Abstract
Purpose
In hepatobiliary surgery, preoperative three-dimensional reconstruction based on CT or MRI can be provided externally or by local, semi-automatic software. We analyzed the time expense and quality of external versus local three-dimensional reconstructions.
Methods
Three first-year residents reconstructed data from 20 patients with liver pathologies using a local, semi-automatic, server-based program. Initially, five randomly selected patient datasets were segmented, with the visualization of an established external company available for comparison at all times (learning phase). The other fifteen cases were compared with the external datasets after completing local reconstruction (control phase). Total time expense/case and for specific manual and semi-automated reconstruction steps were recorded. Segmentation quality was analyzed by testing the equivalence for liver and tumor volumes, portal vein sectors, and hepatic vein territories.
Results
The median total reconstruction time was reduced from 2.5 h (learning phase) to 1.5 h (control phase) (− 42%; p < 0.001). Comparing the total and detailed liver volumes (sectors and territories) as well as the tumor volumes in the control phase equivalence was proven. In addition, a highly significant correlation between the external and local analysis was obtained over all analyzed segments with a very high ICC (median [IQR]: 0.98 [0.97; 0.99]; p < 0.01).
Conclusion
Local, semi-automatic reconstruction performed by inexperienced residents was feasible with an expert level time expense and the quality of the three-dimensional images was comparable with those from an external provider.
Keywords: Liver surgery, 3D segmentation, Surgical planning, Virtual surgery, Surgical education
Introduction
During hepatobiliary surgery, knowledge of the tumor’s localization, relationship to vascular structures, and the functional liver remnant after resection are essential [1, 2]. For complex resections such as extended hepatectomy or major vessel reconstruction, where the limits of resectability are approached, the visualization of critical structures that must be preserved can be demanding, especially since only 29% of patients have standard liver anatomy [3]. In laparoscopic liver resection, knowledge of individual variability is of even higher importance. The imagination of the intraoperative situation from preoperative two-dimensional imaging can be quite difficult in these situations.
Three-dimensional liver reconstruction is a useful tool for visualizing critical structures for surgical planning and intraoperative orientation [3, 4]. Furthermore, it is the basis for three-dimensional printing and computer-assisted navigation [5, 6].
Usually, established external providers create such complete segmentations [7]. However, there is an increasing availability of local server-based segmentation programs. The aim of this study was to evaluate the time expense and the quality of local reconstruction, performed by inexperienced residents, with the external reconstructions.
Methods
Study design
We included 20 consecutive patients with an external preoperative three-dimensional reconstruction available independently from the current study. The same established provider (MeVis Medical Solutions AG, Bremen, Germany) performed all external reconstructions. The three-dimensional images were based on computed tomography (CT) scans. All patients had liver tumors that were resected using an open or laparoscopic approach.
For local reconstruction, a semi-automatic, server-based program (Synapse 3D, FUJIFILM Europe GmbH, Düsseldorf) was used. Three first-year residents of the surgical department, who had no expert knowledge in the fields of liver surgery, radiology, or informatics prior to the study, performed these reconstructions. In preparation, the residents completed a half-day training session at the radiology department’s CT unit under the direction of a radiological consultant to train export functions and the general handling of two-dimensional datasets. The distributor provided a one-time, 2-h training session on the planning software tool.
Patient identifiers (1–20) were assigned randomly. The first five patients’ datasets were reconstructed, with the external visualization available for comparison at all times (learning phase). The other cases (n = 15) were compared with the corresponding external reconstruction after the three-dimensional image was completed (control phase).
External reconstruction
MeVis Medical Solutions is an internationally recognized research and development center for medical image computing closely collaborating with Frauenhofer MEVIS. Several studies have proven the accuracy of the visualization by this provider [8–11].
The CT datasets for each patient were exported as DICOM files, anonymized, and sent to the company. Scans had to be delivered with a quality of 0.8- to 1.5-mm slice increment and with contrasted portal and hepatic veins that were at least 30 Hounsfield units above liver parenchyma. Regular priority visualization takes 3 days, and data were presented as a three-dimensional PDF file that displayed calculated volumes for the liver parenchyma, tumor, portal vein (PV), and hepatic vein (HV) systems.
Local reconstruction
The authors tested different semi-automatic and completely manual software tools for three-dimensional liver reconstruction prior to this study. The mentioned program was acquired for the department independently from the current study.
The patients’ exported DICOM files were uploaded to a local server. The program automatically extracts the raw circumference of the liver parenchyma and the vessels (PV and HV), which needed to be checked and corrected manually. The circumference of the inferior vena cava (IVC) must be traced manually in several slices, and the program interpolates in between. For tumor reconstruction, the largest diameter was marked by hand. Before the three-dimensional model was created, the direction of blood flow in the PV and HV was defined by setting a root point. The feeding vessels were selected manually for the division of the liver parenchyma into different segments and sectors. The program then calculated the volumes automatically.
Segmentation time analysis
The time of reconstruction was recorded for each individual case (total reconstruction time). Furthermore, the duration of each step in visualizing the liver was measured. This included manual editing of the automatically extracted liver parenchyma, IVC, PV, HV, and tumor (step 1, semi-automatic). In addition, the time needed to complete individual segment and territory analysis with optimization of blood vessel visualization was recorded (step 2, fully manual).
Liver volume analysis
We compared the volumes of the liver, tumor, segments (PV), sectors (PV), and territories (HV) of the local and external three-dimensional analyses to objectify one aspect of the quality of the three-dimensional reconstruction.
Statistical analysis
For case number planning, the SAS software (SAS Institute GmbH, Heidelberg, Germany) was used. A minimum case number of 13 was calculated for the control phase applying a lower and upper equivalence bound of 7.5% and a power of 80% based on available literature [12, 13]. Further statistical analyses were performed with IBM SPSS Statistics 23 (IBM, Armonk, NY, USA). The standardized difference was calculated using the following equation:
For equivalence testing, the 95% confidence level of the one sample t test was calculated. A deviation of more than 0.075 was set as equivalence limit. The intraclass correlation coefficient (ICC) was used to correlate volumes. Nonparametric Mann-Whitney U test was performed to analyze time differences. P values were adjusted for multiple testing using the Benjamini and Hochberg (FDR) method. P values < 0.05 were considered statistically significant. Data are presented as the median and interquartile range (IQR).
Results
Patient’s characteristics are displayed in Table 1. The CT data used were from patients with various resectable pathologies and represent very different types of hepatectomy.
Table 1.
Characteristics of selected patients (n = 20)
| Characteristics | Value |
|---|---|
| Age (years, mean (range)) | 62 (22–79) |
| Gender | |
| Male | 13 (65%) |
| Female | 7 (35%) |
| Underlying disease | |
| Colorectal liver metastases | 10 (50%) |
| Hepatocellular carcinoma | 7 (35%) |
| Focal nodular hyperplasia | 1 (5%) |
| Breast cancer metastases | 1 (5%) |
| Echinococcus multilocularis | 1 (5%) |
| Operation type | |
| Laparoscopic | 8 (40%) |
| Open | 12 (60%) |
| Type of resection | |
| Minor resection | |
| Non-anatomical resection | 4 (20%) |
| Segmentectomy | 10 (50%) |
| Bisegmentectomy | 2 (10%) |
| Major resection | |
| Left hemihepatectomy | 1 (5%) |
| Extended left hemihepatectomy | 3 (15%) |
Segmentation time analysis
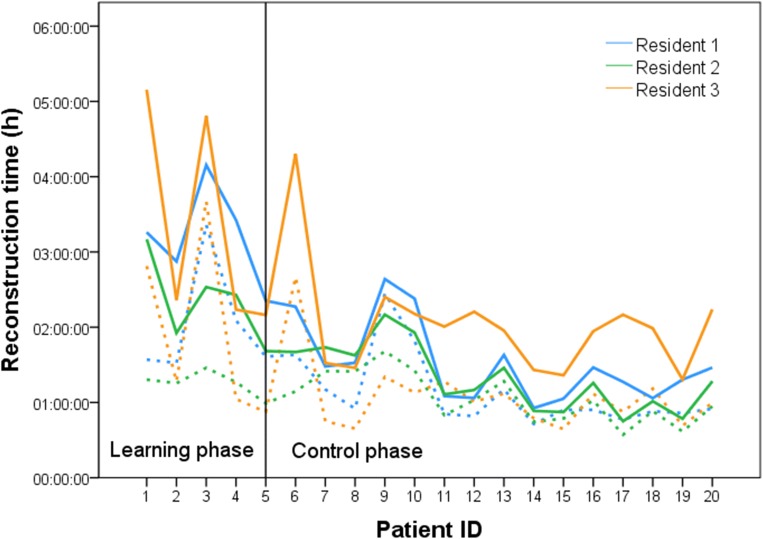
Figure 1 displays the time for step 1 and the total reconstruction time required for editing the data of all patients for each resident. After an initial median total reconstruction time of 2.5 h (learning phase), the duration was reduced to a median level of 1.5 h (control phase) (− 42%; P < 0.001). The median time and IQR for the different reconstruction steps in total and for each resident are shown in Table 2. Between learning and control phase timesavings up to 81% (resident 2) could be achieved for reconstruction step 2.
Fig. 1.
Reconstruction time of the CT scans from all patients. Solid lines represent total reconstruction time. Dashed lines represent the time for reconstruction step 1
Table 2.
Reconstruction time
| Learning phase | Control phase | Difference | ||
|---|---|---|---|---|
| Time (min) | Time (min) | Time (min) | ||
| Median (IQR) | Median (IQR) | Median (%) | P1 | |
| Resident 1 | ||||
| Reconstruction step 1 | 97 (93;163) | 54 (51;70) | 43 (44) | 0.019* |
| Reconstruction step 2 | 80 (46;91) | 27 (13;34) | 53 (66) | 0.001* |
| Total reconstruction time | 196 (157;227) | 88 (64;158) | 108 (55) | 0.002* |
| Resident 2 | ||||
| Reconstruction step 1 | 75 (62;77) | 62 (47;85) | 13 (17) | 0.407 |
| Reconstruction step 2 | 70 (41;100) | 13 (9;19) | 57 (81) | 0.001* |
| Total reconstruction time | 146 (108;171) | 76 (53;100) | 70 (48) | 0.003* |
| Resident 3 | ||||
| Reconstruction step 1 | 68 (58;123) | 61 (45;68) | 7 (10) | 0.182 |
| Reconstruction step 2 | 77 (68;181) | 50 (44;74) | 27 (35) | 0.035* |
| Total reconstruction time | 141 (132;299) | 119 (88;132) | 22 (15) | 0.023* |
| Total | ||||
| Reconstruction step 1 | 77 (64;97) | 57 (48;71) | 20 (26) | 0.002* |
| Reconstruction step 2 | 77 (48;102) | 29 (13;45) | 48 (62) | < 0.001* |
| Total reconstruction time | 152 (134;206) | 88 (68;120) | 64 (42) | < 0.001* |
Learning phase: external reconstruction available for comparison; control phase: external reconstruction not available for comparison. IQR, interquartile range
1Adjusted for multiple testing
*Significant
Liver volume analysis
Comparing the total and detailed liver volumes during the control phase, a high intraclass correlation coefficient was achieved over all analyzed volumes (median ICC [IQR]: 0.98 [0.97; 0.99]; p < 0.01) (Table 3). Equivalence was proven for all three residents in the control phase for tumor volume, total liver volume, left and right portal vein, and the lateral and medial sectors. Liver segment and right sector analysis revealed a higher variability and did thus reach different levels of equivalence for the three residents (Table 3).
Table 3.
Quality assessment of the control phase (n = 15): comparison of liver volume results
| Correlation | One sample t test | ||||
|---|---|---|---|---|---|
| ICC | P1 | Resident 1 | Resident 2 | Resident 3 | |
| 95% CI | 95% CI | 95% CI | |||
| Liver | 0.998 | < 0.01* | [− 0.022;− 0.008] | [− 0.030;0.012] | [− 0.017;− 0.004] |
| Tumor | 0.998 | < 0.01* | [− 0.019;0.060] | [− 0.062;0.013] | [− 0.061;0.026] |
| Left PV | 0.994 | < 0.01* | [− 0.039;− 0.003] | [− 0.054;0.058] | [− 0.050;0.028] |
| Right PV | 0.990 | < 0.01* | [− 0.011;0.036] | [− 0.017;0.062] | [− 0.025;0.013] |
| Lateral PV | 0.997 | < 0.01* | [− 0.046;0.014] | [− 0.075;0.043] | [− 0.040;0.030] |
| Medial PV | 0.992 | < 0.01* | [− 0.058;0.039] | [− 0.069;0.060] | [− 0.096;0.025] |
| Anterior PV | 0.972 | < 0.01* | [− 0.001;0.069] | [− 0.068;0.137] | [− 0.047;0.030] |
| Posterior PV | 0.981 | < 0.01* | [− 0.084;0.042] | [− 0.106; 0.053] | [− 0.045;0.033] |
| Segment 1 | 0.976 | < 0.01* | [− 0.134;0.146] | [− 0.303;− 0.032] | [− 0.194;0.127] |
| Segment 2 | 0.997 | < 0.01* | [− 0.146;− 0.013] | [− 0.255;− 0.021] | [− 0.053;0.010] |
| Segment 3 | 0.988 | < 0.01* | [− 0.143;0.097] | [− 0.040;0.141] | [− 0.046;0.060] |
| Segment 4a | 0.933 | < 0.01* | [− 0.215;0.029] | [− 0.034;0.338] | [− 0.108;0.043] |
| Segment 4b | 0.922 | < 0.01* | [− 0.335;0.153] | [− 0.829;0.002] | [− 0.076;0.081] |
| Segment 5 | 0.937 | < 0.01* | [− 0.039;0.131] | [− 0.424;0.400] | [− 0.197;0.116] |
| Segment 6 | 0.958 | < 0.01* | [− 0.174;0.048] | [− 0.272;0.129] | [− 0.061;0.074] |
| Segment 7 | 0.976 | < 0.01* | [− 0.100;0.049] | [− 0.411;0.169] | [− 0.063;0.064] |
| Segment 8 | 0.976 | < 0.01* | [− 0.031;0.048] | [− 0.128;0.057] | [− 0.109;0.002] |
| Left HV | 0.992 | < 0.01* | [0.065;0.014] | [− 0.075;0.063] | [− 0.048;0.012] |
| Middle HV | 0.976 | < 0.01* | [− 0.012;0.041] | [− 0.014;0.025] | [− 0.035;0.010] |
| Right HV | 0.995 | < 0.01* | [− 0.045;0.011] | [− 0.040;0.058] | [− 0.032;0.020] |
CI, confidence interval. Values in italics are proven equivalence
1Adjusted for multiple testing
*Significant
Liver segmentation
Due to previous resections (right hemihepatectomy) in one case segments 5 to 8 could not be reconstructed. In three cases, segment vessels were displaced by the tumor (twice segment 1, once segment 4a, and three times segment 4b). Poor vascular contrast was responsible for not identifying all segments in three cases: twice segment 1, once segment 4b, and once segment 5. In addition, resident 1 could not identify the feeding vessel once for segment 2 and for segment 5. The later was also not identified by resident 2. In the two additional cases with the local software, the tumor was closely adjacent to the missing segment.
Discussion
The benefit of three-dimensional visualizations, especially for preoperative planning of complex and extended liver resections, has been analyzed previously [4, 14]. This is the first study describing the feasibility and quality of a semi-automatic, local, server-based program for three-dimensional liver reconstruction under the premise that first-year residents performed all reconstructions.
One aspect of this study was to investigate the residents’ time expense in comparison with highly specialized experts for generating the reconstruction. The median time to complete the reconstruction of the patients’ CT scan data was 2,5 h in the learning phase, but was reduced to 1.5 h during the control phase. This is comparable with the work of Endo et al. [1], with a similar total reconstruction time of about 2 h. Two hours is also considered an expert level by Lang et al. [4]. Sboarina et al. stated that with improved skill, the time for liver and vessel segmentation can be reduced by 30–40% [15]. This can be confirmed in our study, when comparing the learning phase to the control phase. The editing time needed for step 1, step 2, and total reconstruction was reduced by 26%, 62%, and 42% respectively. The greatest time saving could be shown for step 2 (fully manual), representing a steep learning curve for all residents. Step 1 (semi-automatic) shows the least time reduction and variations in total reconstruction time are highly dependent on this part of the process. In three patients (3, 9, and 10), the time required for editing during step 1 was longer for all residents due to a necessary alteration in the extracted liver anatomy. In patient 3, the liver parenchyma was not automatically extracted, resulting in the manual editing of the liver data and a longer total reconstruction time, respectively. Furthermore, intraparenchymal HV was traced as the PV, making manual alterations necessary. The reason may be that the patient had steatosis hepatis, which is known to attenuate Hounsfield units [16, 17]. In patient 9, a large tumor influenced the contrasting of the PV, HV, and IVC. In addition, an adjacent lymph node was registered as liver parenchyma. Patient 10 had multiple intrahepatic cystic lesions next to breast cancer metastases, which required extensive manual alteration of the liver data (Fig. 2). To sum up, we could demonstrate that residents can perform three-dimensional liver reconstruction within a comparable time expense to experts after a reasonable time expense for training.
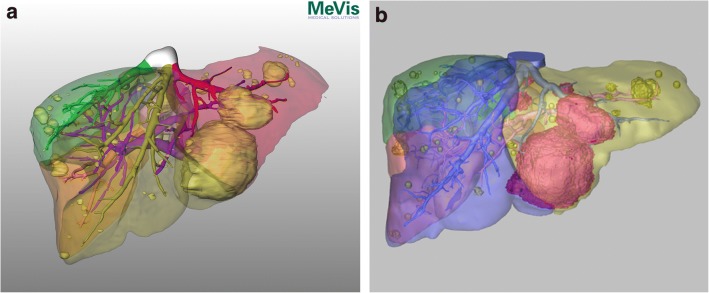
Fig. 2.
Liver reconstruction (Pat. ID 10). External reconstruction (a): Three-dimensional visualization of a patient’s liver with five breast cancer metastases and multiple intrahepatic cystic lesions (yellow). Portal vein (pink) and hepatic vein (color coded) are visualized. In this figure, hepatic vein territories including an inferior right hepatic vein are color coded.
Image courtesy of © 2019 MeVis Medical Solutions AG, Bremen, Germany.
Local reconstruction (b): Three-dimensional visualization of a patient’s liver with five breast cancer metastases (purple) and multiple intrahepatic cystic lesions (yellow). Portal vein (pink) and hepatic vein (blue) are visualized. In this figure, hepatic vein territories including an inferior right hepatic vein are color coded.
Furthermore, we focused on the quality of the liver reconstruction. This is in contrast to the previously cited studies, which measured the general time expense without analyzing the quality of the reconstruction. A very high ICC above 0.9 for all analyzed volumes and a proven equivalence for all sectors and territories demonstrated a good quality of liver segmentation images in the current study. The variance in segmental volumes can be explained by the naturally smooth transitions between segments in general and very thin vessels at the end of the branches [18]. In areas with very small vessels that are difficult to reconstruct especially segment 1 and segment 4a/b, a higher variation was observed. The selection of the feeding vessel and the associated liver volume can also be subjected to human errors and subjective interpretation [18]. Especially for the right hepatic lobe, the assignment of smaller vessels at the borders of the segments can be demanding. On the other hand, the calculation of the parenchyma volume to the feeding vessel is depending on the underlying software and can therefore variate between the external and local reconstructions. The clinical relevance of this variation is questionable.
A limitation of this study is the fact that only surgical residents from one center used the program. Further studies should investigate if quality and time expenses of three-dimensional reconstruction vary by participants with different medical specialties and from different centers. The comparison between several local software providers may also be of interest.
From an economical point of view, annual service costs aside from the initial acquisition price must be considered when using a local segmentation program. In our setting, 45–50 cases are needed to amortize the initial costs. Annual costs will be equalized at about 10 reconstructions per year. In addition, the calculations must include around 2–3 days of a residents working time for the training phase and around 2 h per case. In our opinion, this results in approximately one additional case for the initial costs amortization and one additional case per year to create financial compensation. However, the benefits of a profound knowledge of patients’ individual liver anatomy by performing the reconstruction locally are difficult to put into numbers. The general benefit of a local segmentation program is that the reconstruction is available faster than the usual 3-day time expense for external visualization that is invoiced per case. Thus, a center must evaluate whether the investment of a local reconstruction program is suitable and economic according to its caseload for complex liver surgery.
Conclusion
In conclusion, this study reveals that local, semi-automatic reconstruction is feasible and comparable with an external provider regarding the total and detailed volumes when performed by an inexperienced resident after a reasonable amount of training. Variation in the time expense for reconstructions is currently due to necessary manual corrections of the automated part of the analysis. The software may be optimized to offer different choices in uncertain cases that may improve the automated reconstruction process in the future (deep learning).
Funding information
Open access funding provided by Projekt DEAL. Intramural funding from the Medical Center of the Johannes Gutenberg University of Mainz provided financial support for the project. FUJIFILM Europe GmbH provided no funding or financial benefits.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Markus Paschold and Florentine Huettl shared first authorship.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Endo K, Sata N, Ishiguro Y, Miki A, Sasanuma H, Sakuma Y, Shimizu A, Hyodo M, Lefor A, Yasuda Y. A patient-specific surgical simulator using preoperative imaging data: an interactive simulator using a three-dimensional tactile mouse. J Comput Surg. 2014;1:1–8. doi: 10.1186/s40244-014-0010-5. [DOI] [Google Scholar]
- 2.Lang H, Radtke A, Liu C, Fruhauf NR, Peitgen HO, Broelsch CE. Extended left hepatectomy--modified operation planning based on three-dimensional visualization of liver anatomy. Langenbeck’s Arch Surg. 2004;389(4):306–310. doi: 10.1007/s00423-003-0441-z. [DOI] [PubMed] [Google Scholar]
- 3.Radtke A, Nadalin S, Sotiropoulos GC, Molmenti EP, Schroeder T, Valentin-Gamazo C, Lang H, Bockhorn M, Peitgen HO, Broelsch CE, Malago M. Computer-assisted operative planning in adult living donor liver transplantation: a new way to resolve the dilemma of the middle hepatic vein. World J Surg. 2007;31(1):175–185. doi: 10.1007/s00268-005-0718-1. [DOI] [PubMed] [Google Scholar]
- 4.Lang H, Radtke A, Hindennach M, Schroeder T, Fruhauf NR, Malago M, Bourquain H, Peitgen HO, Oldhafer KJ, Broelsch CE. Impact of virtual tumor resection and computer-assisted risk analysis on operation planning and intraoperative strategy in major hepatic resection. Arch Surg. 2005;140(7):629–638. doi: 10.1001/archsurg.140.7.629. [DOI] [PubMed] [Google Scholar]
- 5.Huber T, Baumgart J, Peterhans M, Weber S, Heinrich S, Lang H. Computer-assisted 3D-navigated laparoscopic resection of a vanished colorectal liver metastasis after chemotherapy. Z Gastroenterol. 2016;54(1):40–43. doi: 10.1055/s-0041-107542. [DOI] [PubMed] [Google Scholar]
- 6.Madurska MJ, Poyade M, Eason D, Rea P, Watson AJ. Development of a patient-specific 3D-printed liver model for preoperative planning. Surg Innov. 2017;24(2):145–150. doi: 10.1177/1553350616689414. [DOI] [PubMed] [Google Scholar]
- 7.Damrah O, Canelo R, Zacharoulis D, Tait P, Jiao LR, Habib N. Three dimensional (3D) computed tomography images reconstruction in liver surgery. In: Karaliotas CC, Broelsch CE, Habib NA, editors. Liver and biliary tract surgery: embryological anatomy to 3D-imaging and transplant innovations. Vienna: Springer Vienna; 2006. pp. 333–338. [Google Scholar]
- 8.Hogemann D, Stamm G, Shin H, Oldhafer KJ, Schlitt HJ, Selle D, Peitgen HO. Individual planning of liver surgery interventions with a virtual model of the liver and its associated structures. Radiologe. 2000;40(3):267–273. doi: 10.1007/s001170050668. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann KS, Ritz JP, Valdeig S, Schenk A, Holmer C, Peitgen HO, Buhr HJ, Frericks BB. Portal vein segmentation of a 3D-planning system for liver surgery--in vivo evaluation in a porcine model. Ann Surg Oncol. 2008;15(7):1899–1907. doi: 10.1245/s10434-008-9934-x. [DOI] [PubMed] [Google Scholar]
- 10.Frericks BB, Caldarone FC, Nashan B, Savellano DH, Stamm G, Kirchhoff TD, Shin HO, Schenk A, Selle D, Spindler W, Klempnauer J, Peitgen HO, Galanski M. 3D CT modeling of hepatic vessel architecture and volume calculation in living donated liver transplantation. Eur Radiol. 2004;14(2):326–333. doi: 10.1007/s00330-003-2161-8. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Zhang Y, Peitgen HO, Schenk A, Yuan L, Wei G, Sun Y. Precise local resection for hepatocellular carcinoma based on tumor-surrounding vascular anatomy revealed by 3D analysis. Dig Surg. 2012;29(2):99–106. doi: 10.1159/000336289. [DOI] [PubMed] [Google Scholar]
- 12.Prodhomme O, Seguret F, Martrille L, Pidoux O, Cambonie G, Couture A, Rouleau C. Organ volume measurements: comparison between MRI and autopsy findings in infants following sudden unexpected death. Arch Dis Child Fetal Neonatal Ed. 2012;97(6):F434–F438. doi: 10.1136/fetalneonatal-2011-301309. [DOI] [PubMed] [Google Scholar]
- 13.Hiroshige S, Shimada M, Harada N, Shiotani S, Ninomiya M, Minagawa R, Soejima Y, Suehiro T, Honda H, Hashizume M, Sugimachi K. Accurate preoperative estimation of liver-graft volumetry using three-dimensional computed tomography. Transplantation. 2003;75(9):1561–1564. doi: 10.1097/01.tp.0000053755.08825.12. [DOI] [PubMed] [Google Scholar]
- 14.Hallet J, Gayet B, Tsung A, Wakabayashi G, Pessaux P. Systematic review of the use of pre-operative simulation and navigation for hepatectomy: current status and future perspectives. J Hepatobiliary Pancreat Sci. 2015;22(5):353–362. doi: 10.1002/jhbp.220. [DOI] [PubMed] [Google Scholar]
- 15.Sboarina A, Foroni RI, Minicozzi A, Antiga L, Lupidi F, Longhi M, Ganau M, Nicolato A, Ricciardi GK, Fenzi A, Gerosa M, De Simone A, Fracastoro G, Guglielmi A, Cordiano C. Software for hepatic vessel classification: feasibility study for virtual surgery. Int J Comput Assist Radiol Surg. 2010;5(1):39–48. doi: 10.1007/s11548-009-0380-4. [DOI] [PubMed] [Google Scholar]
- 16.Karcaaltincaba M, Akhan O. Imaging of hepatic steatosis and fatty sparing. Eur J Radiol. 2007;61(1):33–43. doi: 10.1016/j.ejrad.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Oliva MR, Mortele KJ, Segatto E, Glickman JN, Erturk SM, Ros PR, Silverman SG. Computed tomography features of nonalcoholic steatohepatitis with histopathologic correlation. J Comput Assist Tomogr. 2006;30(1):37–43. doi: 10.1097/01.rct.0000193818.31749.84. [DOI] [PubMed] [Google Scholar]
- 18.Alirr Omar Ibrahim, Rahni Ashrani Aizzuddin Abd. Survey on Liver Tumour Resection Planning System: Steps, Techniques, and Parameters. Journal of Digital Imaging. 2019;33(2):304–323. doi: 10.1007/s10278-019-00262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]