2015-12-684399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plantone D, Koudriavtseva T. Current and future use of chloroquine and hydroxychloroquine in infectious, immune, neoplastic, and neurological diseases: a mini-review. Clin Drug Investig. 2018;38:653–671. doi: 10.1007/s40261-018-0656-y. [DOI] [PubMed] [Google Scholar]
- 58.Vincent MJ, Bergeron E, Benjannet S. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang M, Cao R, Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shippey EA, Wagler VD, Collamer AN. Hydroxychloroquine: an old drug with new relevance. Cleve Clin J Med. 2018;85:459–467. doi: 10.3949/ccjm.85a.17034. [DOI] [PubMed] [Google Scholar]
- 61.Ruiz-Irastorza G, Olivares N, Ruiz-Arruza I, Martinez-Berriotxoa A, Egurbide MV, Aguirre C. Predictors of major infections in systemic lupus erythematosus. Arthritis Res Ther. 2009;11:R109. doi: 10.1186/ar2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sisó A, Ramos-Casals M, Bové A. Previous antimalarial therapy in patients diagnosed with lupus nephritis: influence on outcomes and survival. Lupus. 2008;17:281–288. doi: 10.1177/0961203307086503. [DOI] [PubMed] [Google Scholar]
- 63.Sperber K, Louie M, Kraus T. Hydroxychloroquine treatment of patients with human immunodeficiency virus type 1. Clin Ther. 1995;17:622–636. doi: 10.1016/0149-2918(95)80039-5. [DOI] [PubMed] [Google Scholar]
- 64.Gautret P, Lagier JC, Parola P. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105949. published online March 20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Chen Z, Hu J, Zhang Z. Efficacy of hydroxychloroquine in patients with COVID-19: results of a randomized clinical trial. MedRxiv. 2020 doi: 10.1101/2020.03.22.20040758. published online April 10. (preprint). [DOI] [Google Scholar]
- 66.Tang W, Cao Z, Han M. Hydroxychloroquine in patients mainly with mild to moderate COVID-19: an open-label, randomized, controlled trial. MedRxiv. 2020 doi: 10.1101/2020.04.10.20060558. published online May 7. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mahevas M, Tran V-T, Roumier M. No evidence of clinical efficacy of hydroxychloroquine in patients hospitalized for COVID-19 infection with oxygen requirement: results of a study using routinely collected data to emulate a target trial. MedRxiv. 2020 doi: 10.1101/2020.04.10.20060699. published online April 14. (preprint). [DOI] [Google Scholar]
- 68.Geleris J, Sun Y, Platt J. Observational study of hydroxychloroquine in hospitalized patients with COVID-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2012410. published online May 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Magagnoli J, Narendran S, Pereira F. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with Covid-19. MedRxiv. 2020 doi: 10.1101/2020.04.16.20065920. published online April 23. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fihn SD, Perencevich E, Bradley SM. Caution needed on the use of chloroquine and hydroxychloroquine for coronavirus disease 2019. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.9035. [DOI] [PubMed] [Google Scholar]
- 71.Borba MGS, Val FFA, Sampaio VS. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hardy RS, Raza K, Cooper MS. Therapeutic glucocorticoids: mechanisms of actions in rheumatic diseases. Nat Rev Rheumatol. 2020;16:133–144. doi: 10.1038/s41584-020-0371-y. [DOI] [PubMed] [Google Scholar]
- 73.Meduri GU, Headley AS, Golden E. Effect of prolonged methylprednisolone therapy in unresolving acute respiratory distress syndrome: a randomized controlled trial. JAMA. 1998;280:159–165. doi: 10.1001/jama.280.2.159. [DOI] [PubMed] [Google Scholar]
- 74.Meduri GU, Golden E, Freire AX. Methylprednisolone infusion in early severe ARDS: results of a randomized controlled trial. Chest. 2007;131:954–963. doi: 10.1378/chest.06-2100. [DOI] [PubMed] [Google Scholar]
- 75.Steinberg KP, Hudson LD, Goodman RB. Efficacy and safety of corticosteroids for persistent acute respiratory distress syndrome. N Engl J Med. 2006;354:1671–1684. doi: 10.1056/NEJMoa051693. [DOI] [PubMed] [Google Scholar]
- 76.Tang NL, Chan PK, Wong CK. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin Chem. 2005;51:2333–2340. doi: 10.1373/clinchem.2005.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arabi YM, Balkhy HH, Hayden FG. Middle East Respiratory Syndrome. N Engl J Med. 2017;376:584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.So LK, Lau AC, Yam LY. Development of a standard treatment protocol for severe acute respiratory syndrome. Lancet. 2003;361:1615–1617. doi: 10.1016/S0140-6736(03)13265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arabi YM, Mandourah Y, Al-Hameed F. Corticosteroid therapy for critically ill patients with Middle East Respiratory Syndrome. Am J Respir Crit Care Med. 2018;197:757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 80.Xu X, Shen J, Mall JW. In vitro and in vivo antitumor activity of a novel immunomodulatory drug, leflunomide: mechanisms of action. Biochem Pharmacol. 1999;58:1405–1413. doi: 10.1016/s0006-2952(99)00228-2. [DOI] [PubMed] [Google Scholar]
- 81.Siemasko K, Chong AS, Jack HM, Gong H, Williams JW, Finnegan A. Inhibition of JAK3 and STAT6 tyrosine phosphorylation by the immunosuppressive drug leflunomide leads to a block in IgG1 production. J Immunol. 1998;160:1581–1588. [PubMed] [Google Scholar]
- 82.Manna SK, Mukhopadhyay A, Aggarwal BB. Leflunomide suppresses TNF-induced cellular responses: effects on NF-kappa B, activator protein-1, c-Jun N-terminal protein kinase, and apoptosis. J Immunol. 2000;165:5962–5969. doi: 10.4049/jimmunol.165.10.5962. [DOI] [PubMed] [Google Scholar]
- 83.Kremer JM. Methotrexate and leflunomide: biochemical basis for combination therapy in the treatment of rheumatoid arthritis. Semin Arthritis Rheum. 1999;29:14–26. doi: 10.1016/s0049-0172(99)80034-1. [DOI] [PubMed] [Google Scholar]
- 84.Olsen NJ, Stein CM. New drugs for rheumatoid arthritis. N Engl J Med. 2004;350:2167–2179. doi: 10.1056/NEJMra032906. [DOI] [PubMed] [Google Scholar]
- 85.Leca N, Muczynski KA, Jefferson JA. Higher levels of leflunomide are associated with hemolysis and are not superior to lower levels for BK virus clearance in renal transplant patients. Clin J Am Soc Nephrol. 2008;3:829–835. doi: 10.2215/CJN.03930907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chacko B, John GT. Leflunomide for cytomegalovirus: bench to bedside. Transpl Infect Die. 2012;14:111–120. doi: 10.1111/j.1399-3062.2011.00682.x. [DOI] [PubMed] [Google Scholar]
- 87.John GT, Manivannan J, Chandy S. A prospective evaluation of leflunomide therapy for cytomegalovirus disease in renal transplant recipients. Transplant Proc. 2005;37:4303–4305. doi: 10.1016/j.transproceed.2005.10.116. [DOI] [PubMed] [Google Scholar]
- 88.Teschner S, Burst V. Leflunomide: a drug with a potential beyond rheumatology. Immunotherapy. 2010;2:637–650. doi: 10.2217/imt.10.52. [DOI] [PubMed] [Google Scholar]
- 89.Williams JW, Javaid B, Kadambi PV. Leflunomide for polyomavirus type BK nephropathy. N Engl J Med. 2005;352:1157–1158. doi: 10.1056/NEJM200503173521125. [DOI] [PubMed] [Google Scholar]
- 90.Nguyen L, McClellan RB, Chaudhuri A. Conversion from tacrolimus/mycophenolic acid to tacrolimus/leflunomide to treat cutaneous warts in a series of four pediatric renal allograft recipients. Transplantation. 2012;94:450–455. doi: 10.1097/TP.0b013e318264351e. [DOI] [PubMed] [Google Scholar]
- 91.Chikura B, Lane S, Dawson JK. Clinical expression of leflunomide-induced pneumonitis. Rheumatology (Oxford) 2009;48:1065–1068. doi: 10.1093/rheumatology/kep050. [DOI] [PubMed] [Google Scholar]
- 92.Sawada T, Inokuma S, Sato T. Leflunomide-induced interstitial lung disease: prevalence and risk factors in Japanese patients with rheumatoid arthritis. Rheumatology (Oxford) 2009;48:1069–1072. doi: 10.1093/rheumatology/kep052. [DOI] [PubMed] [Google Scholar]
- 93.Zhu H, Shi X, Ju D, Huang H, Wei W, Dong X. Anti-inflammatory effect of thalidomide on H1N1 influenza virus-induced pulmonary injury in mice. Inflammation. 2014;37:2091–2098. doi: 10.1007/s10753-014-9943-9. [DOI] [PubMed] [Google Scholar]
- 94.Keddie S, Bharambe V, Jayakumar A. Clinical perspectives into the use of thalidomide for central nervous system tuberculosis. Eur J Neurol. 2018;25:1345–1351. doi: 10.1111/ene.13732. [DOI] [PubMed] [Google Scholar]
- 95.Wen H, Ma H, Cai Q. Recurrent ECSIT mutation encoding V140A triggers hyperinflammation and promotes hemophagocytic syndrome in extranodal NK/T cell lymphoma. Nat Med. 2018;24:154–164. doi: 10.1038/nm.4456. [DOI] [PubMed] [Google Scholar]
- 96.Haraf R, Flora AS, Assaly R. Thalidomide as a cough suppressant in idiopathic pulmonary fibrosis. Am J Ther. 2018;25:e687–e688. doi: 10.1097/MJT.0000000000000695. [DOI] [PubMed] [Google Scholar]
- 97.Chen C, Qi F, Shi K. Thalidomide combined with low-dose glucocorticoid in the treatment of COVID-19 pneumonia. Preprints. 2020 doi: 10.1101/202002.0395.v1. published online February 26. (preprint). [DOI] [Google Scholar]
- 98.Chan MC, Kuok DI, Leung CY. Human mesenchymal stromal cells reduce influenza A H5N1-associated acute lung injury in vitro and in vivo. Proc Natl Acad Sci USA. 2016;113:3621–3626. doi: 10.1073/pnas.1601911113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang BX, El Farran CA, Guo HC. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simonson OE, Mougiakakos D, Heldring N. In vivo effects of mesenchymal stromal cells in two patients with severe acute respiratory distress syndrome. Stem Cells Transl Med. 2015;4:1199–1213. doi: 10.5966/sctm.2015-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng G, Huang L, Tong H. Treatment of acute respiratory distress syndrome with allogeneic adipose-derived mesenchymal stem cells: a randomized, placebo-controlled pilot study. Respir Res. 2014;15:39. doi: 10.1186/1465-9921-15-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Winkler AM, Koepsell SA. The use of convalescent plasma to treat emerging infectious diseases: focus on Ebola virus disease. Curr Opin Hematol. 2015;22:521–526. doi: 10.1097/MOH.0000000000000191. [DOI] [PubMed] [Google Scholar]
- 103.Behrens EM, Koretzky GA. Review: cytokine storm syndrome: looking toward the precision medicine era. Arthritis Rheumatol. 2017;69:1135–1143. doi: 10.1002/art.40071. [DOI] [PubMed] [Google Scholar]
- 104.White NJ, Miller KD, Churchill FC. Chloroquine treatment of severe malaria in children. Pharmacokinetics, toxicity, and new dosage recommendations. N Engl J Med. 1988;319:1493–1500. doi: 10.1056/NEJM198812083192301. [DOI] [PubMed] [Google Scholar]
- 105.White NJ, Watt G, Bergqvist Y, Njelesani EK. Parenteral chloroquine for treating falciparum malaria. J Infect Dis. 1987;155:192–201. doi: 10.1093/infdis/155.2.192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


