We read the Article by Lescure and colleagues1 with great interest. During the ongoing coronavirus disease 2019 (COVID-19) pandemic, monitoring patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using viral kinetics or viral loads in various sample types by real-time RT-PCR has become essential. However, understanding whether the RT-PCR test results are interpreted as quantitative, qualitative, or semi-quantitative is important. Since the cycle threshold (Ct) values from RT-PCR can be affected by batch effect, variations among different runs need to be closely monitored by laboratories—especially for quality control in quantitative RT-PCR. Unfortunately, several papers on COVID-19 use the naive Ct values from qualitative RT-PCR as a quantitation unit or use the ΔCt values with incorrect quantitation unit.2, 3 Quantitative RT-PCR is entirely different from qualitative RT-PCR. Ct value itself cannot be directly interpreted as viral load without a standard curve using reference materials. Thorough evaluation of the reliability and robustness of the standard curve is the key to accurately quantify the expected viral copy number.
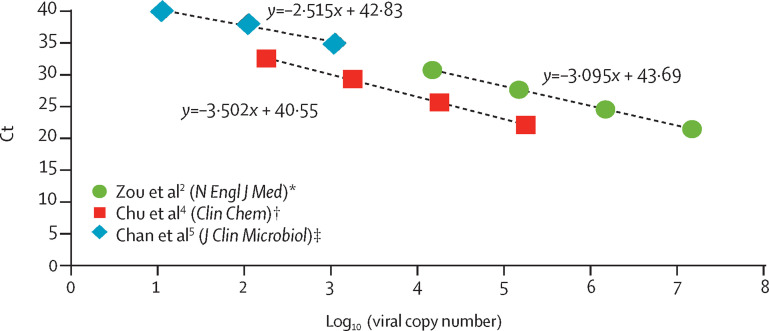
There is wide heterogeneity and inconsistency of the standard curves calculated from studies that provided Ct values from serial dilution samples and the estimated viral loads (figure ).2, 4, 5 An appropriate standard curve with adequate limit of detection is required for viral load quantification to correctly track the viral titre kinetics. A two-step approach using qualitative RT-PCR (for detection) and quantitative RT-PCR (for viral load quantification) is highly recommended for studies focusing on viral loads, as clearly presented by Lescure and colleagues.1 Furthermore, using appropriate quantification units according to different sample types—ie, copies per 1000 cells (for respiratory samples), copies per mL (for plasma), and copies per g (for stool)—should be followed by clinicians, as outstandingly shown by Lescure and colleagues.1
Figure.
Standard curves drawn from papers providing serial dilution factors and corresponding Ct in patient samples
All Ct values were derived from clinical samples targeting RdRp/Orf1b sequence of SARS-CoV-2. Ct=cycle threshold. RdRp=RNA-dependent RNA polymerase. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. *Ct values provided in the figure 1 legend were used. †Ct values of Orf1b in patient 2 provided in table 1 were used. ‡Ct values of RdRp in test 2 of clinical specimen were used.
When interpreting the results of SARS-CoV-2 RT-PCR, the validity of the standard curve using reference materials or in-house plasmid controls with known viral copy numbers should be confirmed first to interpret Ct values as viral loads. In conclusion, precautions are needed when interpreting the Ct values of SARS-CoV-2 RT-PCR results shown in COVID-19 publications to avoid misunderstanding of viral load kinetics for comparison across different studies.
Acknowledgments
We declare no competing interests.
MSH and JHR contributed to study design, data collection, data analysis, data interpretation, literature search, and writing of the Correspondence. J-HB and YC contributed to data collection, data analysis, and data interpretation. All authors reviewed and approved the final version of the Correspondence.
References
- 1.Lescure FX, Bouadma L, Nguyen D, et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30200-0. published online March 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen C, Wang Z, Zhao F, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020 doi: 10.1001/jama.2020.4783. published online March 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JF, Yip CC, To KK, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58:e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]