Abstract
Mate choice behaviors are among the most important reproductive isolating barriers in many animals. Little is known about the genetic basis of reproductively isolating behaviors, but examples to date provide evidence that they can have a simple genetic basis. However, it is unclear if these results indicate that individual genes with large effects are common, or are instead due to ascertainment biases. Here, we present the results of a QTL mapping study for the most important behavioral isolating barrier between Drosophila simulans and D. sechellia: male mate choice. Our QTL results initially suggested that differences in male mate choice may be due to a couple loci with large effects. However, as we divided the largest-effect QTL using stable introgression strains, we found evidence of multiple interacting loci. We further find that separate regions of the genome control different aspects of male choice. Taken together, our results suggest that the genetic architecture of mate choice behavior, in this case, is more complex than QTL mapping suggested, highlighting potential challenges to future mapping studies. We discuss the implications of these results as they relate to signal–receiver coevolution, mate choice, and reproductive isolation.
Subject terms: Evolutionary genetics, Behavioural genetics
Introduction
Behaviors can act as important barriers to reproduction (Coyne and Orr 1997, 2004), preventing gene flow between many taxa (Jiggins et al. 2001; Ligon et al. 2018; Mendelson and Shaw 2002; Pröhl et al. 2006). While behaviors are important in this context, we still have relatively few examples of the genetic underpinnings explaining species’ differences in behavior. As a result, we are unable to draw broad conclusions regarding the proximate and ultimate causes of behavioral evolution. However, the limited results to date have shown that single loci can have large effects on behavioral divergence (Andersson et al. 2012; Arbuthnott 2009; Auer et al. 2020; Cande et al. 2013; Ding et al. 2016; Fanara et al. 2002; Konopka and Benzer 1971; Leary et al. 2012; McGrath et al. 2009, 2011; Prince et al. 2017; Yalcin et al. 2004). Further, many examples highlight the potential of sensory receptor genes to explain these effects (Auer et al. 2020; Cande et al. 2013; Fanara et al. 2002; Leary et al. 2012; McGrath et al. 2011). Such findings imply that substantial changes to animal behavior can arise via the evolution of a single, large-effect locus, and that means that behavioral isolation could evolve more frequently (Hayashi et al. 2007) and quickly (Gavrilets et al. 2007; Gavrilets and Vose 2007) when populations experience some gene flow. Because large-effect genes are easier to detect, however, they are also likely overrepresented in the literature relative to their occurrence in nature (Rockman 2012). Thus, we need more studies of a variety of comparable behavioral traits to better test these hypotheses.
Drosophila courtship behavior provides an excellent genetic model to begin filling in these blanks (Sokolowski 2001). In Drosophila, males perform a complex courtship ritual that can involve tactile, chemosensory, auditory, and visual signals (Greenspan and Ferveur 2000). Males do not court indiscriminately, however. Instead they use signals expressed by females to identify the most suitable mates (Arbuthnott et al. 2017). Thus, courtship behavior in Drosophila represents coordinated communication among signalers and receivers using a variety of sensory stimuli and behavioral responses. Differences in courtship behaviors and signals compose important barriers to interspecific hybridization. For example, in the Drosophila melanogaster subgroup, female gustatory pheromones differ between species (Jallon 1984). The biggest difference in this group of flies is the expression of the cuticular hydrocarbon (CHC) gustatory pheromone, 7,11-heptacosadiene (7,11-HD). While Drosophila express a blend of CHCs, Drosophila melanogaster and D. sechellia females primarily express 7,11-HD. In their sister species, D. simulans, females primarily express a different pheromone, 7-tricosene. Differences in male response to divergent female CHC expression constitute significant barriers to reproduction among these species (Billeter et al. 2009; Shahandeh et al. 2018). D. melanogaster and D. sechellia males preferentially court females that express 7,11-HD, while D. simulans males are deterred from courting by the presence of 7,11-HD (Billeter et al. 2009; Shahandeh et al. 2018). Recent work has begun to illuminate the physiological changes in the nervous system underlying divergent male response to 7,11-HD expression (Clowney et al. 2015; Seeholzer et al. 2018). However, little is known about the genetic loci that underlie this phenotypic evolution.
Here we report the results of a large quantitative trait locus (QTL) study seeking to identify loci underlying natural differences in male mate choice behavior between D. simulans and D. sechellia. Our study identifies three large-effect QTL explaining a significant amount of the phenotypic difference. When we manipulate female CHC expression, we find that different regions of the genome control separate aspects of male mate choice. Contrasting with the simplicity of our initial QTL results, a fine-scale dissection of the largest effect QTL reveals the presence of multiple, nonadditively interacting loci. Ultimately, our findings underscore that the evolution of behaviors may be more complexly regulated (Mackay 2004; MacKay 2009; Zwarts et al. 2011) than found in some case studies thus far (Andersson et al. 2012; Ding et al. 2016; Konopka and Benzer 1971; McGrath et al. 2009; Okhovat et al. 2015; Yalcin et al. 2004). Such a complex genetic architecture has important implications on furthering our understanding of the evolution of animal communication, reproductive isolation, and the speciation process (Kondrashov and Kondrashov 1999; Shaw and Parsons 2002).
Materials and methods
Fly strains and maintenance
We maintained all fly strains as described in Shahandeh et al. (2018). For QTL mapping, we used a single D. simulans parent strain, simC167.4 (Stock #: 14021-0251.199), and a single D. sechellia parent strain, synA, (Stock #: 14021-0248.28) that we will refer to as “D. simulans” and “D. sechellia.” To create introgression strains to fine-map a single QTL, we used two additional strains: sim2133 and sim933 (Stern et al. 2016). We selected these strains because they harbor transgenically inserted fluorescent proteins under the control of the eyeless promotor (see below). We collected all male and female flies used in the experiments described below from these strains, or crosses of these strains, as virgins within 6 h of eclosion on the 11th day following oviposition under light CO2 anesthesia.
Hybrid crosses and backcrosses
To create reciprocal hybrid males, we combined 10 virgin females, collected just 2–3 h following eclosion, with 15 virgin males that had been aged for 7–10 days. D. sechellia females crossed with D. simulans males yield sechXF1 hybrid males (Fig. 1a), and D. simulans females crossed with D. sechellia males yield simXF1 hybrid males (Fig. 1b). These hybrids have an identical background with the exception of their sex chromosomes and maternal inheritance. To generate backcross males for QTL mapping (Fig. 1c), we crossed virgin D. simulans-D. sechellia hybrid females to D. sechellia (BCsech) or D. simulans males (BCsim) using the same crossing method.
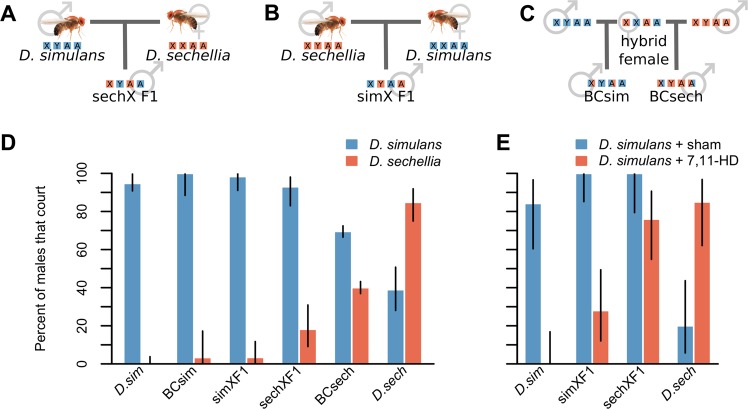
Fig. 1. Courtship behavior of parent strains and hybrids.
a The crosses used to create hybrids with D. sechellia chrX (sechXF1) and b hybrids with D. simulans chrX (simXF1). c The crosses used to create backcross progeny. Autosome and sex chromosome genotypes are represented by colored squares (A autosome, X&Y sex chromosomes, orange D. sechellia chromosome, blue D. simulans chromosome). d CF for each male type when paired with D. simulans females (N = 76, N = 30, N = 70, N = 57, N = 952, and N = 77, for blue bars from left to right) or with D. sechellia females (N = 96, N = 30, N = 59, N = 55, N = 952, and N = 79, orange bars from left to right). e CF for each male type when paired with sham-perfumed D. simulans (N = 19, N = 23, N = 16, and N = 20, for blue bars from left to right) or 7,11-HD-perfumed D. simulans females (N = 20, N = 25, N = 25, and N = 20, orange bars from left to right). Whiskers represent 95% confidence intervals from a binomial test.
Generating recombinant introgression males for fine-mapping
We chose to fine-map a single QTL on chr3 with the largest effect on male courtship behavior toward both species females (see “Results”). To do so, we employed a marker-assisted introgression of chr3 from D. simulans into a D. sechellia background. We first crossed sim2133 and sim933 to create a D. simulans strain harboring a red fluorescent protein marker on chr3 and a yellow fluorescent protein marker on chr2 (strain 3R2Y). We then crossed this strain to D. sechellia. We took the resulting hybrid females (hybrid males are sterile) and crossed them back to the D. sechellia parent strain. We then selected male progeny that had lost the yellow fluorescent protein marker (i.e. chr2 of D. simulans), but retained the red fluorescent protein marker (i.e. chr3 of D. simulans). We crossed these males, a small fraction of which had regained fertility, to D. sechellia females for at least 5 more generations to purify the genetic background, while always selecting for males with red fluorescent protein expression to maintain D. simulans chr3. We always collected and crossed red fluorescent protein positive male offspring because recombination only occurs during female gametogenesis in Drosophila. By only crossing males, the introgressed D. simulans chr3 region remains fully intact. Once we created an initial introgression strain (3R.1), we used females from this strain in a cross to D. sechellia males to harness recombination to create recombinant introgression strains, each with a unique recombination event reducing the size of the original introgression. We identified unique recombinant progeny from this cross first, roughly by polymerase chain reaction using two species-specific alleles flanking the QTL region (You et al. 2008, Table S1), then, precisely by whole genome sequencing. We maintained these lines without recombination, as above. For both the initial introgression and recombinant introgression strains, the D. simulans genomic segment is heterozygous. Thus, each cross produces two types of males: introgression males (marked by red fluorescent protein) and control males, which resemble the D. sechellia parent strain. Introgression and control males are reared in the same vial, controlling for environment, so any difference we observe between the two is directly attributable to the presence or absence of the D. simulans chr3 fragment.
Two-day courtship assays
To measure courtship behavior of D. simulans, D. sechellia, simXF1, and sechXF1 males, we followed the methods first described by Shahandeh et al. (2018). In brief, we observed individual male genotypes with two different female genotypes on consecutive days in a full factorial design collecting minute-by-minute courtship data. As previously reported, for the parent strains, we observed no effect of assay day or female order (Shahandeh et al. 2018). For the hybrids, we also detected no effect of female order on courtship frequency (CF) for either sechXF1 or simXF1 males (all p = 1 after Bonferroni correction). Thus, we analyzed courtship data collected for the same males on each day independently. For males that received the same species female on both days, we only included data from the first day to avoid pseudoreplication. For backcross and chr3 introgression males, we observed all males with D. sechellia females on day 1, and D. simulans females on day 2 because we needed to observe each male with both female types, and there was no detectable effect of female order in the parent or hybrid male strains. Still, for backcross males, there is a small chance of an interaction between genotype and female order for courtship with D. simulans females, although our functional follow-up (observed in single day assays) suggest that this is unlikely (see “Results”).
Perfuming assays
To measure male response to the presence of 7,11-HD on the female cuticle, we perfumed D. simulans females, which lack 7,11-HD expression, with synthetic 7,11-HD (Cayman chemical CAS #10012567) using a protocol modified from Thistle et al. (2012). First, we added 400 μg of synthetic 7,11-HD dissolved in 40 μl 200 proof ethanol to a 20 mm plastic Drosophila rearing vial. We swirled the liquid around the vial, and let the ethanol evaporate for at least 1 h. After the ethanol had evaporated, we added 20 4-day-old virgin D. simulans females to each vial. We vortexed each vial for three 20-s intervals separated by 20 s of rest. We allowed females to recover in these vials at 25 °C and 50% humidity for 30 min before assay. For controls, we created a sham treatment where just 40 μl 200 proof ethanol was added to the vial and the process was repeated. We conducted these assays on a single day because we were concerned that synthetic 7,11-HD might transfer to the courtship assay vials, confounding the data from a sequential observation.
Courtship data analysis
We collected minute-by-minute courtship data over a 30-min period, counting each minute where courtship was observed. From the courtship data we extracted three parameters of male courtship, one of which we will focus on in the main text: CF. CF is a measure of what proportion of a male genotype courts a female type. We calculated CF as the number of males of a single genotype that we observed courting a female genotype divided by the total number of potentially courting pairs. For CF, we only counted males that displayed courtship behavior for more than 10% of the assay time (i.e., males that were scored for courtship in more than 3 min of the 30-min assay) to account for a potential low rate of observer error. We compared CFs between male and female pairings using Fisher’s exact tests with post hoc correction for multiple comparisons (Holm 1979). We estimated 95% confidence intervals (CIs) using a binomial test. We tested for correlations between CF for D. simulans and D. sechellia females among males from our mapping population using Pearson’s correlation test. Analyses for the second and third parameters we collected, courtship effort and courtship latency, are described at length in Supplementary document 1.
Genotyping and linkage map construction
We collected whole genome sequence from five sources: each parent strain, 382 BCsech males, 89 males resulting from a separate third generation backcross, and a set of lines where segments of chr3 from D. simulans were introgressed into a D. sechellia background. We isolated genomic DNA from individual flies using the Qiagen DNEasy blood and tissue kit (catalog #69504). We prepared libraries for illumina sequencing using a modified Nextera library preparation protocol (Baym et al. 2015) and sequenced using the illumina HiSeq3000. We sequenced the parent strains to deep coverage (~40× average), and the backcross male progeny to lesser coverage (~4× average). We aligned reads to a D. simulans reference genome (Hu et al. 2013) using BWA v0.6.2 (Li and Durbin 2009). We called high-confidence single nucleotide variants between our parent strains in the variant call format (Danecek et al. 2011) using SAMtools v1.1 (Li et al. 2009) and BCFtools v1.1 (Li 2011). We then scanned the aligned reads of the 382 backcross males to estimate recombination breakpoints using custom sliding window scripts (Fig. S1).
QTL analysis
Our sliding window analysis produced ~1200 genotype markers across the genome (roughly 200 per autosome arm) for 382 individuals. We imported the list of individual marker genotypes and phenotypes into R/qtl (Broman et al. 2003). We performed single QTL scans using the “scanone()” function with a binomial model for CF (court/did not court). This method identifies the single most likely QTL per chromosome. We additionally used the “scantwo()” function to formally test for epistasis between QTL. Because the single QTL scan for CF produced additional lesser peaks on chr3 (Fig. 2a), we also performed multiple QTL mapping to test for the presence of potential additional QTL at these loci. We first used the “stepwiseqtl()” function to identify the most parsimonious multiple QTL model (Zhou 2010). We then used “fitqtl()” and “refineqtl()” to identify markers associated with QTL peaks. We estimated a genome-wide log of odds (LOD) significance threshold for single and multiple QTL models using 10,000 and 1000 permutations, respectively (multiple QTL permutation is computationally demanding). We calculated CIs as the region surrounding a QTL peak encompassed by a drop of 1.5 in LOD score. For single QTL scans, we calculated QTL effect sizes as the proportion of phenotypic difference displayed by heterozygous and homozygous genotypes relative to the total difference observed among the parent strains (ex: (CFAB−CFAA)/(CFDsim−CFDsech)) using the individuals from our mapping population (Broman and Sen 2010). To calculate combined effect sizes, we compared individuals heterozygous at both QTL to individuals homozygous at both QTL. Effect sizes are often overestimated when using the same data to identify QTL and estimate effects (Beavis 1998). To independently measure QTL effects we phenotyped and genotyped 89 progeny from a hybrid backcross to D sechellia for three generations. These 89 males did not court D. sechellia, but did court D. simulans females. We calculate effect sizes for CF with D. simulans females using the above method with these 89 individuals.
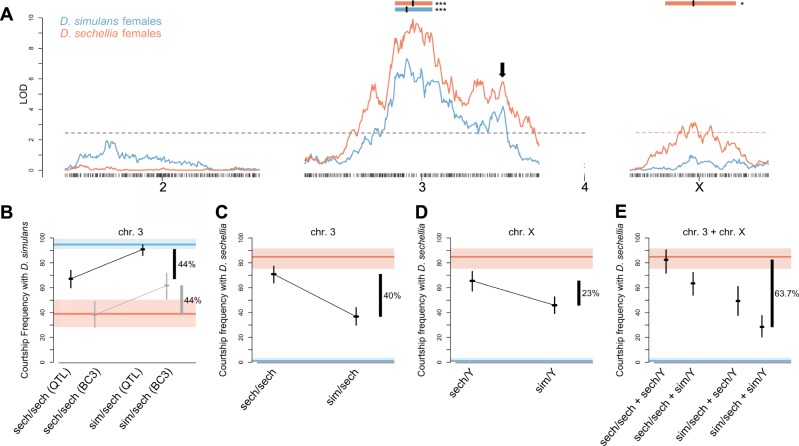
Fig. 2. Single QTL scans for courtship frequency.
a Significant loci for CF toward D. simulans (blue) and D. sechellia females (orange). Significant loci have LOD scores above the dashed lines, which represent alpha = 0.05. Peaks (vertical black lines) and confidence intervals (colored rectangles) are shown above QTL. Asterisks denote significance (*p < 0.05 and ***p < 0.001). The black arrow depicts a secondary peak in LOD score that leads us to hypothesize about the presence of additional QTL on chr3. b The effect of chr3 QTL on CF toward D. simulans females. Black data represents individuals from the mapping population and gray data represents individuals from the separate advanced backcross (N = 186, N = 34, N = 196, and N = 55, from left to right). c The effect of the chr3 QTL on CF toward D. sechellia females (N = 151 for sech/Y and N = 231 for sim/Y). d The effect of chrX QTL on CF toward D. sechellia females (N = 151 for sech/Y and N = 231 for sim/Y). e The combined effect of chr3 and chrX on courtship frequency towards D. sechellia females (N = 74, N = 115, N = 77, and N = 116, from left to right). For all, orange lines represent the phenotype of the D. sechellia parent strain and blue lines represent the phenotype of the D. simulans parent strain. Rectangles of respective colors and whiskers represent the 95% confidence intervals from a binomial test. Effect sizes are estimated on the right.
Results
D. simulans courtship preference alleles are largely autosomal dominant
Drosophila simulans and D. sechellia males preferentially court females of their own species. CF is higher for males paired with their own females vs. females of the other species (Fig. 1d), as previously shown (Shahandeh et al. 2018). We also investigated mating behavior in hybrid males (Fig. 1d). Hybrid males court D. simulans females at high frequencies, regardless of whether the D. simulans parent was their mother (simXF1) or their father (sechXF1, Table S2). However, there was one difference between simXF1 and sechXF1 hybrid males, consistent with some courtship loci residing on chrX (though cytoplasmic inheritance or an effect of chrY cannot be ruled out). A hybrid male is more likely to court a D. sechellia female if chrX is inherited from D. sechellia (p < 0.05, Fig. 1d and Table S2).
BCsim males inherit an average of 25% of their genome from D. sechellia, but behave indistinguishably from simXF1 and D. simulans males (Fig. 1d). Thus, it appears that D. simulans alleles affecting CF are largely autosomal dominant to D. sechellia alleles. BCsech males display increased CF toward D. sechellia females and decreased CF toward D. simulans females relative to sechXF1 males (p < 0.00001 for both, Table S2). These differences in BCsech behavior reflect the effect of making regions of the genome homozygous for D. sechellia sequence, revealing the effects of recessive D. sechellia loci, leading us to select 382 BCsech individuals for QTL mapping (Fig. 1c).
An effect of the X chromosome
We next tested whether these putative X-linked loci are responding to the presence of the 7,11-HD pheromone. This pheromone is present on the D. sechellia female cuticle but absent in D. simulans. Previous results point to this pheromone as the most important cause of male courtship differences between Drosophila species (Billeter et al. 2009). To do so, we observed males with 7,11-HD-perfumed and sham-perfumed D. simulans females.
As expected, males of the parent species respond strongly to the presence of this pheromone (Fig. 1e). D. sechellia males courted 7,11-HD-perfumed females more frequently (CF = 85%) than sham-perfumed D. simulans (CF = 20%, p < 0.001). Like when paired with D. sechellia females, D. simulans males never courted 7,11-HD-perfumed D. simulans females, but they did court sham-perfumed D. simulans females (CF = 84%, p < 0. 00001). simX hybrid males were similar to D. simulans males in that they courted 7,11-HD perfumed less frequently (CF = 28%) than sham-perfumed D. simulans females (CF = 100%, p < 0.00001). These hybrid males are significantly more likely to court 7,11-HD perfumed D. simulans than the D. simulans parent strain, however, indicating some effect of the autosomal D. sechellia genome (p < 0.05). sechXF1 males behave differently from D. sechellia males, in that they do not discriminate between the two treatments (p = 0.0655). Like simXF1 hybrids, 100% of sechXF1 males courted sham-perfumed D. simulans, but 76.0% also courted 7,11-HD-perfumed D. simulans. Thus, sechXF1 males are significantly more willing to court sham-perfumed D. simulans than the D. sechellia parent strain (p < 0.00001), but are also equally willing to court 7,11-HD perfumed D. simulans (p = 0.7095). Thus, males harboring chrX of D. sechellia are not 7,11-HD adverse, like males harboring chrX of D. simulans, indicating the presence of 7,11-HD aversion loci on the D. simulans X chromosome. The fact that both simXF1 and sechXF1 hybrids court sham-perfumed D. simulans females at high frequencies again suggests the likelihood of autosomal dominant D. simulans loci affecting CF toward D. simulans females.
Single QTL scans reveal regions with large effects on CF
A QTL scan for CF toward D. simulans females produced a single, highly significant QTL on the right arm of chr3 (Fig. 2a, blue; peak = 30.55 Mb, CI = 20.60–34.45 Mb). Using the 382 backcross individuals in the mapping population, we estimate this QTL explains 44.3% of the total phenotypic difference between parent strains (Fig. 2b, orange). Our independent estimation of effect size using 89 advanced backcross males not used in the QTL analysis is highly congruent with this measure, at 44.2% of the total phenotypic difference (Fig. 2b, gray). A QTL scan for CF toward D. sechellia identifies a highly overlapping peak on chr3 (Fig. 2a, orange; peak = 31.05 Mb, CI = 20.60–34.45 Mb), with a comparable effect size (40% of the total phenotypic variance, Fig. 2c). These QTL have opposing effects: males with a D. simulans allele in this region court D. simulans females with high frequency, and D. sechellia females infrequently. This result is further supported by the fact that CF toward D. simulans and D. sechellia females among BCsech males was significantly negatively correlated (Pearson’s corr. coefficient = −0.4745, p < 0.00001). Thus, males that court one female type, are significantly less likely to court the other female type.
Our QTL scan for CF toward D. sechellia also identifies a second, less significant QTL on chrX (Fig. 2a; peak = 9.60 Mb, CI = 6.40–15.30 Mb), as expected from the behavior of reciprocal hybrid males (simXF1 and sechXF1). We estimate this QTL explains 23% of the total phenotypic difference between species (Fig. 2d). Individuals that inherit D. sechellia alleles within this region court D. sechellia females more frequently than individuals that inherit D. simulans alleles at the same locus, but there is no effect of this locus on courtship toward D. simulans females. This result is congruent with a 7,11-HD aversion locus on chrX (as demonstrated by our perfuming experiments). Males inheriting D. sechellia alleles within this region are more willing to court D. sechellia females, but not less willing to court D. simulans females, as indicated by a lack of overlapping QTL like we observed on chr3. This implies that these same males do not require 7,11-HD to stimulate courtship, and thus likely are not inheriting 7,11-HD preference alleles at this locus. When we calculate the combined effect of both QTL, we estimate that they explain 63.7% of the total phenotypic difference between species (Fig. 2e), indicating a purely additive relationship between these QTL. Two-dimensional QTL scans detect no evidence of epistasis between individual QTL, supporting this result (Table S3).
A multiple QTL model for CF identifies additional QTL
We also performed multiple QTL mapping to estimate the likelihood of additional QTL on chr3. This method includes QTL detected with a single QTL scan as covariates in its initial model, and then scans for additional QTL and interactions that improve the model fit, allowing us to detect additional QTL on the same chromosome (Broman and Sen 2010).
A multiple QTL model for CF toward D. sechellia identified one additional QTL. This locus is also on the right arm of chr3 (Fig. S2A, peak = 44.25 Mb, CI = 41.25–47.85 Mb), aligning with the secondary peak in LOD score from the single QTL scan (Fig. 2a, black arrow). For males paired with D. simulans females, we identified two additional QTL (Fig. S2B). One is on the right arm of chr3 (peak = 44.25 Mb, CI = 44.15–44.45 Mb) also aligning with the additional QTL for courtship toward D. sechellia females and the secondary peak in LOD score from the single QTL scan (Fig. 2a, black arrow). These additional QTL on the right arm of chr3 highlight this region as a potential hotspot of courtship behavior loci. The second additional QTL was detected on the left arm of chr2 (peak = 8.50 Mb, CI = 3.30–10.20 Mb).
Chr3 introgression validates the largest effect CF QTL
We next sought to fine-map the largest effect QTL with opposing effects: the QTL on ch3. To begin, we used marker-assisted introgression to transfer a portion of D. simulans chr3 into a D. sechellia background (Fig. 3a). To create this strain, we used a parent strain of D. simulans, 3R2Y (see “Materials and methods”), with fluorescent transgenes that allowed us to track the transmission of autosomes across generations (see “Materials and methods,” Fig. 3a). The behavior of 3R2Y was indistinguishable from the D. simulans parent strain we used for QTL mapping (95% court D. simulans females and 0% court D. sechellia females, p = 1 for both, Table S2).
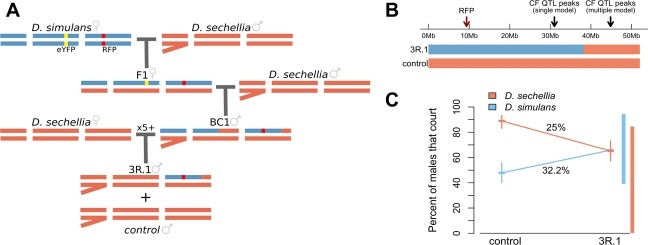
Fig. 3. The effect of D. simulans chr3.
a The crosses used to introgress D. simulans chr3 into D. sechellia. b The introgressed region. The red arrow denotes the position of RFP. The black arrow denotes the position of the CF QTL peaks. c The effect of the introgressed segment (3R.1, N = 161) compared with control males (N = 163) on CF toward D. simulans and D. sechellia females. Whiskers represent 95% confidence intervals from a binomial test. Rectangles to the right show the difference observed between the parent strains. The total percent of the species difference is shown.
The resulting introgression strain, 3R.1, is heterozygous at the major CF QTL on chr3 (Fig. 3b). This strain, however, does not harbor D. simulans alleles at the locus of the additional 3R QTL detected by our multiple QTL model (Fig. 3b). When we compared the behavior of 3R.1 introgression males to their control siblings (who are raised in the same vials but harbor no D. simulans alleles; Fig. 3a), we found two distinguishing patterns of behavior. First, relative to control males, we observed a significant increase in the frequency of 3R.1 introgression males courting D. simulans (p < 0.01). Second, we also observed a significant decrease in the frequency of 3R.1 introgression males courting D. sechellia relative to their control siblings (p < 0.0001, Fig. 3c). When we compare the difference in CF between control and 3R.1 males to that of the parent strains, we find that the introgressed segment explains 25% and 32.2% of the species difference for CF toward D. sechellia and D. simulans females respectively, a notable decrease from our original estimate of ~42%. This is likely due to the exclusion of the additional QTL detected by our multiple QTL model. In this case, the effects of multiple QTL could not be entirely separated in either a single QTL scan or advanced backcross due to linkage between multiple QTL on the right arm of chr3, leading us to overestimate an individual QTL’s effect.
Fine-mapping a large-effect QTL reveals a complicated genetic architecture
We next allowed recombination to randomly break up the introgressed region of 3R.1 to create additional introgression lines with smaller fragments of D. simulans chr3. Our aim was to use markers to identify flies harboring useful recombination to map the causal locus to a small interval, but we instead found that introgression strains demonstrate that species differences result, in part, from epistatic interactions between loci contained within the chr3 QTL.
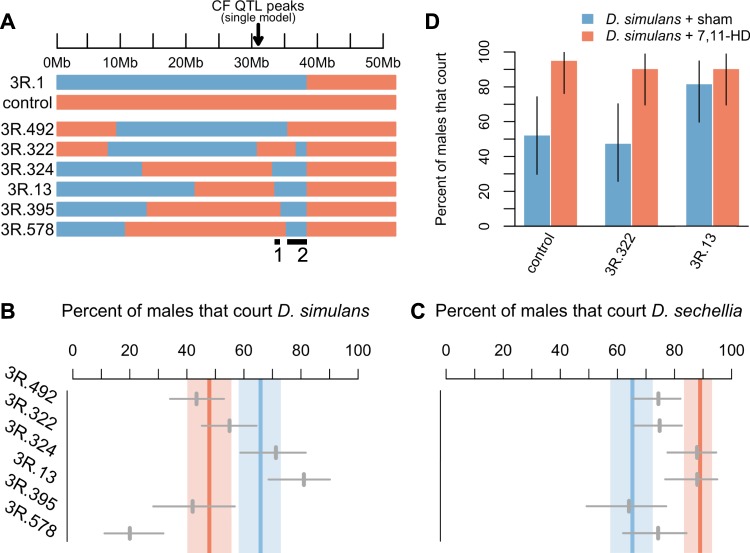
After creating six recombinant sublines with portions of the original introgression (Fig. 4a), we compared the behavior of these lines in two ways. First, we compared these strains by considering CF toward D. simulans females. Recombinant introgression strains that still include the causal loci should court D. simulans females significantly more frequently than control males. For CF with D. simulans females, we identified two such strains: 3R.324 and 3R.13 (Fig. 4b). Males from these strains court D. simulans females at frequencies much greater than control males (p < 0.01 for 3R.324; p < 0.0001 for 3R.13). Importantly, 3R.324 and 3R.13 males also court D. simulans females at frequencies comparable to 3R.1 males (71.2% for 3R.324, p = 0.5331; 81.0% for 3R.13, p = 0.0939). Intriguingly, the introgressed regions of these two strains, while largely overlapping themselves, are also contained in part by each of the other introgression strains, such that the entire overlapping region of 3R.13 and 3R.324 is represented in smaller parts among the other four strains (Fig. 4a). The only plausible explanation for the difference in behavior between 3R.13, 3R.324, and the other strains, is that these two strains harbor at minimum two D. simulans loci that nonadditively interact, while none of the other strains carry both of these loci, and thus do not court D. simulans at high frequencies. There are two regions of chr3 present in both 3R.13 and 3R.324 that are not present together in any of the other introgression strains (Table 1 and Fig. 4a), and thus must act in epistasis to affect CF toward D. simulans females.
Fig. 4. Recombinant introgression males.
a The genotype of recombinant introgression males within the QTL region. Blue denotes heterozygous and orange denotes homozygous D. sechellia regions. The numbered regions mark the proposed locations of epistatic loci that increase courtship toward D. simulans females. These regions are both fully contained in 3R.13 and 3R.324, but not in any of the other strains (Table 1). b CF of recombinant introgression males with D. simulans. c CF of recombinant introgression males with D. sechellia. Thick blue and orange bold lines represent the behavior of 3R.1 males (blue) and control males (orange) surrounded by their 95% CIs. N = 113 for 3R.492, N = 111 for 3R.322, N = 66 for 3R.324, N = 58 for 3R.13, N = 50 for 3R.395, and N = 66 for 3R.578. d The CF of three male genotypes when paired with sham-perfumed (N = 21, N = 21, and N = 22, for blue bars from left to right) and 7,11-HD-perfumed D. simulans females (N = 21, N = 21, and N = 21, for orange bars from left to right). Whiskers represent 95% confidence intervals from a binomial test.
Table 1.
Two regions within the chr3 QTL interact nonadditively to drive courtship toward D. simulans females.
| Male genotype | Courts D. simulans | Genotype region 1 (chr3 33.5–34.4 Mb) | Genotype region 2 (chr3 35.3–38.2 Mb) |
|---|---|---|---|
| Control | Like D. sechellia | D. sechellia | D. sechellia |
| 3R.1 | More than control males | D. simulans | D. simulans |
| Recombinant introgression strains | |||
| 3R.492 | Like control males | D. simulans | D. sechellia |
| 3R.322 | Like control males | D. sechellia |
D. sechellia (3R: 35.3–36.75 Mb) D. simulans (3R: 36.8–38.2 Mb) |
| 3R.324 | More than control males | D. simulans | D. simulans |
| 3R.13 | More than control males | D. simulans | D. simulans |
| 3R.395 | Like control males | D. sechellia | D. simulans |
| 3R. 578 | Less than control males | D. sechellia | D. simulans |
3R.1 introgression males court D. simulans females significantly more than control males. Two strains, 3R.324 and 3R.13, replicate this pattern. These two strains are the only strains that also contain two regions of introgressed D. simulans genome, regions 1 and 2 (see Fig. 4). The remaining introgression strains do not contain D. simulans genome at both regions.
Second, we compared CF of recombinant introgression strains with D. sechellia females to that of 3R.1 and control males. 3R.324 and 3R.13 courted D. sechellia at frequencies indistinguishable from control males (82.0% and 81.2%, respectively, p = 1 for both; Fig. 4c), but significantly higher than 3R.1 males (p < 0.01 for both), suggesting that the alleles affecting attraction to D. simulans females and the alleles affecting attraction to D. sechellia females are genetically separable. The remaining four lines all court D. sechellia at frequencies significantly lower than control males (p < 0.001 for 3R.395, p < 0.01 for 3R.492, and p < 0.05 for 3R.578 and 3R. 322). These four lines also court D. sechellia at frequencies statistically indistinguishable from 3R.1 males (p > 0.40 for all). However, three of these four lines, 3R.322, 492, and 3R.578, still displayed significantly greater CFs with D. sechellia females than with D. simulans females. In this way, these three lines still behave more like control males, which also court D. sechellia females with significantly higher frequencies (p < 0.0001).
The role of 7,11-HD in courtship behavior
The previous results identify two introgression strains that court D. simulans females at higher frequencies than their control male counterparts. To test if the absence of 7,11-HD on the D. simulans cuticle drives attraction to D. simulans, we selected one introgression strain that courts D. simulans females significantly more than control males, 3R.13, and one that does not, 3R.322, to observe with perfumed D. simulans females (Fig. 4d). We found that 3R.13 males court 7,11-HD-perfumed D. simulans just as frequently as sham-perfumed D. simulans females (90.5% and 81.8% of males court, respectively; p = 0.6640). Alternatively, we found that 3R.322 males greatly prefer 7,11-HD-perfumed D. simulans females, courting them 90.5% of the time, compared with just 47.6% of the time when paired with sham-perfumed females (p < 0.05). 3R.322 males behave indistinguishably from control males in this respect (p = 1 when comparing CF with both 7,11-HD and sham-perfumed females). Therefore, 3R.322 males are attracted to 7,11-HD on the female cuticle while 3R.13 males already court D. simulans females at higher frequencies than control males, and thus do not require 7,11-HD to stimulate courtship (Fig. 4b). Importantly, control males behave like D. sechellia males when comparing CF with both 7,11-HD and sham-perfumed females (p = 0.6866 for both).
Discussion
A single CHC pheromone signal isolates D. simulans and D. sechellia
CHC pheromones and corresponding preferences vary and can act as mating signals both among and between Drosophila species (Cobb and Jallon 1990; Jallon and David 1987; Jallon 1984; Pardy et al. 2018; Pischedda et al. 2014) as well as for other insects (Tregenza and Wedell 1997; Zhang et al. 2014). We have previously shown that, broadly, differences in CHCs drive male-mediated reproductive isolation between D. simulans and D. sechellia (Shahandeh et al. 2018). For D. melanogaster and D. simulans, male-mediated reproductive isolation is driven largely by a single CHC expressed on the D. melanogaster female cuticle: 7,11-HD (Billeter et al. 2009). D. sechellia females also express 7,11-HD (Coyne et al. 1994). Here, through the direct manipulation of 7,11-HD expression on the D. simulans cuticle, we show that this specific pheromone is also the primary driver of male-mediated reproductive isolation between these species for the first time. Thus, the evolution of a CHC signal is an important reproductive isolating barrier between D. simulans and D. sechellia, which are sympatric in the Seychelles archipelago (Matute and Ayroles 2014; Shahandeh et al. 2018).
The genetic architecture underlying the evolution of the 7,11-HD signal is well understood. QTL analysis identifies three loci affecting 7,11-HD expression (Gleason 2005). Two of these QTL encompass the enzymes desaturaseF and elongaseF (Gleason et al. 2009), both of which are essential for 7,11-HD biosynthesis (Chertemps et al. 2006, 2007; Combs et al. 2018; Shirangi et al. 2009). This simple genetic architecture suggests that this signal may have diverged quickly, via mutation to just a few large-effect loci. However, in order for reproductive isolation to occur, male receivers must be able to discriminate based on the presence or absences of 7,11-HD on the female cuticle. Until now, little was known about the genetic architecture of 7,11-HD receiver preference evolution. Thus, this information is an important step toward understanding the tempo and mode of the evolution of reproductive isolation among these species and understanding the process of speciation as a whole (Shaw and Parsons 2002).
The genetic architecture of male mate discrimination
Hybrid offspring behavior largely resembles a single parent, D. simulans, highlighting the potential for a simple genetic architecture with largely dominant D. simulans autosomal alleles. The comparison of reciprocal hybrids also suggests a putative role of chrX in male preference, however. Our single QTL scan results largely reflect this pattern (Fig. 2b, c). Further, when we detect more than one QTL, they appear to act purely additively (Fig. 2e). These initial QTL results are consistent with what many other QTL studies have identified as a simple genetic architecture underlying behavioral phenotypes (Arbuthnott 2009; Merrill et al. 2019; Yalcin et al. 2004), even without refining QTL to a gene level.
The results of a multiple QTL model contrast the above results, highlighting a more complex genetic architecture that was not detected by single QTL scans. In total, we identify three QTL affecting CF with D. simulans females, and three affecting CF with D. sechellia. The results of our chr3 introgression support the results of our multiple QTL models. We introgressed the chr3 QTL detected by our single QTL scan for CF, but not those detected by our multiple QTL model (Fig. 3b). When we use this introgression strain, 3R.1, to estimate effect size, our estimates drop significantly (Fig. 3c).
Our understanding of genetic architecture becomes additionally complicated with the data from recombinant lines made from our original introgression (3R.1). Our observations of these strains suggest the presence of multiple, nonadditively interacting loci underlying the effect of a single QTL affecting CF toward D. simulans females (Table 1 and Fig. 4a), making it clear that the genetic architecture of male mate choice, even within the single largest effect QTL, is not as simple as initially thought. Taken together, our findings underscore the difficulties inherent in determining genetic architecture from QTL mapping alone.
Distinct QTL affect different aspects of male preference
The behavior of reciprocal hybrids with perfumed D. simulans females shows that chrX has a substantial effect on CF with females expressing 7,11-HD (Fig. 1e). The difference between reciprocal hybrids is likely attributable to the X-linked QTL we detected, which affected CF toward D. sechellia females (that express 7,11-HD) but not CF with D. simulans females (that lack 7,11-HD). This QTL appears to include D. simulans loci affecting 7,11-HD aversion, but not 7,11-HD attraction; males with D. simulans genotype at this locus court 7,11-HD-perfumed females less because they are aversive to 7,11-HD, but males with D. sechellia genotype at this locus court 7,11-HD-perfumed females equally to sham-perfumed females (Fig. 1e). However, simXF1 males do court 7,11-HD perfumed D. simulans more frequently than the D. simulans parent strain, suggesting the presence of autosomal 7,11-HD attraction loci as well.
When we observe strains with a part of D. simulans chr3 in a D. sechellia background (3R.13 and 3R.322) with perfumed females, we find evidence for this autosomal locus affecting 7,11-HD attraction. While neither strain is aversive to 7,11-HD, presumably due to their D. sechellia X chromosome (see above), one strain (3R.322) is stimulated to higher CF by it. So, autosomal D. sechellia 7,11-HD preference loci must be contained within the nonoverlapping regions of these strains. This is congruent with the results from our QTL mapping: that this locus harbors alleles of opposing effects. When males inherit D. simulans alleles within this region, they are less attracted to D. sechellia females; when males inherit D. sechellia alleles within this region, they are more attracted to D. sechellia females. Taken together, these results suggest that there are two broad loci affecting 7,11-HD response: an X-linked D. simulans aversion locus, and an autosomal D. sechellia attraction locus. Similarly discrete genetic loci affecting separate aspects of behavior have been described in mice (Stacher Hörndli et al. 2019; Weber et al. 2013).
Genetic complexity and the evolution of reproductive isolation
Considering the coevolution of pheromone signals and preferences raises an important question: if the ancestral preference phenotype is for the original pheromone signal, how can newly evolved signals persist? Such traits are expected to experience stabilizing selection by way of preexisting preferences of signal receivers, unless receiver preferences are able to rapidly track large changes in pheromone signals (Baker 1989). While the genetic architecture of signal evolution implies potentially quick divergence due to a few, large-effect mutations (like the retrotransposition of a single gene, desatF (Fang et al. 2009)), the genetic architecture of the receiver’s response appears to have required many more changes. The epistasis we identify potentially augments the complexity of receiver evolution even further (Karageorgi et al. 2019; Lee et al. 2019; Weinreich et al. 2006). Considering signal–receiver coevolution, the differences in genetic architecture allow us to form new hypotheses about the evolution of this particular reproductively isolating mechanism.
The difference in genetic architectures makes it less likely that receiver preferences could quickly track a shift in primary CHC pheromone signal if that were its ancestral function, causing any new mutations modifying the signal to be selected against. Considering our results, it seems more plausible that a shift in CHCs occurred for another reason, and the phenotype diverged irrespective of mate choice. Once the CHC became different between species, then divergent male preferences may have been favored as a species recognition mechanism in sympatry allowing males to avoid costly interspecific courtship and/or mating (Shahandeh et al. 2018). Species recognition could have evolved at a much slower pace once the CHC difference was in place, co-opting it as a sexual signal. We know that sexual selection on mate choice is most important to the evolution of reproductive isolation in such a scenario, particularly when there is significant postmating isolation, as there is in these species (Hudson and Price 2014; Servedio 2001). Indeed, the evolution of such reproductive isolation via reinforcement is thought to be common in Drosophila (Yukilevich 2012).
This hypothesis is reasonable for CHCs, as they also have important ecological functions in Drosophila. Specifically, CHCs are important to desiccation tolerance in insects (Gibbs 1998). Different CHCs vary in their chemical structure-dependent responses to temperature (Gibbs and Pomonis 1995). Underlining this effect, in Drosophila, increases in CHC chain length correlate with increasing environmental temperatures (Gibbs et al. 1998), and quantitative genetic analyses demonstrate a relationship between CHC composition and desiccation resistance (Foley and Telonis-Scott 2011). Thus, the shift in primary pheromones between these species may have first been an adaptation to differences in environment, going unrecognized by males before being co-opted as a species recognition signal. Eventually, D. simulans evolved (at least) an X-linked locus causing 7,11-HD aversion, while D. sechellia separately evolved an autosomal locus causing 7,11-HD attraction. Indeed, adaptation to divergent environments can lead to subsequent reproductive isolation (William and George 1990), but one study divergently selecting on desiccation resistance for 57 generations without a significant cost to conspecific mating between populations still failed to generate premating isolation (Kwan and Rundle 2010), highlighting that divergent ecological selection alone is not always enough to evolve reproductive barriers. We also cannot rule out the possibility that the shift in primary CHC may have been neutral, and does not necessitate an alternatively adaptive function.
Co-localization of mate choice QTL
Another pattern we observed is the co-localization of many QTL to a single region of the genome: the right arm of chr3. For CF, five of the six QTL map to chr3, with the remaining two falling on chr2 and chrX. We know that two of these QTL, on chr3 and chrX, affect 7,11-HD preference. The multiple loci underlying the chr3 QTL affecting 7,11-HD preference only augments this pattern. The remaining five QTL potentially affect separate aspects of male mate choice. Thus, alleles affecting multiple aspects of male mate choice may be maintained in linkage. Simulation has demonstrated that regions of linkage disequilibrium may serve to reduce gene flow when species co-occur (Barton and De Cara 2009). Real-world examples support this hypothesis. For instance, loci controlling threespine stickleback armor traits cluster within the genome, creating freshwater of marine “supergene” regions that may maintain phenotypic divergence among types in the face of gene flow (Miller et al. 2014). Moreover, regions of multiple preference loci maintained in linkage disequilibrium have been identified in Heliconius, and are thought to be important to the maintenance of species boundaries in sympatry (Sekimura and Nijhout 2017). Drosophila simulans and D. sechellia do occur in sympatry where they exchange a small amount of gene flow (Coyne et al. 1994), so the accumulation of mate choice QTL in a single region may reflect similar selective pressures.
Conclusions
Ultimately, our results highlight the difficulty in determining genetic architecture from QTL mapping results alone. This contradicts a one QTL: one locus ascertainment bias perpetuated in the field (despite mounting evidence to the contrary), where the QTL that are successfully fine-mapped to causal loci represent a potentially small subset that have loci with substantial effects, making them easier to dissect (Rockman 2012). Instead, even for between species comparisons, maybe we should expect genetic complexity, even within relatively small QTL or individual loci. In Drosophila, this has been demonstrated in morphological adaptation (Frankel et al. 2011; Rebeiz et al. 2009; Rebeiz and Williams 2017), intraspecific studies of male mate choice behavior (Moehring and Mackay 2004), and now here with interspecific studies of behavioral evolution (Shahandeh et al. 2020). Although this suggests that the loci of behavioral evolution may be more difficult to identify, as we have discussed, we can still glean valuable insights and generate hypotheses, as we have done here, about the tempo and mode of evolution from studies of genetic architecture. In addition, in Drosophila research, our ever increasing abilities to phenotype behaviors at greater resolutions, genotype organisms in mass, and create transgenic strains in multiple species, will continue to improve our ability to dissect complex genetic architectures.
Supplementary information
Acknowledgements
We are grateful to Cameryn Brock, Kevin Card, Devon Cooper, Jason Rodriguez, and Dr Alison Pischedda for their hours spent observing Drosophila courtship. We thank Jeremy McIntyre for his time spent PCR-genotyping recombinants, Dr Scott A. Hodges for his advisement with illumina sequencing, and Dr William R. Rice for his valuable input with our statistical analysis. We used computational facilities purchased with funds from the National Science Foundation (CNS-1725797) and administered by the Center for Scientific Computing (CSC). The CSC is supported by the California NanoSystems Institute and the Materials Research Science and Engineering Center (MRSEC; NSF DMR 1720256) at UC Santa Barbara. This work was supported by the National Institutes of Health (R01 GM098614).
Data availability
Raw sequence reads are available at the Sequence Read Archive (accession PRJNA558426). Scripts used for sequence alignment and analysis are available at GitHub (https://github.com/mshahandeh/genarch_malechoice_Drosophila.git). Courtship and QTL genotype data are available in the Dryad Digital Repository (10.5061/dryad.6c936j6).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Associate editor: Darren Obbard
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41437-020-0309-9) contains supplementary material, which is available to authorized users.
References
- Andersson LS, Larhammar M, Memic F, Wootz H, Schwochow D, Rubin CJ, et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature. 2012;488(7413):642–646. doi: 10.1038/nature11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuthnott D. The genetic architecture of insect courtship behavior and premating isolation. Heredity. 2009;103(1):15–22. doi: 10.1038/hdy.2009.22. [DOI] [PubMed] [Google Scholar]
- Arbuthnott D, Fedina TY, Pletcher SD, Promislow DEL. Mate choice in fruit flies is rational and adaptive. Nat Commun. 2017;8:13953. doi: 10.1038/ncomms13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer Thomas O., Khallaf Mohammed A., Silbering Ana F., Zappia Giovanna, Ellis Kaitlyn, Álvarez-Ocaña Raquel, Arguello J. Roman, Hansson Bill S., Jefferis Gregory S. X. E., Caron Sophie J. C., Knaden Markus, Benton Richard. Olfactory receptor and circuit evolution promote host specialization. Nature. 2020;579(7799):402–408. doi: 10.1038/s41586-020-2073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TC. Sex pheromone communication in the lepidoptera: new research progress. Experientia. 1989;45(3):248–262. [Google Scholar]
- Barton NH, De Cara M. The evolution of strong reproductive isolation. Evolution. 2009;63(5):1171–1190. doi: 10.1111/j.1558-5646.2009.00622.x. [DOI] [PubMed] [Google Scholar]
- Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. Inexpensive multiplexed library preparation for megabase-sized genomes. PLOS ONE. 2015;10(5):e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis William D. Molecular Dissection of Complex Traits. 2019. QTL Analyses: Power, Precision, and Accuracy; pp. 145–162. [Google Scholar]
- Billeter JC, Atallah J, Krupp JJ, Millar JG, Levine JD. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature. 2009;461(7266):987–991. doi: 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen S. A guide to QTL mapping with R/qtl. Biometrics. 2010;66(2):658–659. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Cande J, Prud’homme B, Gompel N. Smells like evolution: the role of chemoreceptor evolution in behavioral change. Curr Opin Neurobiol. 2013;23:152–158. doi: 10.1016/j.conb.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Chertemps T, Duportets L, Labeur C, Ueda R, Takahashi K, Saigo K, et al. A female-biased expressed elongase involved in long-chain hydrocarbon biosynthesis and courtship behavior in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104(11):4273–4278. doi: 10.1073/pnas.0608142104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertemps T, Duportets L, Labeur C, Ueyama M, Wicker-Thomas C. A female-specific desaturase gene responsible for diene hydrocarbon biosynthesis and courtship behavior in Drosophila melanogaster. Insect Mol Biol. 2006;15(4):465–473. doi: 10.1111/j.1365-2583.2006.00658.x. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, Iguchi S, Bussell JJ, Scheer E, Ruta V. Multimodal chemosensory circuits controlling male courtship in Drosophila. Neuron. 2015;87(5):1036–1049. doi: 10.1016/j.neuron.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb M, Jallon JM. Pheromones, mate recognition and courtship stimulation in the Drosophila melanogaster species sub-group. Anim Behav. 1990;39(6):1058–1067. [Google Scholar]
- Combs PA, Krupp JJ, Khosla NM, Bua D, Petrov D, Levine J, et al. Tissue-specific cis-regulatory divergence implicates a fatty acid elongase necessary forinhibiting interspecies mating in Drosophila. Curr Biol. 2018;28(24):3969–3975.e3. doi: 10.1016/j.cub.2018.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Crittenden AP, Mah K, Maht K. Genetics of a pheromonal difference contributing to reproductive isolation in Drosophila. Science. 1994;265(5177):1461–1464. doi: 10.1126/science.8073292. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Patterns of speciation in Drosophila revisited. Evolution. 1997;51(1):295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA (2004) Speciation. Sinauer Associates, Sunderland, MA
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Berrocal A, Morita T, Longden KD, Stern DL. Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature. 2016;536(7616):329–332. doi: 10.1038/nature19093. [DOI] [PubMed] [Google Scholar]
- Fanara JJ, Robinson KO, Rollmann SM, Anholt RRH, Mackay TFC. Vanaso is a candidate quantitative trait gene for Drosophila olfactory behavior. Genetics. 2002;162(3):1321–1328. doi: 10.1093/genetics/162.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Ting CT, Lee CR, Chu KH, Wang CC, Tsaur SC. Molecular evolution and functional diversification of fatty acid desaturases after recurrent gene duplication in Drosophila. Mol Biol Evol. 2009;26(7):1447–1456. doi: 10.1093/molbev/msp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley BR, Telonis-Scott M. Quantitative genetic analysis suggests causal association between cuticular hydrocarbon composition and desiccation survival in Drosophila melanogaster. Heredity. 2011;106(1):68–77. doi: 10.1038/hdy.2010.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474(7353):598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S, Vose A. Case studies and mathematical models of ecological speciation. 2. Palms Ocean Isl Mol Ecol. 2007;16(14):2910–2921. doi: 10.1111/j.1365-294X.2007.03304.x. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Vose A, Barluenga M, Salzburger W, Meyer A. Case studies and mathematical models of ecological speciation. 1. Cichlids in a crater lake. Mol Ecol. 2007;16(14):2893–2909. doi: 10.1111/j.1365-294X.2007.03305.x. [DOI] [PubMed] [Google Scholar]
- Gibbs AG. Water-proofing properties of cuticular lipids. Am Zool. 1998;38(3):471–482. [Google Scholar]
- Gibbs AG, Louie AK, Ayala JA. Effects of temperature on cuticular lipids and water balance in a desert Drosophila: is thermal acclimation beneficial? J Exp Biol. 1998;201(Pt 1):71–80. doi: 10.1242/jeb.201.1.71. [DOI] [PubMed] [Google Scholar]
- Gibbs AG, Pomonis JG. Physical properties of insect cuticular hydrocarbons: the effects of chain length, methyl-branching and unsaturation. Comp Biochem Physiol Part B Biochem. 1995;112(2):243–249. [Google Scholar]
- Gleason JM. Quantitative trait loci for cuticular hydrocarbons associated with sexual Isolation between Drosophila simulans and D. sechellia. Genetics. 2005;171(4):1789–1798. doi: 10.1534/genetics.104.037937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason JM, James RA, Wicker-Thomas C, Ritchie MG. Identification of quantitative trait loci function through analysis of multiple cuticular hydrocarbons differing between Drosophila simulans and Drosophila sechellia females. Heredity. 2009;103(5):416–424. doi: 10.1038/hdy.2009.79. [DOI] [PubMed] [Google Scholar]
- Greenspan RJ, Ferveur JF. Courtship in Drosophila. Annu Rev Genet. 2000;34(1):205–232. doi: 10.1146/annurev.genet.34.1.205. [DOI] [PubMed] [Google Scholar]
- Hayashi TI, Vose M, Gavrilets S. Genetic differentiation by sexual conflict. Evolution. 2007;61(3):516–529. doi: 10.1111/j.1558-5646.2007.00059.x. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat Scand J Stat. 1979;6(6):65–70. [Google Scholar]
- Hu TT, Eisen MB, Thornton KR, Andolfatto P. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 2013;23(1):89–98. doi: 10.1101/gr.141689.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson EJ, Price TD. Pervasive reinforcement and the role of sexual selection in biological speciation. J Hered. 2014;105:821–833. doi: 10.1093/jhered/esu041. [DOI] [PubMed] [Google Scholar]
- Jallon JM, David JR. Variation in cuticular hydrocarbons among the eight species of the Drosophila melanogaster subgroup. Evolution. 1987;41(2):294–302. doi: 10.1111/j.1558-5646.1987.tb05798.x. [DOI] [PubMed] [Google Scholar]
- Jallon JM. A few chemical words exchanged by Drosophila during courtship and mating. Behav Genet. 1984;14(5):441–478. doi: 10.1007/BF01065444. [DOI] [PubMed] [Google Scholar]
- Jiggins CD, Naisbit RE, Coe RL, Mallet J. Reproductive isolation caused by colour pattern mimicry. Nature. 2001;411(6835):302–305. doi: 10.1038/35077075. [DOI] [PubMed] [Google Scholar]
- Karageorgi M, Groen SC, Sumbul F, Pelaez JN, Verster KI, Aguilar JM, et al. Genome editing retraces the evolution of toxin resistance in the monarch butterfly. Nature. 2019;574:1–4. doi: 10.1038/s41586-019-1610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS, Kondrashov FA. Interactions among quantitative traits in the course of sympatric speciation. Nature. 1999;400(6742):351–354. doi: 10.1038/22514. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68(9):2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan L, Rundle HD. Adaptation to desiccation fails to generate pre- and postmating isolation in replicate Drosophila melanogaster laboratory populations. Evolution. 2010;64(3):710–723. doi: 10.1111/j.1558-5646.2009.00864.x. [DOI] [PubMed] [Google Scholar]
- Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE, Macallister IE, et al. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci USA. 2012;109:14081–14086. doi: 10.1073/pnas.1204661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Coradini ALV, Shen A, Ehrenreich IM. Layers of cryptic genetic variation underlie a yeast complex trait. Genetics. 2019;211(4):1469–1482. doi: 10.1534/genetics.119.301907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinforma. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon RA, Diaz CD, Morano JL, Troscianko J, Stevens M, Moskeland A, et al. Evolution of correlated complexity in the radically different courtship signals of birds-of-paradise. PLoS Biol. 2018;16(11):e2006962. doi: 10.1371/journal.pbio.2006962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC. The genetic architecture of quantitative traits: lessons from Drosophila. Curr Opin Genet Dev. 2004;14(3):253–257. doi: 10.1016/j.gde.2004.04.003. [DOI] [PubMed] [Google Scholar]
- MacKay TFC. The genetic architecture of complex behaviors: lessons from Drosophila. Genetica. 2009;136(2):295–302. doi: 10.1007/s10709-008-9310-6. [DOI] [PubMed] [Google Scholar]
- Matute DR, Ayroles JF. Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J Evolut Biol. 2014;27(6):1057–1068. doi: 10.1111/jeb.12391. [DOI] [PubMed] [Google Scholar]
- McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, et al. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61(5):692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, Bargmann CI. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature. 2011;477(7364):321–325. doi: 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson Tamra C., Shaw Kerry L. Contemporary Issues in Genetics and Evolution. Dordrecht: Springer Netherlands; 2002. Genetic and behavioral components of the cryptic species boundary between Laupala cerasina and L. kohalensis (Orthoptera: Gryllidae) pp. 301–310. [PubMed] [Google Scholar]
- Merrill RM, Rastas P, Martin SH, Melo MC, Barker S, Davey J, et al. Genetic dissection of assortative mating behavior. PLoS Biol. 2019;17(2):e2005902. doi: 10.1371/journal.pbio.2005902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Glazer AM, Summers BR, Blackman BK, Norman AR, Shapiro MD, et al. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait loci. Genetics. 2014;197(1):405–420. doi: 10.1534/genetics.114.162420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring AJ, Mackay TFC. The quantitative genetic basis of male mating behavior in Drosophila melanogaster. Genetics. 2004;167(3):1249–1263. doi: 10.1534/genetics.103.024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okhovat M, Berrio A, Wallace G, Ophir AG, Phelps SM. Sexual fidelity trade-offs promote regulatory variation in the prairie vole brain. Science. 2015;350(6266):1371–1374. doi: 10.1126/science.aac5791. [DOI] [PubMed] [Google Scholar]
- Pardy JA, Rundle HD, Bernards MA, Moehring AJ. The genetic basis of female pheromone differences between Drosophila melanogaster and D. simulans. Heredity. 2018;122:93–109. doi: 10.1038/s41437-018-0080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischedda A, Shahandeh MP, Cochrane WG, Cochrane VA, Turner TL. Natural variation in the strength and direction of male mating preferences for female pheromones in Drosophila melanogaster. PLoS ONE. 2014;9(1):e87509. doi: 10.1371/journal.pone.0087509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince DJ, O’Rourke SM, Thompson TQ, Ali OA, Lyman HS, Saglam IK, et al. The evolutionary basis of premature migration in Pacific salmon highlights the utility of genomics for informing conservation. Sci Adv. 2017;3(8):e1603198. doi: 10.1126/sciadv.1603198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröhl H, Koshy RA, Mueller U, Rand AS, Ryan MJ. Geographic variation of genetic and behavioral traits in northern and southern tüngara frogs. Evol; Int J Org Evol. 2006;60(8):1669–1679. [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a drosophila population. Science. 2009;326(5960):1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Williams TM. Using Drosophila pigmentation traits to study the mechanisms of cis-regulatory evolution. Curr Opin Insect Sci. 2017;19:1–7. doi: 10.1016/j.cois.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution. 2012;66(1):1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeholzer LF, Seppo M, Stern DL, Ruta V. Evolution of a central neural circuit underlies Drosophila mate preferences. Nature. 2018;559(7715):564–569. doi: 10.1038/s41586-018-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimura T, Nijhout HF (2017) Diversity and evolution of butterfly wing patterns: an integrative approach. Springer Nature, Singapore
- Servedio MR. Beyond reinforcement: the evolution of premating isolation by direct selection on preferences and postmating, prezygotic incompatibilities. Evolution. 2001;55(10):1909–1920. doi: 10.1111/j.0014-3820.2001.tb01309.x. [DOI] [PubMed] [Google Scholar]
- Shahandeh MP, Pischedda A, Rodriguez JM, Turner TL. The genetics of male pheromone preference difference between drosophila melanogaster and Drosophila simulans. G3 Genes Genomes Genet. 2020;10(1):401. doi: 10.1534/g3.119.400780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahandeh MP, Pischedda A, Turner TL. Male mate choice via cuticular hydrocarbon pheromones drives reproductive isolation between Drosophila species. Evolution. 2018;72(1):123–135. doi: 10.1111/evo.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KL, Parsons YM. Divergence of mate recognition behavior and its consequences for genetic architectures of speciation. Am Naturalist. 2002;159(S3):S61–S75. doi: 10.1086/338373. [DOI] [PubMed] [Google Scholar]
- Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7(8):e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski MB. Drosophila: genetics meets behavior. Nat Rev Genet. 2001;2(11):879–890. doi: 10.1038/35098592. [DOI] [PubMed] [Google Scholar]
- Stacher Hörndli CN, Wong E, Ferris E, Bennett K, Steinwand S, Rhodes AN, et al. Complex economic behavior patterns are constructed from finite, genetically controlled modules of behavior. Cell Rep. 2019;28(7):1814–1829.e6. doi: 10.1016/j.celrep.2019.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Crocker J, Ding Y, Frankel N, Kappes G, Kim E, et al. Genetic and transgenic reagents for Drosophila simulans, D. mauritiana, D. yakuba, D. santomea and D. virilis. G3 Genes Genomes Genet. 2016;7(4):1339–1347. doi: 10.1534/g3.116.038885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell. 2012;149(5):1140–1151. doi: 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregenza T, Wedell N. Definitive evidence for cuticular pheromones in a cricket. Anim Behav. 1997;54(4):979–984. doi: 10.1006/anbe.1997.0500. [DOI] [PubMed] [Google Scholar]
- Weber JN, Peterson BK, Hoekstra HE. Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice. Nature. 2013;493(7432):402–405. doi: 10.1038/nature11816. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312(5770):111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- William R, George S. The evolution of reproductive isolation as a correlated character under sympatric conditions: experimental evidence. Evolution. 1990;44(5):1140–1152. doi: 10.1111/j.1558-5646.1990.tb05221.x. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Willis-Owen SAG, Fullerton J, Meesaq A, Deacon RM, Rawlins JNP, et al. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat Genet. 2004;36(11):1197–1202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
- You FM, Huo N, Gu YQ, Luo MC, Ma Y, Hane D, et al. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukilevich R. Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution. 2012;66(5):1430–1446. doi: 10.1111/j.1558-5646.2011.01534.x. [DOI] [PubMed] [Google Scholar]
- Zhang B, Xue HJ, Song KQ, Liu J, Li WZ, Nie RE, et al. Male mate recognition via cuticular hydrocarbons facilitates sexual isolation between sympatric leaf beetle sister species. J Insect Physiol. 2014;70:15–21. doi: 10.1016/j.jinsphys.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Zhou Q. A guide to QTL mapping with R/qtl. J Stat Softw. 2010;32(1):1–3. [Google Scholar]
- Zwarts L, Magwire MM, Carbone MA, Versteven M, Herteleer L, Anholt RRH, et al. Complex genetic architecture of Drosophila aggressive behavior. Proc Natl Acad Sci USA. 2011;108(41):17070–17075. doi: 10.1073/pnas.1113877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads are available at the Sequence Read Archive (accession PRJNA558426). Scripts used for sequence alignment and analysis are available at GitHub (https://github.com/mshahandeh/genarch_malechoice_Drosophila.git). Courtship and QTL genotype data are available in the Dryad Digital Repository (10.5061/dryad.6c936j6).