Abstract
Alteration of various metabolites has been linked to type 2 diabetes (T2D) and insulin resistance. However, identifying significant associations between metabolites and tissue-specific phenotypes requires a multi-omics approach. In a cohort of 42 subjects with different levels of glucose tolerance (normal, prediabetes and T2D) matched for age and body mass index, we calculated associations between parameters of whole-body positron emission tomography (PET)/magnetic resonance imaging (MRI) during hyperinsulinemic euglycemic clamp and non-targeted metabolomics profiling for subcutaneous adipose tissue (SAT) and plasma. Plasma metabolomics profiling revealed that hepatic fat content was positively associated with tyrosine, and negatively associated with lysoPC(P-16:0). Visceral adipose tissue (VAT) and SAT insulin sensitivity (Ki), were positively associated with several lysophospholipids, while the opposite applied to branched-chain amino acids. The adipose tissue metabolomics revealed a positive association between non-esterified fatty acids and, VAT and liver Ki. Bile acids and carnitines in adipose tissue were inversely associated with VAT Ki. Furthermore, we detected several metabolites that were significantly higher in T2D than normal/prediabetes. In this study we present novel associations between several metabolites from SAT and plasma with the fat fraction, volume and insulin sensitivity of various tissues throughout the body, demonstrating the benefit of an integrative multi-omics approach.
Subject terms: Metabolomics, Data integration, Type 2 diabetes, Positron-emission tomography, Magnetic resonance imaging
Introduction
High-throughput metabolomics promises a deeper understanding of type 2 diabetes (T2D) mellitus1. According to the World Health Organization’s global report on diabetes, the age-standardized global prevalence of diabetes has nearly doubled from 1980 to 20142. Motivated by the necessity of progress to halt the diabetes epidemic, researchers have made significant discoveries to widen our understanding of T2D. A novel classification of T2D has been suggested, comprising of five clusters and reported to better correlate with disease progression and risk of complications3. Further development of precision medicine in T2D will help individualize treatments, predict outcomes and prevent adverse consequences4.
A more precise understanding of T2D is currently being shaped with the rapid development of new methods such as high-throughput metabolomics1,5, single-cell RNA-sequencing6–8 and machine learning (ML)-based approaches for precision medicine9,10. However, integration of multi-dimensional “omics” data from different platforms is challenging. Fortunately, in recent years numerous tools have been made accessible to facilitate this process11,12. In addition to the tools, various studies have provided integration strategies for bulk and single-cell technologies13, with single-cell metabolomics being “around the corner”14.
Previous work on metabolomics in T2D highlights the important role of amino acid profiling, particularly branched-chain amino acids (BCAA) for diabetes risk assessment15–17. Several other metabolites have been reported as significantly associated with diabetes prevalence; these include reduced levels of glycine, lysophosphatidylcholine (LPC) (18:2) as well as increased levels of acetylcarnitine18. Increased levels of deoxycholic acid and monacylglyceride (18:2), as well as lower levels of cortisol, have also been reported as positively associated with T2D risk19.
In parallel with the advances in bioinformatics, imaging techniques have undergone vast improvements in the last years. Improvements from unimodal imaging to bimodal- or even trimodal imaging20 now enable researchers to capture a vast amount of accurate phenotypical information in a reasonable time. Integrated imaging platforms such as positron emission tomography (PET)/magnetic resonance imaging (MRI)21,22 or PET/computerized tomography (CT)23 provide methods to extract quantitatively and spatially accurate tissue-specific information in human subjects.
In this study, we combined data from high-throughput mass spectrometry (MS) with tissue-specific characteristics ([18F]FDG-influx rate during hyperinsulinemia, tissue fat content and volume) from whole-body integrated [18F]FDG-PET/MRI. Our main aim was to discover novel associations of metabolites from subcutaneous adipose tissue (SAT) and plasma with the volume, the fat fraction (FF) and the insulin sensitivity (Ki for [18F]FDG) of tissues throughout the body. We used[18F]FDG PET/MRI in subjects with different degrees of glucose tolerance. An additional aim was to compare non-targeted metabolomics profiling for SAT and blood plasma between normal/prediabetes and T2D subjects.
Results
Baseline characteristics of the subjects are shown in Table 1. Body mass index (BMI) and waist-hip ratio (WHR) were significantly associated with the homeostatic model assessment for insulin resistance (HOMA-IR) and whole-body insulin sensitivity (M-value), while age and sex ratio did not show any biases (Table 1; Supplementary Table S1). Metabolic markers such as hemoglobin A1c (HbA1c), oral glucose tolerance test area under the curve (OGTT AUCglucose), HOMA-IR and M-value were significantly different between the controls, prediabetes and T2D groups (Table 1; Supplementary Table S1).
Table 1.
Baseline characteristics of the 42 subjects in the cohort.
| Parameter | Controls N = 12 | Prediabetes N = 16 | T2D N = 14 | p |
|---|---|---|---|---|
| Age | 60 ± 6 | 64 ± 6 | 62 ± 7 | 0.234 |
| BMI | 30.1 ± 4.7 | 30.4 ± 3.7 | 30.3 ± 3.6 | 0.852 |
| Sex | 6F/6M | 9F/7M | 6F/8M | — |
| WHR | 0.93 ± 0.10 | 1.00 ± 0.09 | 1.01 ± 0.04 | 0.061 |
| HbA1c | 34.2 ± 2.6 | 36.9 ± 3.1 | 55.1 ± 12.1 | 0.000 |
| OGTT AUCglucose | 819.8 ± 138.2 | 1073.1 ± 178.8 | 1731.9 ± 393.5 | 0.000 |
| HOMA-IR | 1.9 ± 1.1 | 2.8 ± 1.5 | 5.1 ± 2.7 | 0.003 |
| M-value | 10.2 ± 2.9 | 8.0 ± 3.8 | 5.3 ± 2.5 | 0.002 |
The mean value and the standard deviation are shown for anthropometric and diabetes metabolic markers. The following units were used: age (years), BMI (kg/m2), sex (proportion of females (F) and males (M)), WHR (ratio of waist- to hip circumference), HbA1c (mmol/mol), OGTT AUCglucose (mmol/L*min) and M-value (mg/kg LBM/min). Statistical significance among controls, prediabetes and T2D for the baseline characteristics was calculated from a Kruskal-Wallis rank sum test.
Metabolite composition in non-diabetes and T2D
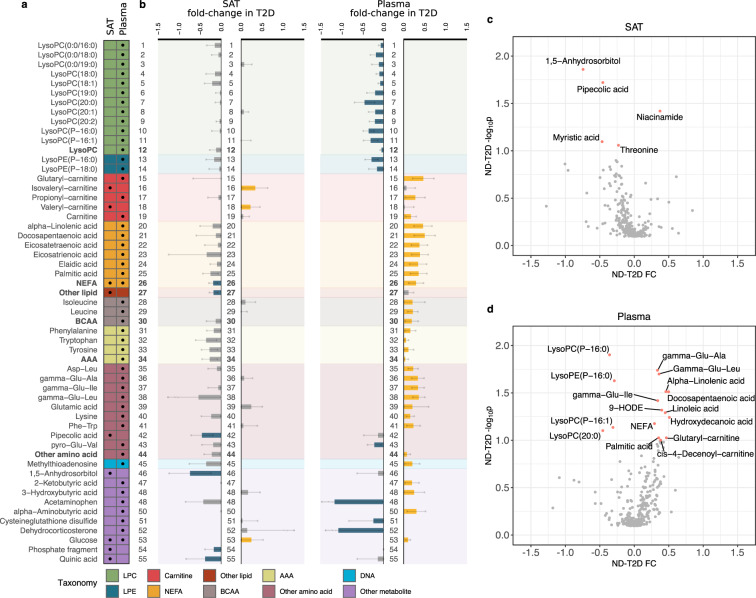
The computational identification that followed the non-targeted profiling identified 259 unique metabolites in the adipose tissue samples and 272 metabolites in plasma (Supplementary Table S2). Levels of 55 unique metabolites were significantly associated (p < 0.1) with at least one T2D metabolic biomarker after correction for BMI, WHR, sex and age (Fig. 1a; Supplementary Tables S3 and S4). We also pooled groups of metabolites belonging to the same taxa and explored their associations with T2D (Fig. 1a; Supplementary Table S2).
Figure 1.
Overview of metabolites associated with T2D. (a) Set of 55 unique metabolites and pools of metabolites significantly associated with at least one diabetes metabolic marker (M-value, OGTT AUCglucose, HbA1c or HOMA-IR) in at least one tissue. Table columns represent SAT and plasma. A black dot implies statistical significance in the corresponding tissue (Mann-Whitney U test permuted p < 0.1; Methods - Statistical analysis). The color-coding of the table refers to the classes of metabolites explained at the bottom legend. (b) Bar plots represent the fold-changes between compound intensities in ND and T2D. The order of the bar plots and the background color matches that of a. Error bars represent 90% confidence intervals (Methods - Statistical analysis). Yellow bars imply statistical significance and increase, blue bars statistical significance and decrease, while grey bars did not cross the statistical significance threshold. The numbering helps in tracing the variation of metabolite across tissues. (c,d) Volcano plots of the differential analysis in adipose tissue (c) and plasma (d). Y-axis shows -log10p and x-axis fold change (FC) of the metabolites. Significant metabolites are marked in pink and are named.
Taxonomy enrichment analysis showed that lysophosphatidylcholines (lysoPCs) (p < 0.001), non-esterified fatty acids (NEFAs) (p < 0.001) and amino acids (AAs) (p < 0.001) in plasma were overrepresented in T2D (Supplementary Table S5). The pools of branched-chain AAs (BCAAs) and aromatic AAs (AAAs) in plasma were inversely associated with the M-value (β = −0.25, p < 0.1 and β = −0.12, p < 0.1, respectively) (Fig. 1a; Supplementary Fig. S1b). In addition to their pools, pathways of BCAAs and AAAs were enriched in plasma of T2D (p < 0.001) (Supplementary Table S6). Additionally, various dipeptides and gamma-dipeptides in plasma were higher in T2D than in ND, and positively associated with OGTT AUCglucose (βmean = 0.39, p < 0.1) (Figs. 1a and 1d; Supplementary Fig. S1b). Plasma levels of lysoPCs and lysophosphatidylethanolamines (lysoPEs) were significantly decreased in T2D and were mainly associated with OGTT AUCglucose and HOMA-IR (βmean = −0.47, p < 0.09 and βmean = −0.53, p < 0.1) (Figs. 1a, 1b and 1d; Supplementary Fig. S1b). Various plasma carnitines and NEFAs were significantly higher in T2D than ND and were associated with OGTT AUCglucose (βmean = 0.43, p < 0.1). The pool of NEFAs in plasma was positively associated with OGTT AUCglucose (β = 0.42, p < 0.1) (Fig. 1b).
In general, the alteration patterns observed in SAT samples and in plasma differed largely (Fig. 1b). In adipose tissue, valeryl- and isovaleryl-carnitine were significantly higher in T2D than ND and positively associated with HOMA-IR (βmean = 0.49, p < 0.05) (Figs. 1a and 1b; Supplementary Fig. S1a). Adipose tissue glucose levels were significantly higher in T2D than ND and were significantly associated with HOMA-IR (β = 0.34, p < 0.1) (Figs. 1a and 1b; Supplementary Fig. S1a). 1,5-anhydrosorbitol was significantly lower in adipose tissue samples of T2D than ND (FC = −0.74, p < 0.05), with a similar trend in plasma that did not cross the threshold of statistical significance (Fig. 1; Supplementary Fig. S1). 1,5-anhydrosorbitol was negatively associated with HOMA-IR (β = −0.74, p < 0.05), HbA1c (β = −0.77, p < 0.001) and OGTT AUCglucose (β = −0.64, p < 0.05). Pipecolic acid was the only adipose tissue amino acid that was significantly associated with HbA1c (β = −0.58, p < 0.05) and was confirmed to be significantly lower in T2D than in ND (FC = −0.46, p < 0.05) (Fig. 1c; Supplementary Fig. S1a).
Associations between metabolites and PET/MRI
Plasma metabolites
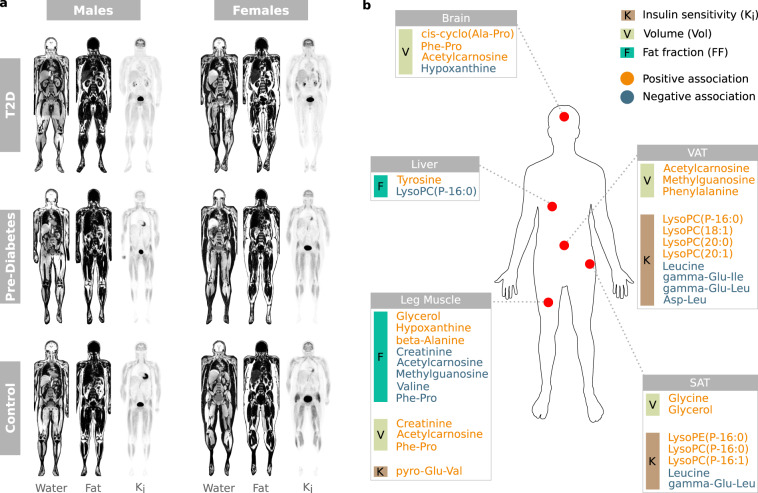
Of the AAAs, phenylalanine was positively associated with VAT volume (β = 0.12, p < 0.1) and tyrosine was positively associated with hepatic fat content (β = 0.25, p < 0.1). Two of the gamma-glutamyl amino acids (isoleucine, leucine) were inversely associated with insulin sensitivity in adipose tissue compartments (SAT Ki and/or VAT Ki) (βmean = −0.48, p < 0.1). Leucine was associated with both SAT Ki (β = −0.53, p < 0.1) and VAT Ki (β = −0.43, p < 0.1). Glycine was positively associated with SAT volume (β = 0.8, p < 0.1) and cis-cyclo(alanine-proline) was positively associated with brain volume (β = 0.52, p < 0.05). Valine was inversely associated with inter-/intramuscular fat in the leg (β = −0.71, p < 0.1) and beta-alanine was positively associated with inter-/intramuscular fat in the leg (β = 0.36, p < 0.1). Pyroglutamylvaline was positively associated with leg muscle Ki (β = 0.56, p < 0.1) and phenylalanylproline was positively associated with both brain volume(β = 0.14, p < 0.05) and leg muscle volume (β = 0.51, p < 0.01) as well as inversely associated with inter-/intramuscular fat in the leg (β = −0.37, p < 0.05) (Fig. 2; Supplementary Fig. S2).
Figure 2.
Representative PET/MR images and associations between tissue-specific image parameters and metabolites. (a) Illustration of water- and fat separated MR images, and [18F]FDG-influx rate (Ki) PET images in each group. (b) Significant positive- and negative associations between plasma metabolites and body composition/glucose uptake.
LysoPC(P-16:0), lysoPC(P-16:1) and lysoPE(P-16:0) were positively associated with SAT Ki (β = 0.5, p < 0.1; β = 0.6, p < 0.05; β = 0.44, p < 0.1, respectively). LysoPC(P-16:0) was also inversely associated with hepatic fat content (β = −0.62, p < 0.1) and positively associated with VAT Ki (β = 0.47, p < 0.1) as were 18:1, 20:0 and 20:1 (βmean = 0.56, p < 0.1) (Fig. 3; Supplementary Fig. S3). Glycerol was positively associated with inter-/intramuscular fat in the leg (β = 0.2, p < 0.1), SAT FF (β = 0.21, p < 0.05) and SAT volume (β = 0.2, p < 0.1). Acetylcarnosine exhibited a positive association with brain volume, leg muscle volume and VAT volume (βmean = 0.4, p < 0.1), as well an inverse association with inter-/intramuscular fat in the leg (β = −0.36, p < 0.05) (Fig. 2; Supplementary Fig. S2).
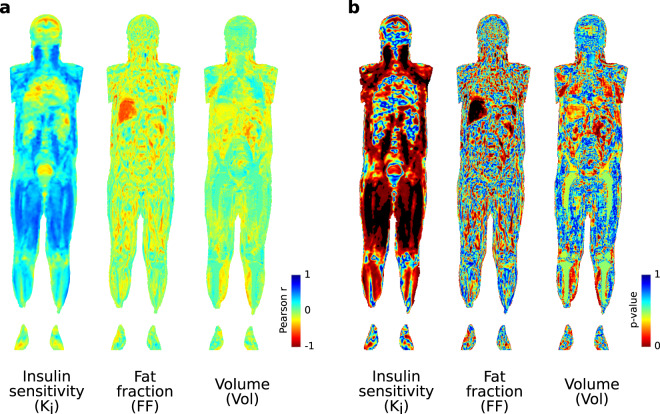
Figure 3.
Voxel-wise correlation maps between lysoPC(P-16:0) and tissue parameters were generated with Imiomics corrected for BMI, WHR, sex and age. The results from the correlation maps for this single example included in the present study show a large overlap with traditional analysis illustrating the usefulness of direct voxel-wise association screening with Imiomics47. (a) Pearson’s r-coefficient maps for [18F]FDG-influx rate (Ki), tissue fat fraction (FF) and tissue volume (Vol) respectively. (b) Corresponding p-value maps. Interpretation: LysoPC (P-16:0) is inversely associated with hepatic fat content and positively associated with retroperitoneal adipose Ki, subcutaneous adipose tissue Ki and Ki in the thigh muscle, psoar major muscle and neck muscles. See Supplementary Fig. S3 for thresholded p-value maps.
Hypoxanthine and methylguanosine showed the strongest associations of the DNA metabolites. Hypoxanthine was inversely associated with brain volume (β = −0.68, p < 0.05) and positively associated with inter-/intramuscular fat in the leg (β = 0.58, p < 0.1). Discordantly, methylguanosine was inversely associated with inter-/intramuscular fat in the leg (β = −0.25, p < 0.1) as well as positively associated with VAT volume (β = 0.46, p < 0.05) (Fig. 2; Supplementary Fig. S2).
Adipose tissue metabolites
Valeryl-carnitine was inversely associated with VAT Ki (β = −0.37, p < 0.1) and tetradecadienyl-carnitine was positively associated with VAT FF (β = 0.48, p < 0.1). The overall class of NEFAs was positively associated with both liver Ki (β = 0.47, p < 0.05) and VAT Ki (β = 0.29, p < 0.1), NEFAs were also inversely associated with VAT FF (β = −0.39, p < 0.05). The specific metabolite associations within the NEFA class are displayed in Supplementary Fig. S4. LysoPE(0:0/20:5) was inversely associated with VAT FF (β = −0.26, p < 0.1). Azelaic acid was inversely associated with both VAT volume (β = −0.63, p < 0.05) and VAT FF (β = −0.59, p < 0.05). Cholic acid was inversely associated with brain volume (β = −0.44, p < 0.1). There was a positive association between glycodeoxycholic acid and VAT volume as well as VAT FF (β = 0.22, p < 0.1; β = 0.3, p < 0.05, respectively) while VAT Ki was inversely associated with glycodeoxycholic acid (β = −0.26, p < 0.1) (Supplementary Fig. S4).
Discussion
We performed cross-sectional metabolic profiling for SAT and plasma in a cohort of 42 subjects that underwent OGTT and whole-body PET/MRI. To the best of our knowledge, this is the first attempt towards exploring T2D by integrating whole-body imaging data and metabolomics. We discovered several interesting associations between metabolites and glucose uptake, fat content and volume of various tissues.
There was a significant increase of plasma metabolites within the classes of BCAAs, AAAs, NEFAs and carnitines in T2D, while lysoPCs and lysoPEs significantly decreased in T2D (Fig. 1; Supplementary Fig. S1; Supplementary Table S5). The enrichment of the pathways for the biosynthesis of unsaturated fatty acids, BCAAs and AAAs, in combination with the significant positive associations of their respective pools with OGTT AUCglucose confirmed their elevated levels in T2D as demonstrated in various other studies (Fig. 1; Supplementary Fig. S1; Supplementary Table S6)24,25. Interestingly, we detected two short-chain carnitine species, namely valeryl- and isovaleryl carnitine, that were significantly higher in SAT of T2D than ND, and strongly associated with the insulin resistance index HOMA-IR. The pattern of metabolic alterations in SAT was divergent when compared to plasma, with the latter demonstrating an enrichment of various associations of metabolites with biomarkers of T2D. We employed MoDentify to explore the association of intra- and inter-tissue modules of metabolites with M-value26. Interestingly, we identified two modules that were associated with the M-value at FDR < 0.1, consisting of metabolites predominantly from SAT rather than plasma (Supplementary Note - S1. MoDentify; Supplementary Fig. S5). While more studies are needed to address the driving metabolic pathways of insulin resistance, our association-based results suggest several interactions of phospholipids and amino acids in SAT that could be relevant for insulin resistance (Supplementary Fig. S5).
Amino acid imbalance commonly manifests in patients with liver disease27. As reported in the seminal paper by Soeters & Fischer, the ratio of BCAAs (leucine, valine, and isoleucine) to AAAs (phenylalanine, tyrosine) decreases with the severity of liver disease28. In the present study, we detected a positive association between plasma levels of tyrosine and liver fat content. This agrees with previous studies that have reported increasing tyrosine levels according to the fibrosis staging in liver diseases27 and in patients with non-alcoholic fatty liver disease (NAFLD)29. The exact underlying mechanism for this increase is not yet clear, but plasma tyrosine levels have been further associated to insulin resistance, although we could not identify any significant association between tyrosine and liver Ki. We also detected an inverse association between plasma levels of lysoPC(P-16:0) and liver fat content. This is consistent with the notion that phosphatidylcholines exert a protective effect against liver damage30 and that choline-deficient diets results in an accumulation of hepatic lipids31. Interestingly, lysoPCs have been proposed as biomarkers for identifying a subset of NAFLD patients with a benign phenotype, where plasma levels of lysoPCs are positively associated with insulin sensitivity32.
Most species of lysoPCs and lysoPEs significantly decrease with obesity33 and insulin resistance18. In the present study, we detected a strong association between multiple species of plasma lysophospholipids and SAT- and VAT Ki (Figs. 2 and 3; Supplementary Fig. S3). In murine models, lysoPCs have been shown to regulate blood glucose levels by enhancing adipocyte glucose uptake through several different mechanisms including increasing the levels of GLUT434. Our results suggested that this is specific for adipose tissue as we found no other significant association between tissue-specific insulin sensitivity and plasma levels of lysophospholipids. A large fraction of these lysoPCs were also associated with T2D (Figs. 1 and 2; Supplemenatry Table S6). Contrary to lysophospholipids, BCAAs and its intermediates were inversely associated with both SAT- and VAT Ki. These findings suggest that BCAAs are upregulated with increasing insulin resistance in adipose tissue as has been confirmed in multiple studies35–37. We found that plasma levels of glycine were positively associated with SAT volume but not VAT volume. On the contrary, VAT volume was positively associated with phenylalanine while SAT volume was not. This could be indicative of different metabolic functions in the two regional fat depots38,39. This is further corroborated by our results from the adipose tissue metabolomics, where we found the strongest associations between NEFAs and VAT Ki and fat fraction, but no significant association with SAT measurements. Overall, the adipose tissue metabolomics revealed a downregulation of NEFAs with increasing VAT FF as well as an inverse association between carnitines and bile acids with VAT Ki.
Skeletal muscle fat content was significantly associated with several metabolites in plasma (Fig. 2). The positive association between hypoxanthine and leg muscle fat fraction suggests an increase of purine catabolism in fatty infiltrated muscle compartments, and similar findings has been reported in subjects with myopathy40–42. A disturbed energy balance in the skeletal muscles of T2D is then reflected by an increase of purine catabolism, resulting in an increase of the final product, uric acid, and possibly all related micro-/macrovascular complications42. In the visual analysis with Imiomics (Fig. 3) we show significant associations between insulin sensitivity in neck- and thigh muscles and lysoPC(P-16:0). These significant findings were not detected when merging leg compartments together (Fig. 2) highlighting the potential benefit of unbiased voxel-wise analysis of the whole-body.
In summary, we proposed a novel approach for integrating data from metabolomics and imaging studies. Our results suggested a large collection of promising associations for plasma and adipose tissue metabolites with the tissue charactheristics of various organs in the body. The results from the present study should be interpreted with caution due to the limited sample size. We did not perform any analyses in the three included groups separately due to the low statistical power that would results from a stratified analyses, and we could therefore not exclude any potential confounding effects related to the groups. Furthermore, the time difference between the image acquisition and tissue sampling for metabolomics must be noted, however, the phenotypical measurements from the imaging data should not differ significantly in that short time-span. The presented multi-omics approach can be used to generate hypotheses for the pathobiology of T2D development, including different disease stages and patient phenotypes.
Methods
Subjects and experimental design
12 healthy controls (6F/6M), 16 subjects with prediabetes (9F/7M), and 14 T2D subjects (6F/8M) were matched for age and BMI. The details of the study design and inclusion and exclusion criteria have been published previously43. Briefly, subjects had been diagnosed according to the American Diabetes Association (ADA) criteria, and the T2D subjects were on a stable dose of metformin as their only antidiabetic medication. Subjects with type 1 diabetes, endocrine disorders, cancer liver dysfunction, renal impairment, alcohol or drug abuse, pregnancy or any serious clinical condition were excluded. The study was approved by the Regional Ethics Review Board at Uppsala University, and all participants provided their written informed consent (Dnr: 2014/313). The whole study was conducted in full accordance with the regional standard practices and the Swedish law. No organs/tissues were procured from prisoners.
Details of the sample collection and the experimental procedures for this cohort have been described earlier44. In summary, the study consisted of two visits. During the first visit, we recorded phenotypic characteristics of the subjects such as anthropometrical measurements and general medical status. Subjects were instructed to fast overnight (>10 h), and blood samples were collected for analysis of HbA1c, plasma glucose, lipids, serum insulin and C-peptide at the Department of Clinical Chemistry, Uppsala University Hospital. A subcutaneous adipose tissue biopsy was taken from the lower part of the abdomen after local epidermal anaesthesia with lidocaine (Xylocaine; AstraZeneca, Södertälje, Sweden) and samples were snap-frozen in liquid nitrogen and stored at −80 °C until metabolic profiling analysis. Next, a 2-h OGTT was performed for analysis of glucose and insulin. During the second visit, 2–3 weeks later, a hyperinsulinemic-euglycemic clamp combined with whole-body integrated PET/MRI was performed during which the [18F]FDG uptake rate in individual tissues and M-value were measured.
PET/MR imaging
PET/MR imaging was performed using an integrated 3.0 T PET/MR scanner (Signa PET/MR, GE Healthcare, Waukesha, WI) as previously described45. The average injected dose of [18F]FDG was 353 MBq (range: 271–445, 4MBq/kg bodyweight). Early tracer dynamics of [18F]FDG were captured in an initial 10-minute dynamic scan of the thorax. Subsequently, five whole-body (covering head-to-toe) PET scans (voxel size = 4.69 × 4.69 × 2.78 mm) were acquired. The PET scans were acquired in time-of-flight mode, and each scan was immediately preceded by a 3D dual-echo water-fat separated MR attenuation correction scan (MRAC; TR = 4.0, Flip = 5, voxel size = 1.95 × 1.95 × 2.6 mm) used for attenuation correction. Water- and fat images from the last MRAC were used and all corrections necessary for quantitative PET were performed. Both PET scans and MRAC images were acquired in free breathing.
Matlab (Matlab 2015b, The Mathworks Inc, Natick, MA) was used for PET data analysis. The Patlak method46 was applied to calculate [18F]FDG influx rate (Ki) utilizing an image-derived input function corrected for blood-cell bound radioactivity45. The Imiomics method47 was used to normalize both MRAC and PET data from all subjects to a common reference coordinate system. Briefly, single-atlas image registration was performed using the MRAC data starting with the bones, followed by non-adipose tissue and lastly adipose tissue with gradually increasing elasticity. The deformation applied along with the original signal intensity was stored in each voxel, enabling calculation of differences in tissue volume, fat content and Ki between subjects. The PET data was registered using the same deformation as the MRAC data.
In this cohort the reference person was a 64-year-old male with a BMI of 26.8, belonging to the control group. Organ segmentation in the reference person was performed by a radiologist with over 5 years of experience and involved areas such as brain, heart, liver, visceral adipose tissue (VAT), SAT, gluteal-/thigh-/calf muscle.
Metabolic profiling
Metabolic profiling by gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS) was performed at the Swedish Metabolomics Center (SMC) in Umeå, Sweden. SAT and plasma MS analyses for the 42 subjects were performed in two batches of 24 and 18 samples, respectively. Detailed description of the sample preparation and metabolic profiling is provided in (Supplementary Note – S2. Metabolic Profiling).
Preprocessing of MS metabolomics data
For the GC-MS data, all non-processed MS files from the metabolic analysis were exported from the ChromaTOF software in NetCDF format to MATLAB R2016a (Mathworks, Natick, MA, USA), where all data pre-treatment procedures, such as base-line correction, chromatogram alignment, data compression and Multivariate Curve Resolution (MCR) were performed using custom scripts. The extracted mass spectra were identified by comparing their retention indices and mass spectra with libraries of retention time indices and mass spectra48. Mass spectra and retention index comparisons were performed using the NIST MS 2.0 software. Annotation of mass spectra was based on reverse and forward searches in the library. Masses and ratio between masses indicative of a derivatized metabolite were especially notified. If the mass spectrum according to SMC’s experience was indicative of a metabolite and the retention index between the sample and library for the suggested metabolite was ±5 (usually less than 3) the deconvoluted “peak” was annotated as a metabolite49. We accepted only peaks that were detected in both GC platforms.
For LC-MS all the data processing was performed using the Agilent Masshunter Profinder version B.08.00 (Agilent Technologies Inc., Santa Clara, CA, USA). The computational processing was performed both in a targeted and a non-targeted fashion. For targeted computational processing, a pre-defined list of metabolites commonly found in plasma and serum were searched for using the Batch Targeted feature extraction in Masshunter Profinder. An-in-house LC-MS library built up by authentic standards run on the same system with the same chromatographic and mass-spec settings were used for the targeted computational processing. The identification of the metabolites was based on MS, MS/MS and retention time information. For the non-targeted computational processing, each tissue group was processed individually using the Batch Recursive Feature Extraction algorithm within Masshunter Profinder.
Metabolites missing in more than 20% of the samples were filtered out, while the remaining missing metabolites were assumed not to exceed the detectable threshold and imputed with 1.00001 (Supplementary Table S2). We calculated the total intensity (TI) for each sample as the sum of metabolite intensities, we computed pools of metabolites belonging to the same class or super class (Supplementary Table S2) and we log2-transformed dataset containing TI, internal standards (IS), metabolites and metabolite pools to better approximate a normal distribution and avoid outlier biases50.
Next, we explored associations among the MS running order, the sample weight, the TI and the IS, and the set of metabolite intensities. We performed a principal component analysis (PCA) on the set of metabolite intensities and considered the first 10 principal components (PCs). We explored correlations of the PCs with the MS running order, the sample weight, the TI and the IS. Significance of the correlations was assessed using the function cor.test from R based on the Pearson correlation coefficient (p < 0.05). We built a linear model using the significantly correlated factors (MS running order, sample weight, TI or IS) as predictors and computed the residuals of the model that would discard confounders’ biases and represent the final set of metabolites (Supplementary Figs. S6 and S7). For the latter we used the function lmFit and residuals.MArrayLM from the package limma in R, that additionally to computing the residuals would center metabolite intensities around 0. Finally, we explored the effects of imputation (Supplementary Figs. S1, S2 and S4) and batch (Supplementary Figs. S6 and S8) to confirm that the downstream analysis would not be biased.
Statistical analysis
Non-parametric Mann-Whitney U Monte Carlo resampling permutation test was used to perform the differential analysis on the intensities of the metabolites for the binary phenotypic groups, non-diabetes (ND) and T2D. ND consisted of control and prediabetes subjects. We performed 100K permutations while correcting for BMI, WHR, age and sex. P-values were Benjamini-Hochberg corrected and the significance threshold was set to 0.1. Fold-changes and 90% confidence intervals were computed.
We explored linear regression models among metabolites and various diabetes biomarkers and imaging measurements. Specifically, metabolites were used as response variables (Supplementary Table S2) while diabetes or imaging measurements as predictors (Supplementary Tables S1 and S7) of the regression models. All models were corrected for confounders (BMI, WHR, age and sex). We used linear regression models that were based on permutation tests (100K permutations) to build a background distribution for the estimation of statistical significance. P-values were Benjamini-Hochberg corrected and the significance threshold was set to 0.1 (Supplementary Table S3 and S4). We performed pathway and taxonomy enrichment analysis for the sets of metabolites significantly associated with T2D in each tissue (Supplementary Note – S3. Pathway and taxonomy enrichment analysis).
The computational tool that performed the bulk of the analysis was used in our former study, and is here reused with some minor updates. It is publicly available under https://github.com/klevdiamanti/MS_targeted.51
Supplementary information
Acknowledgements
We would like to acknowledge the Swedish Metabolomics Centre (www.swedishmetabolomiccentre.se) for the MS and the computational identification of the compounds. We thank the Wikimedia commons users Strogoff and Mikael Häggström for the human silhouette picture used in Fig. 2b. This study was funded by AstraZeneca, the Swedish Research Council 2016-01040, the Swedish Heart and Lung Foundation 20170492, the Swedish Diabetes Foundation (DIA 2017-269 CW; DIA 2017-267 JWE), EXODIAB, the ALF grants, the Ernfors Foundation and the Institute of Computer Science PAN, Poland.
Author contributions
K.D. and R.V. analysed the data, interpreted the results and wrote the first draft of the manuscript. M.C. and G.P. prepared the samples for metabolomics. U.R. provided comments on the manuscript. M.J.P. and J.W.E. recruited the subjects, collected the samples, interpreted the results and wrote the manuscript. C.K. and S.S. wrote the manuscript. J.Ko. assisted in the study design, assisted in the data analysis, interpreted the results and wrote the manuscript. J.Ku., C.W. and H.A. contributed with regards to study design, writing the manuscript and interpreting the results. H.A was the P.I. of the study. All authors read and approved the manuscript.
Data availability
“The metabolomics datasets generated and analysed during the current study are available in MetaboLights (https://www.ebi.ac.uk/metabolights/) under the accession number MTBLS1051. Relevant metadata of study participants are provided in the Supplementary Information file 3”.
Competing interests
C.K. and S.S. are employees of AstraZeneca. H.A. and J.Ku. are employees and shareholders of Antaros Medical. All other authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained errors in the Data availability section and Supplementary Information 3 was omitted. Full information regarding the correction made can be found in the correction for this Article.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Klev Diamanti and Robin Visvanathar.
Change history
11/28/2024
A Correction to this paper has been published: 10.1038/s41598-024-78537-6
Supplementary information
is available for this paper at 10.1038/s41598-020-64524-0.
References
- 1.Sas, K. M., Karnovsky, A., Michailidis, G. & Pennathur, S. Metabolomics and Diabetes: Analytical and Computational Approaches. Diabetes64, 718–732 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Global report on diabetes. (World Health Organization, 2016).
- 3.Ahlqvist, E. et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol.6, 361–369 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Fitipaldi, H., McCarthy, M. I., Florez, J. C. & Franks, P. W. A Global Overview of Precision Medicine in Type 2 Diabetes. Diabetes67, 1911–1922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein, M. S. & Shearer, J. Metabolomics and Type 2 Diabetes: Translating Basic Research into Clinical Application. Journal of Diabetes Researchhttps://www.hindawi.com/journals/jdr/2016/3898502/, 10.1155/2016/3898502 (2016). [DOI] [PMC free article] [PubMed]
- 6.Li, L. et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci. Transl. Med. 7, 311ra174-311ra174 (2015). [DOI] [PMC free article] [PubMed]
- 7.Segerstolpe, Å. et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab.24, 593–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods6, 377–382 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Razavian, N. et al. Population-Level Prediction of Type 2 Diabetes From Claims Data and Analysis of Risk Factors. Big Data3, 277–287 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Zou, Q. et al. Predicting Diabetes Mellitus With Machine Learning Techniques. Front. Genet. 9 (2018). [DOI] [PMC free article] [PubMed]
- 11.Karczewski, K. J. & Snyder, M. P. Integrative omics for health and disease. Nat. Rev. Genet.19, 299–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misra, B. B., Langefeld, C., Olivier, M. & Cox, L. A. Integrated omics: tools, advances and future approaches. J. Mol. Endocrinol.62, R21–R45 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Angelidis, I. et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun.10, 963 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fessenden, M. Metabolomics: Small molecules, single cells. Nature540, 153–155 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Newgard, C. B. et al. A Branched-Chain Amino Acid-Related Metabolic Signature that Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metab.9, 311–326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebholz, C. M. et al. Serum metabolomic profile of incident diabetes. Diabetologia61, 1046–1054 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, T. J. et al. Metabolite profiles and the risk of developing diabetes. Nat. Med.17, 448–453 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang‐Sattler, R. et al. Novel biomarkers for pre‐diabetes identified by metabolomics. Mol. Syst. Biol. 8 (2012). [DOI] [PMC free article] [PubMed]
- 19.Fall, T. et al. Non-targeted metabolomics combined with genetic analyses identifies bile acid synthesis and phospholipid metabolism as being associated with incident type 2 diabetes. Diabetologia59, 2114–2124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gálisová, A. et al. A Trimodal Imaging Platform for Tracking Viable Transplanted Pancreatic Islets In Vivo: F-19 MR, Fluorescence, and Bioluminescence Imaging. Mol. Imaging Biol.21, 454–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quick, H. H. Integrated PET/MR. J. Magn. Reson. Imaging39, 243–258 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Schlemmer, H.-P. W. et al. Simultaneous MR/PET Imaging of the Human Brain: Feasibility Study. Radiology248, 1028–1035 (2008). [DOI] [PubMed] [Google Scholar]
- 23.von Schulthess, G. K., Steinert, H. C. & Hany, T. F. Integrated PET/CT: Current Applications and Future Directions. Radiology238, 405–422 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Chen, T. et al. Tryptophan Predicts the Risk for Future Type 2 Diabetes. PLOS ONE11, e0162192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giesbertz, P. & Daniel, H. Branched-chain amino acids as biomarkers in diabetes. Curr. Opin. Clin. Nutr. Metab. Care19, 48–54 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Do, K. T., Rasp, D. J. N.-P., Kastenmüller, G., Suhre, K. & Krumsiek, J. MoDentify: phenotype-driven module identification in metabolomics networks at different resolutions. Bioinformatics, 10.1093/bioinformatics/bty650. [DOI] [PMC free article] [PubMed]
- 27.Michitaka, K. et al. Amino acid imbalance in patients with chronic liver diseases. Hepatol. Res.40, 393–398 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Soeters, P. & Fischer, J. INSULIN, GLUCAGON, AMINOACID IMBALANCE, AND HEPATIC ENCEPHALOPATHY. The Lancet308, 880–882 (1976). [DOI] [PubMed] [Google Scholar]
- 29.Kawanaka, M. et al. Tyrosine levels are associated with insulin resistance in patients with nonalcoholic fatty liver disease. Hepatic Med. Evid. Res.7, 29–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang, H. et al. Identification of key metabolic changes during liver fibrosis progression in rats using a urine and serum metabolomics approach. Sci. Rep.7, 11433–11433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherriff, J. L., O’Sullivan, T. A., Properzi, C., Oddo, J.-L. & Adams, L. A. Choline, Its Potential Role in Nonalcoholic Fatty Liver Disease, and the Case for Human and Bacterial Genes. Adv. Nutr.7, 5–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann, R. et al. Circulating Lysophosphatidylcholines Are Markers of a Metabolically Benign Nonalcoholic Fatty Liver. Diabetes Care36, 2331–2338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Bas, J. M. et al. Impairment of lysophospholipid metabolism in obesity: altered plasma profile and desensitization to the modulatory properties of n–3 polyunsaturated fatty acids in a randomized controlled trial. Am. J. Clin. Nutr.104, 266–279 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Yea, K. et al. Lysophosphatidylcholine Activates Adipocyte Glucose Uptake and Lowers Blood Glucose Levels in Murine Models of Diabetes. J. Biol. Chem.284, 33833–33840 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Susan et al. Metabolite Profiling Identifies Pathways Associated With Metabolic Risk in Humans. Circulation125, 2222–2231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felig, P., Marliss, E. & Cahill, G. F. Plasma Amino Acid Levels and Insulin Secretion in Obesity. N. Engl. J. Med.281, 811–816 (1969). [DOI] [PubMed] [Google Scholar]
- 37.Lackey, D. E. et al. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am. J. Physiol.-Endocrinol. Metab.304, E1175–E1187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lustgarten, M. S., Price, L. L., Phillips, E. M. & Fielding, R. A. Serum Glycine Is Associated with Regional Body Fat and Insulin Resistance in Functionally-Limited Older Adults. PLOS ONE8, e84034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin, F.-P. J. et al. Topographical Body Fat Distribution Links to Amino Acid and Lipid Metabolism in Healthy Non-Obese Women. PLOS ONE8, e73445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukui, H. et al. Enhanced Activity of the Purine Nucleotide Cycle of the Exercising Muscle in Patients with Hyperthyroidism. J. Clin. Endocrinol. Metab.86, 2205–2210 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Varadaiah, Y. G. C., Sivanesan, S., Nayak, S. B. & Thirumalarao, K. R. Purine metabolites can indicate diabetes progression. Arch. Physiol. Biochem.0, 1–5 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Katsiki, N., Papanas, N., Fonseca, V. A., Maltezos, E. & Mikhailidis, D. P. Uric Acid and Diabetes: Is there a Link? Curr. Pharm. Des.19, 4930–4937 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Boersma, G. J. et al. Altered Glucose Uptake in Muscle, Visceral Adipose Tissue, and Brain Predict Whole-Body Insulin Resistance and may Contribute to the Development of Type 2 Diabetes: A Combined PET/MR Study. Horm. Metab. Res.50, e10–e10 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Pereira, M. J. et al. Impaired adipose tissue lipid storage, but not altered lipolysis, contributes to elevated levels of NEFA in type 2 diabetes. Degree of hyperglycemia and adiposity are important factors. Metabolism65, 1768–1780 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Johansson, E. et al. Whole-Body Imaging of Tissue-specific Insulin Sensitivity and Body Composition by Using an Integrated PET/MR System: A Feasibility Study. Radiology286, 271–278 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Patlak, C. S., Blasberg, R. G. & Fenstermacher, J. D. Graphical Evaluation of Blood-to-Brain Transfer Constants from Multiple-Time Uptake Data. J. Cereb. Blood Flow Metab.3, 1–7 (1983). [DOI] [PubMed] [Google Scholar]
- 47.Strand, R. et al. A concept for holistic whole body MRI data analysis, Imiomics. PLOS ONE12, e0169966 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schauer, N. et al. GC–MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett.579, 1332–1337 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Afzal, M. et al. Integrated Univariate, Multivariate, and Correlation-Based Network Analyses Reveal Metabolite-Specific Effects on Bacterial Growth and Biofilm Formation in Necrotizing Soft Tissue Infections. J. Proteome Res., 10.1021/acs.jproteome.9b00565 (2019). [DOI] [PubMed]
- 50.Nagle, C. A., Klett, E. L. & Coleman, R. A. Hepatic triacylglycerol accumulation and insulin resistance. J. Lipid Res.50, S74–S79 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diamanti, K. et al. Intra- and inter-individual metabolic profiling highlights carnitine and lysophosphatidylcholine pathways as key molecular defects in type 2 diabetes. Sci. Rep.9, 9653 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
“The metabolomics datasets generated and analysed during the current study are available in MetaboLights (https://www.ebi.ac.uk/metabolights/) under the accession number MTBLS1051. Relevant metadata of study participants are provided in the Supplementary Information file 3”.