Abstract
Friedreich’s ataxia (FRDA) is an autosomal recessive neurodegenerative disorder caused by expansion of GAA repeats in intron 1 of the frataxin (FXN) gene, leading to significant decreased expression of frataxin, a mitochondrial iron-binding protein. We previously reported that syngeneic hematopoietic stem and progenitor cell (HSPC) transplantation prevented neurodegeneration in the FRDA mouse model YG8R. We showed that the mechanism of rescue was mediated by the transfer of the functional frataxin from HSPC-derived microglia/macrophage cells to neurons/myocytes. In this study, we report the first step toward an autologous HSPC transplantation using the CRISPR-Cas9 system for FRDA. We first identified a pair of CRISPR RNAs (crRNAs) that efficiently removes the GAA expansions in human FRDA lymphoblasts, restoring the non-pathologic level of frataxin expression and normalizing mitochondrial activity. We also optimized the gene-editing approach in HSPCs isolated from healthy and FRDA patients’ peripheral blood and demonstrated normal hematopoiesis of gene-edited cells in vitro and in vivo. The procedure did not induce cellular toxic effect or major off-target events, but a p53-mediated cell proliferation delay was observed in the gene-edited cells. This study provides the foundation for the clinical translation of autologous transplantation of gene-corrected HSPCs for FRDA.
Keywords: CRISPR/Cas9, gene editing, Friedreich's ataxia, hematopoietic stem cells, ex vivo gene therapy, P53, frataxin, mitochondrial disorder, neurodegenerative disorder
Graphical Abstract

After demonstrating that hematopoietic stem and progenitor cell (HSPC) transplantation prevented disease progression in a mouse model for Friedreich’s ataxia (FRDA), this study presents the manufacturing feasibility of the human product for an autologous HSPC transplantation by removing the diseased-causing GAA expansion in the frataxin gene using CRISPR-Cas9 in patients’ HSPCs. Keywords: CRISPR/Cas9, gene editing, Friedreich’s ataxia, hematopoietic stem cells, ex vivo gene therapy, P53, frataxin, mitochondrial disorder, neurodegenerative disorder
Introduction
Friedreich’s ataxia (FRDA) is an autosomal recessive genetic disorder caused predominantly by a GAA expansion mutation in the first intron of the gene encoding for the mitochondrial protein frataxin.1 This hyper-expansion leads to reduced expression of frataxin by interfering with transcription initiation and elongation.2 Frataxin is involved in iron homeostasis, and its reduction is associated with mitochondrial iron accumulation, leading to oxidative stress.3 The prevalence of FRDA is approximately 1 in 40,000 people with a typical age of onset between 10 and 15 years. FRDA is a progressive, neurodegenerative movement disorder, often associated with cardiomyopathy and diabetes mellitus. FRDA’s pathogenesis is primarily caused by degeneration of the sensory nerve fibers within dorsal root ganglia (DRG), resulting in devolution of coordinated voluntary movement. Ultimately patients require the use of a wheelchair for mobility.4 There is no effective treatment for FRDA.
We previously reported that transplantation of wild-type mouse hematopoietic stem and progenitor cells (HSPCs) into young mice that recapitulate FRDA disease progression (YG8R)5 prevented coordination and locomotion deficits and vacuolar degeneration of the DRG neurons as well as muscle weakness.6 We demonstrated that transplanted HSPCs migrated and engrafted abundantly in affected tissues such as brain, spinal cord, DRG, heart, and skeletal muscle, where they differentiated into microglial cells and macrophages. The mechanism of rescue involves the transfer of the mitochondrial protein frataxin from the HSPC-derived phagocytic cells to the neurons and myocytes. These findings demonstrated for the first time the potential use of HSPC transplantation for the treatment of FRDA. However, there are significant risks associated with allogeneic HSPC transplantation; immunosuppression and graft-versus-host disease are some of the most prominent. Thus, the objective of this new study was to develop an autologous HSPC transplantation approach for FRDA.
Frataxin expression has to be well regulated, downregulation leading to the FRDA phenotype, and overexpression being associated with toxicity.7,8 In addition, 95% of patients carry the GAA expansion in the frataxin (FXN) gene intron 1 locus as the causative mutation. Therefore, removing the hyper-expansion seems the optimal approach as a gene correction strategy for FRDA. Targeted gene correction using artificial endonucleases, such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and RNA-guided nucleases CRISPR (clustered regularly interspaced short palindromic repeat)-Cas9 (CRISPR-associated 9), brings the possibility of gene targeting to the forefront for clinical application.9 Removal of the hyper-expansion for FRDA has previously been explored by two groups using ZFNs or CRISPR-Cas9 in human FRDA lymphoblasts and fibroblasts, as well as murine YG8R-derived fibroblasts.10,11 Both groups showed that excision of the GAA expansion within the first intron of the frataxin gene is repaired by non-homologous end joining (NHEJ) and leads to increased frataxin expression. Li et al.10 also demonstrated an increase in aconitase activity and ATP content in neuronal cells reprogrammed from corrected FRDA fibroblasts. However, the percentage of gene editing and FXN expression was evaluated in clones derived from single cells, and the efficiency was too low to be clinically relevant.
The objective of this study is to efficiently correct human FRDA HSPCs with minimal manipulation to generate vehicles of functional frataxin that will be delivered to diseased tissues. In our previous study, we showed that 30% engraftment of donor wild-type (WT) HSPCs was sufficient to prevent the development of the disease in the YG8R mouse model.6 In addition, because heterozygous individuals are asymptomatic, correcting one allele should recue the cellular phenotype. Finally, introducing mutations by NHEJ-mediated repair within the intronic sequence should have limited consequences to frataxin expression.
Herein, we report the use of the CRISPR-Cas9 system to remove the expansion causing FRDA, thus restoring physiological expression of frataxin in patients’ CD34+ cells. Additionally, we demonstrate that despite a p53-dependant delay in cell proliferation, CRISPR-Cas9 double-strand breaks (DSBs) do not induce toxicity, and corrected CD34+ cells were able to engraft and differentiate in immunodeficient mice. This study represents an efficient and specific gene therapy approach that will generate the cell product for a future ex vivo HSPC gene therapy clinical trial for FRDA.
Results
Optimization of CRISPR-Cas9-Mediated Gene Editing at the FXN Intron 1 Locus in FRDA Fibroblasts and Lymphoblasts
Six guide CRISPR RNAs (crRNAs) were designed following rule set 2 (RS2)12 to remove the GAA expansion within the first intron of the frataxin gene (Figure 1A) and tested in FRDA fibroblasts. Three days post-transfection with different combinations of pre-assembled ribonucleoprotein (RNP) complex long-range PCR was performed to amplify the region containing GAA repeats (∼5 kb). The UP4/DN4 guide pair (4RNP) displayed the greatest gene-editing efficiency excising an ∼2-kb DNA fragment containing the expansion (Figures 1B and 1C). Sequencing of the ∼2-kb resected fragment confirmed directed deletion of the repeats (Figure S1).
Figure 1.
Validation of CRISPR-Cas9-Mediated Gene Editing at the FXN Intron 1 Locus in Human FRDA Fibroblasts
(A) List of the best six crRNAs designed following the rule set 2 surrounding the FXN intron 1 GAA expansion. (B) Position of the crRNAs and regulatory elements surrounding the FXN intron 1 GAA expansion. E-box, enhancer box; mt-binding site, microtubule-binding site. (C) Agarose gel showing the long-range PCR amplification of the region of the FXN intron 1 containing the GAA expansion after gene editing with different pairs of crRNA precomplexed. Optimal gene-editing efficiency was found with the UP4/DN4 pair represented in line 5.
We then optimized the intronic repeat excision protocol using 4RNP and electroporation in non-adherent hematopoietic lymphoblastic cell lines as a relevant model for CD34+ cells from healthy donors, FRDA patients, and related carriers (Table S2), and in the presence or absence of an electroporation enhancer (single-stranded DNA oligonucleotide designed in silico to possess no homology with human, mouse, or rat genomes) to increase RNP uptake. We evaluated FXN editing efficiency by droplet digital PCR (ddPCR) using reference primers at the 5′ end of FXN intron 1 and experimental primers flanking the expected deletion (Figure 2A). Gene-editing efficiency was twice as robust in the three patients’ cell lines when electroporation of the 4RNP was performed in the presence of the enhancer (39.8%–61.9% for FRDA/4RNPenh versus 17%–29.9% for FRDA/4RNP; Figure 2B, p < 0.05). These data represent an optimal approach to remove the GAA hyperexpansion causing FRDA.
Figure 2.
GAA Gene-Editing Optimization in Human FRDA Lymphoblasts Using the UP4/DN4 cRNA Pair
(A) Schematic representing the ddPCR strategy to determine GAA gene-editing efficiency from genomic DNA. Red primers can only amplify the intronic region when GAA gene editing occurs. (B) GAA gene-editing percentage measured by ddPCR in three different FRDA lymphoblastic cell lines 3 weeks post-electroporation with 4RNP or 4RNPenh. Data are means ± SEM. ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0005 (Student’s t test). (C) GAA gene-editing percentage measured by ddPCR in two different healthy lymphoblastic cell lines 3 weeks post-electroporation with 4RNPenh. Data are means ± SEM. (D) Quantification of human frataxin mRNA in healthy and healthy/4RNPenh lymphoblasts normalized to human TBP 3 weeks post-electroporation by ddPCR (n = 3). Data are means ± SEM. NS, not significant (Student’s t test). (E) Representative western blot showing human frataxin protein expression in healthy and healthy/4RNPenh lymphoblasts 3 weeks post-electroporation. The bar graph represents the quantification of human frataxin protein in healthy and healthy/4RNPenh lymphoblasts normalized to actin 3 weeks post-electroporation (n = 3). Data are means ± SEM. NS, not significant (Student’s t test). (F) Quantification of human frataxin mRNA in healthy, carrier, FRDA, FRDA/4RNP, and FRDA/4RNPenh lymphoblasts 3 weeks post-electroporation by ddPCR (n = 3). Data are represented as fold change relative to Healthy1 normalized to human TBP. Data are means ± SEM. ∗p < 0.05 (one-way ANOVA). (G) Representative western blot showing human frataxin protein expression in Healthy1, Carrier1, FRDA1, FRDA1/4RNP, and FRDA1/4RNPenh lymphoblasts 3 weeks post-electroporation. The bar graph represents the quantification of human frataxin protein in healthy, carrier, FRDA, FRDA/4RNP, and FRDA/4RNPenh lymphoblasts normalized to actin 3 weeks post-electroporation (n = 3). Data are means ± SEM. ∗p < 0.05 (one-way ANOVA). (H) Mitochondrial activity measured in healthy, carrier, FRDA, and FRDA/4RNPenh lymphoblasts in the presence of succinate. Data are means ± SEM. ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0005 (one-way ANOVA).
CRISPR-Cas9-Mediated GAA Gene Editing Restores Frataxin Expression and Mitochondrial Function in FRDA Lymphoblasts
Because regulatory elements are included in the excised DNA fragment after 4RNP-mediated gene editing, we first ensured that normal frataxin expression was maintained in modified healthy cells. Healthy lymphoblasts with 60% of gene-editing efficiency (Figure 2C) had no difference in FXN expression at both the mRNA and protein levels (Figures 2D and 2E). In contrast, significant increases in frataxin expression was seen at both the transcriptional and translational levels in the three 4RNPenh-modified FRDA lymphoblast cell lines compared to untreated cells (Figures 2F and 2G), leading to frataxin expression comparable to the asymptomatic carrier and healthy cell lines.
Biolog’s MitoPlate I-1 was utilized to assess mitochondrial function by measuring the rates of electron flow into and through the electron transport chain in response to metabolic substrates. First, the specificity of our assay was verified by measuring mitochondrial activity in the presence of specific complex I, II, or III inhibitors in response to succinate (Figure S2). We then quantified basal mitochondrial activity in all of the cell lines without inhibitors. Results exhibit a defect in electron flow rate for complex II/III in the FRDA cell lines (∼40% less when compared to healthy cell lines). Removal of the hyper-expansion was able to partially restore mitochondrial function in the edited FRDA cells compared to the controls (Figure 2H). These data suggest that our genetic-editing approach not only restores frataxin expression but also improves cellular mitochondrial functions in FRDA patient cells.
Gene Editing Triggers a p53-Mediated DNA Damage Response That Delays Cell Proliferation
To assess whether the GAA removal was inducing any toxicity in the edited lymphoblastic cell lines, proliferation rate and viability were determined during 7 days post-electroporation. A significant but limited decrease in cell viability was observed at 24 and 48 h post-electroporation in FRDA/4RNP and FRDA/4RNPenh compared to FRDA control, with viability still greater than 75% (Figure 3A). However, proliferation was significantly impacted in both FRDA/4RNP and FRDA/4RNPenh cells, exhibiting no proliferative activity during the initial 24 h post-electroporation, resulting in lower number of cells thereafter (Figure 3B).
Figure 3.
Impact of GAA Gene Editing on FRDA Lymphoblast Viability and Proliferative Capacity
(A) Percentage of live FRDA, FRDA/4RNP, and FRDA/4RNPenh lymphoblasts over time post-electroporation (n = 3 for the three FRDA lymphoblastic cell lines). Data are means ± SEM. ∗p < 0.05 and ∗∗p < 0.005 (one-way ANOVA). (B) Fold expansion of FRDA, FRDA/4RNP, and FRDA/4RNPenh lymphoblasts (n = 3 for the three FRDA lymphoblastic cell lines) over time post-electroporation. Data are means ± SEM. (C) Western blot representing the time course expression of p53 and p21 after 4RNPenh electroporation in FRDA lymphoblasts. (D) Western blot representing the time course expression of p53 and p21 after RNP/control guide electroporation in FRDA lymphoblasts.
A recent study reported that DSBs created by CRISPR-Cas9 led to a transient p53-mediated DNA damage response, which delayed cell proliferation.13 We confirmed that p53 and p21 expression were transiently induced by 4RNPenh in FRDA lymphoblasts, with a peak at 24 h post-electroporation (Figure 3C). The same experiment carried with a control crRNA did not affect p53/p21 expression, suggesting that their overexpression is specifically due to DSBs (Figure 3D). To demonstrate that p53 transient overexpression is not due to the disease status of the FRDA cells, healthy lymphoblasts were also analyzed showing that p53/p21 were also transiently overexpressed (Figure S3A). To emphasize the involvement of p53 in this delay, HL-60, a lymphoblastic cell line characterized by a p53 deficiency,14 was similarly electroporated with 4RNPenh (63.7% ± 3.1% GAA gene editing, data not shown). No delay in proliferation was observed in gene-modified cells compared to non-electroporated or Cas9-only control cells (Figure S3B). Finally, the FRDA GM15850 lymphoblastic cell line was knocked down for p53 using a short hairpin RNA (shRNA) anti-p53 (Figure S3C, left) and subjected to 4RNPenh electroporation. This new cell line exhibited the same proliferation profile as that of the control GM15850KD/Cas9, as opposed to GM15850/4RNPenh, which was delayed compared to GM15850/Cas9 (Figure S3C, right). Taken together, these data confirmed that the delay in cell growth observed in the edited cells was due to p53 expression induced by DSBs.
Optimization and In Vitro and In Vivo Assessments of CRISPR-Cas9-Mediated FXN GAA Excision in Peripheral Blood CD34+ Cells
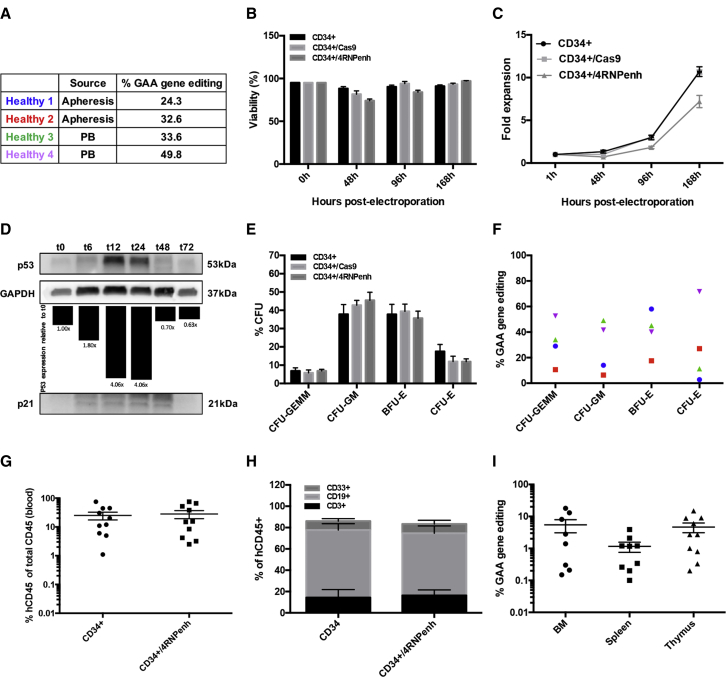
We next optimized 4RNPenh-mediated gene editing in healthy human CD34+ HPSCs isolated from two different sources: frozen granulocyte colony-stimulating factor (G-CSF)-mobilized apheresis bags and fresh peripheral blood (Figure 4A). Gene editing efficiency in apheresis and peripheral derived cells reached 32.6% and 49.8%, respectively (Figure 4A). Cell viability of CD34+/4RNPenh was significantly lower at 48 and 96 h post-electroporation compared to CD34+ cells (CD34+) and Cas9-electroporated cells (CD34+/Cas9), but still above 74% (Figure 4B). As expected, significant delay in cell proliferation was observed in CD34+/4RNPenh compared to CD34+ and CD34+/Cas9 controls (Figure 4C), which was shown to be due to p53 transient overexpression (Figures 4D; Figure S4).
Figure 4.
GAA Gene Editing in Healthy CD34+ Cells and Consequence on Hematopoiesis Reconstitution Capacity
(A) GAA gene-editing percentage measured by ddPCR in four different healthy donors from two different sources (apheresis bags or fresh peripheral blood) 1 week post-electroporation. (B) Percentage of live CD34+, CD34+/Cas9, and CD34+/4RNPenh cells over time post-electroporation (n = 4 for the three CD34+ cell lines). Data are means ± SEM. ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0005 (one-way ANOVA). (C) Fold expansion of CD34+, CD34+/Cas9, and CD34+/4RNPenh cells (n = 4 for the three CD34+ cell lines) over time post-electroporation. Data are means ± SEM. (D) Western blot representing the time course expression of p53 and p21 after 4RNPenh electroporation in CD34+ cells. (E) Colony-forming unit assay showing the percentages of the different colony types formed (n = 4). CFU-GEMM, CFU for granulocytes/erythroid cells/macrophages/megakaryocytes; CFU-GM, CFU for granulocytes/macrophages; BFU-E, burst-forming units for erythroid cells; CFU-E, CFU for erythroid cells. Data are means ± SEM. Not significant (one-way ANOVA). (F) GAA gene-editing percentage measured by ddPCR in each colony type for each healthy donor. (G) Human engraftment of gene-modified CD34+ cells in peripheral blood of transplanted NSG mice 3 months post-transplantation by flow cytometry (n = 10 for CD34+ and n = 10 for CD34+/4RNPenh), determined as the percentage of human CD45+ cells of all human and murine CD45+ cells. Data are means ± SEM. Not significant (Student’s t test). (H) Lineage distribution of human cells engrafted in NSG mice in the bone marrow 3 months post-transplantation determined by flow cytometry using fluorescent-labeled antibodies to human T cells (CD3), human B cells (CD19), and human myeloid cells (CD33+). Data are means ± SEM. Not significant (Student’s t test). (I) GAA gene-editing percentage measured by ddPCR in bone marrow (BM), spleen, and thymus of transplanted NSG mice 3 months post-transplantation. Data are means ± SEM.
The differentiation ability of individual CD34+ cells was verified using colony-forming unit (CFU) assays showing that hematopoietic lineage colony distribution in 4RNPenh-edited CD34+ cells was similar to controls (Figure 4E). Heterogeneous rates of gene editing were measured in each colony type for the different donor cells, illustrating an unbiased distribution among the different hematopoietic cell lineages: 10.6%–52.6% in CFU for granulocytes/erythroid cells/macrophages/megakaryocytes (CFU-GEMM), 6.4%–49% in CFU for granulocytes/macrophages (CFU-GM), 17.5%–58% in burst-forming units for erythroid cells (BFU-E), and 2.8%–71.7% in CFU for erythroid cells (CFU-E) (Figure 4F). A single colony analysis also showed that all of the clones presented with a monoallelic GAA deletion (Figure S5).
The repopulating capacity of edited CD34+ cells was also evaluated in vivo through xenotransplantation in NSG mice. Control and 4RNPenh-edited CD34+ cells were transplanted into sub-lethally irradiated newborn NSG mice via intrahepatic injection. Engraftment and hematopoiesis reconstitution assessments displayed substantial numbers of human hematopoietic CD45+ cells in peripheral blood of all transplanted mice in both groups (24.93% ± 7.461% in CD34+ mice, and 27.99% ± 8.734% in CD34+/4RNPenh mice) (Figure 4G). Enumeration of T cells (CD3), B cells (CD19), and myeloid progenitors (CD33) by flow cytometry showed comparable lineage distribution in the bone marrow of both CD34+ and CD34+/4RNPenh mice (Figure 4H). The input cells had a gene-editing rate ranging from 24.3% to 49.8%, and at 3 months post-transplant, gene editing ranged from 0.15% to 18% with a mean of 5.49% in bone marrow, from 0.1% to 3.90% with a mean of 1.16% in spleen, and from 0.2% to 15% with a mean of 4.65% in thymus (Figure 4I).
Overall, the data obtained from CD34+ cells demonstrated a high percentage of GAA repeat excision and normal hematopoietic lineage differentiation capacity in vivo and in vitro.
FXN GAA Expansion Editing in Peripheral Blood CD34+ Cells from FRDA Patients
Directed removal of FXN GAA expansion was then tested in CD34+ cells isolated from fresh peripheral blood collected from five FRDA patients, with three healthy parents (carrier) and three healthy donors used as controls (Table S3). Gene-repair rates of the input cells ranged from 12.1% to 55.9% with a mean of 29.66% (Figure 5A). Cell viability of 4RNPenh-edited FRDA CD34+ cells was significantly decreased at 6, 48, 72, and 96 h post-electroporation compared to controls, but it was consistently more than 70% (Figure 5B). As expected, 4RNPenh-edited FRDA CD34+ cells exhibited a delay in cell proliferation (Figure 5C).
Figure 5.
GAA Gene Editing in FRDA Patient CD34+ Cells and Impact on FXN Expression and Hematopoiesis Reconstitution
(A) GAA gene-editing percentage measured by ddPCR in five different FRDA patient donors from fresh peripheral blood 1 week post-electroporation. (B) Viability of healthy, carrier, FRDA, and FRDA/4RNPenh CD34+ cells over time post-electroporation (n = 3 for healthy, n = 3 for carrier, n = 5 for FRDA, and n = 5 for FRDA/4RNPenh). Data are means ± SEM. ∗p < 0.05 and ∗∗p < 0.005 (one-way ANOVA). (C) Fold expansion of healthy, carrier, FRDA, and FRDA/4RNPenh cells over time post-electroporation (n = 3 for healthy, n = 3 for carrier, n = 5 for FRDA, and n = 5 for FRDA/4RNPenh). Data are means ± SEM. (D) Quantification of human frataxin mRNA in healthy, carrier, FRDA, and FRDA/4RNPenh CD34+ cells 1 week post-electroporation by ddPCR (n = 3 for healthy, n = 3 for carrier, n = 5 for FRDA, and n = 5 for FRDA/4RNPenh). Data are normalized to human TBP. Data are means ± SEM. ∗p < 0.05 and ∗∗p < 0.05 (one-way ANOVA). (E) Correlation curve showing the significant relationship between the percentage of GAA gene editing and the increased expression of human frataxin mRNA in gene-modified CD34+ cells from FRDA patient donors. (F) Colony-forming unit assay showing the percentages of the different colony types formed (n = 3 for healthy, n = 3 for carrier, n = 5 for FRDA, and n = 5 for FRDA/4RNPenh). CFU-GEMM, CFU for granulocytes/erythroid cells/macrophages/megakaryocytes; CFU-GM, CFU for granulocytes/macrophages; BFU-E, burst-forming units for erythroid cells); CFU-E, CFU for erythroid cells. Data are means ± SEM. Not significant (one-way ANOVA). (G) GAA gene-editing percentage measured by ddPCR in each colony type for each FRDA donor. (H) Quantification of the human mitochondrial (mt) complex subunit mRNA mtDN6 (complex I), mtCO2 (complex II), and mtATP6 (complex V) by qRT-PCR in healthy, FRDA, and FRDA/4RNPenh CD34+ cells 1 week post-electroporation. Data are represented as fold change relative to healthy and normalized to human tubulin. Data are means ± SEM. ∗p < 0.05 and ∗∗p < 0.005 (Student’s t test).
We then assessed frataxin transcriptional expression in the different donor groups. An increase in FXN mRNA expression was observed in the edited cells from all of the patients with a mean expression of 0.334 for FRDA cells versus 1.25 for FRDA/4RNPenh cells (Figure 5D). While this increase was not significant, there was a direct correlation between the proportion of gene correction and level of FXN expression (R2 = 0.73) (Figure 5E). A CFU assay demonstrated colony lineages from FRDA/4RNPenh CD34+ cells comparable to FDRA, healthy, and carrier CD34+ cell controls (Figure 5F). Gene-editing rates in the different colony lineages confirmed that CRISPR-Cas9-mediated expansion removal was retained and unbiasedly distributed within the different hematopoietic progeny (Figure 5G).
To investigate functional rescue using a limited number of cells, we tested the expression of three mitochondrial (mt) complex subunit genes mtDN6 (complex I), mtCO2 (complex II), and mtATP6 (complex V), which were shown to be affected in whole-blood cells from FRDA patients.15 A decrease of expression of these markers was observed in FRDA CD34+ cells compared to healthy cells. An increased expression of these markers was observed in gene-edited FRDA CD34+, cells although it did not reach significance (Figure 5H). These data demonstrate that CRISPR-Cas9-mediated GAA excision from the frataxin gene is feasible in the targeted CD34+ HSPCs from FRDA patients, leading to an increase in FXN expression and an improvement of mitochondrial biomarkers.
Assessment of Nuclease Specificity
The specificity of 4RNPenh-mediated gene editing was assessed in FRDA CD34+ cells using in silico COSMID tool (https://crispr.bme.gatech.edu).16 Results showed six potential off-target sites within the following genes’ introns: AGAP1, UNC5D, LRP1B, RARB, EPHX2, and DGKG (Table S4). Indel formation was tested via a T7 endonuclease 1 (T7E1) mismatch detection assay using guide DNA (gDNA) isolated from the edited cells. No detectable off-target activity was found in these six regions for all of the patients (Figure S6). To confirm this result, we also sequenced PCR products and compared them to PCR amplicons from corresponding non-edited FRDA CD34+ cells. Deconvolution analysis using the ICE Synthego software (https://ice.synthego.com) exhibited no indel formation in any of the edited gDNA compared to the non-edited gDNA (Table S5). Finally, these same amplicons have been analyzed by next-generation sequencing (NGS) for a more precise reading. For the six intronic areas, more than 98.66% of reads were without indels (Figure S7). Collectively, these data show that UP4 and DN4 crRNA are very specific to the target region of frataxin gene.
Discussion
Herein, we report preliminary data for the preclinical study of targeted gene editing of HSPCs for an autologous transplantation strategy for FRDA by removing the GAA expansion in intron 1 of the frataxin gene using CRIPSR-Cas9. The efficiency of gene repair in CD34+ cells isolated from peripheral blood from FRDA patients could reach 55%. We previously showed that a proportion of 30% of the cell product using WT HSPCs could rescue FRDA.6 In addition, increased frataxin expression could be achieved in CD34+ cells isolated from FRDA patients, as well as presence and stable rates of gene editing in the hematopoietic progeny in vitro and in vivo, and no off-target activity was detected in the final cell products. Thus, our study supports the development of this potential therapeutic strategy for FRDA. Recent results of ongoing clinical trials using CRISPR-Cas9-mediated gene-edited HSPC therapy for transfusion-dependent β-thalassemia and sickle cell disease support the efficacy and safety of this approach.17
Optimization, safety, and efficacy of our targeted gene-editing approach were obtained through a series of functional assays using FRDA patients’ cell lines. Because of the presence of an mt-binding site and E-box regulation sequences only 98 bp upstream of the GAA repeats, excision of the expansion made their removal inevitable. Although these regulatory sequences are known to contribute to the frataxin promoter activity,18 their deletion did not impact frataxin expression in healthy cells. This result confirms previous data obtained in K562 cells.10 Initial optimization of RNP transfection via electroporation in patient-derived lymphoblasts showed that inclusion of a carrier DNA enhancer dramatically increased the percentage of gene-editing efficiency. This is the first demonstration that this enhancer could increase targeted genome efficiency in CD34+ cells, and validation of its use in clinical trials should be the next critical step. Physiologic levels of frataxin, comparable to those of carriers and healthy controls, were thus achieved, both at the mRNA and protein levels.
Deficits of frataxin are associated with mitochondrial iron accumulation, deficient Fe-S cluster biogenesis, increased sensitivity to oxidative stress, and deficits in respiratory chain complex activity.19,20 However, FRDA patients’ blood cells do not exhibit major defects in mitochondrial function.21 Hence, functional rescue was difficult to demonstrate using lymphoblasts, and regular mitochondrial and oxidative stress assays failed to show impairment. However, Biolog’s MitoPlate I-1 assay, which is highly strenuous on the respiratory chain pathway, revealed significant differences between healthy and FRDA lymphoblasts. Partial functional rescue in gene-corrected FRDA lymphoblasts was then demonstrated.
While our approach did not establish a deleterious cytotoxic effect, it notably delayed cell proliferation during the first 48 h post-electroporation. We confirmed that p53 overexpression, in response to the presence of DNA DSBs, was responsible for this cell cycle block.13 Indeed, the ribonuclease complex 4RNPenh generates four potential DSBs to remove the GAA expansion in FXN, and we showed overexpression of p53 at 24-h post-electroporation in healthy and FRDA lymphoblasts as well as in CD34+ cells only when electroporated with 4RNPenh but not with a crRNA control. In contrast, cell proliferation was not affected in p53-knockout HL60 lymphoblasts or FRDA lymphoblasts with p53 knockdown. Because human CD34+ cells were transplanted in NSG mice only 24 h after electroporation, the p53-mediated proliferation delay observed after 4RNPenh-mediated gene editing could explain the drop of efficiency rate between the input cells and the in vivo bone marrow cells. This reduction in gene-repair proportions from in vitro to in vivo have been reported by other groups.22, 23, 24, 25, 26 Efforts to expand CD34+ cells in culture in an undifferentiated state27 or using p53 inhibitors13 should help to significantly increase gene-editing efficiency in vivo.
Among the five FRDA patients tested, gene-editing efficiency was variable. Although size of the trinucleotide repeat expansion correlates with the disease severity,28 we did not find any correlation between expansion size and gene-editing efficiency. However, abnormal secondary DNA structures by long GAA tracts29, 30, 31, 32 could presumably be responsible for challenging the access of crRNA to the targeting site and explain the different genome editing rates obtained in the different patients’ lymphoblasts and CD34+ cells.
Our results support the use of the CRISPR-Cas9 to remove the GAA expansion in FRDA patients’ CD34+ cells, leading to physiological rescue of frataxin expression, when the percentage of gene editing is sufficiently high, without cytotoxic effects in vitro or in vivo, and maintaining HSPC engraftment ability and clonogenicity. This work represents a step toward the clinical translation of autologous transplantation of gene-corrected HSPCs for FRDA.
Materials and Methods
Human Blood Cells
CD34+ HSPCs from healthy donors were obtained from two different sources: leukapheresis bags of G-SCF-mobilized cells kindly provided by Dr. J. Bui (University of California, San Diego), and 500 mL of freshly withdrawn peripheral blood from the Scripps Research’s Normal Blood Donor Service. For the study including FRDA patients, up to 100 mL of peripheral blood was withdrawn from FRDA patients, carriers, or healthy donors after written informed consent under Institutional Review Board-approved protocol #161762. The conduct of these studies conformed to the Declaration of Helsinki protocols and all US federal regulations required for the protection of human subjects.
Mouse Studies
Use of non-obese diabetic (NOD) severe combined immunodeficiency (SCID) Il2rg−/− (NSG) mice (Jackson Laboratory, Bar Harbor, ME, USA) for xenotransplantation studies was approved by the National Institute of Allergy and Infectious Diseases (NIAID) Institutional Animal Care and Use Committee under protocol S14042. The conduct of these studies conformed to Association for Assessment and Accreditation of Laboratory Animal Care International guidelines and all US federal regulations governing the protection of research animals.
RNP Protein Complex Assembly
Single Alt-R crRNA and Alt-R trans-activating crRNA (tracrRNA) (100 μM, Integrated DNA Technologies, San Diego, CA, USA) oligonucleotides were mixed at equimolar concentrations to a final concentration of 44 μM and complexed together at 95°C for 5. The Alt-R S.p. HiFi Cas9 nuclease V3 protein (62 μM, IDT, San Diego, CA, USA) was diluted in electroporation buffer (buffer R, Invitrogen, Carlsbad, CA, USA) or Opti-MEM to 36 μM. Equivalent volumes of crRNA-tracrRNA and diluted HiFi Cas9 nuclease were mixed together and incubated at room temperature for 10–20 min.
Transfection
Human FRDA fibroblasts (GM03816) from the Coriell Institute were cultured in Eagle’s minimal essential medium (EMEM)/15% fetal bovine serum (FBS) at 37°C, 5% CO2.. Transfection was carried out using the Lipofectamine CRISPRMAX Cas9 transfection reagent kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Briefly, RNP complex and CRISPRMAX reagent were each diluted in 50 μL of Opti-MEM medium and mixed. The RNP complex solution was then immediately added to the CRISPRMAX reagent solution and mixed. After a 5- to 10-min incubation, the solution was added to the cells.
Long-Range PCR
At 72 h post-transfection, gDNA was isolated using QuickExtract DNA extraction solution (Lucigen, Middleton, WI, USA) and PCR was carried out using 10 ng of gDNA, 2× Super Master Mix (BioTool, Houston, TX, USA), and primers listed in Table S1. The PCR program was set up as follows: (94°C for 20 s, 65.6°C for 2 min 30 s) × 20, (94°C for 20 s, 65.6°C for 2 min 30 s + 15 s/cycle) × 17. PCR products were then run on a 0.7% agarose gel. Band intensity was quantified using ImageJ.
Lymphoblast and CD34+ HSPC Electroporation
Human FRDA lymphoblasts from the Coriell Institute (Table S2) were cultured in RPMI 1640/15% FBS at 37°C, 5% CO2. 1 × 105 lymphoblasts were resuspended in 9 μL of electroporation buffer (buffer R, Invitrogen, Carlsbad, CA, USA); 1 μL of RNP complex was added as well as 1.8 μM Alt-R Cas9 electroporation enhancer (IDT, San Diego, CA, USA) when required. 10 μL was then pipetted into the 10-μL Neon tip (Invitrogen, Carlsbad, CA, USA), and the cell/RNP complex mixture was electroporated at 1,600 V, 10 ms, for three pulses using the Neon electroporator (Invitrogen, Carlsbad, CA, USA). Cells were immediately returned to pre-equilibrated cultured media in 24-well plates. The same protocol and number of cells were used for CD34+ HSPCs for in vitro experiments while 1 × 106 CD34+ HSPCs (resuspended in 90 μL of buffer R) were electroporated with 10 μL of RNP complex for transplantation in one NSG mouse.
ddPCR
gDNA from cells was isolated using QuickExtract (Lucigen, Middleton, WI, USA) and from tissues using a DNeasy Blood & Tissue kit (QIAGEN, Germantown, MD, USA). To measure GAA gene-editing efficiency, 100 ng of gDNA, HindIII (NEB, Ipswich, MA, USA), and 1× ddPCR supermixes for probes (no dUTP [deoxyuridine triphosphate]) (Bio-Rad, Hercules, CA, USA) were used in combination with two sets of primers/probes (Table S1) to generate the droplets using the QX200 droplet generator (Bio-Rad, Hercules, CA, USA). Next, the droplets were transferred to a 96-well plate and the following PCR was carried out: 95°C for 10-min ramp at 2°C/s, (94°C for 30-s ramp at 2°C/s, 60°C for 1-min ramp at 2°C/s) × 39, 98°C for 10-min ramp at 2°C/s. The 96-well plate was then read by the QX200 Droplet reader (Bio-Rad, Hercules, CA, USA). For frataxin expression, the protocol was similar but using cDNA and the following sets of primers: human frataxin (Bio-Rad, Hercules, CA, USA, #10031252) and human TBP (Bio-Rad, Hercules, CA, USA, #10031255).
Western Blot
Proteins were isolated from lymphoblasts and transferred to a polyvinylidene fluoride membrane as previously described.33 The following antibodies were used: mouse anti-frataxin antibody (Abcam, Cambridge, MA, USA, ab110328), mouse anti-p53 antibody (Santa Cruz Biotechnology [SCBT], Santa Cruz, CA, USA, sc-126), mouse anti-p21 antibody (SCBT, Santa Cruz, CA, USA, sc-6246), rabbit anti-PAN actin antibody (Cell Signaling Technology, Danvers, MA, USA, #4968S), and mouse anti-GAPDH antibody (SCBT, Santa Cruz, CA, USA, sc-365062) followed by goat anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies. Signal was revealed using the Azure 600 imager, and bands were quantified using the AzureSpot analysis software (Azure Biosystems, Dublin, CA, USA).
MitoPlate I-1
Mitochondrial activity within lymphoblasts was measured using the MitoPlate I-1 (Biolog, Hayward, CA, USA, #14104) following the manufacturer’s instructions. Briefly, wells containing the different mitochondrial inhibitors were rehydrated with a solution containing redox dye mix, saponin, and succinate for 1 h at 37°C. Lymphoblasts were washed with PBS (1×) and resuspended at a density of 105 cells/30 μL using 1× Biolog MAS (Biolog, Hayward, CA, USA) and added to each well. The MitoPlate I-1 was then loaded into the OmniLog PM-M system (Biolog, Hayward, CA, USA) for kinetic reading.
Lymphoblast p53 Knockdown
GM15850 lymphoblasts (600 viable cells/mL) were incubated overnight with RPMI 1640/15% FBS, Polybrene (5 μg/mL), and p53 shRNA (h) lentivirus particles (SCBT, Santa Cruz, CA, USA, #sc-29435-V) at an MOI of 10. On the next day, media were replaced with RPMI 1640/15% FBS and cells were incubated during two nights. Selection occurred through incubation of transduced cells with puromycin concentrations of 0, 2, 5, and 10 μg/mL to produce stable cell lines, which were analyzed for p53 knockdown via western blot.
CD34+ HSPC Isolation and In Vitro Differentiation
Peripheral blood was first diluted in PBS (1×) and white blood cells were separated from red blood cells and plasma using Ficoll, while leukapheresis bags were directly thawed and diluted in Miltenyi buffer. Then, CD34+ HSPCs were isolated using the Miltenyi Biotec MACS human CD34 MicroBead kit (Miltenyi Biotec, San Diego, CA, USA) following the manufacturer’s instructions. With 100 mL of peripheral blood, an average of 1.5 × 106 CD34+ HSPCs were obtained. Cells were cultured in complete medium consisting of Iscove’s modified Dulbecco’s medium (IMDM) supplemented with FBS, BSA, glutamine, penicillin/streptomycin, human interleukin (hIL)-3, hIL-6, and human stem cell factor (h-SCF) (PeproTech, Rocky Hill, NJ, USA) at 37°C. Cell proliferation and viability were determined using an automated cell counter (Bio-Rad, Hercules, CA, USA). CFU assays were performed using MethoCult H4434 enriched methylcellulose (STEMCELL Technologies, Cambridge, MA, USA). Two days after electroporation, 3,000 cells from each condition were mixed with 3 mL of MethoCult and plated in triplicate into 35-mm gridded cell culture dishes. After 12–14 days of culture at 37°C, 5% CO2, the different types of hematopoietic colonies were identified and counted. CFU (∼15/type) were plucked for genomic DNA isolation using QuickExtract (Lucigen, Middleton, WI, USA).
Allelic Deletion Detection
Single CD34+ colonies were isolated and individually placed in QuickExtract (Lucigen, Middleton, WI, USA) for DNA isolation. PCR was carried out using 25 ng of gDNA, PCR Master Mix (2×) (Thermo Fisher Scientific, West Columbia, SC, USA), 4% DMSO, and primers listed in Table S1. The PCR program was set up as follows: (95°C for 30 s, 56°C for 30 s, and 72°C for 1 min) × 35. PCR products were then run on a 1% agarose gel. Colony genotypes were then binned based on positive bands into non-allelic, mono-allelic, or bi-allelic deletion.
NSG Mouse Transplantation
The NOD SCID Il2rg−/− (NSG) mice (Jackson Laboratory, Bar Harbor, ME, USA) were housed in a pathogen-free colony in a biocontainment vivarium and handled in laminar flow hoods. Newborn pups at 3–7 days of life of both sexes were injected with 1 × 106 cells/pup via intrahepatic injection of unmodified or gene-edited human CD34+ cells 1 day after conditioning with 1.25 Gy of sub-lethal body irradiation from an X-ray energy irradiator, and allowed to engraft during 12–16 weeks.34
Flow Cytometry Analysis of Hematopoiesis
Human engraftment in NSG mice was determined 2 months post-transplantation from peripheral blood using a fluorescein isothiocyanate (FITC) anti-human CD45 (BioLegend, San Diego, CA, USA) and a phycoerythrin (PE) anti-mouse CD45 (BioLegend, San Diego, CA, USA). Hematopoiesis reconstitution was determined at the time of sacrifice, 12–16 weeks post-transplantation, from peripheral blood using the following conjugated antibodies: allophycocyanin (APC)/Cyanine7 anti-human CD19, FITC anti-human CD3, PE anti-human CD33, and APC anti-human CD45 (BioLegend, San Diego, CA, USA). All of the flow cytometric analyses were performed using the BD LSRFortessa flow cytometer (BD Biosciences, San Jose, CA, USA).
qRT-PCR
Total RNA was prepared from lymphoblasts or CD34+ cells using an RNeasy kit (QIAGEN, Germantown, MD, USA) according to the manufacturer’s instructions. cDNA was then prepared using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA), and primers listed in Table S1 and SYBR Green master mix (Bio-Rad, Hercules, CA, USA) were used following the manufacturer’s instructions.
Off-Target Assessment
Potential off-target regions were predicted using COSMID software. An Alt-R genome editing detection kit (IDT, San Diego, CA, USA) and primers listed in Table S1 were used to detect the presence of potential indels within edited gDNA. Fragments were Sanger sequenced and Amplicon-EZ next-generation sequenced by Genewiz (La Jolla, CA, USA). Sanger chromatograms were compared to controls via Synthego ICE deconvolution, and single-nucleotide polymorphisms were detected from NGS.
Author Contributions
C.J.R. designed, collected, and analyzed the data and wrote the manuscript. J.N.R., Y.S., and J.H.H. collected and analyzed the data. J.S. isolated HSPCs from apheresis bags and peripheral blood and counted CFU colonies. J.L. and P.M. designed the crRNA and sorted the off-target sequences. S.C. conceived and supervised the study and wrote the manuscript.
Conflicts of Interest
S.C. is inventor on a patent titled “Methods of treating mitochondrial disorders” (#20378-201301) and is a cofounder, shareholder, and a member of both the Scientific Board and Board of Directors of GenStem Therapeutics. S.C. serves as a consultant for AVROBIO and receives compensation for these services. S.C. also serves as a member of the Scientific Review Board and Board of Trustees of the Cystinosis Research Foundation. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies.
Acknowledgments
We thank Jack Bui for the leukapheresis bags. We thank Laura Hernandez and Jose Cano for blood withdrawing. We thank all of the blood donor volunteers for participating in this study. This work was supported by National Institutes of Health (NIH) RO1-DK090058 and R01-NS108965, the Friedreich's Ataxia Research Alliance (FARA), the California Institute of Regenerative Medicine (CIRM, CLIN-09230), and by the Cystinosis Research Foundation. J.N.R. received a fellowship from CIRM (CIRM Bridges to Stem Cell Research Program, EDUC2-08388). Finally, we thank the summer students who have been involved in this project, Avery Mubarak and Soline Grimbert.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.04.018.
Supplemental Information
References
- 1.Campuzano V., Montermini L., Moltò M.D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 2.Ohshima K., Montermini L., Wells R.D., Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;273:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 3.Pandolfo M., Pastore A. The pathogenesis of Friedreich ataxia and the structure and function of frataxin. J. Neurol. 2009;256(Suppl 1):9–17. doi: 10.1007/s00415-009-1003-2. [DOI] [PubMed] [Google Scholar]
- 4.Cook A., Giunti P. Friedreich’s ataxia: clinical features, pathogenesis and management. Br. Med. Bull. 2017;124:19–30. doi: 10.1093/bmb/ldx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anjomani Virmouni S., Ezzatizadeh V., Sandi C., Sandi M., Al-Mahdawi S., Chutake Y., Pook M.A. A novel GAA-repeat-expansion-based mouse model of Friedreich’s ataxia. Dis. Model. Mech. 2015;8:225–235. doi: 10.1242/dmm.018952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocca C.J., Goodman S.M., Dulin J.N., Haquang J.H., Gertsman I., Blondelle J., Smith J.L.M., Heyser C.J., Cherqui S. Transplantation of wild-type mouse hematopoietic stem and progenitor cells ameliorates deficits in a mouse model of Friedreich’s ataxia. Sci. Transl. Med. 2017;9:eaaj2347. doi: 10.1126/scitranslmed.aaj2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navarro J.A., Llorens J.V., Soriano S., Botella J.A., Schneuwly S., Martínez-Sebastián M.J., Moltó M.D. Overexpression of human and fly frataxins in Drosophila provokes deleterious effects at biochemical, physiological and developmental levels. PLoS ONE. 2011;6:e21017. doi: 10.1371/journal.pone.0021017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vannocci T., Notario Manzano R., Beccalli O., Bettegazzi B., Grohovaz F., Cinque G., de Riso A., Quaroni L., Codazzi F., Pastore A. Adding a temporal dimension to the study of Friedreich’s ataxia: the effect of frataxin overexpression in a human cell model. Dis. Model. Mech. 2018;11:dmm032706. doi: 10.1242/dmm.032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song M., Ramakrishna S. Genome editing in stem cells for disease therapeutics. Mol. Biotechnol. 2018;60:329–338. doi: 10.1007/s12033-018-0072-9. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., Polak U., Bhalla A.D., Rozwadowska N., Butler J.S., Lynch D.R., Dent S.Y., Napierala M. Excision of expanded GAA repeats alleviates the molecular phenotype of Friedreich’s ataxia. Mol. Ther. 2015;23:1055–1065. doi: 10.1038/mt.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouellet D.L., Cherif K., Rousseau J., Tremblay J.P. Deletion of the GAA repeats from the human frataxin gene using the CRISPR-Cas9 system in YG8R-derived cells and mouse models of Friedreich ataxia. Gene Ther. 2017;24:265–274. doi: 10.1038/gt.2016.89. [DOI] [PubMed] [Google Scholar]
- 12.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiroli G., Conti A., Ferrari S., Della Volpe L., Jacob A., Albano L., Beretta S., Calabria A., Vavassori V., Gasparini P. Precise gene editing preserves hematopoietic stem cell function following transient p53-mediated DNA damage response. Cell Stem Cell. 2019;24:551–565.e8. doi: 10.1016/j.stem.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf D., Rotter V. Major deletions in the gene encoding the p53 tumor antigen cause lack of p53 expression in HL-60 cells. Proc. Natl. Acad. Sci. USA. 1985;82:790–794. doi: 10.1073/pnas.82.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jasoliya M.J., McMackin M.Z., Henderson C.K., Perlman S.L., Cortopassi G.A. Frataxin deficiency impairs mitochondrial biogenesis in cells, mice and humans. Hum. Mol. Genet. 2017;26:2627–2633. doi: 10.1093/hmg/ddx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol. Ther. Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vertex . 2019. CRISPR therapeutics and vertex announce positive safety and efficacy data from first two patients treated with investigational CRISPR/Cas9 gene-editing therapy CTX001® for severe hemoglobinopathies.https://investors.vrtx.com/news-releases/news-release-details/crispr-therapeutics-and-vertex-announce-positive-safety-and [Google Scholar]
- 18.Greene E., Mahishi L., Entezam A., Kumari D., Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandolfo M. Iron metabolism and mitochondrial abnormalities in Friedreich ataxia. Blood Cells Mol. Dis. 2002;29:536–547, discussion 548–552. doi: 10.1006/bcmd.2002.0591. [DOI] [PubMed] [Google Scholar]
- 20.Rötig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 21.Selak M.A., Lyver E., Micklow E., Deutsch E.C., Onder O., Selamoglu N., Yager C., Knight S., Carroll M., Daldal F. Blood cells from Friedreich ataxia patients harbor frataxin deficiency without a loss of mitochondrial function. Mitochondrion. 2011;11:342–350. doi: 10.1016/j.mito.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoban M.D., Cost G.J., Mendel M.C., Romero Z., Kaufman M.L., Joglekar A.V., Ho M., Lumaquin D., Gray D., Lill G.R. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125:2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Ravin S.S., Reik A., Liu P.Q., Li L., Wu X., Su L., Raley C., Theobald N., Choi U., Song A.H. Targeted gene addition in human CD34+ hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 2016;34:424–429. doi: 10.1038/nbt.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiroli G., Ferrari S., Conway A., Jacob A., Capo V., Albano L., Plati T., Castiello M.C., Sanvito F., Gennery A.R. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 2017;9:eaan0820. doi: 10.1126/scitranslmed.aan0820. [DOI] [PubMed] [Google Scholar]
- 26.Kuo C.Y., Long J.D., Campo-Fernandez B., de Oliveira S., Cooper A.R., Romero Z., Hoban M.D., Joglekar A.V., Lill G.R., Kaufman M.L. Site-specific gene editing of human hematopoietic stem cells for X-linked hyper-IgM syndrome. Cell Rep. 2018;23:2606–2616. doi: 10.1016/j.celrep.2018.04.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tajer P., Pike-Overzet K., Arias S., Havenga M., Staal F.J.T. Ex vivo expansion of hematopoietic stem cells for therapeutic purposes: lessons from development and the niche. Cells. 2019;8:169. doi: 10.3390/cells8020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filla A., De Michele G., Cavalcanti F., Pianese L., Monticelli A., Campanella G., Cocozza S. The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am. J. Hum. Genet. 1996;59:554–560. [PMC free article] [PubMed] [Google Scholar]
- 29.Herman D., Jenssen K., Burnett R., Soragni E., Perlman S.L., Gottesfeld J.M. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 30.Al-Mahdawi S., Pinto R.M., Ismail O., Varshney D., Lymperi S., Sandi C., Trabzuni D., Pook M. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum. Mol. Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 31.Saveliev A., Everett C., Sharpe T., Webster Z., Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 32.van Rensburg R., Beyer I., Yao X.Y., Wang H., Denisenko O., Li Z.Y., Russell D.W., Miller D.G., Gregory P., Holmes M. Chromatin structure of two genomic sites for targeted transgene integration in induced pluripotent stem cells and hematopoietic stem cells. Gene Ther. 2013;20:201–214. doi: 10.1038/gt.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lobry T., Miller R., Nevo N., Rocca C.J., Zhang J., Catz S.D., Moore F., Thomas L., Pouly D., Bailleux A. Interaction between galectin-3 and cystinosin uncovers a pathogenic role of inflammation in kidney involvement of cystinosis. Kidney Int. 2019;96:350–362. doi: 10.1016/j.kint.2019.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huey D.D., Niewiesk S. Production of Humanized Mice through Stem Cell Transfer. Curr. Protoc. Mouse Biol. 2018;8:17–27. doi: 10.1002/cpmo.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.