Clostridioides difficile infection (CDI) is associated with increasing morbidity and mortality posing an urgent threat to public health. Recurrence of CDI after successful treatment with antibiotics is high, thus necessitating discovery of novel therapeutics against this enteric pathogen. Administration of the secondary bile acid ursodeoxycholic acid (UDCA; ursodiol) inhibits the life cycles of various strains of C. difficile in vitro, suggesting that the FDA-approved formulation of UDCA, known as ursodiol, may be able to restore colonization resistance against C. difficile in vivo.
KEYWORDS: C. difficile, FXR, UDCA, bile acids, inflammation, microbiome, ursodiol
ABSTRACT
Clostridioides difficile infection (CDI) is associated with increasing morbidity and mortality posing an urgent threat to public health. Recurrence of CDI after successful treatment with antibiotics is high, thus necessitating discovery of novel therapeutics against this enteric pathogen. Administration of the secondary bile acid ursodeoxycholic acid (UDCA; ursodiol) inhibits the life cycles of various strains of C. difficile in vitro, suggesting that the FDA-approved formulation of UDCA, known as ursodiol, may be able to restore colonization resistance against C. difficile in vivo. However, the mechanism(s) by which ursodiol is able to restore colonization resistance against C. difficile remains unknown. Here, we confirmed that ursodiol inhibits C. difficile R20291 spore germination and outgrowth, growth, and toxin activity in a dose-dependent manner in vitro. In a murine model of CDI, exogenous administration of ursodiol resulted in significant alterations in the bile acid metabolome with little to no changes in gut microbial community structure. Ursodiol pretreatment resulted in attenuation of CDI pathogenesis early in the course of disease, which coincided with alterations in the cecal and colonic inflammatory transcriptome, bile acid-activated receptors nuclear farnesoid X receptor (FXR) and transmembrane G-protein-coupled membrane receptor 5 (TGR5), which are able to modulate the innate immune response through signaling pathways such as NF-κB. Although ursodiol pretreatment did not result in a consistent decrease in the C. difficile life cycle in vivo, it was able to attenuate an overly robust inflammatory response that is detrimental to the host during CDI. Ursodiol remains a viable nonantibiotic treatment and/or prevention strategy against CDI. Likewise, modulation of the host innate immune response via bile acid-activated receptors FXR and TGR5 represents a new potential treatment strategy for patients with CDI.
INTRODUCTION
Clostridioides difficile is a leading nosocomial enteric pathogen that causes significant human morbidity and mortality, resulting in 29,000 deaths per year and costing more than $4.8 billion in health care expenses annually in the United States (1–5). A major risk factor for C. difficile infection (CDI) is the use of antibiotics (6, 7). Antibiotics lead to significant and enduring shifts in the gut microbiota and metabolome (8–10), resulting in loss of colonization resistance against C. difficile. Although the exact mechanisms of colonization resistance against C. difficile remain unclear, there is increasing evidence that microbe-derived secondary bile acids play an important role (10–13).
Antibiotics, either vancomycin or fidaxomicin, are recommended for an initial episode of CDI (14). Unfortunately antibiotic treatment can further disrupt the gut microbial community, and recurrence of CDI after cessation of antibiotics is high, occurring in 20 to 30% of patients (3, 15–18). Even fidaxomicin, a bactericidal antibiotic against C. difficile, alters the intestinal microbial community structure, just to a lesser extent than vancomycin does (19). Fecal microbiota transplantation (FMT) is a highly effective treatment for recurrent CDI; however, the long-term ramifications of this conditionally FDA-approved procedure are unknown (20, 21). A potential mechanism by which FMT leads to resolution of CDI in a mouse model (22, 23) and in humans (13) is by restoring microbe-derived secondary bile acids.
Secondary bile acids, such as deoxycholic acid (DCA), lithocholic acid (LCA), and ursodeoxycholic acid (UDCA; ursodiol), are produced as a collaborative effort by the host (production of primary bile acids) and the gut microbiota (biochemical modification of host primary bile acids into secondary bile acids) (24, 25). Our group and others have demonstrated that numerous microbe-derived secondary bile acids, including UDCA, are able to inhibit spore germination, growth, and toxin activity of various C. difficile strains in vitro (26, 27). We also know that secondary bile acids are associated with colonization resistance against C. difficile in mice and humans (10, 13, 28). Furthermore, oral UDCA administration successfully prevented recurrence of C. difficile ileal pouchitis in a single patient (29), suggesting that exogenous administration of the secondary bile acid UDCA may be able to restore colonization resistance against C. difficile in vivo (29).
UDCA is available in an FDA-approved formulation known as ursodiol (30). Ursodiol is used to treat a variety of hepatic and biliary diseases due to its myriad of beneficial effects, including anticholestatic, antifibrotic, antiproliferative, and anti-inflammatory properties (31–40). Additionally, off-label use of ursodiol is currently in phase 4 clinical trials for the prevention of recurrent CDI (41). It is well established that ursodiol alters the hepatic, biliary, and serum bile acid pools (31, 42). However, it remains unclear whether and how exogenously administered ursodiol alters the indigenous gut microbial community structure and/or bile acid metabolome, which contributes to colonization resistance against C. difficile.
On the basis of our previous work with UDCA (26), we hypothesized that ursodiol would directly inhibit different stages of the C. difficile life cycle and ameliorate disease when administered to mice in a model of CDI. Due to their detergent-like properties, bile acids can also directly alter the gut microbial composition, subsequently altering bile acid metabolism and host physiology (43). Gastrointestinal cross talk between bile acids and the gut microbiota is well recognized (reviewed by Wahlström et al.) (43), but the impact of ursodiol on these interactions remains unknown. Similarly, the central role bile acids play in regulation of the innate immune response is growing (reviewed by Fiorucci et al. [44]). Because of this, we also investigated the indirect effects or the extent to which the impact of ursodiol on CDI correlated with changes in the gut microbiota, bile acid metabolome, and the host inflammatory response.
Here, we show that ursodiol is able to significantly decrease C. difficile spore germination, growth, and toxin activity in a dose-dependent manner in vitro, but not to the same level in vivo. In C. difficile-infected mice, ursodiol pretreatment resulted in attenuation of CDI pathogenesis early in the course of disease, which coincided with alterations in gut bile acids, the host inflammatory transcriptome, and gene expression of bile acid-activated receptors nuclear farnesoid X receptor (FXR) and transmembrane G-protein-coupled membrane receptor 5 (TGR5), which are able to modulate the innate immune response through signaling pathways such as NF-κB. Although ursodiol pretreatment did not restore colonization resistance against C. difficile, it attenuated an overly robust host inflammatory response that is detrimental to the host during CDI. Together, these results suggest that ursodiol remains a viable nonantibiotic treatment and/or prevention strategy against CDI by acting on the FXR-FGR axis via NF-κB signaling. Targeting the bile acid-activated receptors FXR and TGR5 represents a novel approach for the development of therapies for the prevention and treatment of CDI.
RESULTS
Ursodiol directly inhibits different stages of the C. difficile life cycle in vitro.
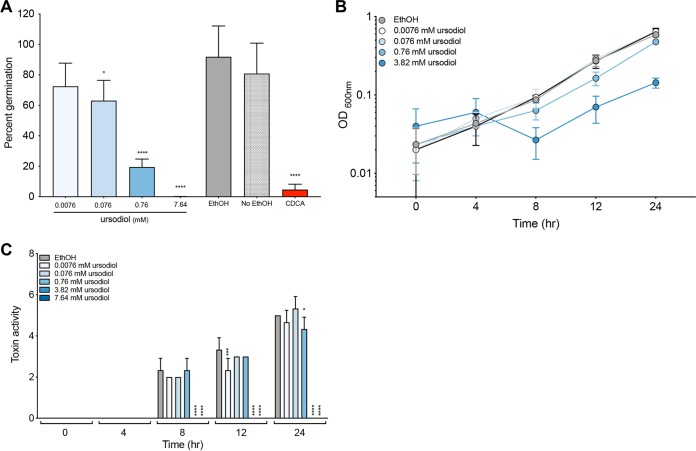
To determine the direct effects of ursodiol on the life cycles of clinically relevant C. difficile strain R20291, in vitro assays to assess spore germination and outgrowth, growth, and toxin activity were performed (Fig. 1). Taurocholic acid (TCA)-mediated spore germination and outgrowth of C. difficile were significantly inhibited by ursodiol in a dose-dependent manner (Fig. 1A, blue bars). No significant difference was observed between positive controls, sterile ethanol, and water supplemented with 0.1% TCA (Fig. 1A, ethanol represented as a solid gray bar). As expected, the negative control, 0.04% chenodeoxycholic acid (CDCA) supplemented with 0.1% TCA resulted in significant inhibition of spore germination and outgrowth (Fig. 1A, red bar). CDCA is a known inhibitor of spore germination (45).
FIG 1.
Ursodiol inhibits different stages of the C. difficile life cycle. (A) In vitro assay to assess whether the addition of ursodiol (0.0076, 0.076, 0.76, and 7.64 mM) (A) inhibits C. difficile R20291 TCA-mediated spore germination and outgrowth compared to the positive control, ethanol (EthOH) (gray bar), and the negative control, 0.04% CDCA (red bar). The data presented represent triplicate experiments. Statistical significance between treatment groups and the positive controls was determined by Student’s parametric t test with Welch’s correction. (B) Growth kinetics of C. difficile over a 24-h period. Growth curves were done in BHI medium with ethanol (gray line) and addition of various concentrations of ursodiol (blue lines). The data presented represent mean OD600 ± standard error of the mean (SEM) (error bars) from triplicate experiments. (C) Culture supernatants, taken throughout a 24-h growth curve in panel B, were used for a Vero cell cytotoxicity assay, and the data are expressed as log10 reciprocal dilution toxin per 100 μl of C. difficile culture supernatant. The data presented represent the results from triplicate experiments. Statistical significance between treatment groups and the positive controls was determined by a two-way ANOVA with Dunnett’s multiple-comparison test (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001).
C. difficile growth decreased when exposed to ursodiol at 8, 12, and 24 h in a dose-dependent manner compared to the ethanol control (Fig. 1B, blue lines). For the 3.82 mM concentration of ursodiol, a significantly lower optical density (OD) compared to the ethanol control was observed at time points 8, 12, and 24 h (P = 0.0316, P = 0.0001, and P = 0.0001, respectively). For the 0.76 mM ursodiol treatment, a significantly lower OD compared to the ethanol control was observed at time points 12 and 24 h (P = 0.0001 for both time points), and for the 0.076 mM ursodiol treatment at time point 24 h (P = 0.0208). No significant difference in OD between the ethanol control and treatments was noted at time point 0 or 4 h. The highest concentration of ursodiol (7.64 mM) interfered with OD measurement and was not included in the analysis.
Since OD measurements do not capture viability and spore formation of C. difficile, we also did differential plating to evaluate the CFU per milliliter at each time point (see Fig. S1 in the supplemental material). At 24 h, ursodiol inhibited C. difficile total vegetative cells and spores compared to the ethanol control. The highest concentration of ursodiol (7.64 mM) had a significant and immediate inhibitory effect on C. difficile viability compared to ethanol at time zero. Ursodiol treatment also significantly decreased C. difficile spores.
Toxin activity was not detected until 8 h of C. difficile growth (Fig. 1C). Higher concentrations of ursodiol resulted in significant decreases in C. difficile toxin activity compared to the ethanol control at 8, 12, and 24 h (Fig. 1C, blue bars). Using a Spearman’s correlation (r), the CFU/milliliter and toxin activity at 8 and 12 h were significantly correlated (8 h, r = 0.6399, P = 0.0042; 12 h, r = 0.6981, P = 0.0013), suggesting that low toxin activity is due to decreased growth.
Ursodiol alters the fecal bile acid metabolome with minimal change to the microbiota.
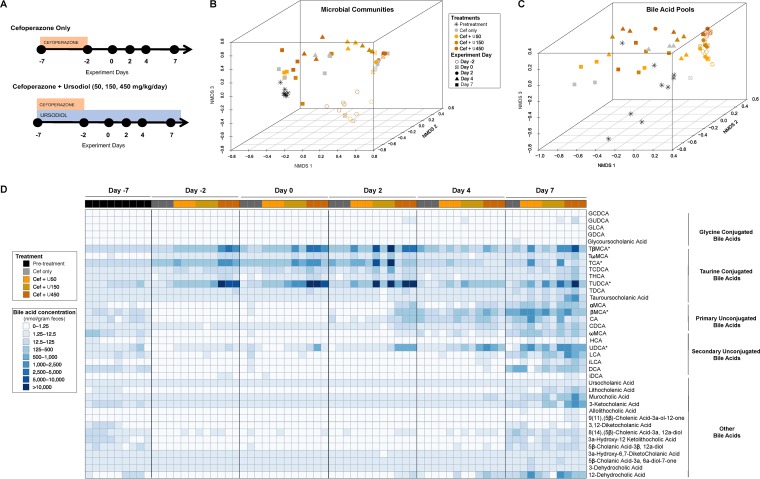
Since the gut microbiota and bile acids mediate colonization resistance against C. difficile, we aimed to determine how different doses of ursodiol administered alone impacts the bacteria in our antibiotic-treated mouse model. C57BL/6J mice were administered three doses of ursodiol (50, 150, and 450 mg/kg of body weight/day) via oral gavage for 14 days (Fig. 2A). Mice were also concurrently administered cefoperazone (in drinking water ad libitum) for 5 days in order to mimic the CDI mouse model. Paired fecal samples were collected from the same mice serially over the 14-day period to evaluate how ursodiol affected the gut microbial community structure and bile acid metabolome.
FIG 2.
Ursodiol does not alter the gut microbiota but does alter the bile acid metabolome. (A) Groups of C57BL/6J mice (eight mice/treatment group) were given cefoperazone in their drinking water ad libitum for 5 days alone (Cef only) and or with ursodiol at three doses (50, 150, and 450 mg/kg) given daily via oral gavage for 14 days. Three mice per treatment group were selected for further microbiome and bile acid metabolomic analysis. (B) NMDS ordination of the fecal microbial communities from mice treated and untreated with ursodiol was calculated from Yue and Clayton dissimilarity metric (θYC) on OTU at a 97% cutoff. (C) NMDS ordination illustrates dissimilarity indices via Horn distances between the bile acid profiles of fecal samples from treated and untreated mice. (D) A heatmap of bile acid concentrations present in nanomoles per gram of feces, ranging from 0 to 38,221.91 (nmol/g feces). Asterisks next to bile acids indicate the bile acids that were significantly altered during ursodiol treatment. Abbreviations: GCDCA, glycochenodeoxycholic acid; GUDCA, glycoursodeoxycholic acid; GLCA, glycolithocholic acid; GDCA, glycodeoxycholic acid; TβMCA, tauro-β-muricholic acid; TωMCA, tauro ω-muricholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholic acid; THCA, taurohyodeoxycholic acid; TUDCA, tauroursodeoxycholic acid; TDCA, taurodeoxycholic acid; αMCA, α-muricholic acid; βMCA, β-muricholic acid; CA, cholic acid; CDCA, chenodeoxycholic acid; ωMCA, ω-muricholic acid; HCA, hyodeoxycholic acid; UDCA, ursodeoxycholic acid; LCA, lithocholic acid; iLCA, isolithocholic acid; DCA, deoxycholic acid; iDCA, isodeoxycholic acid.
The fecal microbial community structures of all treatment groups were significantly different from pretreatment at all time points, based on an analysis of molecular variance (AMOVA) performed for each day (Fig. 2B). There were no significant differences between the fecal microbial community structures based on ursodiol pretreatment. The shifts in the fecal microbiota seen in the nonmetric multidimensional scaling (NMDS) were predominantly due to the cefoperazone treatment and experiment day, and not a result of the ursodiol pretreatment.
Targeted bile acid analysis via ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) detected 38 bile acids in the feces of mice. During the first 9 days of the experiment (day −2 to day 7), the fecal bile acid profiles of mice cluster based on the dose of ursodiol administered (Fig. 2C). Ursodiol dramatically alters the fecal bile acid metabolome compared to pretreatment as seen in Fig. 2D and Table S3 in the supplemental material. Bile acids that significantly changed over the course of the experiment in mice that received ursodiol compared to mice that received only cefoperazone included tauro-β-muricholic acid (TβMCA), taurocholic acid (TCA), tauroursodeoxycholic acid (TUDCA), β-muricholic acid (βMCA), and UDCA (Fig. S2, two-way analysis of variance [ANOVA] with Tukey’s multiple-comparison posthoc test performed on each experiment day, P values ranged from P < 0.0001 to P = 0.0012).
Ursodiol attenuates disease in mice early during C. difficile infection.
Due to the potent inhibitory effect of ursodiol on C. difficile in vitro, and increased gastrointestinal bile acids in vivo, we sought to determine the extent to which pretreatment with ursodiol influences the natural course of CDI in a reproducible mouse model. Since supraphysiologic UDCA intestinal concentrations were achieved in Fig. 2D when mice were dosed with 450 mg/kg/day of ursodiol, we selected this dose (46).
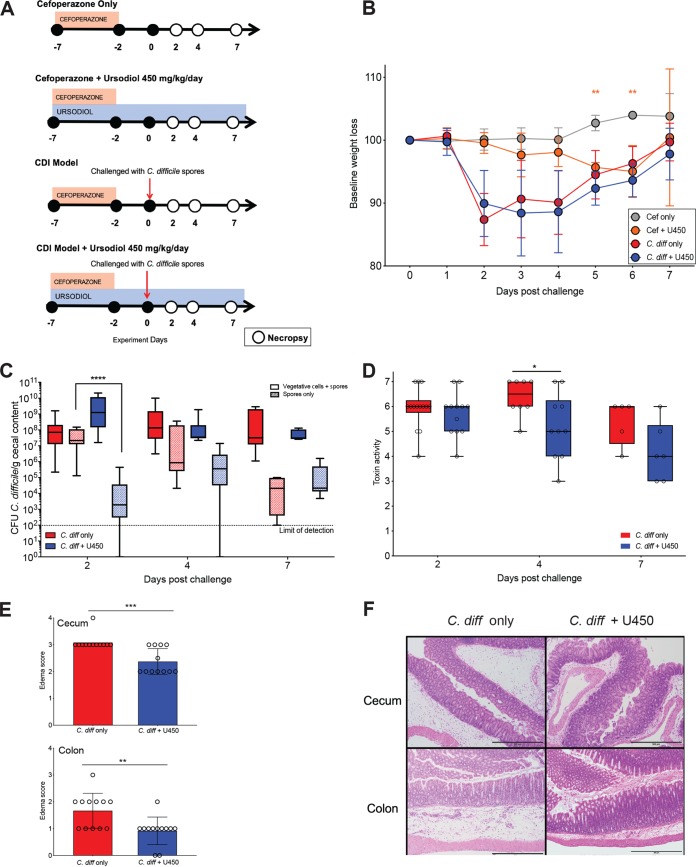
All C57BL/6J mice in each group received cefoperazone in their drinking water from day −7 to day −2 with a 2-day washout on normal drinking water. Control groups included two uninfected groups that received cefoperazone and no ursodiol (Cef only), and cefoperazone with ursodiol (Cef + U450) (Fig. 3A). An additional control group received cefoperazone and no ursodiol, with C. difficile challenge (C. diff only). Mice were orally gavaged with ursodiol (450 mg/kg in corn oil vehicle) daily for 7 days prior to challenge with C. difficile on day 0, and throughout the infection (C. diff + U450) (Fig. 3A).
FIG 3.
Ursodiol attenuates disease early during C. difficile infection. (A) Groups of C57BL/6J mice (12 mice/treatment group) were given cefoperazone in theirdrinking water ad libitum for 5 days alone and or with ursodiol at 450 mg/kg given daily via oral gavage for 14 days. Two groups of mice were then challenged with 105 spores of C. difficile via oral gavage at day 0. Mice were monitored for weight loss and clinical signs of CDI from day 0 to day 7. Necropsy was performed on days 2, 4, and 7 (open circles). (B) Baseline weight loss of mice challenged with and without C. difficile and pretreated with and without ursodiol. Significance was determined by a two-way ANOVA with Sidak’s multiple-comparison posthoc test. (C) C. difficile bacterial load in the cecal content throughout CDI. Solid boxes represent total vegetative cells and spores, and hatched boxes represent spores only. Significance was determined by a two-way ANOVA with Tukey’s multiple-comparison posthoc test. (D) Vero cell cytotoxicity assay from cecal content throughout CDI. Significance was determined by a Student’s t test corrected by the Holm-Sidak method for multiple comparisons. (E) Histopathologic scoring of edema in the murine cecum and colon at day 2 postchallenge. Significance was determined by a Student’s t test corrected by the Holm-Sidak method for multiple comparisons. Error bars represent the standard deviations from the mean. (F) Representative histomicrograph images of H&E-stained cecum (top) and colon (bottom) for groups indicated, shown at 100× total magnification with 500-μm scale bars. Statistical significance for all graphs is indicated as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Two groups of mice (C. diff only and C. diff + U450) were challenged with 105 spores of C. difficile R20291 on day 0 and within 24 h were colonized with greater than 107 CFU per gram of feces (data not shown). Mice challenged with C. difficile with and without ursodiol pretreatment lost a significant amount of their baseline weight by day 2 postchallenge (C. diff only [red circles] and C. diff + U450 [blue circles] [Fig. 3B]). Additionally, there was no significant difference in weight loss between these groups over the course of the experiment (based on two-way ANOVA). However, mice pretreated with ursodiol (C. diff + U450) had a 24- to 36-h delay in clinical signs (lethargy, inappetence, diarrhea, and hunched posture) related to CDI compared to untreated mice (C. diff only). Uninfected control mice (Cef only and Cef + U450) revealed no clinical signs of disease during the experiment and remained C. difficile free. The Cef only group of mice displayed weight gain over the course of the experiment as expected (Fig. 3B, gray circles). The Cef + U450 group did have significant weight loss compared to Cef only mice at day 5 and day 6 of the experiment (two-way ANOVA with Sidak’s multiple-comparison posthoc test; P = 0.0013 and P = 0.0001, respectively; Fig. 3B, orange circles). However, no significant difference was noted between these groups by day 7 (Fig. 3B). An additional control group of mice challenged with C. difficile and pretreated with the vehicle corn oil alone (C. diff + corn oil) displayed no significant difference in disease outcome compared to the C. diff only group (Fig. S3).
Mice pretreated with ursodiol (C. diff + U450) had equivalent total cecal bacterial loads of C. difficile (CFU/gram of cecal content) compared to untreated mice (C. diff only) (Fig. 3C). However, mice pretreated with ursodiol had a significant reduction in C. difficile spores within cecal content at day 2 compared to untreated mice (log transferred CFU/g content, two-way ANOVA with Tukey’s multiple-comparison posthoc test, P < 0.0001; Fig. 3C). The ceca of mice remained persistently colonized with C. difficile throughout the 7-day experiment. At day 4, C. difficile toxin activity within cecal content was significantly reduced in ursodiol-pretreated compared to untreated mice (Student’s t test corrected with the Holm-Sidak method for multiple comparisons, P = 0.014; Fig. 3D).
Histopathologic evaluation of cecal and colonic tissue revealed no significant difference in total histologic score between ursodiol-pretreated and untreated mice (Fig. S4). When assessing individual parameters, there was no significant difference between epithelial damage or inflammatory infiltrate between the treatment groups. At day 2, mice pretreated with ursodiol had a significant reduction in edema score (scored 0 to 4) in both cecal and colonic tissue compared to untreated mice (Student’s t test corrected with the Holm-Sidak method for multiple comparisons, P = 0.00025 and P = 0.0049, respectively; Fig. 3E and F).
Ursodiol alters the host inflammatory transcriptome early during CDI.
Since there were minimal changes in the C. difficile life cycle early during infection to explain the decrease in edema, we wanted to examine the impact of ursodiol pretreatment on the host inflammatory response during CDI. We interrogated transcriptional changes related to the cecal and colonic inflammatory response at day 2 postchallenge using a mouse-specific panel of inflammatory markers (Nanostring nCounter) that included 264 transcripts. After processing the raw data, mRNA log2 normalized z-scores for 261 genes were obtained and genes significantly altered by ursodiol pretreatment compared to untreated mice with CDI were evaluated. Ursodiol-mediated alterations in gene expression fold change were different between cecal and colonic tissue (Fig. 4 and Fig. S5). In cecal tissue, 173 genes had a significant expression fold change when comparing ursodiol-pretreated mice to untreated mice, with increased expression in 74 genes and decreased expression in 99 genes (Fig. 4A and B and Tables S4 and S5). In colonic tissue, 139 genes had a significant expression fold change when comparing ursodiol-pretreated to untreated mice, with increased expression in 58 genes and decreased expression in 81 genes (Fig. S5A and B and Tables S4 and S5).
FIG 4.
Ursodiol alters the host inflammatory transcriptome of the cecum early during CDI. (A) Venn diagram depicting the mRNA expression fold change comparing the ursodiol-pretreated compared to untreated mice in cecal tissue at day 2 postchallenge with C. difficile, from two independent experiments performed with a total of 12 mice/treatment group. Of the 261 genes evaluated, a total of 173 cecal genes had significant gene expression fold changes (increased expression, red; decreased expression, green). (B) Volcano plots highlighting genes whose transcript levels changed by greater than onefold and met the significance threshold of P ≤ 0.05. Genes highlighted in red had increased transcript levels, while those highlighted in green had decreased levels. Blue points represent genes whose results were significant but did not met the specified fold change (FC). Black points represent genes whose results failed to meet the significance threshold. (C) Cecal inflammatory transcriptome heatmap of calculated z-scores for log2-transformed count data of mRNA expression levels (y axis) comparing ursodiol-pretreated (blue) to untreated mice (red). Dendrograms represent unsupervised clustering. (D) mRNA expression fold differences in increased expression (red) and decreased expression (green) of genes involved with NF-κB signaling in cecal tissue comparing ursodiol-pretreated to untreated mice.
Ingenuity Pathway Analysis (IPA) identified NF-κB signaling as an inflammatory canonical pathway in both cecum and colon significantly altered in ursodiol-pretreated mice (P < 0.0001 for both tissues). Based on unsupervised clustering, genes involved with NF-κB signaling in ursodiol-pretreated mice clustered distinctly and separately from untreated mice in both cecal and colonic tissue (Fig. 4C and Fig. S5C). Differences in individual gene expression fold changes were noted in NF-κB signaling between the cecal and colonic tissue (Fig. 4D and Fig. S5D). In both tissues, components important for mediating the host inflammatory response to CDI, such as interleukin 1 receptor 1 (IL-1R1) and various Toll-like receptors (TLRs), were significantly decreased in expression in the ursodiol-pretreated mice compared to untreated mice at day 2 (Fig. 4D and Fig. S5D; see Table S6 for P values). Collectively, these results suggest that ursodiol pretreatment in CDI mice alters the inflammatory transcriptome in cecal and colonic tissue by impacting essential host inflammatory signaling pathways, such as NF-κB, early in the course of CDI.
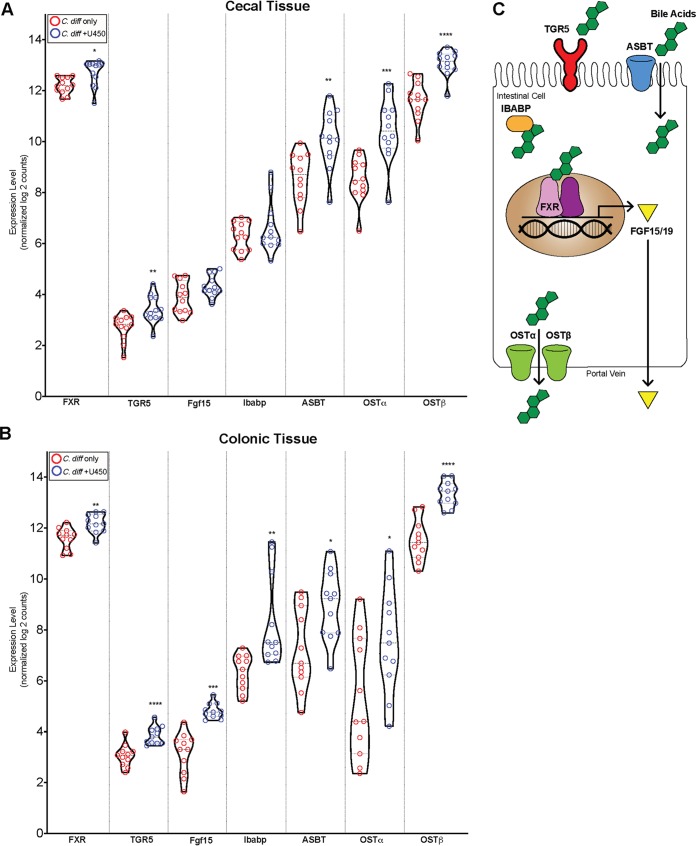
Ursodiol induces bile acid transport and signaling via FXR/FGF15 pathway.
Since ursodiol treatment alters the bile acid metabolome in mice (Fig. 2D) and bile acids are known to modulate signaling via the nuclear bile acid receptor FXR and the plasma membrane receptor TGR5 (47), we sought to define transcriptional changes in bile acid transport and signaling via the FXR/FGF15 (fibroblast growth factor 15) pathway. We selected seven genes in the FXR/FGF15 pathway to interrogate within cecal and colonic tissue using a customized murine panel (Nanostring nCounter). Bile acids within the gastrointestinal tract are transported back to the liver via enterohepatic recirculation by several active transporters, ASBT (apical sodium-dependent bile acid transporter) on the apical surface, and OSTα/β (organic solute transporters) on the basolateral surface of enterocytes (Fig. 5C) (47). Cytoplasmic IBABP (ileal bile acid-binding protein) helps to facilitate bile acid transport (Fig. 5C) (47). Bile acids in ileal enterocytes and colonic L cells bind to FXR and activate the FXR-RXR heterodimer complex, resulting in transcription of target genes, such as FGF15/19. FGF15/19 is secreted into the portal vein and transported back to the liver, where it inhibits expression of CYP7A1 in hepatocytes to directly regulate bile acid synthesis (47).
FIG 5.
Ursodiol alters the FXF-FGF15 transcriptome during CDI. (A and B) Violin plots of cecal (A) and colonic (B) mRNA expression levels of normalized log2counts of seven genes involved with the FXF-FGF15 pathway. Significance was determined by a Student’s parametric t test with Welch’s correction (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). (C) Schematic of seven genes within the FXF-FGF15 pathway. Bile acids modulate signaling via the nuclear bile acid receptor FXR (farnesoid X receptor) and plasma membrane receptor TGR5 (or G-protein-coupled bile acid receptor Gpbar1). Bile acids within the gastrointestinal tract are transported back to the liver via enterohepatic recirculation by several active transporters, ASBT (apical sodium-dependent bile acid transporter) on the apical surface, and OSTα/β (organic solute transporters) on the basolateral surfaces of enterocytes. Cytoplasmic IBABP (ileal bile acid-binding protein) helps to facilitate bile acid transport. Bile acids in ileal enterocytes and colonic L cells bind to FXR and activate the FXR-RXR heterodimer complex, resulting in transcription of target genes, such as FGF15/19. FGF15/19 is secreted into the portal vein and transported back to the liver, where it inhibits expression of CYP7A1 in hepatocytes to directly regulate bile acid synthesis.
In cecal tissue, mRNA expression of receptors FXR and TGR5 and bile acid transporters ASBT and OSTα/β were significantly increased in ursodiol-pretreated mice compared to untreated mice at day 2 postchallenge (Fig. 5A, Student’s parametric t test with Welch’s correction, see Table S7 for P values). In colonic tissue, mRNA expression of the receptors FXR and TGR5, the signaling molecule FGF15, and the bile acid transporters ASBT and OSTα/β were significantly increased in ursodiol-pretreated mice compared to untreated mice (Fig. 5B, Student’s parametric t test with Welch’s correction, see Table S7 for P values). Collectively, these results demonstrate that pretreatment of ursodiol in mice with CDI increases the expression of the FXR/FGF15 pathway transcriptome in both cecal and colonic tissue.
DISCUSSION
Ursodiol, the FDA-approved commercial formulation of UDCA, is used to treat a variety of hepatic and biliary diseases, including cholesterol gallstones, primary biliary cirrhosis, primary sclerosing cirrhosis, cholangitis, nonalcoholic fatty liver disease, chronic viral hepatitis C, and cholestasis of pregnancy (31–40). However, limited evidence is available for use of ursodiol in CDI patients. To date, only a single case report demonstrating successful prevention of recurrent C. difficile ileal pouchitis with oral UDCA administration is available (29), suggesting that exogenous administration of the secondary bile acid UDCA may be able to restore colonization resistance against C. difficile in vivo (29). Although the mechanisms by which ursodiol exerts its impact on C. difficile are unknown, off-label use of ursodiol is currently in phase 4 clinical trials for the prevention of recurrent CDI (41). In the present study, ursodiol pretreatment impacted the life cycle of C. difficile in vivo. Although C. difficile bacterial load and toxin activity were not affected at day 2, pretreatment with ursodiol resulted in a 24- to 36-h delay in the clinical course of CDI, suggesting attenuation of CDI pathogenesis early during infection. Although ursodiol pretreatment did not alter the gut microbiota composition, significant modifications in the bile acid metabolome were observed. Several key bile acid species were significantly increased following ursodiol administration, namely, UDCA, TUDCA, and TβMCA. The tissue inflammatory transcriptome was altered, including gene expression of bile acid-activated receptors nuclear farnesoid X receptor (FXR) and transmembrane G-protein-coupled membrane receptor 5 (TGR5), which are able to modulate the innate immune response through signaling pathways such as nuclear factor κ-B (NF-κB). This is the first study to investigate the impact of innate immune response regulation by bile acid-activated receptors, FXR and/or TGR5, during CDI.
Modulation of bile acid pools to restore colonization resistance against C. difficile is a rational therapeutic option, based on the importance bile acids play in the life cycle of C. difficile (25, 27). Traditional therapeutic approaches to CDI have focused on restoring bile acid pools by reestablishing the entire microbial community, such as with fecal microbiota transplantation (13, 28), or supplementation of key commensals, such as Clostridium scindens, a bacterium capable of synthesizing microbe-derived secondary bile acids, which when supplemented leads to partial restoration of colonization resistance against C. difficile (11). In the present study, we deduced that direct supplementation of bile acids, such as UDCA or ursodiol, which are known to inhibit the life cycle of C. difficile (26), may be sufficient to restore or partially restore colonization resistance against this enteric pathogen by directly impacting C. difficile, but also by modulating the host inflammatory response.
The host innate immune response to CDI is an essential component of C. difficile pathogenesis (1). The acute host innate immune response is initiated by C. difficile toxin-mediated damage to the intestinal epithelial cells, loss of the epithelial barrier, and subsequent translocation of luminal gut microbes (1, 48). C. difficile toxin A (TcdA) and toxin B (TcdB) directly disrupt the cytoskeleton of intestinal epithelial cells, resulting in disassociation of tight junctions and loss of epithelial integrity (1, 49). In response to toxin-mediated destruction, resident immune cells and intoxicated epithelial cells release proinflammatory cytokines and chemokines to recruited circulating innate immune cells and adaptive immune cells (reviewed by Abt et al. [1]). A prominent inflammatory pathway that is activated in intestinal epithelial cells by TcdA and TcdB is the NF-κB signaling pathway through the phosphorylation of mitogen-activated protein kinase (MAPK), which leads to the transcription of proinflammatory cytokines, including gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), interleukin 6 (IL-6), and IL-1β (50–53). In the present study, pretreatment with ursodiol resulted in alterations in the host intestinal inflammatory transcriptome. Genes encoding proteins involved in the NF-κB signaling pathway in both the cecum and colon segregated based on ursodiol pretreatment. This is not surprising given the broad ursodiol-mediated alterations in bile acid metabolome observed and the accumulating evidence that bile acid-activated receptors regulate the host innate immune response (recently reviewed by Fiorucci et al. [44]). In particular, interactions between the gut microbiota and bile acids result in alterations in bile acid pools, which in turn modulate signaling via the FXR and TGR5, both of which can regulate the innate immune response (44, 47, 54). Bile acid activation of TGR5 involves cyclic AMP (cAMP)-protein kinase A (PKA)-mediated inhibition of NF-κB, and bile acid activation of FXR leads to FXR-nuclear receptor corepressor 1 (NCor1)-mediated repression of NF-κB-responsive elements (NRE). Bile acid activation of both pathways blunt NF-κB gene transcription, including proinflammatory cytokines IFN-γ, TNF-α, IL-6, and IL-1β (55, 56). Additionally, TGR5 directly regulates IL-10 expression by a cAMP/PKA/p CAMP-responsive element binding protein pathway, thus further attenuating the inflammatory response (57).
Bile acids differ in their affinity and agonistic/antagonistic effects for FXR and TGR5 (44). We observed global alterations in bile acids that are both FXR and TGR5 agonists and antagonists, lending to the complexity of unravelling the cumulative effects on bile acid-activated receptors and how this impacts the overall innate immune response during CDI. Based on FXR and TGR5 signaling transcriptomics, ursodiol pretreatment in the present study resulted in increased cecal and colonic expression of genes in these pathways during CDI. Collectively, this suggests that ursodiol-induced alterations in the bile acid metabolome result in alterations of host inflammatory transcriptome, including alterations in the NF-κB signaling pathway, likely via the bile acid-activated receptors FXR and TGR5. Furthermore, this principle has also recently been demonstrated in recurrent CDI patients receiving FMTs, where effectiveness of FMT was associated with an upregulation of the FXR-FGF pathway (58).
While the present study demonstrated ursodiol-mediated attenuation of CDI early in the disease course, complete restoration of colonization resistance against this enteric pathogen was not achieved. Several limitations of the use of ursodiol pretreatment prior to CDI need to be considered. First, once daily dosing may be sufficient to obtain supraphysiologic pulses of UDCA, but these concentrations may not be sustained in the gastrointestinal tract throughout the day. Consequently, C. difficile in vivo is exposed to a daily pulse of ursodiol that inhibits its life cycle, similar to our in vitro results, only during a limited window. Alterations in the dosing scheme such as dividing the dose over two or four administration times may ameliorate this potential limitation. Second, alterations in the inflammatory microenvironment during the initial phases of CDI were observed in this study; however, these effects were not sustained throughout CDI. Potentially, longer pretreatment with ursodiol is needed to result in sustained alterations to the host innate immune response. Last, ursodiol may be more effective as a preventative for relapsing CDI, meaning that following successful treatment of CDI with antibiotic treatment, ursodiol could be administered to prevent relapse of infection.
Regardless, our results highlight that ursodiol pretreatment was able to significantly alter the bile acid metabolome and host inflammatory transcriptome during CDI. Although ursodiol pretreatment did not result in colonization resistance against C. difficile, attenuated pathogenesis of C. difficile was noted early in the disease course. Collectively, we theorize that ursodiol-induced alterations within the intestinal bile acid metabolome result in activation of bile acid receptors, such as FXR and TGR5, which are able to modulate the innate immune response through signaling pathways such as NF-κB, thus mitigating an overly robust host inflammatory response that can be detrimental to the host. Given these results and the effectiveness of ursodiol to inhibit the C. difficile life cycle, ursodiol remains a viable nonantibiotic treatment and/or prevention strategy against CDI. The clinical utility of ursodiol to mitigation of an overly robust host inflammatory response may be particularly beneficial for treating patients with severe CDI. Further studies are required to define the optimum dose and duration of therapy in humans, particularly since ursodiol undergoes rapid absorption in the small intestine, thus resulting in low intracolonic concentrations. Moreover, targeting bile acid-activated receptors FXR and TGR5 to attenuate an overly robust detrimental host immune response during CDI merits further consideration as a potential therapeutic intervention for CDI patients.
MATERIALS AND METHODS
Spore preparation.
C. difficile R20291 spores were prepared as described previously (22, 26, 59). Briefly, C. difficile R20291 was grown at 37°C anaerobically overnight in 2 ml Columbia broth and then added to 40-ml culture of Clospore medium in which it was allowed to sporulate for 5 to 7 days. Spores were harvested by centrifugation and subjected to three to five washes with sterile cold water to ensure spore purity. Spore stocks were stored at 4°C in sterile water until use.
C. difficile in vitro spore germination and outgrowth assay.
The spore germination and outgrowth assay performed was modified from Carlson et al. (60), Theriot et al. (22), and Thanissery et al. (26). Purified spores were enumerated and tested for purity before use. The spore stock was resuspended in ultrapure water to achieve an initial spore concentration of approximately 106 spores/ml. The spore suspension was subjected to heat treatment (65°C for 20 min) to eliminate any vegetative cells. The spores were then plated on both brain heart infusion (BHI) with 100 mg/liter l-cysteine and BHI medium supplemented with 0.1% taurocholic acid (TCA) and further incubated at 37°C for 24 h. We observed a lawn of C. difficile colonies on BHI medium supplemented with 0.1% TCA and no visible growth on BHI medium alone, which indicates that the spore suspension was devoid of vegetative cells. Bile acid solutions containing four or five concentrations were used to study a dose response (Ursodiol USP [0.0076, 0.076, 0.76, and 7.64 mM]; CAS 128-13-2 [Spectrum Chemical]). Bile acids were dissolved in ethanol, passed into the anaerobic chamber (Coy Labs), and added to BHI broth with 0.1% TCA. Chendeoxycholate (CDCA) (catalog no. 50328656; Fisher Scientific) at 0.04% is a known inhibitor of TCA-mediated spore germination and was used as a negative control. TCA at 0.1% was made in water, and water and ethanol were used as positive controls. C. difficile R20291 spores were added to BHI broth supplemented with and without TCA (0.1%) and bile acids at the concentrations listed above and allowed to incubate for 30 min at 37°C anaerobically. Bacterial enumeration of the samples was performed on both BHI agar (vegetative cells only) and BHI agar supplemented with 0.1% TCA (germinated spores and vegetative cells). Percent germination was calculated as [(CFU on BHI supplemented with TCA plus SBAs)/(CFU on BHI supplemented with TCA alone)] × 100. All measurements were performed in triplicate for each isolate and expressed as percent germination.
C. difficile in vitro growth kinetics assay.
The growth kinetic assay was modified from Thanissery et al. (26) and performed over a 24-h period. C. difficile R20291 was cultured overnight at 37°C in BHI plus 100 mg/liter l-cysteine broth in an anaerobic chamber. After 14 h of growth, C. difficile R20291 was subcultured 1:10 and 1:5 into BHI plus 100 mg/liter l-cysteine and allowed to grow for 3 h anaerobically at 37°C. The culture was then diluted in fresh BHI to a starting optical density at 600 nm (OD600) of 0.01 within a conical tube at a final volume of 50 ml. Five concentrations of filter-sterilized ursodiol (0.0076, 0.076, 0.76, 3.82, and 7.64 mM dissolved in ethanol) were added to each culture and anaerobically incubated for 72 h at 37°C. The optical density was monitored every 4 h for 12 h and then once at 24 h. Cultures were inverted prior to obtaining the optical density on a cell density meter (WPA Biowave). Due to optical density interference from gelatin formation at 7.64 mM ursodiol, this concentration was assessed only for viability.
To assess C. difficile R20291 viability, culture aliquots (100 μl) were taken at each time point and enumerated on TBHI plates (BHI supplemented with TCA) to obtain total CFU per milliliter, which represents total vegetative cells and spores of C. difficile R20291. At each time point, a second culture aliquot (100 μl) was heat treated at 65°C for 20 min in order to eliminate all vegetative cells. This heat-treated culture aliquot was then enumerated on TBHI plates to obtain total CFU/ml, which represents total spores only.
C. difficile in vitro Vero cell cytotoxicity assay.
The cytotoxicity protocol is modified from the protocol of Winston et al. (61). Vero cells were grown and maintained in Dulbecco modified Eagle medium (DMEM) (catalog no. 11965-092; Gibco Laboratories) with 10% fetal bovine serum (Gibco Laboratories (catalog no. 16140-071; Gibco Laboratories) and 1% penicillin-streptomycin solution (catalog no. 15070-063; Gibco Laboratories). Cells were incubated with 0.25% trypsin (catalog no. 25200-056; Gibco Laboratories) washed with 1× DMEM and harvested by centrifugation at 1,000 rpm for 5 min. Cells were plated at 1 × 104 cells per well in a 96-well flat-bottom microtiter plate (catalog no. 3596; Corning) and incubated overnight at 37°C and 5% CO2. Culture supernatants from the anaerobic growth assay were defrosted on ice, and three 200-μl aliquots from each treatment and time point were then centrifuged at 1,750 rpm for 5 min to pellet vegetative C. difficile. Supernatant was collected from each sample, and 10-fold dilutions, to a maximum of 10−6, were performed. Sample dilutions were incubated 1:1 with phosphate-buffered saline (PBS) (for all dilutions) or antitoxin (performed for 10−1 and 10−4 dilutions only) (catalog no. T5000; TechLabs) for 40 min at room temperature. Following incubation, these admixtures were added to the Vero cells, and the plates were incubated overnight at 37°C 5% CO2. Vero cells were viewed under ×200 magnification for rounding after overnight incubation. The cytotoxic titer was defined as the reciprocal of the highest dilution that produced rounding in 80% of Vero cells for each sample. Vero cells treated with purified C. difficile toxins (A and B) and antitoxin (List Biological Labs [catalog no. 152C and 155C]; TechLabs [catalog no. T5000]) were used as controls.
Ethics statement.
The Institutional Animal Care and Use Committee (IACUC) at North Carolina State University College of Veterinary Medicine (NCSU) approved this study. The NCSU Animal Care and Use policy applies standards and guidelines set forth in the Animal Welfare Act and Health Research Extension Act of 1985. Laboratory animal facilities at NCSU adhere to guidelines set forth in the Guide for the Care and Use of Laboratory Animals (62). The animals’ health statuses were assessed daily, and moribund animals were humanely euthanized by CO2 asphyxiation followed by secondary measures (cervical dislocation). Trained animal technicians or a veterinarian performed animal husbandry in an AAALAC-accredited facility during this study.
Animals and housing.
C57BL/6J mice (females and males) were purchased from Jackson Laboratories (Bar Harbor, ME) and quarantined for 1 week prior to starting the antibiotic water administration to adapt to the new facilities and avoid stress-associated responses. Following quarantine, the mice were housed with autoclaved food, bedding, and water. Cage changes were performed weekly in a laminar flow hood by laboratory staff. Mice had a 12-h cycle of light and darkness.
Ursodiol dosing experiment and sample collection.
Groups of 5-week-old C57BL/6J mice (male and female) were treated with ursodiol at three doses (50, 150, and 450 mg/kg dissolved in corn oil; Ursodiol USP, Spectrum Chemical, CAS 128-13-2) given daily via oral gavage for 14 days. Concurrently mice were administered cefoperazone (0.5 mg/ml), a broad-spectrum antibiotic, in their drinking ad libitum for 5 days. A control group of mice were administered only cefoperazone in their drinking water ad libitum for 5 days. Two independent experiments were performed, with a total of eight mice per treatment group. Fecal pellets were collected twice daily, flash frozen, and stored at –80°C until further analysis.
Murine model of C. difficile infection and sample collection.
Groups of 5-week-old C57BL/6J mice (male and female; 12 mice/treatment group) were given cefoperazone (0.5 mg/ml), in their drinking ad libitum for 5 days, followed by a 2-day wash out with regular drinking water ad libitum (Fig. 3A). A group of cefoperazone-treated mice were pretreated with ursodiol (450 mg/kg dissolved in corn oil; Ursodiol USP, Spectrum Chemical, CAS 128-13-2) daily via oral gavage. Mice were then challenged with 105 spores of C. difficile R20291 via oral gavage at day 0 (C. diff + U450). Mice were monitored for weight loss and clinical signs of CDI (lethargy, inappetence, diarrhea, and hunched posture) for 7 days postchallenge. Fecal pellets were collected daily for C. difficile enumeration. Controls for this experiment included cefoperazone-treated mice, no ursodiol, challenged with C. diff (C. diff only), and cefoperazone-treated mice pretreated with the corn oil vehicle (equivalent volume, 75 μl) daily via oral gavage and then challenged with 105 spores of C. difficile R20291 via oral gavage at day 0 (C. diff + corn oil). Uninfected controls included cefoperazone-treated mice (Cef only) and mice pretreated with ursodiol (450 mg/kg dissolved in corn oil; Cef + U450). Animals were euthanized after losing 20% of initial baseline weight or after developing any severe clinical signs noted above.
Necropsy was performed on days 2, 4, and 7 postchallenge. The contents and tissue from the cecum and colon were collected immediately at necropsy, flash frozen in liquid nitrogen, and stored at –80°C until further analysis. To prevent RNA degradation, snips of cecal and colonic tissue were also stored in RNA-Later at 80°C until further analysis. The cecum and colon were prepared for histology by placing intact tissue into histology cassettes, stored in 10% neutral buffered formalin at a ratio of 1:10 tissue-fixative at room temperature for 72 h, and then transferred to room temperature 70% ethyl alcohol. Tissues were processed, paraffin embedded, sectioned at 4 μm thick, and stained by routine hematoxylin and eosin (H&E) for histopathological examination (LCCC Animal Histopathology Core Facility at the University of North Carolina at Chapel Hill). Histopathologic evaluation was conducted by a board-certified veterinary pathologist (S. Montgomery) in an a priori blind manner in which individuals within groups were masked at time of scoring. For tissue scoring, a 0 to 4 numerical scoring system was employed to separately assess edema, inflammatory cell infiltration, and epithelial cell damage in the cecum and colon based upon a previously published scoring scheme (63).
Bile acid quantitation by UPLC-MS/MS of murine cecal and fecal content.
Targeted analysis of bile acids on fecal pellets (three mice per treatment group were selected) were performed with an Acquity ultraperformance liquid chromatography (UPLC) system using a C8 BEH column (2.1 by 100 mm; 1.7 μm) coupled with a Xevo TQ-S triple quadrupole mass spectrometer equipped with an electrospray ionization (ESI) source operating in negative ionization mode (All Waters, Milford, MA) as previously described (64). The sample was thawed on ice, and 25 mg was added to 1 ml of precooled methanol containing 0.5 μM stable-isotope-labeled bile acids as internal standards (IS), followed by homogenization (Precellys, Bertin Technologies, Rockville, MD) with 1.0-mm-diameter zirconia/silica beads (BioSpec, Bartlesville, OK) and centrifugation (Eppendorf, Hamburg, Germany) at 13,200 × g and 4°C for 20 min. Two hundred microliters of the supernatant was transferred to an autosampler vial. Following centrifugation, the supernatant was transferred to an autosampler vial for quantitation. Bile acids were detected by either multiple reaction monitoring (MRM) (for conjugated bile acid) or selected ion monitoring (SIM) (for nonconjugated bile acid). Mass spectrometry (MS) methods were developed by infusing individual bile acid standards. Calibration curves were used to quantify the biological concentration of bile acids. Bile acid quantitation was performed at Penn State University.
Heatmaps and box and whisker plots of bile acid concentrations and nonmetric multidimensional scaling (NMDS) depicting the dissimilarity indices via Horn distances between bile acid profiles were generated using R packages (http://www.R-project.org).
Illumina MiSeq sequencing of bacterial communities.
Microbial DNA was extracted from murine feces (three mice per treatment group were selected) using the PowerSoil-htp 96-well soil DNA isolation kit (Mo Bio Laboratories, Inc.). The V4 region of the 16S rRNA gene was amplified from each sample using a dual-indexing sequencing strategy (65). Each 20-μl PCR mixture contained 2 μl of 10× Accuprime PCR buffer II (Life Technologies), 0.15 μl of Accuprime high-fidelity Taq (catalog no. 12346094) DNA polymerase (Life Technologies), 2 μl of a 4.0 μM primer set, 1 μl DNA, and 11.85 μl sterile double-distilled water (ddH2O) (free of DNA, RNase, and DNase contamination). The template DNA concentration was 1 to 10 ng/μl for a high bacterial DNA/host DNA ratio. PCR was performed under the following conditions: (i) 2 min at 95°C; (ii) 30 cycles, with 1 cycle consisting of 95°C for 20 s, 55°C for 15 s, and 72°C for 5 min; (iii) 72°C for 10 min. Each 20-μl PCR mixture contained 2 μl of 10× Accuprime PCR buffer II (Life Technologies), 0.15 μl of Accuprime high-fidelity Taq (catalog no. 12346094) DNA polymerase (Life Technologies), 2 μl of 4.0 μM primer set, 1 μl DNA, and 11.85 μl sterile ddH2O (free of DNA, RNase, and DNase contamination). The template DNA concentration was 1 to 10 ng/μl for a high bacterial DNA/host DNA ratio. PCR was performed under the following conditions: (i) 2 min at 95°C; (ii) 20 cycles, with 1 cycle consisting of 95°C for 20 s, 60°C for 15 s, and 72°C for 5 min (with a 0.3°C increase of the 60°C annealing temperature each cycle); (iii) 20 cycles, with 1 cycle consisting of 95°C for 20 s, 55°C for 15 s, and 72°C for 5 min; (iv) 72°C for 10 min. Libraries were normalized using a Life Technologies SequalPrep normalization plate kit (catalog no. A10510-01) following the manufacturer’s protocol. The concentration of the pooled samples was determined using the Kapa Biosystems library quantification kit for Illumina platforms (catalog no. KK4854; Kapa Biosystems). The sizes of the amplicons in the library were determined using the Agilent Bioanalyzer high-sensitivity DNA analysis kit (catalog no. 5067-4626). The final library consisted of equal molar amounts from each of the plates, normalized to the pooled plate at the lowest concentration.
Sequencing was done on the Illumina MiSeq platform, using a MiSeq reagent kit V2 with 500 cycles (catalog no. MS-102-2003) according to the manufacturer’s instructions, with modifications (65). Libraries were prepared according to Illumina’s protocol for preparing libraries for sequencing on the MiSeq (part 15039740 Rev. D) for 2 or 4 nM libraries. The final load concentration was 4 pM (but it can be up to 8 pM) with a 10% PhiX spike to add diversity. Sequencing reagents were prepared according to Illumina’s protocol for 16S sequencing with the Illumina MiSeq personal sequencer (65). (Updated versions of this protocol can be found at http://www.mothur.org/wiki/MiSeq_SOP.) Custom read 1, read 2, and index primers were added to the reagent cartridge, and FASTQ files were generated for paired-end reads.
Microbiome analysis.
Analysis of the V4 region of the 16S rRNA gene was done using mothur (version 1.40.1) (65, 66). Briefly, the standard operating procedure (SOP) at http://www.mothur.org/wiki/MiSeq_SOP was followed to process the MiSeq data. The paired-end reads were assembled into contigs and then aligned to the SILVA 16S rRNA sequence database (release 132) (67, 68) and were classified to the mothur-adapted RDP training set v16 (69) using the Wang method and an 80% bootstrap minimum to the family taxonomic level. All samples with <500 sequences were removed. Chimeric sequences were removed using UCHIME (70). Sequences were clustered into operational taxonomic units (OTU) using a 3% species-level definition. The OTU data were then filtered to include only those OTU that made up 1% or more of the total sequences. The percentage of relative abundance of bacterial phyla and family members in each sample was calculated. A cutoff of 0.03 (97%) was used to define operational taxonomic units (OTU) and Yue and Clayton dissimilarity metric (θYC) was utilized to assess beta diversity. Standard packages in R were used to create NMDS ordination on serial fecal samples.
mRNA isolation and gene expression analysis.
RNA extraction was performed on cecal and colonic tissue using Zymo Research RNA extraction kit (Irvine, CA). After RNA extraction, samples were quantified using an Invitrogen Qubit (Thermo Fisher Scientific) and run on an Agilent Bioanalyzer NanoChip (Santa Clara, CA) to assess the quality of RNA. RNA extraction, quantification, and purity assessment were performed at the Microbiome Shared Resource Lab, Duke University, Durham, NC.
The nCounter Mouse Inflammation v2 gene expression panel and custom code set was purchased from NanoString Technologies (Seattle, WA) and consists of 254 inflammation-related mouse genes, including six internal reference genes (see Table S1 in the supplemental material) and 10 custom genes of interest (Table S2). The nCounter assay was performed using 100 ng of total RNA. Hybridization reactions were performed according to the manufacturer’s instructions with 5-μl diluted sample preparation reaction, and samples were hybridized overnight. Hybridized reactions were purified using the nCounter Prep Station (NanoString Technologies), and data collection was performed on the nCounter Digital Analyzer (NanoString Technologies) following the manufacturer’s instructions to count targets. Nanostring nCounter Max platform was utilized at UNC Translational Genomics Laboratory to perform these assays.
The raw data were normalized to six internal reference genes within each tissue type with the lowest coefficient of variation using nSolver software following the manufacturer’s instructions (NanoString Technologies). Using the nSolver Advanced analysis software, after preprocessing and normalization, the counts were log2 transformed and z-scores were calculated (NanoString Technologies).
To identify highly significant genes and molecular pathways, the gene set was subsequently uploaded to Ingenuity Pathway Analysis (IPA) database (Qiagen, Redwood City, CA) and analyzed in the context of known biological response and regulatory networks. IPA was also used to assess gene expression fold changes comparing ursodiol-pretreated CDI mice to untreated CDI mice.
Statistical analysis.
Statistical tests were performed using Prism version 7.0b for Mac OS X (GraphPad Software, La Jolla, California, USA) or using R packages (http://www.R-project.org). Statistical significance was set at a P value of <0.05 for all analyses. For in vitro experiments, significance between treatments in spore germination was calculated by Student’s parametric t test with Welch’s correction. Two-way analysis of variance (ANOVA) test followed by Dunnett’s multiple-comparison posthoc test was used to calculate significance between treatments for the anaerobic growth assay and toxin activity assay.
For the in vivo experiments, a two-way ANOVA followed by Tukey’s multiple-comparison posthoc test was used to calculate significance between treatment groups in bile acid profiles, individual bile acids, C. difficile load (CFU/gram) cecal content, and total histologic score. A two-way ANOVA followed by Sidak’s multiple-comparison posthoc test was used to calculate significance between treatment groups in baseline weight loss. A Student’s t test corrected for multiple comparisons using the Holm-Sidak method was used to calculate significance between treatment groups in cecal content toxin activity and edema score. Analysis of molecular variance (AMOVA) was used to detect significant microbial community clustering of treatment groups in NMDS plots (71). nSolver software and IPA were used to calculate significance in gene expression fold change comparing mice pretreated with ursodiol to untreated mice.
Availability of data and material.
Raw 16S sequences have been deposited in the Sequence Read Archive (SRA) under accession numbers PRJNA622536 and SAMN14518789 to SAMN14518901. All other data, including bile acid metabolomics and Nanostrings expression data are provided in Tables S3 to S7 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
J.A.W. was funded by the Ruth L. Kirschstein National Research Service Award Research Training grant T32OD011130 by NIH. This research was supported by work performed by The University of Michigan Microbial Systems Molecular Biology Laboratory (microbiome sequencing). The Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center. C.M.T. is funded by the National Institute of General Medical Sciences of the National Institutes of Health under award R35GM119438. This project was also funded by an intramural grant from the North Carolina State University College of Veterinary Medicine.
C.M.T. is a scientific advisor to Locus Biosciences, a company engaged in the development of antimicrobial technologies. C.M.T. is a consultant for Vedanta Biosciences and Summit Therapeutics.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerding DN, Lessa FC. 2015. The epidemiology of Clostridium difficile infection inside and outside health care institutions. Infect Dis Clin North Am 29:37–50. doi: 10.1016/j.idc.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubberke ER, Olsen MA. 2012. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis 55(Suppl 2):S88–S92. doi: 10.1093/cid/cis335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magill SS, O’Leary E, Janelle SJ, Thompson DL, Dumyati G, Nadle J, Wilson LE, Kainer MA, Lynfield R, Greissman S, Ray SM, Beldavs Z, Gross C, Bamberg W, Sievers M, Concannon C, Buhr N, Warnke L, Maloney M, Ocampo V, Brooks J, Oyewumi T, Sharmin S, Richards K, Rainbow J, Samper M, Hancock EB, Leaptrot D, Scalise E, Badrun F, Phelps R, Edwards JR. 2018. Changes in prevalence of health care-associated infections in U.S. hospitals. N Engl J Med 379:1732–1744. doi: 10.1056/NEJMoa1801550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tartof SY, Rieg GK, Wei R, Tseng HF, Jacobsen SJ, Yu KC. 2015. A comprehensive assessment across the healthcare continuum: risk of hospital-associated Clostridium difficile infection due to outpatient and inpatient antibiotic exposure. Infect Control Hosp Epidemiol 36:1409–1416. doi: 10.1017/ice.2015.220. [DOI] [PubMed] [Google Scholar]
- 7.Abou Chakra CN, Pepin J, Sirard S, Valiquette L. 2014. Risk factors for recurrence, complications and mortality in Clostridium difficile infection: a systematic review. PLoS One 9:e98400. doi: 10.1371/journal.pone.0098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antunes LC, Han J, Ferreira RB, Lolic P, Borchers CH, Finlay BB. 2011. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother 55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theriot CM, Koenigsknecht MJ, Carlson PE Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorg JA. 2014. Microbial bile acid metabolic clusters: the bouncers at the bar. Cell Host Microbe 16:551–552. doi: 10.1016/j.chom.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. 2014. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol 306:G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. 2018. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:987–994. doi: 10.1093/cid/ciy149. [DOI] [PubMed] [Google Scholar]
- 15.Kociolek LK, Gerding DN. 2016. Breakthroughs in the treatment and prevention of Clostridium difficile infection. Nat Rev Gastroenterol Hepatol 13:150–160. doi: 10.1038/nrgastro.2015.220. [DOI] [PubMed] [Google Scholar]
- 16.Kelly CP, LaMont JT. 2008. Clostridium difficile–more difficult than ever. N Engl J Med 359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 17.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 364:422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 18.Theriot CM, Young VB. 2015. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu Rev Microbiol 69:445–461. doi: 10.1146/annurev-micro-091014-104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tannock GW, Munro K, Taylor C, Lawley B, Young W, Byrne B, Emery J, Louie T. 2010. A new macrocyclic antibiotic, fidaxomicin (OPT-80), causes less alteration to the bowel microbiota of Clostridium difficile-infected patients than does vancomycin. Microbiology 156:3354–3359. doi: 10.1099/mic.0.042010-0. [DOI] [PubMed] [Google Scholar]
- 20.Gough E, Shaikh H, Manges AR. 2011. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 21.Khoruts A, Sadowsky MJ, Hamilton MJ. 2015. Development of fecal microbiota transplantation suitable for mainstream medicine. Clin Gastroenterol Hepatol 13:246–250. doi: 10.1016/j.cgh.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Theriot C, Bowman A, Young V. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:e00045-15. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theriot CM, Young VB. 2014. Microbial and metabolic interactions between the gastrointestinal tract and Clostridium difficile infection. Gut Microbes 5:86–95. doi: 10.4161/gmic.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridlon JM, Kang DJ, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Winston JA, Theriot CM. 2020. Diversification of host bile acids by members of the gut microbiota. Gut Microbes 11:158–114. doi: 10.1080/19490976.2019.1674124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thanissery R, Winston JA, Theriot CM. 2017. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winston JA, Theriot CM. 2016. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 41:44–50. doi: 10.1016/j.anaerobe.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weingarden AR, Dosa PI, DeWinter E, Steer CJ, Shaughnessy MK, Johnson JR, Khoruts A, Sadowsky MJ. 2016. Changes in colonic bile acid composition following fecal microbiota transplantation are sufficient to control Clostridium difficile germination and growth. PLoS One 11:e0147210. doi: 10.1371/journal.pone.0147210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weingarden AR, Chen C, Zhang N, Graiziger CT, Dosa PI, Steer CJ, Shaughnessy MK, Johnson JR, Sadowsky MJ, Khoruts A. 2016. Ursodeoxycholic acid inhibits Clostridium difficile spore germination and vegetative growth, and prevents the recurrence of ileal pouchitis associated with the infection. J Clin Gastroenterol 50:624–630. doi: 10.1097/MCG.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.US FDA. 2016. Drugs@FDA: FDA-approved drugs. Ursodiol. US Food and Drug Administration, Washington, DC.
- 31.Ridlon JM, Bajaj JS. 2015. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B 5:99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikegami T, Matsuzaki Y. 2008. Ursodeoxycholic acid: mechanism of action and novel clinical applications. Hepatol Res 38:123–131. doi: 10.1111/j.1872-034X.2007.00297.x. [DOI] [PubMed] [Google Scholar]
- 33.Fischer S, Muller I, Zundt BZ, Jungst C, Meyer G, Jungst D. 2004. Ursodeoxycholic acid decreases viscosity and sedimentable fractions of gallbladder bile in patients with cholesterol gallstones. Eur J Gastroenterol Hepatol 16:305–311. doi: 10.1097/00042737-200403000-00010. [DOI] [PubMed] [Google Scholar]
- 34.Tsubakio K, Kiriyama K, Matsushima N, Taniguchi M, Shizusawa T, Katoh T, Manabe N, Yabu M, Kanayama Y, Himeno S. 2002. Autoimmune pancreatitis successfully treated with ursodeoxycholic acid. Intern Med 41:1142–1146. doi: 10.2169/internalmedicine.41.1142. [DOI] [PubMed] [Google Scholar]
- 35.Sinakos E, Marschall HU, Kowdley KV, Befeler A, Keach J, Lindor K. 2010. Bile acid changes after high-dose ursodeoxycholic acid treatment in primary sclerosing cholangitis: relation to disease progression. Hepatology 52:197–203. doi: 10.1002/hep.23631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poupon RE, Bonnand AM, Queneau PE, Trepo C, Zarski J, Vetter D, Raabe JJ, Thieffin G, Larrey D, Grange JD, Capron JP, Serfaty L, Chretien Y, St Marc Girardin MF, Mathiex-Fortunet H, Zafrani ES, Guechot J, Beuers U, Paumgartner G, Poupon R. 2000. Randomized trial of interferon-alpha plus ursodeoxycholic acid versus interferon plus placebo in patients with chronic hepatitis C resistant to interferon. Scand J Gastroenterol 35:642–649. doi: 10.1080/003655200750023624. [DOI] [PubMed] [Google Scholar]
- 37.Serfaty L, De Leusse A, Rosmorduc O, Desaint B, Flejou JF, Chazouilleres O, Poupon RE, Poupon R. 2003. Ursodeoxycholic acid therapy and the risk of colorectal adenoma in patients with primary biliary cirrhosis: an observational study. Hepatology 38:203–209. doi: 10.1053/jhep.2003.50311. [DOI] [PubMed] [Google Scholar]
- 38.Carey EJ, Ali AH, Lindor KD. 2015. Primary biliary cirrhosis. Lancet 386:1565–1575. doi: 10.1016/S0140-6736(15)00154-3. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, Liu XH, Qi HB, Li Z, Fu XD, Chen L, Shao Y. 2015. Ursodeoxycholic acid and S-adenosylmethionine in the treatment of intrahepatic cholestasis of pregnancy: a multi-centered randomized controlled trial. Eur Rev Med Pharmacol Sci 19:3770–3776. [PubMed] [Google Scholar]
- 40.Mueller M, Thorell A, Claudel T, Jha P, Koefeler H, Lackner C, Hoesel B, Fauler G, Stojakovic T, Einarsson C, Marschall HU, Trauner M. 2015. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J Hepatol 62:1398–1404. doi: 10.1016/j.jhep.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.US National Library of Medicine. 2019. C. Difficile and Ursodiol: ClinicalTrials.gov. US National Library of Medicine, National Institutes of Health, Bethesda, MD.
- 42.Copaci I, Micu L, Iliescu L, Voiculescu M. 2005. New therapeutical indications of ursodeoxycholic acid. Rom J Gastroenterol 14:259–266. [PubMed] [Google Scholar]
- 43.Wahlstrom A, Sayin SI, Marschall HU, Backhed F. 2016. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Fiorucci S, Biagioli M, Zampella A, Distrutti E. 2018. Bile acids activated receptors regulate innate immunity. Front Immunol 9:1853. doi: 10.3389/fimmu.2018.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorg JA, Sonenshein AL. 2009. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol 191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamilton JP, Xie G, Raufman JP, Hogan S, Griffin TL, Packard CA, Chatfield DA, Hagey LR, Steinbach JH, Hofmann AF. 2007. Human cecal bile acids: concentration and spectrum. Am J Physiol Gastrointest Liver Physiol 293:G256–G263. doi: 10.1152/ajpgi.00027.2007. [DOI] [PubMed] [Google Scholar]
- 47.Wahlstrom A, Kovatcheva-Datchary P, Stahlman M, Backhed F, Marschall HU. 2017. Crosstalk between bile acids and gut microbiota and its impact on farnesoid X receptor signalling. Dig Dis 35:246–250. doi: 10.1159/000450982. [DOI] [PubMed] [Google Scholar]
- 48.Naaber P, Mikelsaar RH, Salminen S, Mikelsaar M. 1998. Bacterial translocation, intestinal microflora and morphological changes of intestinal mucosa in experimental models of Clostridium difficile infection. J Med Microbiol 47:591–598. doi: 10.1099/00222615-47-7-591. [DOI] [PubMed] [Google Scholar]
- 49.Hunt JJ, Ballard JD. 2013. Variations in virulence and molecular biology among emerging strains of Clostridium difficile. Microbiol Mol Biol Rev 77:567–581. doi: 10.1128/MMBR.00017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popoff MR. 2018. Clostridium difficile and Clostridium sordellii toxins, proinflammatory versus anti-inflammatory response. Toxicon 149:54–64. doi: 10.1016/j.toxicon.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Kim JM, Lee JY, Yoon YM, Oh YK, Youn J, Kim YJ. 2006. NF-kappa B activation pathway is essential for the chemokine expression in intestinal epithelial cells stimulated with Clostridium difficile toxin A. Scand J Immunol 63:453–460. doi: 10.1111/j.1365-3083.2006.001756.x. [DOI] [PubMed] [Google Scholar]
- 52.Lee JY, Park HR, Oh YK, Kim YJ, Youn J, Han JS, Kim JM. 2007. Effects of transcription factor activator protein-1 on interleukin-8 expression and enteritis in response to Clostridium difficile toxin A. J Mol Med 85:1393–1404. doi: 10.1007/s00109-007-0237-7. [DOI] [PubMed] [Google Scholar]
- 53.Jefferson KK, Smith MF Jr, Bobak DA. 1999. Roles of intracellular calcium and NF-kappa B in the Clostridium difficile toxin A-induced up-regulation and secretion of IL-8 from human monocytes. J Immunol 163:5183–5191. [PubMed] [Google Scholar]
- 54.Fiorucci S, Distrutti E. 2015. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol Med 21:702–714. doi: 10.1016/j.molmed.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. 2009. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol 183:6251–6261. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 56.Cipriani S, Mencarelli A, Chini MG, Distrutti E, Renga B, Bifulco G, Baldelli F, Donini A, Fiorucci S. 2011. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One 6:e25637. doi: 10.1371/journal.pone.0025637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biagioli M, Carino A, Cipriani S, Francisci D, Marchiano S, Scarpelli P, Sorcini D, Zampella A, Fiorucci S. 2017. The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J Immunol 199:718–733. doi: 10.4049/jimmunol.1700183. [DOI] [PubMed] [Google Scholar]
- 58.Monaghan T, Mullish BH, Patterson J, Wong GK, Marchesi JR, Xu H, Jilani T, Kao D. 2018. Effective fecal microbiota transplantation for recurrent Clostridioides difficile infection in humans is associated with increased signalling in the bile acid-farnesoid X receptor-fibroblast growth factor pathway. Gut Microbes 10:142–148. doi: 10.1080/19490976.2018.1506667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez J, Springthorpe VS, Sattar SA. 2011. Clospore: a liquid medium for producing high titers of semi-purified spores of Clostridium difficile. J AOAC Int 94:618–626. doi: 10.1093/jaoac/94.2.618. [DOI] [PubMed] [Google Scholar]
- 60.Carlson PE Jr, Kaiser AM, McColm SA, Bauer JM, Young VB, Aronoff DM, Hanna PC. 2015. Variation in germination of Clostridium difficile clinical isolates correlates to disease severity. Anaerobe 33:64–70. doi: 10.1016/j.anaerobe.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winston JA, Thanissery R, Montgomery SA, Theriot CM. 2016. Cefoperazone-treated mouse model of clinically-relevant Clostridium difficile strain R20291. J Vis Exp 2016:54850. doi: 10.3791/54850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 63.Theriot CM, Koumpouras CC, Carlson PE, Bergin II, Aronoff DM, Young VB. 2011. Cefoperazone-treated mice as an experimental platform to assess differential virulence of Clostridium difficile strains. Gut Microbes 2:326–334. doi: 10.4161/gmic.19142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarafian MH, Lewis MR, Pechlivanis A, Ralphs S, McPhail MJ, Patel VC, Dumas ME, Holmes E, Nicholson JK. 2015. Bile acid profiling and quantification in biofluids using ultra-performance liquid chromatography tandem mass spectrometry. Anal Chem 87:9662–9670. doi: 10.1021/acs.analchem.5b01556. [DOI] [PubMed] [Google Scholar]
- 65.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.