Abstract
Evolutionary studies over the last several decades have invoked fitness trade-offs to explain why species prefer some environments to others. However, the effects of population size on trade-offs and ecological specialization remain largely unknown. To complicate matters, trade-offs themselves have been visualized in multiple ways in the literature. Thus, it is not clear how population size can affect the various aspects of trade-offs. To address these issues, we conducted experimental evolution with Escherichia coli populations of two different sizes in two nutritionally limited environments, and studied fitness trade-offs from three different perspectives. We found that larger populations evolved greater fitness trade-offs, regardless of how trade-offs are conceptualized. Moreover, although larger populations adapted more to their selection conditions, they also became more maladapted to other environments, ultimately paying heavier costs of adaptation. To enhance the generalizability of our results, we further investigated the evolution of ecological specialization across six different environmental pairs, and found that larger populations specialized more frequently and evolved consistently steeper reaction norms of fitness. This is the first study to demonstrate a relationship between population size and fitness trade-offs, and the results are important in understanding the population genetics of ecological specialization and vulnerability to environmental changes.
Subject terms: Experimental evolution, Population genetics
Introduction
Adaptation of a biological population to a given environment may not concomitantly increase its fitness in other environments (Kassen 2002; Anderson et al. 2013; Kassen 2014; Cooper 2014; Bono et al. 2017). In many cases, adaptation to one environment can also lead to maladaptation to others (Cooper and Lenski 2000; Kassen 2002; Bataillon et al. 2011; Remold 2012). Such incongruity in fitness changes across environments forms the basis of ecological specialization, which happens when the fittest type in one environment cannot be the fittest type in another environment (Fry 1996), and tends to restrict the niche breadth of populations to a narrow range of environments (Levins 1962, 1968; Futuyma and Moreno 1988; Agrawal et al. 2010). Studies of ecological specialization routinely invoke trade-offs in fitness across environments (Levins 1962), with the latter leading to the intuitive and widespread assumption that the jack-of-all-trades is a master-of-none (MacArthur 1984). Such fitness trade-offs and ecological specialization underlie the evolution of a wide range of biological properties and processes, including (but not restricted to) the composition of ecological communities (Kneitel and Chase 2004; Farahpour et al. 2018), host specificity in several systems (Rausher 1984; Joshi and Thompson 1995; Messina and Durham 2015), virulence (Messenger et al. 1999), resistance to a variety of agents like herbivores (Koricheva 2002), parasites (Boots 2011), and antibiotics (Andersson and Hughes 2010; MacLean et al. 2010).
The term “trade-off” has been used in a variety of contexts in evolutionary studies (Agrawal et al. 2010), with the following three main usages: (1) when a trait that is adaptive in a given environment is costly (maladaptive or detrimental to fitness) in others (Bell and Reboud 1997; Kassen 2014; Rodríguez-Verdugo et al. 2014; Sane et al. 2018); (2) unequal adaptation to alternative environments, wherein populations become adapted to all the environments under consideration, but no single genotype can be the fittest one in all the environments (Bell and Reboud 1997; Remold 2012; Kassen 2014); (3) life-history trade-offs across traits that arise within a single environment due to the systemic properties and constraints of organismal features (Stearns 1989; Prasad et al. 2001; Knops et al. 2007). The first two of the above usages pertain to trade-offs in fitness across environments, which can directly give rise to ecological specialization. Although the physiological and molecular mechanisms of trade-offs and specialization are difficult to decipher in multicellular organisms (Agrawal et al. 2010), there is a fairly detailed understanding of such mechanisms in microbes (Ferenci 2016). However, the role of a key parameter like population size in determining fitness trade-offs, and the resultant specialization across a pair of environments, remains relatively unclear, even in asexual microbes.
Population size is known to shape a large variety of evolutionary phenomena and properties, including rate and extent of adaptation (Desai and Fisher 2007; Desai et al. 2007; Sniegowski and Gerrish 2010; Chavhan et al. 2019a), repeatability of adaptation (Szendro et al. 2013; Lachapelle et al. 2015), biological complexity (LaBar and Adami 2016), and efficiency of natural selection (Ohta 1992; Petit and Barbadilla 2009; Chavhan et al. 2019a). Numerous theoretical and empirical results have indirectly linked population size with the extent of ecological specialization. For example, multiple theoretical and empirical studies have established that larger populations generally adapt faster (Gerrish and Lenski 1998; Desai and Fisher 2007; Desai et al. 2007; Sniegowski and Gerrish 2010). Moreover, larger populations are expected to adapt primarily via rare large-effect beneficial mutations, while relatively smaller populations adapt slower through common beneficial mutations of modest- effect sizes (reviewed in Sniegowski and Gerrish 2010). Interestingly, the link between the size of a beneficial mutation and its deleterious pleiotropic effects has been touched upon by a rich array of theoretical studies. In Fisher’s geometrical model of adaptation on a multidimensional phenotype space where each orthogonal dimension represents a trait, larger mutational effect sizes are associated with greater probabilities of affecting multiple traits deleteriously (Fisher 1930). This was the basis of Fisher’s argument for micromutationism, the idea that adaptation is largely driven by mutations of very small effects. Although Fisher’s original model assumed that all traits are equally important to fitness, it is easy to imagine cases where a few focal trait(s) affect(s) fitness while others do not (Orr and Coyne 1992). In such a case, beneficial mutations with larger effects on the focal trait(s) would not only be selectively favored, but also likely to be associated with large pleiotropic disadvantages to other traits. Along these lines, several theoretical studies assume that larger mutational benefits also have heavier pleiotropic disadvantages (Lande 1983; Orr and Coyne 1992; Otto 2004). When we combine these two insights, a new testable hypothesis emerges: larger asexual populations should show greater specialization by adapting more specifically to their environment of selection, and should also suffer heavier costs in alternative environments. To the best of our knowledge, there are no direct experimental tests of the relationships of such specializations and their underlying trade-offs with population size. To begin with, as pointed out repeatedly in the literature, experimental evolution studies of fitness trade-offs and the resulting specialization have not been conducted at variable population sizes (Kawecki et al. 2012; Bataillon et al. 2013; Cooper 2014; Kraemer and Boynton 2017). Furthermore, several recent evolution experiments with microbes, which have provided important insights in this regard, have focused on the pleiotropic profiles of individual mutations (reviewed in Bono et al. 2017), and not on population-level properties like population size. To address this lacuna, here we use experimental evolution with Escherichia coli populations of different sizes to test if larger populations evolve bigger fitness trade-offs and specialize more across environments.
To avoid semantic ambiguity, we follow Fry (1996) and define specialization across two environments as any case where the reaction norms for fitness intersect with each other. Evolutionary experiments have conventionally studied fitness trade-offs across environments from three major perspectives: (1) as negative correlations in fitness of populations selected in two or more distinct environments (Bell and Reboud 1997; Jessup and Bohannan 2008; Lee et al. 2009); (2) as fitness deficits below the ancestral levels (costs of adaptation) in the alternative environment(s) that accompany adaptation to the environment in which evolution takes place (Lee et al. 2009; Bono et al. 2017); (3) as differences in fitness across environmental pairs (Kassen 2014; Schick et al. 2015). Although these perspectives can potentially be related, they are clearly not equivalent, and therefore might lead to different insights about the process of ecological specialization. For example, ecological specialization can happen with or without costs of adaptation (see Appendix S1 and Fig. S1 in Supplementary Information for details). Therefore, we decided to use all above three perspectives to investigate how population size affects fitness trade-offs and the resulting specialization across environments. To this end, we propagated replicate E. coli populations of two different sizes in two different nutritionally limiting environments (galactose minimal medium and thymidine minimal medium). We note that these experiments can also be contextualized in terms of a combination of two other questions: (A) do pleiotropic effects act reciprocally across a pair of environments in question? (B) Are pleiotropic effects correlated to the corresponding direct effects on fitness? Although several previous studies have investigated these questions separately, they have not combined them in order to look at the effects of population size on fitness trade-offs. With regard to the reciprocity of trade-offs, for example, collateral sensitivity profiles have been shown to be reciprocal across several pairs of antibiotics (Imamovic and Sommer 2013). Contrastingly, other studies have found asymmetric (and not reciprocal) trade-offs across pairs of environments (Travisano 1997; Lee et al. 2009). Along similar lines, previous studies have reported evidence of both high and low correlations of pleiotropic and direct fitness effects (Ostrowski et al. 2005; Sane et al. 2018). Linking the above two questions, we also sought to determine if our experimental populations evolved reciprocal trade-offs, and whether direct fitness gains in one environment led to correlated disadvantages in the other.
We found that, descending from a common ancestor, our experimental populations evolved a strong negative correlation between fitness across the two environments. As expected, the larger populations adapted more to the selection environments. Interestingly, the larger populations also paid heavier costs of adaptation. We further assayed the fitness of the evolved populations in two more nutritionally limiting environments, which enabled us to quantify the extent of specialization across six environmental pairs. Remarkably, we found that the larger populations specialized more, evolving steeper reaction norms of fitness. To the best of our knowledge, this is the first study to directly test the effects of population size on ecological specialization brought about by fitness trade-offs.
Materials and methods
Experimental evolution
We founded 24 populations from a single E. coli MG1655 colony and propagated them for ~480 generations at two different population sizes: large (L) or small (S) (defined below). For each population size, we had two different kinds of environments: thymidine (T) or galactose (G) as the sole carbon source in a M9-based minimal medium (for details, see Appendix S2 in Supplementary information). This 2 × 2 design gave rise to four population types (TL, TS, GL, and GS) where the first letter represented the only carbon source in the selection environment, and the second letter represented the population size. We chose these carbon sources because they have very different metabolic pathways in E. coli (Frey 1996; Loh et al. 2006; Díaz-Mejía et al. 2009; Barupal et al. 2013). Furthermore, galactose and thymidine differ in terms of their uptake within E. coli cells. Specifically, whereas the uptake of thymidine occurs via NupG and NupC proteins (Patching et al. 2005), galactose uptake is mediated by GalP (Henderson et al. 1992). Taken together, based on disparate metabolism and uptake mechanisms of the two carbon sources, we expected E. coli populations to show fitness trade-offs across them. Each population type had six independently evolving replicate populations. We propagated all the 24 populations using the standard batch-culture technique at a volume of 300 µl in 96-well plates shaking continuously at 150 rpm at 37 °C. The large populations (TL and GL) had a periodic bottleneck of 1:10, while the small ones (TS and GS) faced a periodic bottleneck of 1:104. To ensure that our treatments did not spend vastly different amounts of time in the stationary phase, we bottlenecked the large populations every 12 h (every 3.3 generations), and the small ones every 48 h (every 13.3 generations). Overall, L and S corresponded approximately to 9.9 × 107 and 3.9 × 105, respectively, in terms of the harmonic mean population size (Lenski et al. 1991). In terms of the measure of population size relevant for the extent of adaptation in asexual populations (Chavhan et al. 2019a), L and S corresponded approximately to 9.0 × 106 and 2.2 × 103, respectively.
Quantification of fitness and trade-offs across environments
At the end of our evolution experiment, we revived the cryostocks belonging to each experimental population in glucose-based M9 minimal medium, and allowed them to grow for 24 h. Next, we performed automated growth assays on each of the 24 revived populations using a well-plate reader in multiple environments (Synergy HT, BIOTEK® Winooski, VT, USA). Using optical density at 600 nm as the measure of population density, we obtained growth readings every 20 min for 24 h. We ensured that the physical conditions during the assays were identical to culture conditions (96-well plates shaking at 150 rpm at 37 °C). Using a randomized complete block design (RCBD), we conducted the fitness measurements over six different days, assaying one replicate population of each type in both the environments on a given day (Milliken and Johnson 2009). We estimated fitness as the maximum growth rate (R) (Kassen 2014; Ketola and Saarinen 2015; Vogwill et al. 2016), which was computed as the maximum slope of the growth curve over a moving window of ten readings (Leiby and Marx 2014; Karve et al. 2015, 2016, 2018; Chavhan et al. 2019a, b).
We labeled the environment in which selection occurred as “home” and the other (alternative) environment(s) as “away.” The presence of the common ancestor as the reference against which fitness gains or reductions could be tested allowed us to differentiate between ecological specialization and costs of adaptation. We studied trade-offs and ecological specialization from three major conventional perspectives:
(1) If fitness trade-offs exist between two environments, selection is expected to result in strong negative correlations in fitness across them (Kassen 2014). Therefore, we determined if relative fitness in Galactose (henceforth “Gal”) had a significant negative correlation with relative fitness in Thymidine (henceforth “Thy”).
(2) We also determined if our experimental populations paid significant costs of adaptation. To this end, we first established whether our experimental populations had adapted significantly to their home environment (Thy for TL/TS and Gal for GL/GS). Next, we determined if the populations had maladapted significantly to their away environment (Gal for TL/TS and Thy for GL/GS). For this, we normalized all fitness values in a given environment by the ancestral fitness in the corresponding environment, which is equivalent to scaling the ancestral fitness value to 1 (Kassen 2014). We then used single-sample t-tests to ascertain if the fitness of a given population type differed significantly from the ancestor. We corrected for inflations in family-wise error rate using the Holm–Šidák procedure (Abdi 2010). We also computed Cohen’s d to analyse the statistical significance of these differences in terms of effect sizes (Cohen 1988). We concluded that a population had paid a cost of adaptation only if it had adapted significantly to its home environment (i.e., scaled fitness >1), and simultaneously maladapted significantly to its away environment (i.e., scaled fitness <1).
We used a mixed-model ANOVA with RCBD to analyze if population size and the identity of the home environment interacted with each other statistically to shape the fitness in home environment (henceforth Fitnesshome). To this end, we used “population size” (two levels: L or S) and “home environment” (two levels: Gal or Thy) as fixed factors crossed with each other, and “day of assay” (six levels: 1–6) as the random factor. We also analyzed the effect size of the main effects using partial η2, interpreting the latter as representing a small, medium, or large effect for partial η2 < 0.06, 0.06 < partial η2 < 0.14, and 0.14 < partial η2, respectively (Cohen 1988).
Furthermore, we analyzed if the relative extent of fitness loss in the away environment (=1—Fitnessaway) was significantly different for the large and small populations. To this end, we used a mixed-model ANOVA (RCBD) with “population size” (two levels: L or S) and “home-away pair” (two levels: Gal–Thy or Thy–Gal) as fixed factors crossed with each other, and “day of assay” (six levels: 1–6) as the random factor.
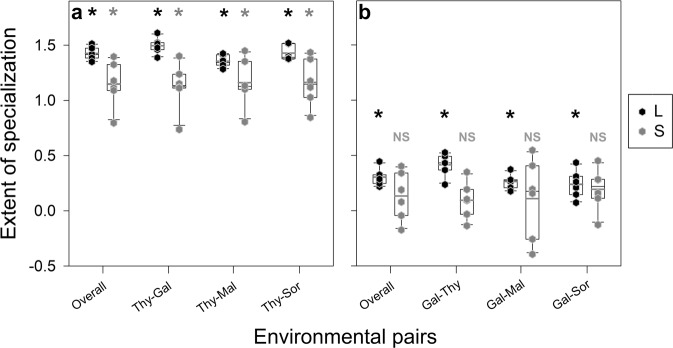
(3) We quantified the environmental specificity of adaptation using differences in the relative fitness of experimental populations across different home-away environmental pairs. This quantity represents the difference in the degrees to which a population adapts to the two environments under consideration (Remold 2012). Such a difference between home- and away-relative fitness values (=Fitnesshome–Fitnessaway) can be represented graphically as slopes of reaction norms of fitness. Since the quantification of reaction norm slopes does not require selection in each assay environment, we enhanced the generalizability of our study by assaying the fitness of all the 24 evolved populations (TL, TS, GL, and GS) in two more nutritionally limited environments (Maltose minimal medium (henceforth “Mal”) and Sorbitol minimal medium (henceforth “Sor”)). This allowed us to compare the reaction norm slopes of large and small populations across six home-away environmental pairs (Thy–Gal, Thy–Mal, and Thy–Sor for TL and TS; Gal–Thy, Gal–Mal, and Gal–Sor for GL and GS).
We first determined if a population type had specialized significantly across a given home-away pair. We followed Fry (1996) to identify specialization across a pair of environments as any case where the population’s reaction norm intersected with the corresponding ancestral reaction norm. This would happen whenever the unambiguous fittest type in one environment is not unambiguously the fittest type in the other environment (Fig. 1). To identify cases of specialization, we first determined if the population type in question had significantly different Fitnesshome relative to the common ancestor. Next, we determined if the population type’s Fitnessaway was significantly different from that of the ancestor. If the population type increased its fitness significantly in both its home and away environments, its reaction norm would not intersect with the ancestral norm. This would imply lack of ecological specialization. On the other hand, if the population type’s fitness increased significantly in its home environment, but failed to do so in its away environment, its reaction norm would intersect with the ancestral norm, revealing significant specialization across the environmental pair in question (Fig. 1). We performed this procedure for each of the four population types to determine if specialization had occurred across the six home-away pairs under consideration.
Fig. 1. Schematic representation of ecological specialization across two environments.
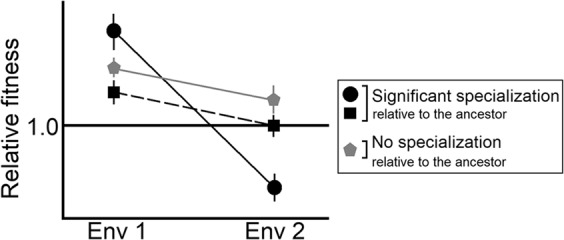
All the fitness values are scaled by the ancestral value (=1 (horizontal black line)). The error bars represent 95% confidence intervals. Significant ecological specialization occurs when the reaction norms of fitness intersect (which happens when the unambiguous fittest type in one environment is not the unambiguous fittest type in the other environment).
We also compared the reaction norm slopes of each of the four population types with those of the ancestor across all the home-away pairs under consideration. To this end, we conducted single-sample t-tests against the ancestral level (ancestral reaction norms have zero slope), followed by correction for family-wise error rates using the Holm–Šidák procedure.
To further study how population size affects the specificity of adaptation, we determined if the reaction norm slopes were significantly different across large and small populations. To this end, we conducted two separate mixed-model ANOVAs (RCBD), one for selection in Thy and the other for selection in Gal. The design of these mixed-model ANOVAs had “population size” (two levels: L or S) and “home-away pair” (three home-away pairs) as fixed factors crossed with each other, and “day of assay” (six levels: 1–6) as the random factor.
Results
Fitness trade-offs as negative correlations: fitness in Gal was negatively correlated with fitness in Thy
We found a strong negative correlation between fitness in Gal and fitness in Thy (Fig. 2; Spearman’s ρ = –0.744; P = 3.04 × 10–5). This negative correlation revealed that the fittest type in Gal was never the fittest type in Thy (compare Fig. 2 with Fig. S1), which implied the occurrence of ecological specialization in our experimental populations.
Fig. 2. Correlation between relative fitness values in galactose and thymidine minimal media after evolution in these environments at two different population sizes.
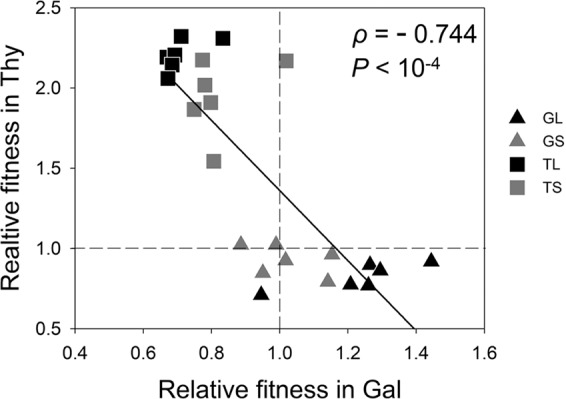
The black line represents the best linear fit (R² = 0.63); the dotted lines represent ancestral levels of fitness.
Since trade-offs based on negative fitness correlations can potentially happen with or without costs of adaptation (Fig. S1; Fry 1996; Kassen 2014), we next determined if our populations had evolved significant costs of adaptation. We also examined whether larger populations had evolved greater costs of adaptation.
Trade-offs as costs of adaptation: larger populations paid more costs of adaptation
Following Kassen (2014), we defined costs of adaptation as the simultaneous occurrence of fitness increase (adaptation) in the home environment and fitness decrease in the away environment (maladaptation). A comparison of Fig. 2 with Fig. S1 shows that the negative correlation between relative fitness values in Gal and Thy was accompanied by costs of adaptation.
To begin with, we found a significant main effect of population size on adaptation to the home environment, with the larger populations showing higher Fitnesshome than the smaller ones (mixed-model ANOVA: population size (F1,15 = 10.998, P = 0.005, η2 = 0.423 (large effect)). We also found a significant main effect of the home environment (F1,15 = 176.969, P = 1.044 × 10–9, η2 = 0.921 (large effect)). Importantly, we did not find a significant population size × home environment interaction (F1,15 = 0.102, P = 0.753).
We found that evolution in Thy resulted in significant costs of adaptation in the case of both large (TL) and small (TS) populations (Table 1), i.e., both TL and TS adapted to Thy, but became maladapted to Gal (Fig. 2; Table 1). However, evolution in Gal incurred significant costs of adaptation only in the case of the large populations (GL); i.e., the GL populations adapted to Gal, but became maladapted to Thy (Fig. 2; Table 1). The small populations that evolved in Gal (GS) neither adapted significantly to Gal nor became significantly maladapted to Thy (Fig. 2; Table 1). Thus, the GS populations did not pay any costs of adaptation.
Table 1.
Occurrence of adaptation and maladaptation events in population types selected in Gal and Thy separately.
| Population type | Fitness change in Gal | Fitness change in Thy | Cost of adaptation |
|---|---|---|---|
| TL | Maladaptation, P = 2.683 × 10–4 | Adaptation, P = 3.240 × 10–6 | Yes |
| TS | Maladaptation, P = 0.007 | Adaptation, P = 7.327 × 10–4 | Yes |
| GL | Adaptation, P = 0.049 | Maladaptation, P = 0.013 | Yes |
| GS | Not significant, P = 0.617 | Not significant, P = 0.122 | None |
The Holm–Sidak corrected P values correspond to single-sample t-tests against the ancestral levels of fitness (=1). P < 0.05 is shown in boldface. All the cases with P < 0.05 were also found to have large-effect sizes (see Table S1).
Hence, larger populations paid significant costs of adaptation in both the environments, but the smaller ones did so only in one of the two environments under consideration.
Next, we determined if larger populations also had a higher magnitude of loss in relative fitness below the ancestral levels in their away environments. Indeed, we found that the larger populations lost significantly greater fitness than the smaller ones in their away environments (Fig. 3, mixed-model ANOVA: population size (main effect) F1,15 = 9.558, P = 0.007, partial η2 = 0.389 (large effect); home environment (main effect) F1,15 = 9.650, P = 0.007, partial η2 = 0.391 (large effect); population size × home environment (interaction) F1,15 = 0.002, P = 0.963). This result also implies that the effect of population size on Fitnessaway would be the opposite of its effects on Fitnesshome. Table 1 also shows that adaptation in Thy (in the case of TL and TS) was always accompanied by maladaptation in Gal; analogously, populations that adapted significantly to Gal showed significant maladaptation in Thy. This demonstrates that the fitness trade-offs were reciprocal across the Thy–Gal environmental pair.
Fig. 3. Loss of fitness below the ancestral levels in the away environments.
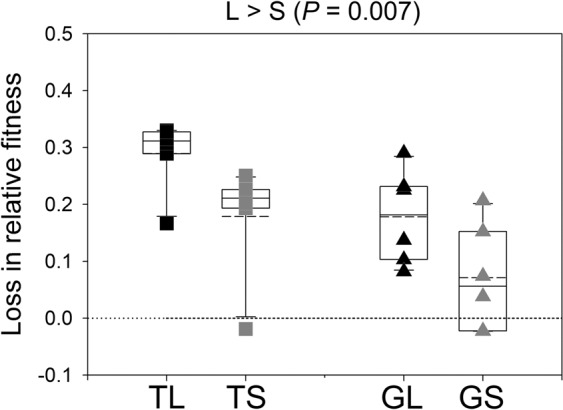
In each case, the loss in relative fitness was computed as the difference between the descendant population’s relative fitness and the ancestor’s relative fitness. L and S represent large and small populations, respectively. The solid lines in the box plots mark the 25th, 50th, and 75th percentiles, while the whiskers mark the 10th and 90th percentiles; the dashed lines represent means (N = 6). The dotted line represents no loss from the ancestral fitness levels. See the text for details.
Overall, larger population adapted more to their home environments while paying greater costs of adaptation. This made them significantly more maladapted to the away environments.
Thus, as compared with the small populations, the large populations lost more Fitnessaway. We note that the magnitudes of such fitness decline cannot reflect the full extent of ecological specialization. This is because the former only represent deficits in Fitnessaway, whereas ecological specialization involves a difference between the extents of adaptation across the home and away environments. Thus, as the next logical step, we set out to measure the extent of specialization in our experimental populations.
Fitness trade-offs as extents of ecological specialization: larger populations specialized more
We used the reaction norm slope as a measure of the specificity of adaptation, computed as the difference between Fitnesshome and Fitnessaway. As described earlier, we assayed the fitness of our experimental populations in two more nutrient-limited environments, which allowed us to compute the reaction norm slope across six different environmental pairs (three for Gal-selected populations and three for Thy-selected populations). Moreover, the normalization of fitness in any given environment with the corresponding ancestral value ensured that the slope of the ancestral reaction norms of relative fitness is zero across all environmental pairs (Kassen 2014).
First, we determined whether our experimental populations had specialized significantly to their home environment. Following a previous study (Fry 1996), we identified the evolution of specialization as the intersection of the reaction norms of the descendant treatments with that of the ancestor (Fig. 1).
On the one hand, the TL and TS populations had significantly greater fitness than the ancestor in their home environment (Table 1). On the other hand, the TL and TS populations had lower fitness than the ancestor in all the three away environments under consideration (Table S1). This reveals that the average reaction norms of both TL and TS populations intersected with the ancestral norms across all the three environmental pairs under consideration (Thy–Gal, Thy–Mal, and Thy–Sor) (Fig. 4a). Hence, both the TL and TS populations had specialized significantly across all the three home-away pairs.
Fig. 4. Reaction norms of fitness across the six home-away environmental pairs used in our study.
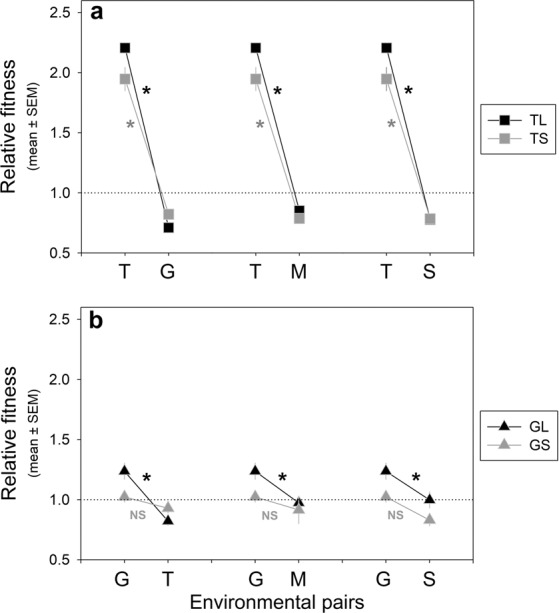
The error bars represent SEM (N = 6). The asterisks represent significant ecological specialization with respect to the ancestor for the corresponding home-away pair; “NS” denotes that the corresponding ecological specialization was not significant (see Table S1 and the text for details). The dotted lines represent ancestral reaction norms. Significant specialization happened when the reaction norms of a treatment population intersected with the ancestral norm. a Reaction norms for populations selected in Thy (TL (large) and TS (small)). b Reaction norms for populations selected in Gal (GL (large) and GS (small)). Also see Fig. 5 for reaction norm slopes across the six environmental pairs.
The large populations evolved in Gal (GL) had adapted significantly to their home environments (Table 1). Furthermore, the relative fitness of GL was not significantly greater than the ancestor in any of the three away environments (Table S1). Combining these pieces of information, GL populations specialized significantly (i.e., the fittest type in home environment was not the unambiguous fittest type in the away environment) across all the three home-away pairs (Gal–Thy, Gal–Mal, and Gal–Sor) (Fig. 4b). Interestingly, the small populations evolved in Gal (GS) did not have significantly different fitness as compared with the ancestor in any of the four environments under consideration (Table S1). This implies that the GS populations did not specialize significantly across any of the three home-away pairs under consideration (Fig. 4b).
Among the Thy-selected populations, we found that both the large (TL) and small (TS) populations evolved significantly greater reaction norm slopes than those of the ancestor (i.e., reaction norm slope = 0) (Fig. 5a and Table S2).
Fig. 5. Slopes of reaction norms of the fitness of our experimental populations across six environmental pairs.
The asterisks represent significant differences (single-sample t-tests (P < 0.05)) from the ancestral slope (=0); “NS” denotes that the corresponding slopes are not significantly different from 0. The solid lines in the box plots mark the 25th, 50th, and 75th percentiles, while the whiskers mark the 10th and 90th percentiles; the thick horizontal lines represent means (N = 6). See the text and Table S2 for details, and Fig. 4 for reaction norms across the six environmental pairs. a Populations evolved in Thy: reaction norm slopes (L > S (P < 10–6)). b Populations evolved in Gal: reaction norm slopes (L > S (P < 10–2)). Overall, the larger populations had steeper reaction norms.
Among the Gal-selected populations, we found that the GL populations had significantly steeper reaction norms than the ancestor across all the three home-away pairs under consideration (Fig. 5b and Table S2). However, the reaction norm slopes of the GS populations were not significantly different from those of the ancestor across any of the three home-away pairs (Fig. 5b and Table S2).
We further determined if larger populations had steeper reaction norms than smaller populations over the six different home-away environmental pairs. Indeed, we found a significant main effect of population size, with the larger populations evolving steeper reaction norms than the smaller ones; importantly, population size and home-away pair did not show significant statistical interaction (Fig. 5: mixed-model ANOVA for Thy-selected lines: population size (main effect) F1,25 = 43.664, P = 6.357 × 10–7, partial η2 = 0.636 (large effect); home-away pair (main effect) F2,25 = 0.566, P = 0.575; population size × home-away pair (interaction) F2,25 = 1.471, P = 0.249; mixed-model ANOVA for Gal-selected lines: population size (main effect) F1,25 = 8.147, P = 0.009, partial η2 = 0.246 (large effect); home-away pair (main effect) F2,25 = 0.428, P = 0.657; population size × home-away pair (interaction) F2,25 = 1.721, P = 0.199).
Overall, the large populations not only specialized more frequently (six home-away pairs for the large populations vs. three for the small ones), but they also evolved higher magnitudes of specificity of adaptation.
Discussion
Fitness trade-offs across environments and the ensuing ecological specialization play key roles in understanding a variety of important phenomena, including the maintenance of biodiversity and local adaptation (reviewed in Futuyma and Moreno 1988; Agrawal et al. 2010). However, little is known about the relationship between fitness trade-offs and population size, even in relatively simple organisms like microbes.
In this study, we conducted experimental evolution to directly test how population size influences fitness trade-offs and the resulting ecological specialization. Inconsistencies in the usage of terms like trade-offs and costs of adaptation in the evolutionary biology literature complicate comparisons across studies. Therefore, here we investigated fitness trade-offs with three different (but not necessarily independent) perspectives. Our primary result is that regardless of how we chose to visualize trade-off, larger populations suffered more fitness trade-offs, and thus evolved higher extents of ecological specialization. To the best of our knowledge, this is the first study to address and experimentally demonstrate this relationship between population size and fitness trade-offs.
The theory of adaptive dynamics in asexual populations predicts that while larger populations adapt primarily via rare large-effect beneficial mutations, such mutations remain largely inaccessible to small populations, which adapt via mutations of relatively smaller effect sizes (Sniegowski and Gerrish 2010; Chavhan et al. 2019a). Moreover, a large body of studies suggests that mutational benefits of large sizes lead to heavier disadvantages due to antagonistic pleiotropy (Orr and Coyne 1992; Otto 2004; Griswold 2007; Hague et al. 2018). Our result that larger population pay higher costs of adaptation can thus be explained by a combination of the above two ideas. In other words, the notion that larger populations adapt primarily via large-effect beneficial mutations, which, in turn, are expected to lead to higher pleiotropic disadvantages, can potentially explain why larger population paid higher costs of adaptation, which resulted in steeper reaction norms (Figs. 5 and S2). Furthermore, since multiple beneficial mutations can simultaneously rise to high frequencies in large populations (Desai and Fisher 2007; Desai et al. 2007), our observations can also be explained by the pleiotropic effects of a higher number of beneficial mutations in the larger experimental populations (TL and GL). Although our observations align with the underlying assumption that larger-effect beneficial mutations should be associated with heavier pleiotropic disadvantages, we have not presented direct genetic evidence for this assumption. We note that the small experimental populations in our study (TS and GS) harbored enough individuals to undergo clonal interference between distinct beneficial mutations, while the larger populations (TL and GL) likely underwent adaptation based on multiple beneficial mutations within 480 generations (Desai and Fisher 2007). Although the timescale of 480 generations is long enough to lead to substantial fitness increase, it is likely to be inadequate for the fixation of multiple beneficial mutations (Cooper 2018). Thus, our experimental populations are expected to be genetically heterogeneous. Hence, any genetic test of the underlying assumption regarding the scaling of pleiotropy with the direct effects of the individual mutations in our experiment would need to account for the genetic background (epistatic interactions) and allelic frequencies at several loci. Although interesting in its own right, such an investigation was out of the scope of this study.
A previous study with a collection of single beneficial mutations had found their pleiotropic effects in terms of carbon usage to be rarely antagonistic (Ostrowski et al. 2005), which contrasts with our observations. Below, we briefly speculate on the basis of these differences. Ostrowski et al. (2005) had determined the pleiotropic effects of single beneficial mutations that had started rising in frequency during selection in glucose minimal medium (Rozen et al. 2002). Glucose is known to be the preferred carbon source for E. coli (Brückner and Titgemeyer 2002; Görke and Stülke 2008). This preference for glucose is a likely outcome of metabolic optimization during the evolutionary history of these bacteria. Thus, the scope for adaptation on glucose should be relatively lower as compared with that on other carbon sources (like Thy and Gal). Since lower scope for adaptation is associated with slower increase in fitness (Couce and Tenaillon 2015), adaptation in glucose minimal medium should be relatively slower. Indeed, it took more than 1000 generations for the populations in Lenski’s long term evolution experiment to increase their growth rate by ~30% on glucose as the sole carbon source (Novak et al. 2006). Contrastingly, even the small populations of our study could increase their growth rate in Thy by >70% within 480 generations (this increase was >100% for the large populations). As compared with Thy, Gal is closer to glucose in terms of metabolic pathways, suggesting that the adaptation in Gal should be slower than that in Thy. Our observations agree with this expectation (~20% increase in GL and no significant increase in GS) (Table 1). Furthermore, we note that the average fitness effect of mutations enriched on glucose for 400 generations reported in Ostrowski et al. (2005) was approximately 10%. Taken together, the direct effects of beneficial mutations that drove adaptation in our study (in TL, TS, and GL, but not in GS) are expected to be greater than those reported in Ostrowski et al. (2005). The small size of direct fitness in their study could be a potential reason behind the lack of detectable antagonistic pleiotropic effects. Thus, the notion that larger direct effects are associated with heavier pleiotropic disadvantages could be enough to explain why our results are different from Ostrowski et al. (2005). Last, since GL and TL populations are large enough to have multiple beneficial mutations within the same lineage rising simultaneously to high frequencies, the substantial pleiotropic costs in these populations are likely to be associated with multiple mutations as opposed to single mutations as reported in Ostrowski et al. (2005).
We briefly note that although maladaptation to the away environments can potentially be caused by the accumulation (via drift in the home environment) of neutral mutations that are contextually deleterious in the away environments, it is unlikely to be an explanation of our observations. There are two key reasons behind this assertion. First, a period of 480 generations is too small for mutation accumulation to show its phenotypic effects in clonally derived bacterial populations with harmonic mean population sizes >104 (which happens to be the case here) (Kassen 2002; Cooper 2018). Second, the effects of accumulation of conditionally neutral mutations are not expected to be different across populations of different sizes (Kimura 1983; Hall and Colegrave 2008).
Our study shows that when the environment remains constant for long periods, adaptation in larger numbers can make populations more specialized to this environment. This relationship between population size and ecological specialization has many important implications. Foremost, owing to their higher extent of specialization, larger populations can become vulnerable to sudden changes in the environment, as predicted by a recent study (Chavhan et al. 2019b). Interestingly, if the environment abruptly shifts between two states that show fitness trade-offs with each other, then populations with a history of evolution at larger numbers would be at a greater disadvantage than historically smaller populations. Microbial populations routinely experience such abrupt shifts across environmental states that are known to show fitness trade-offs with each other. For example, costs of antimicrobial resistance are expected to check the spread of resistant microbes if antimicrobials are removed abruptly from the environments (Andersson and Hughes 2010; Hill et al. 2015). Moreover, pathogens are also expected to experience fitness trade-offs when they migrate across different hosts (Turner and Elena 2000; Smith-Tsurkan et al. 2010).
Our results also predict that in the face of environmental changes, larger populations may not adapt better than smaller ones. Pleiotropy has been routinely invoked to explain why evolution should mostly proceed via small-effect mutations in nature (where the environment is rarely constant, both spatially and temporally) (Lande 1983; Orr and Coyne 1992; Tenaillon 2014; Dillon et al. 2016). Our results are in accordance with this long-held assumption, and lead to the prediction that environmental fluctuations across states that show fitness trade-offs can potentially explain why small populations can be successful in nature. We note that the evolution of ecological specialization may sometimes require thousands of generations (Kassen 2002, 2014; Cooper 2014). It would therefore be particularly interesting to study how specialization evolves in populations of different sizes if the environment fluctuates at much smaller timescales (tens of generations). Our results can act as stepping stones for more complex investigations of the links between population size and trade-offs, particularly in fluctuating environments.
Supplementary information
Acknowledgements
We thank Milind Watve and MS Madhusudhan for their valuable inputs. YC was supported by a Senior Research Fellowship initially sponsored by IISER Pune and then by Council for Scientific and Industrial Research (CSIR), Govt. of India. SM was supported by an INSPIRE undergraduate fellowship, sponsored by the Department of Science and Technology (DST), Govt. of India. This project was supported by an external grant (BT/PR22328/BRB/10/1569/2016) from the Department of Biotechnology, Govt. of India, and internal funding from IISER Pune.
Author contributions
YC and SD designed the study. YC and SM conducted the experiments. YC analyzed the data. YC and SD wrote the paper with inputs from SM.
Data availability
All the data relevant to this study can be found at 10.5061/dryad.bnzs7h46z.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Louise Johnson
Supplementary information
The online version of this article (10.1038/s41437-020-0308-x) contains supplementary material, which is available to authorized users.
References
- Abdi H (2010) Holm’s sequential Bonferroni procedure. In: Salkind N (ed) Encyclopedia of research design, p 1–8. Sage, Thousand Oaks, CA
- Agrawal AA, Conner JK, Rasmann S. Tradeoffs and negative correlations in evolutionary ecology. Evol Darwin First. 2010;150:243–268. [Google Scholar]
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- Anderson JT, Lee C-R, Rushworth CA, Colautti RI, Mitchell-Olds T. Genetic trade-offs and conditional neutrality contribute to local adaptation. Mol Ecol. 2013;22:699–708. doi: 10.1111/j.1365-294X.2012.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barupal DK, Lee SJ, Karoly ED, Adhya S. Inactivation of metabolic genes causes short- and long-range dys-regulation in Escherichia coli metabolic network. PLoS ONE. 2013;8:e78360. doi: 10.1371/journal.pone.0078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillon T, Joyce P, Sniegowski P. As it happens: current directions in experimental evolution. Biol Lett. 2013;9:20120945. doi: 10.1098/rsbl.2012.0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillon T, Zhang T, Kassen R. Cost of adaptation and fitness effects of beneficial mutations in pseudomonas fluorescens. Genetics. 2011;189:939–949. doi: 10.1534/genetics.111.130468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G, Reboud X. Experimental evolution in Chlamydomonas II. Genetic variation in strongly contrasted environments. Heredity. 1997;78:498–506. [Google Scholar]
- Bono LM, Smith LB, Pfennig DW, Burch CL. The emergence of performance trade-offs during local adaptation: insights from experimental evolution. Mol Ecol. 2017;26:1720–1733. doi: 10.1111/mec.13979. [DOI] [PubMed] [Google Scholar]
- Boots M. The evolution of resistance to a parasite is determined by resources. Am Nat. 2011;178:214–220. doi: 10.1086/660833. [DOI] [PubMed] [Google Scholar]
- Brückner R, Titgemeyer F. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett. 2002;209:141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- Chavhan Y, Ali SI, Dey S. Larger numbers can impede adaptation in asexual populations despite entailing greater genetic variation. Evol Biol. 2019;46:1–13. [Google Scholar]
- Chavhan Y, Karve S, Dey S. Adapting in larger numbers can increase the vulnerability of Escherichia coli populations to environmental changes. Evolution. 2019;73:836–846. doi: 10.1111/evo.13700. [DOI] [PubMed] [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. Hillsdale, N.J: L. Erlbaum Associates; 1988. [Google Scholar]
- Cooper VS. The origins of specialization: insights from bacteria held 25 years in captivity. PLOS Biol. 2014;12:e1001790. doi: 10.1371/journal.pbio.1001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS. Experimental evolution as a high-throughput screen for genetic adaptations. mSphere. 2018;3:e00121–18. doi: 10.1128/mSphere.00121-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- Couce A, Tenaillon OA. The rule of declining adaptability in microbial evolution experiments. Front Genet. 2015;6:99. doi: 10.3389/fgene.2015.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MM, Fisher DS. Beneficial mutation–selection balance and the effect of linkage on positive selection. Genetics. 2007;176:1759–1798. doi: 10.1534/genetics.106.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MM, Fisher DS, Murray AW. The speed of evolution and maintenance of variation in asexual populations. Curr Biol. 2007;17:385–394. doi: 10.1016/j.cub.2007.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Mejía JJ, Babu M, Emili A. Computational and experimental approaches to chart the Escherichia coli cell-envelope-associated proteome and interactome. FEMS Microbiol Rev. 2009;33:66–97. doi: 10.1111/j.1574-6976.2008.00141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon MM, Rouillard NP, Dam B, Gallet R, Cooper VS. Diverse phenotypic and genetic responses to short-term selection in evolving Escherichia coli populations. Evolution. 2016;70:586–599. doi: 10.1111/evo.12868. [DOI] [PubMed] [Google Scholar]
- Farahpour F, Saeedghalati M, Brauer VS, Hoffmann D. Trade-off shapes diversity in eco-evolutionary dynamics. eLife. 2018;7:e36273. doi: 10.7554/eLife.36273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T. Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol. 2016;24:209–223. doi: 10.1016/j.tim.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford, UK: Oxford University Press; 1930. [Google Scholar]
- Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J Publ Fed Am Soc Exp Biol. 1996;10:461–470. [PubMed] [Google Scholar]
- Fry JD. The evolution of host specialization: are trade-offs overrated? Am Nat. 1996;148:S84–S107. [Google Scholar]
- Futuyma DJ, Moreno G. The evolution of ecological specialization. Annu Rev Ecol Syst. 1988;19:207–233. [Google Scholar]
- Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102–103:127–144. [PubMed] [Google Scholar]
- Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Griswold CK. The relationship between the pleiotropic phenotypic effects of a mutation fixed by selection. Heredity. 2007;98:232–242. doi: 10.1038/sj.hdy.6800933. [DOI] [PubMed] [Google Scholar]
- Hague MTJ, Toledo G, Geffeney SL, Hanifin CT, Brodie ED, Brodie ED. Large-effect mutations generate trade-off between predatory and locomotor ability during arms race coevolution with deadly prey. Evol Lett. 2018;2:406–416. doi: 10.1002/evl3.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AR, Colegrave N. Decay of unused characters by selection and drift. J Evol Biol. 2008;21:610–617. doi: 10.1111/j.1420-9101.2007.01473.x. [DOI] [PubMed] [Google Scholar]
- Henderson PJ, Baldwin SA, Cairns MT, Charalambous BM, Dent HC, Gunn F, et al. in bacteria. Int Rev Cytol. 1992;137:149–208. [PubMed] [Google Scholar]
- Hill JA, O’Meara TR, Cowen LE. Fitness trade-offs associated with the evolution of resistance to antifungal drug combinations. Cell Rep. 2015;10:809–819. doi: 10.1016/j.celrep.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Imamovic L., Sommer M. O. A. Use of Collateral Sensitivity Networks to Design Drug Cycling Protocols That Avoid Resistance Development. Science Translational Medicine. 2013;5(204):204ra132–204ra132. doi: 10.1126/scitranslmed.3006609. [DOI] [PubMed] [Google Scholar]
- Jessup CM, Bohannan BJM. The shape of an ecological trade‐off varies with environment. Ecol Lett. 2008;11:947–959. doi: 10.1111/j.1461-0248.2008.01205.x. [DOI] [PubMed] [Google Scholar]
- Joshi A, Thompson JN. Trade-offs and the evolution of host specialization. Evol Ecol. 1995;9:82–92. [Google Scholar]
- Karve SM, Bhave D, Dey S. Extent of adaptation is not limited by unpredictability of the environment in laboratory populations of Escherichia coli. J Evol Biol. 2018;31:1420–1426. doi: 10.1111/jeb.13338. [DOI] [PubMed] [Google Scholar]
- Karve SM, Bhave D, Nevgi D, Dey S. Escherichia coli populations adapt to complex, unpredictable fluctuations by minimizing trade-offs across environments. J Evol Biol. 2016;29:2545–2555. doi: 10.1111/jeb.12972. [DOI] [PubMed] [Google Scholar]
- Karve SM, Daniel S, Chavhan YD, Anand A, Kharola SS, Dey S. Escherichia coli populations in unpredictably fluctuating environments evolve to face novel stresses through enhanced efflux activity. J Evol Biol. 2015;28:1131–1143. doi: 10.1111/jeb.12640. [DOI] [PubMed] [Google Scholar]
- Kassen R. The experimental evolution of specialists, generalists, and the maintenance of diversity. J Evol Biol. 2002;15:173–190. [Google Scholar]
- Kassen R (2014) Experimental evolution and the nature of biodiversity. Roberts and Company, Greenwood Village, CO
- Kawecki TJ, Lenski RE, Ebert D, Hollis B, Olivieri I, Whitlock MC. Experimental evolution. Trends Ecol Evol. 2012;27:547–560. doi: 10.1016/j.tree.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Ketola T, Saarinen K. Experimental evolution in fluctuating environments: tolerance measurements at constant temperatures incorrectly predict the ability to tolerate fluctuating temperatures. J Evol Biol. 2015;28:800–806. doi: 10.1111/jeb.12606. [DOI] [PubMed] [Google Scholar]
- Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
- Kneitel JM, Chase JM. Trade-offs in community ecology: linking spatial scales and species coexistence. Ecol Lett. 2004;7:69–80. [Google Scholar]
- Knops JMH, Koenig WD, Carmen WJ. Negative correlation does not imply a tradeoff between growth and reproduction in California oaks. Proc Natl Acad Sci. 2007;104:16982–16985. doi: 10.1073/pnas.0704251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koricheva J. Meta-analysis of sources of variation in fitness costs of plant antiherbivore defenses. Ecology. 2002;83:176–190. [Google Scholar]
- Kraemer SA, Boynton PJ. Evidence for microbial local adaptation in nature. Mol Ecol. 2017;26:1860–1876. doi: 10.1111/mec.13958. [DOI] [PubMed] [Google Scholar]
- LaBar T, Adami C. Different evolutionary paths to complexity for small and large populations of digital organisms. PLoS Comput Biol. 2016;12:e1005066. doi: 10.1371/journal.pcbi.1005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachapelle J, Reid J, Colegrave N. Repeatability of adaptation in experimental populations of different sizes. Proc R Soc Lond B Biol Sci. 2015;282:20143033. doi: 10.1098/rspb.2014.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. The response to selection on major and minor mutations affecting a metrical trait. Heredity. 1983;50:47–65. [Google Scholar]
- Lee M-C, Chou H-H, Marx CJ. Asymmetric, bimodal trade-offs during adaptation of methylobacterium to distinct growth substrates. Evolution. 2009;63:2816–2830. doi: 10.1111/j.1558-5646.2009.00757.x. [DOI] [PubMed] [Google Scholar]
- Leiby N, Marx CJ. Metabolic erosion primarily through mutation accumulation, and not tradeoffs, drives limited evolution of substrate specificity in Escherichia coli. PLoS Biol. 2014;12:e1001789. doi: 10.1371/journal.pbio.1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- Levins R. Theory of fitness in a heterogeneous environment. I. The fitness set and adaptive function. Am Nat. 1962;96:361–373. [Google Scholar]
- Levins R (1968) Evolution in changing environments: some theoretical explorations. Princeton University Press, New Jersey
- Loh KD, Gyaneshwar P, Markenscoff Papadimitriou E, Fong R, Kim K-S, Parales R, et al. A previously undescribed pathway for pyrimidine catabolism. Proc Natl Acad Sci USA. 2006;103:5114–5119. doi: 10.1073/pnas.0600521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur RH. Geographical ecology: patterns in the distribution of species. New Jersey: Princeton University Press; 1984. [Google Scholar]
- MacLean RC, Hall AR, Perron GG, Buckling A. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat Rev Genet. 2010;11:405–414. doi: 10.1038/nrg2778. [DOI] [PubMed] [Google Scholar]
- Messenger SL, Molineux IJ, Bull JJ. Virulence evolution in a virus obeys a trade-off. Proc R Soc Lond B Biol Sci. 1999;266:397–404. doi: 10.1098/rspb.1999.0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina FJ, Durham SL. Loss of adaptation following reversion suggests trade-offs in host use by a seed beetle. J Evol Biol. 2015;28:1882–1891. doi: 10.1111/jeb.12704. [DOI] [PubMed] [Google Scholar]
- Milliken GA, Johnson DE (2009) Analysis of messy data, volume I: designed experiments. Chapman and Hall/CRC, Boca Raton, FL
- Novak M, Pfeiffer T, Lenski RE, Sauer U, Bonhoeffer S. Experimental tests for an evolutionary trade-off between growth rate and yield in E. coli. Am Nat. 2006;168:242–251. doi: 10.1086/506527. [DOI] [PubMed] [Google Scholar]
- Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- Orr HA, Coyne JA. The genetics of adaptation: a reassessment. Am Nat. 1992;140:725–742. doi: 10.1086/285437. [DOI] [PubMed] [Google Scholar]
- Ostrowski EA, Rozen DE, Lenski RE. Pleiotropic effects of beneficial mutations in Escherichia Coli. Evolution. 2005;59:2343–2352. [PubMed] [Google Scholar]
- Otto Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc R Soc Lond B Biol Sci. 2004;271:705–714. doi: 10.1098/rspb.2003.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patching SG, Baldwin SA, Baldwin AD, Young JD, Gallagher MP, Henderson PJF, et al. The nucleoside transport proteins, NupC and NupG, from Escherichia coli: specific structural motifs necessary for the binding of ligands. Org Biomol Chem. 2005;3:462–470. doi: 10.1039/b414739a. [DOI] [PubMed] [Google Scholar]
- Petit N, Barbadilla A. Selection efficiency and effective population size in Drosophila species. J Evol Biol. 2009;22:515–526. doi: 10.1111/j.1420-9101.2008.01672.x. [DOI] [PubMed] [Google Scholar]
- Prasad NG, Shakarad M, Anitha D, Rajamani M, Joshi A. Correlated responses to selection for faster development and early reproduction in drosophila: the evolution of larval traits. Evolution. 2001;55:1363–1372. doi: 10.1111/j.0014-3820.2001.tb00658.x. [DOI] [PubMed] [Google Scholar]
- Rausher MD. Tradeoffs in performance on different hosts: evidence from within- and between-site variation in the beetle Deloyala guttata. Evolution. 1984;38:582–595. doi: 10.1111/j.1558-5646.1984.tb00324.x. [DOI] [PubMed] [Google Scholar]
- Remold Susanna. Understanding specialism when the jack of all trades can be the master of all. Proceedings of the Royal Society B: Biological Sciences. 2012;279(1749):4861–4869. doi: 10.1098/rspb.2012.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Verdugo A, Carrillo-Cisneros D, González-González A, Gaut BS, Bennett AF. Different tradeoffs result from alternate genetic adaptations to a common environment. Proc Natl Acad Sci. 2014;111:12121–12126. doi: 10.1073/pnas.1406886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen DE, de Visser JAGM, Gerrish PJ. Fitness effects of fixed beneficial mutations in microbial populations. Curr Biol. 2002;12:1040–1045. doi: 10.1016/s0960-9822(02)00896-5. [DOI] [PubMed] [Google Scholar]
- Sane M, Miranda JJ, Agashe D. Antagonistic pleiotropy for carbon use is rare in new mutations. Evolution. 2018;72:2202–2213. doi: 10.1111/evo.13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick A, Bailey SF, Kassen R. Evolution of fitness trade-offs in locally adapted populations of pseudomonas fluorescens. Am Nat. 2015;186:S48–S59. doi: 10.1086/682932. [DOI] [PubMed] [Google Scholar]
- Smith-Tsurkan SD, Wilke CO, Novella IS. Incongruent fitness landscapes, not tradeoffs, dominate the adaptation of vesicular stomatitis virus to novel host types. J Gen Virol. 2010;91:1484–1493. doi: 10.1099/vir.0.017855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ. Beneficial mutations and the dynamics of adaptation in asexual populations. Philos Trans R Soc B Biol Sci. 2010;365:1255–1263. doi: 10.1098/rstb.2009.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC. The evolutionary significance of phenotypic plasticity. BioScience. 1989;39:436–445. [Google Scholar]
- Szendro IG, Franke J, JAGM de Visser, Krug J. Predictability of evolution depends nonmonotonically on population size. Proc Natl Acad Sci. 2013;110:571–576. doi: 10.1073/pnas.1213613110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O. The utility of Fisher’s geometric model in evolutionary genetics. Annu Rev Ecol Evol Syst. 2014;45:179–201. doi: 10.1146/annurev-ecolsys-120213-091846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano M. Long-term experimental evolution in Escherichia coli. VI. environmental constraints on adaptation and divergence. Genetics. 1997;146:471–479. doi: 10.1093/genetics/146.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PE, Elena SF. Cost of host radiation in an RNA virus. Genetics. 2000;156:1465–1470. doi: 10.1093/genetics/156.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogwill T, Phillips RL, Gifford DR. Divergent evolution peaks under intermediate population bottlenecks during bacterial experimental evolution. Proc R Soc B. 2016;283:20160749. doi: 10.1098/rspb.2016.0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data relevant to this study can be found at 10.5061/dryad.bnzs7h46z.