Abstract
ETS1 has shown dichotomous roles as an oncogene and a tumor suppressor gene in diverse cancers, but its functionality in breast cancer tumorigenesis still remains unclear. We utilized the Cancer Genome Atlas (TCGA) database to analyze comprehensive functions of ETS1 in human breast cancer (BRCA) patients by investigating its expression patterns and methylation status in relation to clinical prognosis. ETS1 expression was significantly diminished by hyper-methylation of the ETS1 promoter region in specimens from BRCA patients compared to a healthy control group. Moreover, ETS1high BRCA patients showed better prognosis and longer survival compared to ETS1low BRCA patients. Consistent with clinical evidence, comparative transcriptome analysis combined with CRISPR/Cas9 or shRNA based perturbation of ETS1 expression revealed direct as well as indirect mechanisms of ETS1 that hinder tumorigenesis of BRCA cells. Taken together, our study enlightens a novel function of ETS1 as a tumor suppressor in breast cancer cells.
Keywords: ETS1, breast cancer, tumor suppressor, DNA methylation, regulatory elements
Introduction
Breast cancer is the most commonly diagnosed cancer and the second leading cause of death from cancer in women (1). Development and progress of breast cancer are mediated by a complicated process in which many genes and signaling pathways are intertwined (2). Profiling of gene-expression in patient specimen has been used to identify the key molecular switches of tumor development (3). Human breast cancer datasets, including The Cancer Genome Atlas (TCGA) (4) and Curtis (5) provide comprehensive genomic profiles of breast cancer.
ETS1 (ETS proto-oncogene 1, transcription factor) has initially been characterized as the proto-oncogenic transcription factor that contributes to tumor angiogenesis and invasiveness in cancer cells (6–8). Previously, high levels of ETS1 expression have been closely associated with higher chance of metastatic potential and poor prognosis in various types of cancers (9–14). ETS1 is known to enhance the expression of numerous tumorigenic genes involved in tumor angiogenesis, cancer cell invasion, and energy metabolism (15). These include vascular endothelial growth factor (VEGF) and certain proteases such as MMP-1, MMP-3, and MMP-9, as well as urokinase type plasminogen activator (uPA), which is associated with extracellular matrix (ECM) degradation (16–19). Despite the established oncogenic function of ETS1 in human cancers, recent studies have proposed contrasting roles of ETS1 as anti-oncogenes suggesting dichotomous roles of ETS1 for tumorigenesis in context-dependent manner (20, 21). However, the functionality and molecular action mechanisms of ETS1 in BRCA tumorigenesis still remain unclear.
In this study, we revealed ETS1 as the tumor suppressor gene in BRCA cells. In humans, poor prognosis of BRCA patients was negatively correlated with ETS1 expression, repressed by hyper-CpG methylation in ETS1 promoter locus. Furthermore, we showed the direct and indirect mechanisms of ETS1 to hinder tumorigenesis of BRCA cells. Overall, our findings enlighten the novel function of ETS1 as the tumor suppressor gene, which can be the potential target for novel therapeutics in BRCA.
Materials and Methods
Cell Culture, Plasmid, and Reagents
MDA-MB-231 cells were cultured in DMEM (WELGENE: LM 001-05) supplemented with 10% FBS (Gibco: 10099-141) and 100 U/ml of penicillin-streptomycin (Thermo: 15140122). Mutant MDA-MB-231 cells (ΔCRE) harboring deleted promoter region (−540 to −80) of ETS1 were established using the CRISPR/Cas9 method (22). Mutations were confirmed by Sanger sequencing, and the effect of CRE deletion on ETS1 level was tested by immunoblotting. Cells were harvested with 0.05% trypsin-EDTA (Gibco: 25300-054). The following chemicals were used; phorbol 12-myristate 13-acetate (PMA, Calbiochem: 524400) and Ionomycin (Calbiochem: 407950).
Knockdown and Ectopic Expression of ETS1 by Lentiviral Transduction
Gene knockdown was accomplished using the shRNA system with control shRNA (TR30021) or ETS1 targeted shRNA (TL313153) (OriGene Technologies, Rockville, MD). MDA-MB-231 cells were exposed to lentiviral concentrates. Gene overexpression was accomplished using Human cDNA clone ETS1 (RC215203L2) (OriGene Technologies, Rockville, MD). MCF-7 cells were infected with lentiviral particle encoding hETS1. After lentiviral transduction, cells were harvested and sorted with Moflo XDP (Beckman Coulter, Fullerton, CA) for GFP negative and positive cells.
RNA Isolation, cDNA Synthesis, and Quantitative RT-PCR
Total RNA was extracted using a TRI Reagent (Molecular Research Center) following standard protocols. Reverse transcription of 1 μg RNA was performed using oligo (dT) primer (Promega: C1101) with Improm II Reverse Transcription system (Promega) according to the manufacturer's protocols. Quantitative RT-PCR was performed using SYBR Green Dye mix (Takara: RR420) on Rotor-Gene Q (Qiagen, Hilden, Germany). Data was normalized to human hypoxanthine-guanine phosphoribosyl transferase (HPRT). Primer sequences are provided in Supplementary Table 1.
Chromatin Immunoprecipitation (ChIP)-PCR Assay
ChIP-PCR assays were performed by Simple ChIP plus Enzymatic Chromatin IP Kit (Cell Signaling: #9005) according to the manufacturer's protocols. Briefly, cells were cross-linked with 1% of formaldehyde and lysed for nuclei preparation. Nucleus pellet was treated with micrococcal nuclease and sonicated for chromatin fragmentation. Protein-chromatin complex was incubated with antibody targeting anti-ETS1 (Cell Signaling: #14069) at 4°C overnight. Rabbit IgG (Vector Laboratories) was used as negative control. After immuno-precipitation, 50 μ Dynabeads protein G or A (Life technologies) were added and rotated further for 6-h at 4°C. Ab/protein/chromatin complex were reverse-crosslinked at 65°C overnight, and DNA was purified by DNA purification columns (Cell Signaling: #10010). The relative enrichment of specific regions in precipitated DNA was measured by quantitative PCR (qRT-PCR). To quantify protein binding in specific genomic locus, purified DNA was used for qRT-PCR. Primer sequences are listed in Supplementary Table 2.
Immunoblot Assay
Whole cell lysates were extracted using RIPA buffer according to manufacturer's protocols. Protein concentration was measured by Bradford protein assay (Bio-Rad: #5000001), and 20 or 30 μg of proteins were used for SDS-PAGE (10%) and then transferred onto a nitrocellulose membrane (Bio-Rad: 162-0097). The following primary antibodies targeting ETS1 (Santa Cruz Biotechnology: sc-55581) and ACTIN (Abcam: ab3280) were used. Protein expression was visualized with ImageQuant™ LAS 4000 (GE healthcare Life Science, Piscataway, NJ). ACTIN expression was used as a loading control for whole cell lysates.
Flow Cytometric Analysis
MDA-MB-231 (WT) and ΔCRE cells were harvested, washed with PBS, fixed by 2 ml of cold 70% ethanol dropwise, and incubated at −20°C overnight. For checking proliferation by Ki-67, diluted anti-Ki-67 antibody (BioLegend: #652404) was added and incubated at room temperature (RT) for 30 min in the dark. After incubation, cells were washed and re-suspended in 200 μl of PBS. Cells were then analyzed with BD LSRFortessa (BD Biosciences, San Jose, CA) and FlowJo software (Treestar, San Carlos, CA).
Xenograft Cancer Model
Six-week-old female nude mice (Orient Bio) were injected subcutaneously with MDA-MB-231 (5 × 106) or ΔCRE cells (5 × 106) to the left or right abdomen, respectively. In addition, nude mice were injected subcutaneously with MCF-7 GFP negative (ETS1−) or MCF-7 GFP positive (ETS1+) (5 × 106) to the left or right abdomen, respectively. Tumor size was measured every 4 days by caliper measurements. All mice were housed in a specific pathogen-free barrier facility, and allocated and randomized for the experiments by a technician at the animal facility. The experiments were performed in accordance with protocols approved by POSTECH Institutional Animal Care and Use Committee, Korea. All experiments were performed in accordance with relevant guidelines and regulations.
RNA-Sequencing
Total RNA was extracted and purified with RibospinTMII (GeneAll biotechnology: 314-150). RNA was subjected to library preparation with TruSeq Stranded mRNA Sample Preparation Kit (Illumina: RS-122-2101~2), and RNA-sequencing was performed by NextSeq 500 Sequencing System (Illumina, San Diego, CA). Sequences were mapped to hg19 with TopHat (version 2.0.12). Estimated expression level was generated with Cufflinks (version 2.2.1), and differentially expressed genes were selected using Cuffdiff (version 2.2.1). RNA-seq was performed on two biological replicates. The set of differential expressed genes were analyzed using DAVID gene functional classification tool (23).
Dataset for Human Samples and Human Cell Lines
TCGA (The Cancer Genome Atlas) data were analyzed using a web-based program. Wanderer Website (http://gattaca.imppc.org:3838/wanderer/) was used to determine differential expressions of ETS1, target genes, and methylation profiles in the tumor and normal specimens (24). OncoLnc (http://www.oncolnc.org/) was used to calculate survival rate according to ETS1 expression level in tumor samples (25). Top or bottom 10% of total patients in ETS1 expression or methylation level was selected for further analysis. The cBioPortal (http://www.cbioportal.org/) was used to determine the correlation between ETS1 alteration and survival rate in tumor samples.
Statistical Analysis
Error bars indicate standard deviation (SD). All t-tests performed were student two-tailed tests (*P < 0.05, **P < 0.01, and ***P < 0.001).
Data Accessibility
RNA-seq datasets have been deposited in GEO database with accession code GSE106634.
Results
Clinical Relevance of ETS1 Expression in Breast Cancer Patients
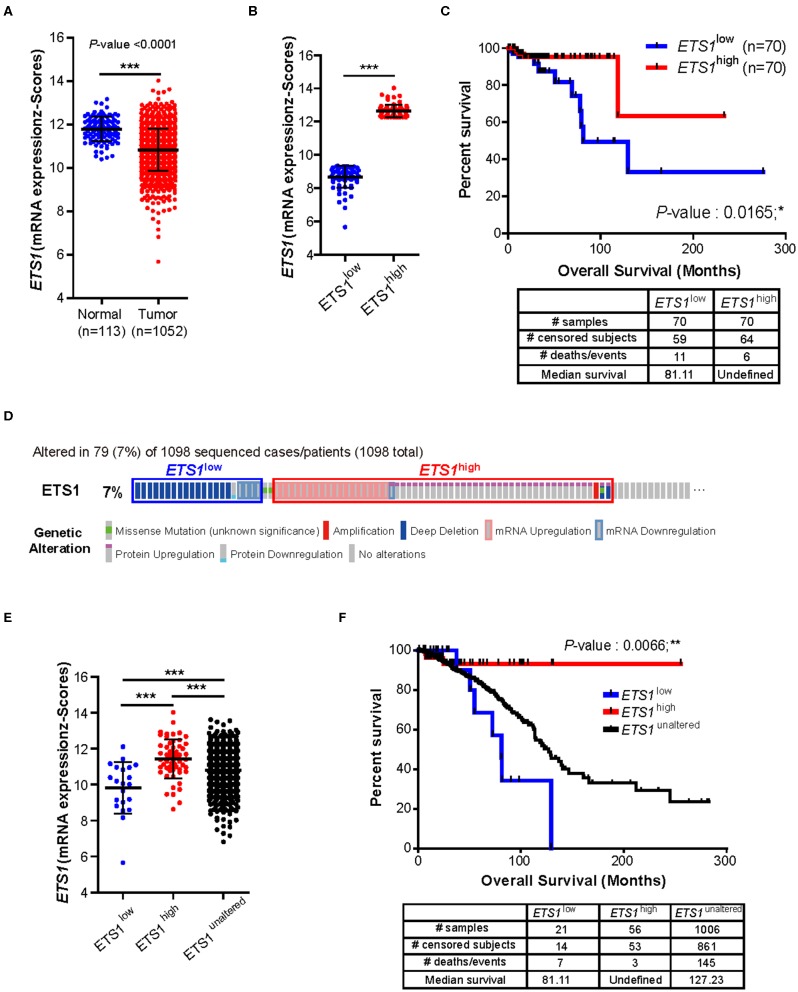
To determine whether ETS1 acts as a pro- or anti-tumorigenic factor in BRCA tumorigenesis, we performed correlative analysis of ETS1 expression with clinical outcome of BRCA patients. Through the analysis of The Cancer Genome Atlas (TCGA) database, we found that, in contrast to previous studies (10, 13, 26–29), breast cancer specimens (n = 1,052) significantly reduced ETS1 expression compared to normal tissues (n = 113, Figure 1A). This observation was further validated by analyzing another large clinical dataset, Curtis, that contained 2,000 breast tumor samples (5). Consistently, significantly curtailed expression of ETS1 in breast cancer specimens (n = 1,992) was observed in BRCA specimens compared to normal specimens (n = 144; Supplementary Figure 1A). Next, to assess whether ETS1 expression level has clinical implications, we classified breast cancer patients into two groups based on ETS1 expression in BRCA specimens (ETS1high and ETS1low; Figure 1B). Intriguingly, we found that BRCA patients in the ETS1low group were significantly more likely to have a poor prognosis in TCGA (P = 0.0165) as well as the Curtis (P = 0.0076) database (Figure 1C and Supplementary Figures 1A,B), compared to BRCA patients in ETS1high group not in triple-negative type specific manners (Supplementary Figures 1C,D) suggesting the potential of ETS1 as the anti-tumorigenic factor in breast cancer. To confirm whether ETS1 has anti-oncogenic function in general or in specific types of tumor, we extended previous analysis to other cancers as well. Intriguingly, significantly reduced expression of ETS1 was observed in not all but specific types of cancers, including Bladder Urothelial Carcinoma (BLCA), colon adenocarcinoma (COAD), and Lung adenocarcinoma (LUAD), compared to normal specimens (Supplementary Figure 2A), which is highly correlated with poor clinical outcomes in general, consistent with BRCA (Supplementary Figure 2B). Collectively, these results suggest anti-tumorigenic functions of ETS1 in a tumor-dependent manner. Next, we questioned whether low levels of ETS1 in BRCA patients is associated with genetic alterations around ETS1 genomic locus compared to healthy counterparts, using cBioPortal (Computational Biology Center, Memorial Sloan Kettering Cancer Center). We found that about 7% of BRCA patients (79 among 1,098) in TCGA dataset showed genetic alterations in the ETS1 gene including amplification or deep deletion, which led to down- or up-regulation of ETS1 levels, respectively (Figure 1D). Hence, we then divided the patients into three groups, based the effect of generic alteration of ETS1 on its expression; ETS1low, ETS1high, and ETS1unaltered (Figure 1E), and assessed the effect of genetic alteration on clinical outcomes in BRCA patients. Compared to ETS1unaltered, genetic alterations triggering diminished ETS1 expression (ETS1low) were highly correlated with poor clinical outcomes, while ETS1high showed increased survival rate (Figure 1F). Altogether, these clinical results indicate ETS1 as a novel tumor suppressor gene in BRCA.
Figure 1.
Clinical implication of ETS1 level in breast cancer patients. (A) Comparison of ETS1 expression between normal (n = 113) and BRCA patient (n = 1052) specimens in publicly available datasets (TCGA). Each symbol represents an individual; horizontal lines indicate the mean. ***P < 0.001 (Student t-test). (B) ETS1 expressions of ETS1low (n = 70) and ETS1high (n = 70) groups. (C) Correlation between ETS1 expression and survival rate. A total of 1,052 samples were divided into ETS1high (n = 70) and ETS1low (n = 70) groups according to ETS1 expression levels. Overall survival analysis was performed based on ETS1 levels in BRCA patients. Blue and red lines indicate patients with ETS1low (n = 70) and ETS1high expression (n = 70), respectively. P-value was calculated using log-rank test (P = 0.0165). (D) Profiles of genetic alterations of ETS1 in the 1,098 human BRCA specimens in TCGA dataset. (E) ETS1 expression levels of three groups (unaltered, group 1, and group 2). (F) Overall survival rates in three groups (unaltered, group 1, and group 2) of breast cancer patients. Black line indicates unaltered group. Blue and red lines indicate patients with ETS1low (P = 0.0551) and ETS1high expression (P = 0.0452), respectively, compared to unaltered group. P-values were calculated by log-rank test. *P < 0.05, **P < 0.005.
The Effects of DNA Methylation Status on ETS1 Promoter Region for Its Expression in BRCA Patients
Next, we questioned how breast tumor cells maintain lower ETS1 expression than normal cells. To answer this, we decided to focus on DNA methylation, which is one of the major epigenetic mechanisms to control gene expression (30, 31), by comparing methylation status of CpG sites on ETS1 genomic locus between BRCA patients (n = 732) and healthy counterparts (n = 84) (24). Intriguingly, we found that tumor specimens showed significantly higher methylation levels in two CpG island regions, located around the promoter of ETS1 (locus #1 and locus #2), compared to normal specimens (Figure 2A and Supplementary Table 3). Then, we determined whether methylation status of ETS1 promoter regions is accompanied with ETS1 expression level. Hyper-methylation in locus #1 but not #2 of ETS1 promoter region was closely associated with decreased ETS1 expression in BRCA specimens, indicating inverse correlation of ETS1 methylation status with its expression (Figures 2B,C). Among 10 CpG sites in locus #1, methylation status of cg26559804, cg26503877, and cg11588197 showed significant inverse correlation with ETS1 expression in multiple types of cancers (Supplementary Figure 3). Moreover, seven remaining CpG methylation sites seemed to be BRCA-specific, since other tumors showed comparable CpG methylation patterns in normal and tumor specimens (Supplementary Figure 3). Next, we examined the methylation level of the ETS1 promoter (Metlow vs. Methigh) and its clinical outcomes in BRCA patients. Consistent with ETS1 expression, enhanced CpG methylation (Methigh) was correlated with low ETS1 level (Figure 2D), resulting in poor prognosis (Figure 2E). Collectively, these results suggest that DNA methylation on ETS1 promoter locus is a key factor to determine ETS1 expression and disease prognosis in BRCA patients.
Figure 2.
Impact of DNA methylations status of ETS1 promoter on its expression in BRCA patients. (A) Mean methylation (β value) on CpG sites of ETS1 locus between normal (n = 98) and BRCA patient (n = 743) specimens by TCGA Wanderer tool. Blue and red lines indicates normal or BRCA patients, respectively. *P < 0.05, **P < 0.01, ***P < 0.001 (two-way ANOVA with Bonferroni test). (B,C) Scatterplots depicting linear regression (black line) and Pearson correlation analysis with corresponding P-values. Correlation between ETS1 expression and methylation status represented by β value on each CpG site of locus #1 (B) and locus #2 (C) in normal and tumor samples. Each symbol represents an individual human specimen. (D) ETS1 expression levels between Metlow vs. Methigh groups in cg26559804. Blue or red dot indicates methylation low (Metlow; n = 100) or methylation high (Methigh; n = 100), respectively. (E) Overall survival rates in two groups (Metlow vs. Methigh) of breast cancer patients. Blue and red lines indicate patients with Metlow and Methigh (P = 0.0191), respectively. P-values were calculated using log-rank test.
ETS1 Regulates Growth and Proliferation of Tumor Cells
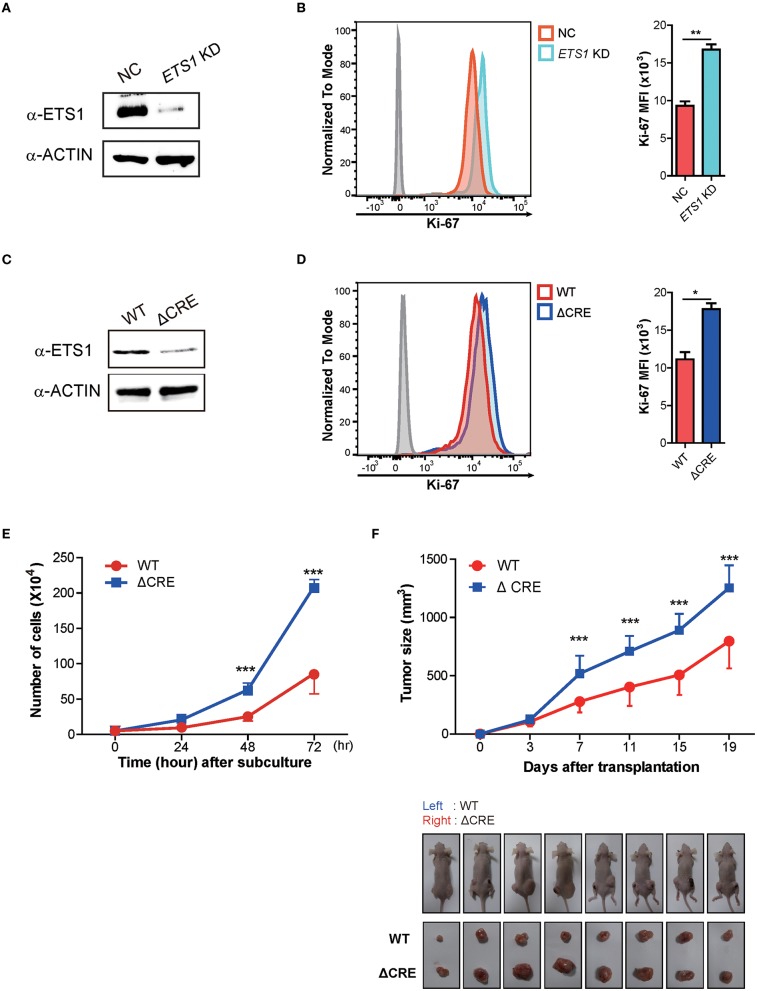
The abovementioned findings on BRCA patients support ETS1 as a tumor suppressor in breast cancer. To elucidate how ETS1 affects tumorigenesis of BRCA, we tested the effects of knockdown of ETS1 on tumorigenicity of breast cancer cells. To achieve this goal, we employed MDA-MB-231 cells, well-characterized breast cancer cell lines with high levels of ETS1 expression (32). MDA-MB-231 cells were transfected with mock and ETS1 targeting shRNA vector tagged with GFP and found around 70% diminished ETS1 protein level in ETS1-shRNA+ cells compared with mock-shRNA+ cells (Figure 3A). Interestingly, knockdown of ETS1 expression significantly increased proliferation capacity of breast cancer cells measured by Ki-67 expression (Figure 3B). To confirm previous observations, we generated mutant MDA-MB-231 cells, in which a core regulatory element (CRE; −540 to −80) of ETS1 promoter was deleted by CRISPR/Cas9 methods, and named as “ΔCRE” (22). Similar to shRNA based knockdown system, ΔCRE cells demonstrated decreased expression of ETS1 with enhanced proliferation and growth rate (Figures 3C–E) suggesting anti-tumorigenic function of ETS1 in BRCA cells. To further validate anti-tumorigenic roles of ETS1 in vivo, WT or ΔCRE cells were subcutaneously transplanted into the left or right sides of flank using nude mice and monitored tumor growth in vivo, respectively. Indeed, mice engrafted with ΔCRE cells developed significantly larger volume of tumor compared to mice injected with WT cells (Figure 3F). Since MDA-MB-231 is a triple negative breast cell line that can only represent a specific subset of breast cancer, we tried to confirm whether ETS1 inhibits the proliferation of tumor cells with MCF-7 cells (ER+, PR+ and HER2− cell; Supplementary Figure 4). Ectopic expression of ETS1 in MCF-7 cells significantly reduced cell proliferation in vitro (Supplementary Figure 4A) and suppressed tumor growth in vivo (Supplementary Figure 4B). Altogether, these results indicate that ETS1 suppresses proliferation and growth of breast cancer cells in vitro as well as in vivo.
Figure 3.
ETS1 inhibits the proliferation and growth of breast cancer cells in vitro as well as in vivo. MDA-MB-231 cells were transfected with negative control (NC) and ETS1 shRNA-GFP reporter vector, and transfected cells were sorted according to GFP-expression. The FACs plots are representative of data from three independent experiments. (A) Immunoblotting analysis for ETS1 protein levels in NC and knockdown cells. (B) Analysis of cell proliferation (Ki-67) comparing NC and ETS1 KD cells. (C) Immunoblotting analysis for levels of ETS1 in MDA-MB-231 (WT) cells and ΔCRE cells. (D) Quantification of cell proliferation based on detection of Ki-67 protein by flow cytometry. (E) Cell count for 72-h culture periods. *P < 0.05, **P < 0.01, ***P < 0.001 (two-way ANOVA with Bonferroni test). (F) Tumor growth curves in nude mice after injection of WT (left) and ΔCRE (right) cells. Tumor volumes were measured at an interval of 4 days (n = 10). Images of representative tumor-bearing mice (top) and isolated tumors (bottom). *P < 0.05, **P < 0.01, ***P < 0.001 (two-way ANOVA with Bonferroni test). Data is representative of three independent experiments. For reasons of clarity, blots were cropped to the bands of interest. Samples derived from the same experiment and gels/blots were processed in parallel. See full-length blots in Supplementary Figure 6.
ETS1 Regulates Expression of Cell Proliferation-Related Factors
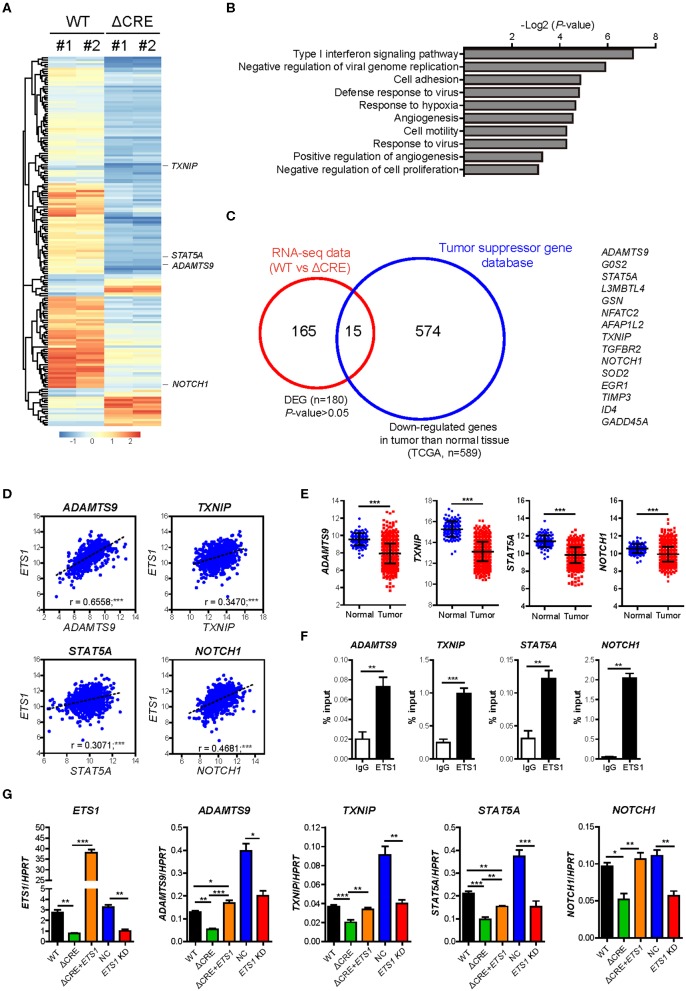
To determine how ETS1 regulates proliferation and growth of breast cancer cells, we performed an RNA-sequencing (RNA-seq) based transcriptome analysis with WT and ΔCRE cells (Figure 4A and Supplementary Table 4). Among 180 differentially expressed genes (DEG), 80% of DEGs were down-regulated in ΔCRE cells compared to WT cells, indicating a major role of ETS1 in BRCA as transcriptional activator. To enlighten down-stream pathways affected by ETS1 in breast cancer cells, we performed Gene Ontology (GO) and found that various tumorigenic pathways, including cell adhesion, angiogenesis, and proliferation, were significantly altered in ΔCRE cells (Figure 4B and Supplementary Table 5). Intriguingly, we noticed that several well-known tumor suppressor genes were significantly under-expressed in ΔCRE cells, of which expression levels were also reduced in BRCA tumor tissue compared to normal tissue from TCGA (Figures 4A,C and Supplementary Tables 6, 7) (33). Furthermore, we found a strong correlative expression between ETS1 and these tumor suppressor genes (ADAMTS9, TXNIP, STAT5A, and NOTCH1) in BRCA specimen samples that were actually under-expressed in BRCA compared to normal tissue (Figures 4D,E), suggesting the potential mechanism of ETS1 as the tumor suppressor through direct activation of other tumor suppressor genes in BRCA cells. Indeed, we found highly conserved putative ETS1 binding sites on the promoter regions of ADAMTS9, TXNIP, STAT5A, and NOTCH1 using ECR browser (Supplementary Figure 5), and the direct binding of ETS1 in these genomic loci was confirmed by ChIP assay (Figure 4F) in BRCA cells. To elucidate the effects of ETS1 binding on these loci, we first knockdowned ETS1 expression and checked its impact on transcriptional change of these tumor suppressor genes in MDA-MB-231 cells. Consistent with ΔCRE cells, knockdown of ETS1 significantly attenuated the expression level of these tumor suppressor genes (Figure 4G). Furthermore, knockdown effects of ETS1 were confirmed by rescuing ETS1 expression in ΔCRE cells that significantly enhanced the expression of ADAMTS9, TXNIP, STAT5A, and NOTCH1 compared to ΔCRE cells (Figure 4G). Altogether, these data indicate dichotomous functions of ETS1 as anti-tumorigenic factors through direct activation of targets but also trans-activating other tumor suppressor genes in breast cancer cells.
Figure 4.
ETS1 resultes muti-turmogeneic pathways in breast cancer cells. Transcriptome datasets from WT and ΔCRE cells were visualized by (A) heatmap analysis and further applied in (B) biological process of Gene Ontology (GO) enrichment analysis. (C) Venn diagram depicting the overlap of differentially expressed genes (DEGs) together with down-regulated genes in tumor compared to normal tissue, based on TSGene (33), a web resource for tumor suppressor genes. (D) Scatterplots depicting linear regression (black dot line) and Pearson correlation analysis with corresponding P-values. Correlation between ETS1 and target genes in BRCA. Each symbol represents an individual human specimen. (E) mRNA expression of ETS1 target genes in normal and tumor specimens. (F) ChIP assay was performed using anti-ETS1 antibody with MDA-MB-231 cells. Relative enrichments were determined by qRT-PCR with primers specific for ETS1 binding sites in the target gene locus. Representative data from three independent experiments. (G) Validation of representative target genes by qRT-PCR. Representative data from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (One-way ANOVA with Bonferroni test). *P < 0.05, **P < 0.01, ***P < 0.001 (Student t-test).
Discussion
In this study, we defined ETS1 as the tumor suppressor in BRCA together with detailed molecular action mechanisms. Analysis of human breast cancer specimens showed a lower level of ETS1 through hyper-DNA methylation on the ETS1 promoter region in BRCA compared to normal specimens, which was closely correlated with poor prognosis in a large number of BRCA patients. Furthermore, we elucidated the action mechanisms of ETS1 as a tumor suppressor not only by directly activating down-stream targets linked with tumor cell proliferation/growth but also trans-activation of other tumor suppressor genes, such as ADAMTS9, TXNIP, STAT5A, and NOTCH1.
Previous studies have shown dual functions of ETS1 as both pro-oncogenic (9–13) and anti-oncogenic (19, 20, 31) factors. How does ETS1 have paradoxical roles in tumorigeneisis? Interestingly, we found a distinct expression pattern of ETS1 depending on tumor types. For example, unlike BRCA, high level of ETS1 was observed in cancer specimens from GBM, HNSC, KIRC, PCPG, SARC, and THCA; while other tumor types, including BRCA, showed low levels of ETS1 compared to normal specimens (Supplementary Figure 2A, data not shown). In addition, we have shown that ETS1 directly activates several invasiveness factors, such as ENG and MMP14 (Supplementary Table 5) enhancing invasive phenotypes in vitro as well as in vivo (22). Similarly to ETS1, various factors are thought to have dual roles, as tumor suppressor and activator, in a cell type-dependent manner; however, how they switch between the two functionalities has never been established. For instance, PGC-1α, a master regulator of energy metabolism, has recently been shown to exert anti-metastatic effects in cancer through inhibition of EMT, but it also plays the opposite role in specific cancer subtypes by providing growth advantages (34). In addition, TGFβ has shown its anti-tumorigenic function at early stage of cancer, while it supported tumor metastasis in later stages of cancer (35). Furthermore, it is intriguing that ΔCRE cells decreased the expression of gene sets involved in immune responses, such as Type I interferon signaling (OAS-1, -3 and IFIT-1, -2, -3, etc.) and anti-viral responses (TREM183, BNIP3 etc.) which suggest potential roles of ETS1 to modulating immunogenicity of BRCA cells. Hence, it is essential to understand how ETS1 has dichotomous roles in tumorigenesis, which is currently under investigation.
Our comparative transcriptome analysis between MDA-MB-231 cells (WT) and ΔCRE cells has clearly revealed that ETS1 is directly involved in multiple steps to hinder tumorigenesis including angiogenesis, cell survival, proliferation, and even cell adhesion (Figure 4B). To take a step forward from this analysis, we have identified a unique feature of ETS1 to inhibit tumorigenesis of BRCA cells by directly trans-activating core regulators of tumorigenesis as tumor suppressor genes in BRCA cells. ADAMTS9 (36, 37) and TXNIP (38) have been shown to inhibit cancer cell proliferation, and STAT5A and NOTCH1 are well-known for their versatile roles to suppress proliferation, survival, differentiation, or senescence of cancer cells (39–42). ETS1 directly bound in genomic loci of these regulators (Figure 4F) and activated their transcription in breast cancer cells (Figure 4G). Hence, our results enlightened that ETS1 exerts its anti-tumorigenic function in at least two distinct ways: (i) direct activation of gene-sets involved in BRCA tumorigenesis, and (ii) indirect inhibition of tumorigenesis through trans-activation of canonical tumor suppressor genes in BRCA cells.
Previously, transcription factors (TFs) were implicated in a majority of human diseases, such as cancer and autoimmune diseases. In addition, TFs were considered to be “undruggable” targets, except for ligand-inducible nuclear receptors (43). However, numerous recent cases have successfully targeted TFs, suggesting TFs to be feasible targets for drug development (43). Since ETS1 level is strikingly correlated with a patient's prognostic status, ETS1-targeted therapy seems attractive. Previously, our group and others have identified up-stream signaling pathways and major transcriptional activators for ETS1 transcription in breast cancer cells (22, 44). In addition, previous studies have shown the importance of post-translational modification (phosphorylation, acetylation, sumoylation, and ubiquitination) and co-factor interaction (45). In this context, our result proposed potential therapeutic approach targeting cis-regulatory elements of ETS1 to modulate the overall level in cancer cells. Therefore, in the future, we will direct our studies to investigate these mechanisms for the development of therapeutics targeting ETS1 in BRCA.
In this study, we defined ETS1 as a tumor suppressor that inhibits growth and proliferation of breast cancer cells in both humans and mice. Our study indicated that ETS1 might be a good therapeutic target for BRCA due to its intrinsic and extrinsic propensities to inhibit tumorigenesis. Moreover, given the intense interest in understanding the biomarkers for predicting breast cancer prognosis, our findings indicate that evaluating ETS1 level in tumors may be an important predicator for BRCA patients.
Data Availability Statement
RNA-seq datasets have been deposited in the NCBI Gene Expression Omnibus with accession code GSE106634.
Ethics Statement
This study was carried out in accordance with the principles of the Basel Declaration and recommendations of International Association of Veterinary Editors guidelines, Pohang University of Science and Technology (POSTECH) Institutional Animal Care and Use Committee. The protocol was approved by POSTECH Institutional Animal Care and Use Committee.
Author Contributions
S-HI, G-CK, and H-KK designed the study and wrote the paper. G-CK, C-GL and RV performed the experiments. TK and KK analyzed RNA sequencing data. YK and JN provided intellectual contributions. S-HI, DR, C-GL, and H-KK edited the paper.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Haejin Jung for technical assistance in cell sorting.
Footnotes
Funding. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1A6A1A03032869, 2019R1F1A1060415, 2019M3C9A6091949), a new faculty research seed money grant of Yonsei University College of Medicine for 2019 (2019-32-0022) and a faculty research grant of Yonsei University College of Medicine (6-2019-0113), and Korea Institute of Science and Technology intramural grant (2Z06220 and 2Z06130).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.00642/full#supplementary-material
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. (2016) 66:7–30. 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Feng Y, Spezia M, Huang S, Yuan C, Zeng Z, Zhang L, et al. Breast cancer development and progression: risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. (2018) 5:77–106. 10.1016/j.gendis.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Güler EN. Gene expression profiling in breast cancer and its effect on therapy selection in early-stage breast cancer. Eur J Breast Health. (2017) 13:168–74. 10.5152/ejbh.2017.3636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Network CGA. Comprehensive molecular portraits of human breast tumors. Nature. (2012) 490:61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. (2012) 486:346–52. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimoto J, Aoki I, Toyoki H, Khatun S, Tamaya T. Clinical implications of expression of ETS-1 related to angiogenesis in uterine cervical cancers. Ann Oncol. (2002) 13:1598–604. 10.1093/annonc/mdf248 [DOI] [PubMed] [Google Scholar]
- 7.Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. (2015) 35:20–38. 10.1016/j.semcancer.2015.09.010 [DOI] [PubMed] [Google Scholar]
- 8.Hsing M, Wang Y, Rennie PS, Cox ME, Cherkasov A. ETS transcription factors as emerging drug targets in cancer. Med Res Rev. (2020) 40:413–30. 10.1002/med.21575 [DOI] [PubMed] [Google Scholar]
- 9.Fujimoto J, Aoki I, Toyoki H, Khatun S, Sato E, Sakaguchi H, et al. Clinical implications of expression of ETS-1 related to angiogenesis in metastatic lesions of ovarian cancers. Oncology. (2004) 66:420–8. 10.1159/000079491 [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi E, Nakayama T, Nanashima A, Matsumoto K, Yasutake T, Sekine I, et al. Ets-1 proto-oncogene as a potential predictor for poor prognosis of lung adenocarcinoma. Tohoku J Exp Med. (2007) 213:41–50. 10.1620/tjem.213.41 [DOI] [PubMed] [Google Scholar]
- 11.Rothhammer T, Hahne J, Florin A, Poser I, Soncin F, Wernert N, et al. The Ets-1 transcription factor is involved in the development and invasion of malignant melanoma. Cell Mol Life Sci. (2004) 61:118–28. 10.1007/s00018-003-3337-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama T, Ito M, Ohtsuru A, Naito S, Sekine I. Expression of the ets-1 proto-oncogene in human colorectal carcinoma. Mod Pathol. (2001) 14:415–22. 10.1038/modpathol.3880328 [DOI] [PubMed] [Google Scholar]
- 13.Span PN, Manders P, Heuvel JJ, Thomas CM, Bosch RR, Beex LV, et al. Expression of the transcription factor Ets-1 is an independent prognostic marker for relapse-free survival in breast cancer. Oncogene. (2002) 21:8506–9. 10.1038/sj.onc.1206040 [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Zhou R, Zhou H, Li Q, Hong J, Meng R, et al. ETS-1 induces endothelial-like differentiation and promotes metastasis in non-small cell lung cancer. Cell Physiol Biochem. (2018) 45:1827–39. 10.1159/000487874 [DOI] [PubMed] [Google Scholar]
- 15.Verschoor ML, Verschoor CP, Singh G. Ets-1 global gene expression profile reveals associations with metabolism and oxidative stress in ovarian and breast cancers. Cancer Metab. (2013) 1:17. 10.1186/2049-3002-1-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behrens P, Mathiak M, Mangold E, Kirdorf S, Wellmann A, Fogt F, et al. Stromal expression of invasion-promoting, matrix-degrading proteases MMP-1 and-9 and the Ets 1 transcription factor in HNPCC carcinomas and sporadic colorectal cancers. Int J Cancer. (2003) 107:183–8. 10.1002/ijc.11336 [DOI] [PubMed] [Google Scholar]
- 17.Nakada M, Yamashita J, Okada Y, Sato H. Ets-1 positively regulates expression of urokinase-type plasminogen activator (uPA) and invasiveness of astrocytic tumors. J Neuropathol Exp Neurol. (1999) 58:329–34. 10.1097/00005072-199904000-00003 [DOI] [PubMed] [Google Scholar]
- 18.Park YH, Jung HH, Ahn JS, Im Y-H. Ets-1 upregulates HER2-induced MMP-1 expression in breast cancer cells. Biochem Biophys Res Commun. (2008) 377:389–94. 10.1016/j.bbrc.2008.09.135 [DOI] [PubMed] [Google Scholar]
- 19.Nazir SU, Kumar R, Singh A, Khan A, Tanwar P, Tripathi R, et al. Breast cancer invasion and progression by MMP-9 through Ets-1 transcription factor. Gene. (2019) 711:143952. 10.1016/j.gene.2019.143952 [DOI] [PubMed] [Google Scholar]
- 20.Suzuki H, Romano-Spica V, Papas TS, Bhat NK. ETS1 suppresses tumorigenicity of human colon cancer cells. Proc Natl Acad Sci. (1995) 92:4442–6. 10.1073/pnas.92.10.4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fry EA, Inoue K. Aberrant expression of ETS1 and ETS2 proteins in cancer. Cancer Rep Rev. (2018) 2:1–10. 10.15761/CRR.1000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim G-C, Kwon H-K, Lee C-G, Verma R, Rudra D, Kim T, et al. Upregulation of Ets1 expression by NFATc2 and NFKB1/RELA promotes breast cancer cell invasiveness. Oncogenesis. (2018) 7:91. 10.1038/s41389-018-0101-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. (2009) 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 24.Díez-Villanueva A, Mallona I, Peinado MA. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenet Chromatin. (2015) 8:22. 10.1186/s13072-015-0014-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ PrePrints. (2016) 4:e1780v1 10.7287/peerj.preprints.1780 [DOI] [Google Scholar]
- 26.Zhang Y, Yan L-X, Wu Q-N, Du Z-M, Chen J, Liao D-Z, et al. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. (2011) 71:3552–62. 10.1158/0008-5472.CAN-10-2435 [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee T, Kumar A, Mathur M, Chattopadhyay TK, Ralhan R. Ets-1 and VEGF expression correlates with tumor angiogenesis, lymph node metastasis, and patient survival in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. (2003) 129:430–6. 10.1007/s00432-003-0457-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson B, Reich R, Goldberg I, Gotlieb WH, Kopolovic J, Berner A, et al. Ets-1 messenger RNA expression is a novel marker of poor survival in ovarian carcinoma. Clin Cancer Res. (2001) 7:551–7. Available online at: https://clincancerres.aacrjournals.org/content/7/3/551 [PubMed] [Google Scholar]
- 29.Takanami I, Takeuchi K, Karuke M. Expression of ETS-1 is correlated with urokinase-type plasminogen activator and poor prognosis in pulmonary adenocarcinoma. Tumor Biol. (2001) 22:205–10. 10.1159/000050617 [DOI] [PubMed] [Google Scholar]
- 30.Locke WJ, Guanzon D, Ma C, Liew YJ, Duesing KR, Fung KY, et al. DNA methylation cancer biomarkers: translation to the clinic. Front Genet. (2019) 10:1150. 10.3389/fgene.2019.01150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. (2016) 76:3446–50. 10.1158/0008-5472.CAN-15-3278 [DOI] [PubMed] [Google Scholar]
- 32.Barrett JM, Puglia MA, Singh G, Tozer RG. Expression of Ets-related transcription factors and matrix metalloproteinase genes in human breast cancer cells. Breast Cancer Res Treat. (2002) 72:227–32. 10.1023/A:1014993006190 [DOI] [PubMed] [Google Scholar]
- 33.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. (2012) 41:D970–D6. 10.1093/nar/gks937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan Z, Luo X, Xiao L, Tang M, Bode AM, Dong Z, et al. The role of PGC1α in cancer metabolism and its therapeutic implications. Mol Cancer Ther. (2016) 15:774–82. 10.1158/1535-7163.MCT-15-0621 [DOI] [PubMed] [Google Scholar]
- 35.Lebrun J-J. The dual role of TGF in human cancer: from tumor suppression to cancer metastasis. ISRN Mol Biol. (2012) 2012:381428 10.5402/2012/381428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du W, Wang S, Zhou Q, Li X, Chu J, Chang Z, et al. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. (2013) 32:3319–28. 10.1038/onc.2012.359 [DOI] [PubMed] [Google Scholar]
- 37.Lung HL, Lo PHY, Xie D, Apte SS, Cheung AKL, Cheng Y, et al. Characterization of a novel epigenetically-silenced, growth-suppressive gene, ADAMTS9, and its association with lymph node metastases in nasopharyngeal carcinoma. Int J Cancer. (2008) 123:401–8. 10.1002/ijc.23528 [DOI] [PubMed] [Google Scholar]
- 38.Morrison JA, Pike LA, Sams SB, Sharma V, Zhou Q, Severson JJ, et al. Thioredoxin interacting protein (TXNIP) is a novel tumor suppressor in thyroid cancer. Mol Cancer. (2014) 13:62. 10.1186/1476-4598-13-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferbeyre G, Moriggl R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochim Biophys Acta. (2011) 1815:104–14. 10.1016/j.bbcan.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 40.Nowell CS, Radtke F. Notch as a tumour suppressor. Nat Rev Cancer. (2017) 17:145–59. 10.1038/nrc.2016.145 [DOI] [PubMed] [Google Scholar]
- 41.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. (2003) 3:756–67. 10.1038/nrc1186 [DOI] [PubMed] [Google Scholar]
- 42.Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J Clin Oncol. (2004) 22:2053–60. 10.1200/JCO.2004.11.046 [DOI] [PubMed] [Google Scholar]
- 43.Bushweller JH. Targeting transcription factors in cancer-from undruggable to reality. Nat Rev Cancer. (2019) 19:611–24. 10.1038/s41568-019-0196-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tetsu O, McCormick F. ETS-targeted therapy: can it substitute for MEK inhibitors? Clin Transl Med. (2017) 6:16. 10.1186/s40169-017-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. (2003) 2:29. 10.1186/1476-4598-2-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets have been deposited in GEO database with accession code GSE106634.
RNA-seq datasets have been deposited in the NCBI Gene Expression Omnibus with accession code GSE106634.