Abstract
The wide use of pesticides in agriculture expose microbiota to stressful conditions that require the development of survival strategies. The bacterial response to many pollutants has not been elucidated in detail, as well as the evolutionary processes that occur to build adapted communities. The purpose of this study was to evaluate the bacterial population structure and adaptation strategies in planktonic and biofilm communities in limited environments, as tanks containing water used for washing herbicide containers. This biodiversity, with high percentage of nonculturable microorganisms, was characterized based on habitat and abiotic parameters using molecular and bioinformatics tools. According to water and wastewater standards, the physicochemical conditions of the tank water were inadequate for survival of the identified bacteria, which had to develop survival strategies in this hostile environment. The biodiversity decreased in the transition from planktonic to biofilm samples, indicating a possible association between genetic drift and selection of individuals that survive under stressful conditions, such as heating in water and the presence of chlorine, fluorine and agrochemicals over a six-month period. The abundance of Enterobacter, Acinetobacter and Pseudomonas in biofilms from water tanks was linked to essential processes, deduced from the genes attributed to these taxonomic units, and related to biofilm formation, structure and membrane transport, quorum sensing and xenobiotic degradation. These characteristics were randomly combined and fixed in the biofilm community. Thus, communities of biofilm bacteria obtained under these environmental conditions serve as interesting models for studying herbicide biodegradation kinetics and the prospects of consortia suitable for use in bioremediation in reservoirs containing herbicide-contaminated wastewater, as biofilters containing biofilm communities capable of degrading herbicides.
Keywords: Microbiology, Adaptation, Biodiversity, Bacteria, Microbial genomics, Biofilms, Microbial biotechnology, Environmental microbiology, Xenobiotics, Herbicide biodegradation, Bacterial diversity, Bioremediation, Bacterial adaptation
Microbiology; Adaptation; Biodiversity; Bacteria; Microbial genomics; Biofilms; Microbial biotechnology; Environmental microbiology; Xenobiotics; Herbicide biodegradation; Bacterial diversity; Bioremediation; Bacterial adaptation.
1. Introduction
The field of environmental microbiology, including the composition, structure and function of microbial communities in nature, has been gaining interest in academia over the last few years (Zhou et al., 2015). Notably, contaminated environments expose microbiota to stressful and selective conditions, and microorganisms can then undergo structural and metabolic adaptations to survive under these conditions (Olson et al., 2017). These adaptive strategies are linked at the cellular level to specific microbial populations, which results in different responses to compounds such as pesticides (Prione et al., 2016; Dobrzanski et al., 2018) and allows their use in biotechnological processes to help degrade them in the environment (Martins et al., 2007; Silva et al., 2007; Olchanheski et al., 2014).
Great microbial diversity, ineffective culture techniques, and the large percentage of microorganisms that are nonculturable has limited the ability to understand microbial degradation in the environment (Deng et al., 2018; Oueriaghli et al., 2018). Therefore, new research tools are needed for the characterization of microbiological biodiversity, i.e., molecular microbial ecology, using high-throughput sequencing technologies combined with bioinformatics data analysis techniques (Gutarowska et al., 2015; Ranjan et al., 2016; Zhou et al., 2015; Battin et al., 2016).
The use of molecular technologies is fundamental to understanding the structure and functions of microbiota and nonculturable microorganisms (Valeriani et al., 2019). Advances in genomics and metagenomics, marker genes, 16S rRNA gene sequencing and other molecular approaches are essential for predicting soil microbiome functions and improving agriculture production (Fierer, 2017). These approaches are fundamental to the creation of genomic libraries that allow the identification and isolation of enzymes with different biocatalytic activities from culturable strains and thereby allow their biotechnological exploitation (Al-Amoudi et al., 2016; Madhavan et al., 2017). For example, using these techniques, Krohn-Molt et al. (2017) evaluated the algal genes that are essential to the carbon cycle and linked to a unique and specific microbiome. Knowledge of the taxonomic structure of a microbiome increases the reliable handling of its latent biotechnological potential (Ji et al., 2017; Ofaim et al., 2017).
The composition and relative abundance of the soil microbiomes in the Amazon forest in Brazil have been correlated with different land use types, such as deforestation, which leads to carbon cycle disturbances and, consequently, climate change (Kroeger et al., 2018). Therefore, microbiomes, which play key roles in biogeochemical cycles, are affected by anthropogenic action, particularly the use of xenobiotic compounds, and these effects lead to changes in sustainability and environmental quality (Egea et al., 2017; Kroeger et al., 2018). High-density DNA microarray and 16S rDNA amplicon next-generation sequencing have been used to study the impacts of the insecticide chlorpyrifos, the herbicide isoproturon and the fungicide tebuconazole on the soil bacterial community, and significant differences have been found in field experiments but not in soil microcosms exposed to these pesticides (Storck et al., 2018). These methods are considered a guide for more specific studies in this field. A molecular approach has also been used to analyze the effects of combinations of glyphosate with different herbicides on the rhizobacterial community of transgenic plants, and the results indicate that glyphosate alone appears to be less aggressive than the tested combinations (Valverde et al., 2014). Some studies using molecular techniques have led to the discussion about the non-interference of herbicides on microbiotes, such as the approach using high-throughput phylogenetic marker gene sequencing that led Dennis et al. (2018) to the conclusion that the herbicides glyphosate, glufosinate, paraquat and paraquat-diquat do not significantly interfere in the soil richness, evenness and composition of bacterial and archaeal communities under laboratory conditions.
In aqueous environments, which are also subject to contamination by agrochemicals, microbiomes exhibit two possible lifestyles: planktonic and biofilm. The first term refers to the cells of microorganisms suspended in water (e.g., in an ocean water column) (Sunagawa et al., 2015). However, one of the main adaptive strategies identified in microbiomes in contaminated or extreme environments is the formation of biofilms; during biofilm formation, cells adhere to a substrate or one another by secreting a gelatinous matrix of extracellular polymeric substances (EPS), which is configured as an extension of the cell itself (Decho and Gutierrez, 2017; Maunders and Welch, 2017). The community can thus capture organic compounds for metabolization by extracellular enzymes through organized gene expression mediated by cell-cell communication or quorum sensing. This structure is also responsible for increased adaptability, stability, and cellular integrity (Zhao et al., 2014; Lerch et al., 2017; Anutrakunchai et al., 2018). Molecular approaches as 16S rRNA gene amplicon sequencing has been used to verify the impacts of herbicides and fertilizers on planktonic marine microbial communities in the Great Barrier Reef. However, water characteristics, such as salinity, rainfall, temperature and quality, have a notable influence on the composition of microbial communities (Angly et al., 2016). Thus, aquatic environments are complex and subject to various physicochemical interferences, which makes it difficult to understand the impact of agrochemicals on microbiomes (Wang et al., 2017; Chen et al., 2018a, Chen et al., 2018b).
The described environments usually involve complex biotic and abiotic interactions over time and space. The in situ bacterial response system to pollutants has not been elucidated in detail, as well as the sequence of evolutionary processes necessary to build communities more adapted to the impacted environment. Thus, the objective of this study was to identify the population structure of planktonic and biofilm bacterial communities in an environment that is more limited in terms of habitat and abiotic parameters, e.g., highly contaminated wastewater (WW) in tanks used for storing water obtained from washing the containers of herbicides, building a theoretical model for predicting adaptive systems to large-scale contaminated environments and potential biotechnological utilization of these bacterial communities.
2. Materials and methods
2.1. Study area
The object of this study was the residual water obtained after washing containers of 30 agrochemicals (Table A.1). The clean water to carry out these washes came from an artesian well and used as an experimental control. After washing, the water was kept in two 10,000-L tanks (Figure 1) for 6 months at BASF – Brasil, Experimental Station located at the Capão da Onça Farm School, State University of Ponta Grossa, State of Paraná, Brazil (25°05′31.3″S; 50°03′28.0″W) (altitude: 1000 m). One of the duties of this company, in association with the State University of Ponta Grossa, is to assist in cleaning containers of pesticides used by farmers to reduce environmental contamination, but only the brands produced by this company. Due to the high level of contamination of this water and the need for transportation to other centers to properly dispose of this environmental liability, it is necessary to decrease the volume of this water, by evaporation, to lower freight costs. The first tank (T1) contained water obtained after the washing of agrochemical containers that had lost some volume via evaporation process during incubation at 100 °C for 190 min, and this water was then stored in a second tank (T2) until being transported and discarded.
Figure 1.
Evaporation process of water obtained from the washing of pesticide containers used for DNA collection in this study. S: sanitary sink; TR: tractor spray washing trough; SH: shed where the two tanks (T1 and T2) are located; T1: tank 1; T2: tank 2; W: water filters; TI: timer for controlling the flow of water; TC: solar heating temperature control panel; and So: solar heating system.
2.2. Physicochemical analyses of water
The following physicochemical characteristics of the water samples collected from the tanks and the artesian well were analyzed according to the Standard methods for the examination of water and wastewater (1998): chlorine, conductivity, color, fluorine, pH, total dissolved solids, turbidity, biochemical oxygen demand (BOD) and chemical oxygen demand (COD).
2.3. Collection of samples and extraction of DNA
Four liters of water were collected from T1 and T2 to obtain planktonic samples. The water was passed through autoclaved glass fiber membranes in 750-mL triplicates; specifically, the samples were first passed through a 5-μm-thick prefilter, then through a 1.2-μm filter and subsequently through a 0.45-μm filter (Merck Millipore, Carrigtohill, Ireland) to obtain the DNA samples used in this study. The final filter porosity of 0.45 μM for bacterial collection was selected based on the difficulty of filtration due to the presence of particulate matter in the water samples. The samples were denoted M1 (preheating) and M2 (postheating) and stored at -80 °C.
Biofilm samples were collected in triplicate from the wall surface of the tanks in contact with the water line using sterile swabs (one swab was used for each triplicate sample). The samples were named B1 (preheating) and B2 (postheating). Similarly, biofilms were collected from the 4-L flasks from which water samples were collected after being kept immobile for 5 days. The samples were denoted G1 (preheating) and G2 (postheating), and the material, which was collected in triplicate, was stored at -80 °C.
For alpha diversity analyzes, each triplicate was represented by the letters A, B and C placed after the letters and numbers that define the planktonic and biofilm communities, as explained above.
The samples of planktonic origin (retained in the membranes) were macerated in liquid nitrogen using a pestle. The swabs used for biofilm collection were submerged in 1 mL of saline buffer (0.9% NaCl) and homogenized for 5 min to extract the biological material from the cells in the buffer. DNA was extracted using the PowerSoil DNA extraction kit (MO BIO Laboratories Inc., Carlsbad, CA, USA), and the material obtained after extraction was stored at -80 °C.
2.4. Sequencing
The samples were sequenced using the 515F/806R primers, which are specific for the V4 region of the 16S rRNA gene (Caporaso et al., 2012). Amplification and sequencing were performed using the dual index method reported by the University of Minnesota Genomics Center (Minneapolis, MN, USA) (Gohl et al., 2016). A negative control containing sterile water was included in each amplified and sequenced sample plate. Sequencing was performed on the Illumina MiSeq platform using an iSeq 100 System (Illumina, Inc. San Diego, CA, USA) (Caporaso et al., 2012). The sequence data are available from the Sequence Read Archive of the National Center for Biotechnology Information under the accession number SRA:PRJNA528924.
2.5. Bioinformatics
Sequence processing was performed using mothur software (version 1.29.2). The prokaryotic sequence data were trimmed to the first 160 nucleotides (nt), and end pairing was performed using fastq-join software (Aronesty, 2013). To obtain high-quality data, the sequences were trimmed according to a previous description for V5–V6 data (Staley et al., 2015). The overall alignment was performed in reference to the SILVA database (version 119) (Pruesse et al., 2007), the sequences were subjected to 2% preclustering to remove sequencing errors (Huse et al., 2010), and chimeric sequences were identified and removed using UCHIME software (Edgar et al., 2011). Operational taxonomic units (OTUs) were assigned through complete linkage clustering based on ≥97% identity. Taxonomic attributions were obtained by comparing the Ribosomal Database Project (version 14) to a bootstrap limit of 60%, as previously described (Wang et al., 2007).
2.6. Predicted metagenomics bioinformatics analysis
After sequence processing described at section 2.5, bacterial communities were ordered based on OTU abundance using multidimensional scaling. The relative abundance of the taxonomic groups and the functional abundances inferred in PICRUSt were regressed in relation to the ordering scores to determine significant correlations with the ordering scores of the bacterial communities (Langille et al., 2013). The 16S rDNA data were analyzed as indicated by the PICRUSt genome prediction software [http://picrust.github.io/picrust/] from raw sequence reads in the following environment: NumPy (1.7.1), biom-format (1.3.1), PyCogent (1.5.3), PICRUSt (1.0.0-dev), and PICRUSt script (1.0.0-dev). Functional predictions were assigned up to KO tier 3 for all genes. Results from the MG-RAST QIIME report were compared with predictions from PICRUSt using the “compare_biom.py” subroutine with normalization and observations not in the “expected data” file ignored. Results were compared using both shotgun metagenomic and PICRUSt predictions as the expected data. Statistical analyses were performed using the vegan package in R (R Foundation for Statistical Computing, 2007).
3. Results and discussion
Three types of information were analyzed to obtain insights regarding the adaptive strategies used by different bacterial communities in response to the contaminated environments used in this study: physicochemical characteristics of the environment, taxonomic composition at the time of collection, and survey of characteristics associated with survival of the identified taxonomic units.
3.1. Physicochemical characteristics and selection pressure
Physicochemical data were analyzed because they provide information on the conditions under which the microbiome adapts to the environment. The data on artesian well, T1 and T2 water samples are shown in Table 1. The average water temperature of these samples was 24 ± 1.4 °C.
Table 1.
Physicochemical analyses of tanks 1 (T1) and 2 (T2) and the artesian well (AW).
| Parameters | T1 | T2 | AW | WHO |
|---|---|---|---|---|
| Chlorine (mg/L) | 1.7 ± 0.1A | 1.6 ± 0.0A | 0 ± 0.0B | 0.7C |
| Conductivity (μS/cm) | 418.3 ± 2.0A | 413.0 ± 0.7A | 13.6 ± 0.6B | 10–100B |
| Color (Pt/Co) | 245.0 ± 1.5A | 250.0 ± 0.0B | 0.0 ± 0.0C | 0C |
| Fluorine (mg/L) | 0.8 ± 0.0A | 0.8 ± 0.0A | 0.1 ± 0.0B | 1.5C |
| pH at 25 °C | 5.5 ± 0.1A | 5.3 ± 0.0A | 4.8 ± 0.1B | 6.5–8.5C |
| TDS (ppm/NaCl) | 186.3 ± 1.0A | 183.2 ± 0.3A | 0.0 ± 0.0B | 100–600A |
| Turbidity (NTU) | 204.0 ± 5.8A | 206.3 ± 3.6A | 0.3 ± 0.0B | 1C |
| BOD | 80.3 ± 1.5A | 69.0 ± 4.6B | ND | 0C |
| COD | 1352.3 ± 29.0A | 1105.0 ± 35.5B | ND | 0C |
The parameter values are presented as the means from triplicate measurements ±standard deviation. WHO: World Health Organization; TDS: total dissolved solids; BOD: biochemical oxygen demand; COD: chemical oxygen demand; and ND: not detected. The letters A, B, C, and D indicate significant differences within each row.
According to the parameters shown in Table 1, the water characteristics of tanks 1 and 2 were not significantly different from each other, except for color, BOD and COD.
The data obtained in this study indicate that the heating process did not interfere with most physicochemical parameters of the water between the tanks. As discussed later in this article, changes in the microbiota did not shift these parameters, except BOD and COD, the most relevant to the discussion on metabolism and degradation of xenobiotics. Nevertheless, significant differences in all tested parameters were found between the water from the tanks and the water from the artesian well (Table 1), and these differences were mainly due to the presence of pesticides (Table A.1).
The chlorine and fluorine contents of T1 and T2 are noteworthy because these specific values are close to the acceptable concentrations for drinking water provided by the WHO (World Health Organization, 2011), even though these residues were not added to the water from the artesian well and that from the abovementioned tanks (Table 1). Moreover, according to the WHO, the pH was also inadequate for bacterial growth because at low pH values the disinfection efficiency of free chlorine is increased. Differently, the highest levels of chlorine were found in hot water spring in Siloam, South Africa, but also the highest pH, which must have decreased the effectiveness of chlorine as disinfectant (Table 2, Tekere et al., 2011).
Table 2.
Comparison of the physicochemical parameters of tanks 1 (T1) and 2 (T2) and the artesian well (AW) with those of different environments.
| Environment | Chlorine | Fluorine | BOD | COD | TDS (ppm/NaCl) | pH |
|---|---|---|---|---|---|---|
| T1 | 1.66B | 0.82C | 80.33C | 1352.33A | 186.27D | 5.45E |
| T2 | 1.57B | 0.81C | 69.00D | 1105.00B | 183.17D | 5.32E |
| AW | 0.00D | 0.05D | 0.00F | 0.00D | 0.00D | 4.75F |
| OC | 0.00D | - | - | - | - | 7.78C |
| DW | 0.70C | 1.50B | - | - | 600.00A | 7.50C |
| RS | 0.94C | - | - | - | 39826.47D | - |
| SHS | 44.35A | 6.11A | - | - | 197.32D | 9.50B |
| WW | - | - | 4886.26A | 308.91C | 2593.69D | 7.66C |
| WT | - | - | 12547.50B | 395.00C | 9470.50C | 10.40A |
| YRB | - | - | 3.28E | - | - | 7.61C |
| GLF | - | - | - | - | 23602.07B | - |
| FAZ | - | - | - | - | - | 7.09D |
Data for the following environments were obtained from the literature: oceanic transition zone (OC), drinking water (DW), Red Sea (RS), Siloam Hot Spring (SHS), wastewater (WW), WW tributary (WT), Yeongsan River Basin (YRB), Gulf of Mexico (GLF) and a freshwater aquaculture system (FAS). A dashed line (-) indicates that no information is available. The letters A, B, C, D, E, and F indicate significant differences within each column.
The BOD data indicated biological activity, even in an environment unfavorable to the development of microorganisms. COD values are usually higher than BOD values because compounds can be more easily oxidized via the chemical route than by microorganisms (Gholizadeh et al., 2016). The BOD and COD values obtained for T1 were higher than those found for T2, which indicated that T1 likely contained greater amounts of contaminants and microorganisms. For the same reason, the BOD and COD values obtained for T1 and T2 were higher than those found for the water collected directly from the artesian well. The increases in conductivity and COD found in the water from both tanks was most likely related to the nonbiological oxidation processes of the pesticides and indicated important changes to the physicochemical characteristics of the water from the artesian well.
The physicochemical data obtained in our study were compared with those of water with different origins and from environments with different characteristics. These environments were drinking water (World Health Organization, 2011), the interior transition zone of the ocean (Mende et al., 2017), the Red Sea (Thompson et al., 2017), the southwest part of the Gulf of Mexico (Godoy-Lozano et al., 2018), a hot water spring in Siloam, South Africa (Tekere et al., 2011), an integrated system of anaerobic-aerobic reactors for the treatment of WW in Ethiopia (Desta et al., 2014) and the Yeongsan River Basin of South Korea (Jang et al., 2014). These articles describe environments with different conditions of environmental stress and variations in physicochemical parameters which were compared with T1 and T2, in an attempt to categorize the type of environmental stress found in these tanks. The two tanks did not exhibit significant differences in the chlorine level, but these levels were significantly higher than those found in seawater and potable water. However, the levels found in the well water were not significantly different from those found in seawater. The fluorine levels in both tanks did not exhibit significant differences but were lower than those found drinking water and hot water spring in Siloam, South Africa, and higher than those found in the water from the artesian well. The pH of the water in the tanks did not exhibit significant differences but was significantly more acidic than those of the water from the artesian well, residual tributaries, and ocean, fresh water and drinking water. In general, the analysis of total dissolved solids (ppm/NaCl, corresponding to salinity) revealed that the values obtained in this study did not significantly differ from the levels found in WW, drinking water and a hot water spring in Siloam, South Africa. The tanks exhibited a lower BOD and a higher COD than WW and a WW tributary (WT), probably because the methods used for the treatment of these effluents contain microorganisms that increase the BOD but not the COD compared with the values found for the tanks used in this study (Table 2).
These comparisons indicate that the tank environments investigated in this study generally present quite stressful conditions for bacteria. Based on the findings from this analysis, T1 and T2 were characterized as environments with inadequate levels of chlorine and fluorine for the survival of the genera identified in this study. However, according to the BOD and COD results, these genera were probably able to tolerate these conditions, perhaps due to inactivity in planktonic form. It is important to remember that these physicochemical analyses more directly reflect the planktonic community compared with the biofilm community. These structures, even though they interface with the environment, most likely represent modifications that improve the metabolism of the microorganisms and their adaptability to the studied environment (Lerch et al., 2017) and can function as a sanctuary for metabolically integrated bacterial species in a hostile environment (Lewenza et al., 2018), as will be discussed in the following section.
3.2. Biodiversity, bacterial taxonomy and adaptation
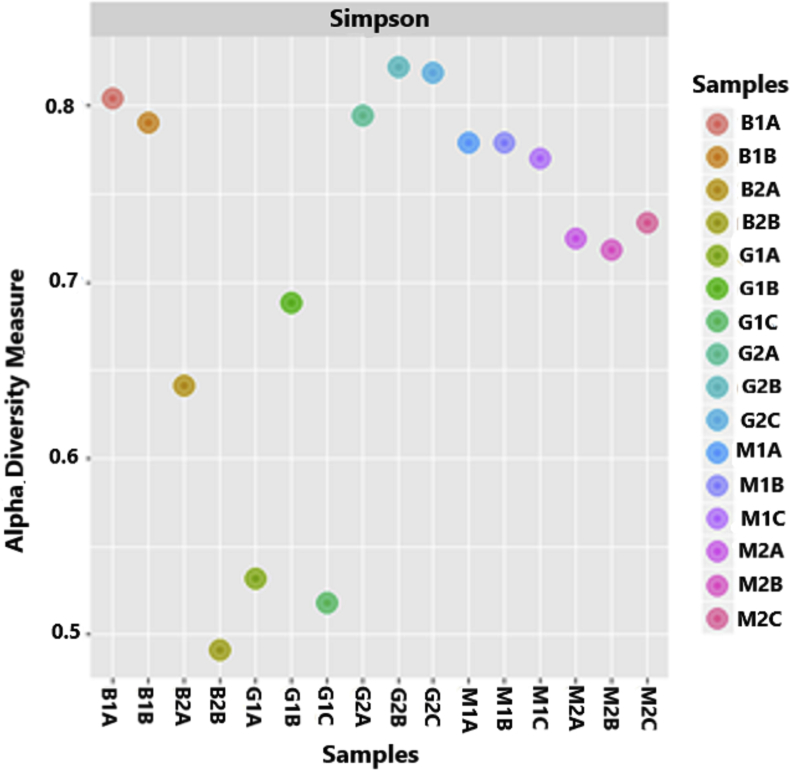
The taxonomic identity of groups found in the planktonic habitats (M1, preheating and M2, postheating), tank biofilm habitats (B1, preheating and B2, postheating), and flask biofilm habitats (G1, preheating and G2, postheating), might indicate changes in the bacterial population structures that adapt to the stress induced by the physicochemical conditions and contamination. Sample coverage data in terms of the Simpson indices from the 16S rRNA gene sequencing and diversity calculations are shown in Figure 2.
Figure 2.
Measurements of alpha diversity in the triplicate planktonic samples (M1A/M1B/M1C and M2A/M2B/M2C) and triplicate biofilm samples from the tanks (B1A/B1B and B2A/B2B) and flasks (G1A/G2B/G2C and G2A/G2B/G2C). Each triplicate was represented by the letters A, B and C placed after the letters and numbers that define the planktonic and biofilm communities.
Simpson's diversity index demonstrates the distribution of alpha diversity (Figure 2). The genera of the planktonic communities (M1/M2) identified based on the obtained sequences are represented in the biofilm communities (B1/B2 and G1/G2), but present different diversity indices depending on selection and adaptation to the environment.
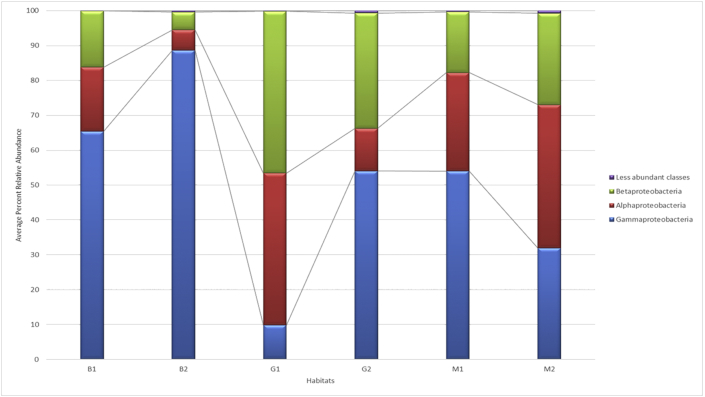
The analyses of the sequences obtained in all the communities studied in this work showed that the most representative classes were Gammaproteobacteria, Betaproteobacteria and Alphaproteobacteria, as shown in Figure 3.
Figure 3.
Abundance of bacterial classes in the biofilm and planktonic samples. B1 and B2: taxonomic units identified from the pre- and postheating biofilms, respectively; M1 and M2: taxonomic units identified from the pre- and postheating planktonic samples, respectively; and G1 and G2: taxonomic units identified from the biofilms that formed in flasks with pre- and postheating water, respectively.
A search of literature on the National Center for Biotechnology Information PubMed website, on February 27, 2020, with the keywords " agrochemical " and “biodegradation”, in the last five years, showed an exclusivity of articles published with these classes, with no articles on this subject in relation to other classes of Proteobacteria. There is a preponderance of studies with Gammaproteobacteria in relation to Betaproteobacteria and Alphaproteobacteria, which reflects the distribution found for these classes in the biofilm communities (Figure 3), indicating the importance of the formation of these structures for adaptation to environments with the presence of xenobiotics. However, the relationship between adaptation to stressful environments and structural and metabolic characteristics needs to be analyzed in more detail, observing the characterization in bacterial families.
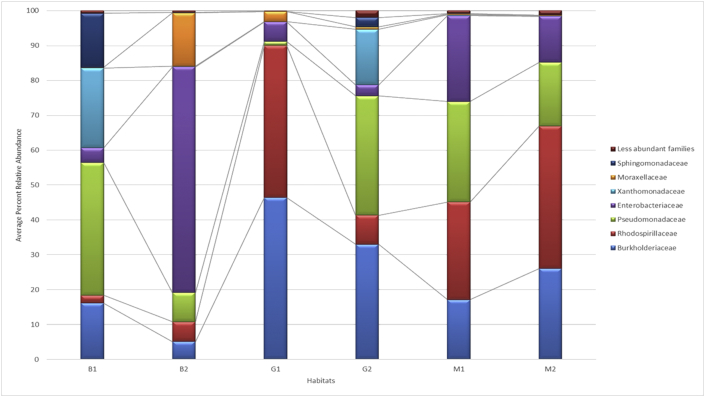
The most representative families, as Sphingomonadaceae, Moraxellaceae, Xanthomonadaceae, Enterobacteriaceae, Pseudomonadaceae, Rhodospirillaceae and Burkholderiaceae, were identified from the sequences obtained in all the communities (Figure 4).
Figure 4.
Abundance of bacterial families in planktonic and biofilm samples. B1 and B2: taxonomic units identified from the pre- and postheating biofilms, respectively; M1 and M2: taxonomic units identified from the pre- and postheating planktonic samples, respectively; and G1 and G2: taxonomic units identified from the biofilms that formed in flasks with pre- and postheating water, respectively.
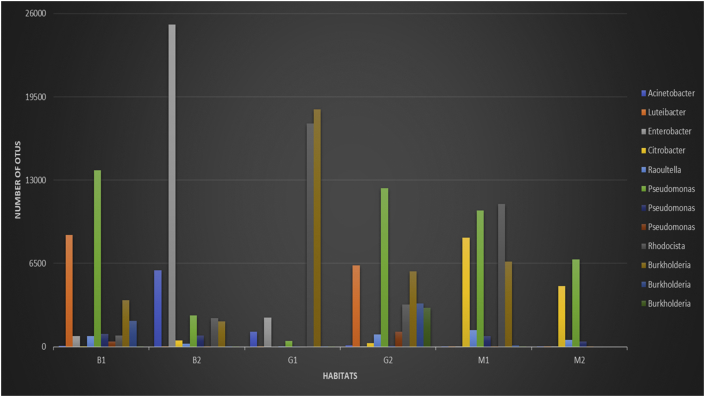
The most representative genus, as Acinetobacter, Luteibacter, Enterobacter, Citrobacter, Raoultella, Pseudomonas, Rhodocista, and Burkholderia, identified from the sequences obtained in all the communities, are shown in Figure 5.
Figure 5.
Most frequent genera identified in planktonic and biofilm samples. M1: preheating planktonic community; M2: postheating planktonic community; B1: preheating biofilm community; B2: postheating biofilm community; G1: preheating container biofilm community; and G2: postheating container biofilm community.
The data shown in Figures 3, 4, and 5 indicates the existence of high variation in the population structure among different communities in relation to classes and families. Water heating is a nondirectional selective process because none of the taxonomic groups identified in this study exhibit resistance responses to high temperatures; however, this process changed the number of taxonomic units, as reflected by decreased diversity in all the communities in the postheating samples, and more intense changes were observed in the biofilm communities than in the planktonic communities. The effect of temperature can be observed in Figure 2 and leads to lower diversity and abundance among the communities that settled in the biofilm structure in the tanks and in the flasks after the heating process. The planktonic community exerts a direct influence on biofilm formation (Hallatschek et al., 2007). The communities are formed in the following chronological order: the community in T1 is formed first, and this community is followed by that in B1 and then that in G1. This chronological order is valid for postheating communities, with the proviso that the T2 community is formed after the T1 community.
A more obvious abundance changes were observed in the very dissimilar biofilm population structures in G1 and G2, which were derived from similar planktonic strains (Figures 2, 3, 4, and 5). This finding was likely obtained because these communities had less time to consolidate adapted population structures through processes of selection and functional redundancy, that probably occurred in B1 and B2. The concept of functional redundancy describes the overlap between functional capacities of populations in a given habitat and their taxonomic composition (Comte et al., 2013).
A lower population structure variation was observed in the planktonic samples than in the biofilm samples in tanks and flasks, which might indicate an association between the genetic drift of individuals who survived the heating process and the subsequent selection of differentiated characteristics that allow individuals to adapt to the tank environment. Water contaminated with 30 different agrochemicals presents physicochemical conditions that are unfavorable to the survival of microorganisms and is thus considered a stressful environment (Tables 1 and Table A.1). The organization of bacteria in biofilms in tanks and flasks, with their cooperative or antagonistic interactions, might lead to a specific combination of species with increased adaptability to the environment (Herschend et al., 2017). Thus, the most dominant sequences in B2, which correspond to the genera Enterobacter and Acinetobacter, are poorly represented in the other communities studied, including the biofilms in B1, G1 and G2 (Figures 2 and 5).
Theoretically, the transition from planktonic to biofilm life involves changes in the regulation of different groups of genes and can be considered a transition phase prior to colonization of a new environment (Berlanga and Guerrero, 2016). In our study, there was probably this kind of transition, since the tanks were filled with water first, after that there was the formation of biofilms. In addition, the same 16S rRNA gene sequences were identified in all communities, even though showing significant changes in the abundance of these sequences (Figures 3, 4, and 5). Throughout the establishment of a community, the most abundant genera tend to retain most of their genes related to basic cellular processes, leading to the survival of individuals of different species that share the same function rather than survival of the fittest (Stephens et al., 2015).
The reduction in diversity observed after water heating possibly led to genetic drift, resulting in alteration of the fine structure of the M2, B2 and G2 communities in relation to that of the M1, B1 and G1 communities and the retention of genes involved in central adaptive processes. If the genetic drift and functional diversity of different bacterial communities were less preponderant, more similar diversity indices would be found between the B1 and B2 communities, the B2 and G2 communities, and G1 and G2 communities (Hallatschek et al., 2007) than those observed in Figure 2. In addition, the data shown in Figure 5 indicate that species belonging to the same genus but containing different 16S rRNA gene sequences might contribute to the different communities observed after adaptation to the environments studied in this work (Łukasik et al., 2018). Nevertheless, these hypotheses were established based on the sequences identified and not the taxonomic units.
Aquatic environments are complex (Angly et al., 2016); therefore, the analysis of changes in the structure of communities of microorganisms during adaptation to environmental contaminants is difficult. According to Zeglin (2015), various studies have revealed the connections among drivers of ecosystem process and microbial diversity in stream environments, and these findings highlight the within-stream spatial differences, such as metal concentrations, and conclude that microorganism taxonomic turnover could mediate ecosystem-scale responses to altered environmental parameters. Confined environments with a high number of agrochemicals, such as those investigated in this study, might allow the safe proposal of fundamental hypotheses that can then be tested at a larger environmental scale.
3.3. Adaptative characteristics of identified genera
The genera that were previously found to show a close relationship to agrochemical biodegradation are the same as those that dominate the biofilm communities B1 and B2. The survey of possible characteristics related to adaptation to the biofilm environment described in this study was related to the genera of the most prevalent bacteria in these communities identified by 16S rRNA gene sequencing (Figures 2 and 5) and supported by Predicted Metagenomics Analysis (section 3.4).
A model was proposed to explain a possible range of characteristics that could confer adaptive advantages over the stressful environment of the water tanks used for washing agrochemical containers. The population fluctuations observed in this study might be the result of genetic drift and the capacity for biofilm formation, which involves membrane transport, the interaction of signaling molecules with signal transduction, and functional bacterial versatility (Roller et al., 2013). The biofilm structure depends on the presence of EPS, which involves the production of glycans and lipid transportation in catabolism. Biofilms confer genomic plasticity and phenotypic flexibility to the species present in the films and thus allow successful reproduction in stressful environments (Decho and Gutierrez, 2017). Enterobacter, Acinetobacter and Pseudomonas genera, belonging to B2 community, have the potential to harbor genes that codify for these characteristics. Our results indicate a high functional similarity in biofilm communities due to environmental uniformity, with strong changes in population structure (B1, B2, G1 and G2 biofilm communities), suggesting functional redundancy in these communities (Comte et al., 2013).
A search on the National Center for Biotechnology Information PubMed website, on October 25, 2019, with the keywords "degradation" or “biofilm” associated with the most frequent genera identified in each community (Figure 5) recovered data that can corroborate the hypotheses presented here. These two characteristics were evaluated as being considered fundamental for the survival of bacteria identified in the stressful environments studied in this work. For the Acinetobacter genus, second most abundant in B2, 5933 articles on degradation and 893 on biofilm were retrieved. Luteibacter is the second most abundant genus in B1 and G2, and 24 articles on degradation and only 1 on biofilm were retrieved. Enterobacter is the most abundant genus in B2 and the third in G1, 5822 articles retrieved on degradation and 254 on biofilm. Citrobacter was an abundant genus in planktonic communities, being the third in M1 and the second in M2, presenting 2438 articles recovered on degradation but only 82 on biofilm, which may explain its low representativeness in biofilm communities. Raoultella was a genus with relative abundance in planktonic communities too, with only 142 articles on degradation and 15 articles on biofilm, not being prevalent in biofilm communities. Pseudomonas was an abundant genus in planktonic and biofilm communities, with the exception of G1, and also with the largest number of articles retrieved, with 52614 on degradation and 7228 on biofilm, showing its adaptive potential for different environmental conditions. Rhodocista was the second most abundant genus in G1 and the first in M1, with only 10 articles on degradation and 1 on biofilm, showing a relatively low adaptive potential. Burkholderia, a genus phylogenetically close to Pseudomonas, was abundant in both planktonic and biofilm communities, with 4091 articles on degradation and 494 on biofilm, also showing, like Pseudomonas, its functional versatility.
Thus, the more plastic genera, such as Pseudomonas and Burkholderia could remain abundant in most communities studied. For biofilm communities it seems that there was a selection for genera, probably with functional complementarity, or for degradation, or biofilm, or both. However, only after a possible event of genetic drift and selection for a longer time, the B2 community was composed with more abundance of genera with characteristics that theoretically would allow a better adaptation to the studied environments: Enterobacter, Acinetobacter and Pseudomonas.
The availability of nutrients, even if xenobiotics, has an important role in determining the population structure, as may have been the case of agrochemicals as possible sources of nutrients for the microbiota identified in T1 and T2 and also for some environments mentioned in Table 2. This is the case of bacteria and archaea that employ a variety of strategies to minimize cellular demand for limiting nutrients in the interior transition zone of the ocean (Mende et al., 2017). Bacterial community structure is enriched with hydrocarbon-degrading bacteria in the shallow zone with higher risk of large-scale oil spills at southwest part of the Gulf of Mexico (Godoy-Lozano et al., 2018). Clostridia, Betaproteobacteria, Bacteroidia, Deltaproteobacteria and Gammaproteobacteria are the dominant bacterial classes implicated in the removal of various carbon containing pollutants of natural and synthetic origins in the system of anaerobic-aerobic reactors for the treatment of WW in Ethiopia (Desta et al., 2014).
However, only the composition of species fails to explain the adaptive potential of microorganism communities to stressful environments, as is the case of the lack of tolerance to high temperatures presented by the predominant classes in communities M2, B2 and G2. This is also true for the marine microbial communities across environmental gradients in the Red Sea, in which one-quarter of taxonomic and functional variation could not be explained by temperature, nitrate and chlorophyll (Thompson et al., 2017). In hot water spring in Siloam, South Africa, was not found thermophiles as the most abundant organisms, but the genera Stenotrophomonas, Zavarzinella and Aquaspirillum (Tekere et al., 2011). The work of Jang et al. (2014) among Escherichia coli populations from Yeongsan River Basin of South Korea has shown that strains of this species could be clustered based on their genotypes and environment conditions, and the distributional differences of phylogenetic groups in different environments may be caused by different genomic adaptability and plasticity. The discussions presented in these articles alongside our results are compatible with the hypotheses of functional redundancy (Comte et al., 2013) and functional stability of communities under environmental stress, even with changes in the population structure (Jiang et al., 2020).
The higher diversity in the planktonic community compared with the biofilm community (Figure 2) has good potential to lead to species fixation over time, even if the less efficient species become extinct. Some mutations might be randomly lost by genetic drift; however, significant advantageous mutations are more likely to undergo fixation in the population. A strong selection pressure in a community increases the frequencies of genotypes that improve competence in the population (Brooks et al., 2011). This model can be applied for mutations in populations with one species or populations of different species but with gene flow potential, as might occur in the B2 community.
The selection of different taxonomic groups but with redundant functions suitable for survival in stressful environments, as may have occurred in the B2 community, is similar to that reported by Yao et al. (2019), who compared giant panda gut microbiomes by 16S rRNA gene sequencing and metagenomics analysis. In this article was showed a large variation in bacterial population structure, even with similarity in animal diets. Dietary conditions, such as the presence of toxins, may have led to selective pressure in favor of distinct bacterial genera with redundant functions important for survival in the panda's intestinal environment, such as cellulose and xenobiotic degradation. Jiang et al. (2020) described the hypothesis of functional stability in soil microbial communities under long-term stress of heavy metals and subsequent herbicide siduron. The results show that the alpha diversity and composition of microbial communities are affected significantly after these treatments but does not appear to impair the functions of microbial communities or their functional stability. The authors conclude that the structural composition of soil microbial community is more important than the diversity of microbial species to explain the functional stability of microbial communities under chemical stresses, and that a number of soil properties and the disturbance history also play a decisive role.
The environments studied in this work underwent a sequence of physicochemical changes involving chemical contamination and heating over six months. During these events, the genera with the capacity to develop useful functions (Roller et al., 2013) for biofilm consolidation, such as Enterobacter, Acinetobacter and Pseudomonas, dominated the B2 community (Hallatschek et al., 2007; Komarova, 2014), which was formed after the M1, M2 and B1 communities by selection and probably by genetic drift. The genera identified in this study have agrochemical degradation potential and the capacity for structuring biofilm communities, as shown literature review, which suggests the viability of searching for promising strains among the culturable collection obtained in this study. The high temperature is considered bactericidal for all planktonic genera identified in this work (World Health Organization, 2011). Therefore, the nondirectional selective effect induced by high temperature was independent of the genomes of the identified genera.
Environmental biofilm communities have higher metabolic complexity than those of their individual components (Flemming and Wuertz, 2019). Thus, the study of biofilm formation in environments contaminated with pesticides, which can be stressful to microorganisms, can potentially provide information regarding the systems involved in adaptation. This information could provide a strategy for exploiting enzymatic potentials, particularly those related to the biodegradation and bioremediation of herbicides (Berlanga and Guerrero, 2016). The predictable metagenomics by the identification of bacterial diversity via sequencing of the 16S rRNA gene and the identification of the genes and their functions linked to these sequences can allow the creation of a theoretical model in which the most appropriate biotechnological use can be predicted for the different communities of an environment under stress. These studies might provide knowledge that can be used to exploit the enzymatic potential of bacterial communities under these environmental conditions, particularly that related to the biodegradation kinetics of herbicides in reservoirs containing herbicide-contaminated wastewater. Based on these results, a research program has been started with the construction of biofilters composed of different matrices colonized by bacterial communities from tanks biofilms, specifically postheating, with the objective of bioremediation of the washing water of herbicide containers, an important environmental liability.
3.4. Predicted Metagenomics Analysis
The comparative data of clustering 16S rRNA genes are shown in Figure 6. This analysis was carried out as a survey of biotechnological potential (Imfeld and Vuilleumier, 2012; Kroeger et al., 2018) mainly related to biodegradation in the storage tanks of water contaminated with agrochemicals, which are considered a significant environmental liability.
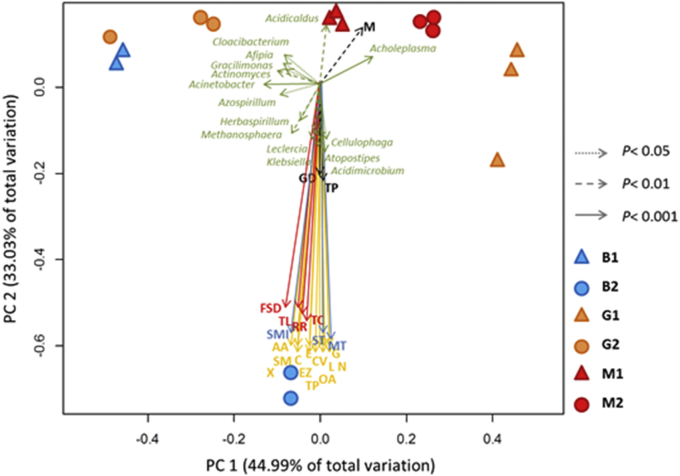
Figure 6.
Ordering the sites based on OTU abundance by multidimensional scaling. Only the bacterial genus (green), genetic process (red), cellular process (black), environmental process (blue) and metabolic pathways (yellow) with p < 0.05 after 999 permutations are displayed. Each vector shows the direction of increase for a given variable, and its length indicates the strength of the correlation between the variable and the ordination scores. Legend: M: Motility; GD: Growth and Death; TP: Transportation in Catabolism; MT: Membrane Transport; SMI: Signaling Molecule Interaction; ST: Signal Transduction; TL: Translation; TC: Transcription; RR: Replication and Repair; FSD: Folding, Sorting and Degradation; AA: Amino Acid; SM: Secondary Metabolites; C: Carbohydrate; E: Energy; EZ: Enzyme; G: Glycan; L: Lipid; CV: Cofactors and Vitamins; OA: Other Amino Acids; TP: Terpenoids and Polyketides; N: Nucleotide; XB: Xenobiotic Biodegradation.
Nonmetric multidimensional scaling (NMDS) shows the correlations between the number and types of 16S rRNA gene sequences grouped by the 6 different communities. Genes related to bacterial motility have a high correlation with the two planktonic communities M1 and M2 (Table 3).
Table 3.
Correlations of the functional characteristics identified in our analyses of the studied communities. M1: preheating planktonic community; M2: postheating planktonic community; B2: postheating biofilm community; r2: coefficient of determination; p: coefficient of significance.
| Functional characteristics |
r2 |
P |
|---|---|---|
| Communities M1 and M2 | ||
| Motility | 0.6196 | <0.002 |
| Community B2 | ||
| Cellular processes | ||
| Growth and death | 0.8670 | <0.001 |
| Transportation in catabolism | 0.8977 | <0.001 |
| Environmental processes | ||
| Membrane transport | 0.9827 | <0.001 |
| Interaction of signaling molecules | 0.9422 | <0.001 |
| Signal transduction | 0.9838 | <0.001 |
| Genetic information processes | ||
| Translation | 0.9880 | <0.001 |
| Transcription | 0.9863 | <0.001 |
| Repair and replication | 0.9887 | <0.001 |
| Folding, sorting and degradation | 0.9888 | <0.001 |
| Metabolic processes | ||
| Enzymes | 0.9838 | <0.001 |
| Amino acids | 0.9872 | <0.001 |
| Secondary metabolites | 0.9883 | <0.001 |
| Carbohydrates | 0.9874 | <0.001 |
| Energy | 0.9864 | <0.001 |
| Glycans | 0.9825 | <0.001 |
| Lipids | 0.9850 | <0.001 |
| Cofactors and vitamins | 0.9872 | <0.001 |
| Other amino acids | 0.9883 | <0.001 |
| Terpenoids, polyketides | 0.9845 | <0.001 |
| Nucleotides | 0.9865 | <0.001 |
| Xenobiotic biodegradation | 0.9402 | <0.001 |
On the other hand, genes associated with other cellular processes, environmental processes, gene information processes and metabolic processes have a strong correlation with the biofilm community B2 (Table 3), which is predominantly composed by the genera and Enterobacter, Acinetobacter and Pseudomonas.
4. Conclusions
In this work, bacterial communities were characterized based on their population structure and functional profile in a type of environment that was define as stressful. The communities were characterized as tank biofilm, flask biofilm, and planktonic communities, and all of these communities were obtained from the pre- and postheating samples and formed at different times. The water heating process might have induced a genetic drift phenomenon, which decreased the diversity of the bacterial biofilm community in the planktonic communities. The genera Enterobacter, Acinetobacter and Pseudomonas, which were dominant in the tank biofilm community after water heating, have metabolic potential for survival in this environment, even in the presence of inadequate levels of chlorine, fluorine and agrochemicals, probably due to redundancy in functions related to biofilm formation and structure, membrane transport, quorum sensing and xenobiotic degradation, well-documented characteristics in the genera described in this paper. Biofilm bacterial communities obtained under these environmental conditions constitute interesting models for studying the kinetics of herbicide biodegradation and the prospects of strains suitable for use in bioremediation in reservoirs containing herbicide-contaminated wastewater, as the construction of biofilters containing postheating biofilm communities capable of degrading xenobiotics.
Declarations
Author contribution statement
Jhenifer Yonara Lima: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Cassiano Moreira, Paloma Nathane Nunes Freitas: Performed the experiments.
Luiz Ricardo Olchanheski: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Sonia Alvim Veiga Pileggi, Rafael Mazer Etto: Analyzed and interpreted the data; Wrote the paper.
Christopher Staley: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Michael Jay Sadowsky: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Marcos Pileggi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Coordination for the Improvement of Higher Level Personnel (CAPES), the National Council of Technological and Scientific Development (CNPq), and the Foundation for Research Support of the State of Paraná (Fundação Araucária).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors thank Maria Janina Pinheiro Diniz and Gessica da Costa for assisting in the preparation and execution of the experiments, and Bruno César do Espírito Santo for assistance with the resolution of the figures.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Table A1
References
- Al-Amoudi S., Razali R., Essack M., Amini M.S., Bougouffa S., Archer J.A.C. Metagenomics as a preliminary screen for antimicrobial bioprospecting. Gene. 2016;594:248–258. doi: 10.1016/j.gene.2016.09.021. [DOI] [PubMed] [Google Scholar]
- Angly F.E., Heath C., Morgan T.C., Tonin H., Rich V., Schaffelke B. Marine microbial communities of the Great Barrier Reef lagoon are influenced by riverine floodwaters and seasonal weather events. PeerJ. 2016;4 doi: 10.7717/peerj.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anutrakunchai C., Bolscher J.G.M., Krom B.P., Kanthawong S., Chareonsudjai S., Taweechaisupapong S. Impact of nutritional stress on drug susceptibility and biofilm structures of Burkholderia pseudomallei and Burkholderia thailandensis grown in static and microfluidic systems. PloS One. 2018;13 doi: 10.1371/journal.pone.0194946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E. Comparison of sequencing utility programs. Open Bioinf. J. 2013;7:1–8. [Google Scholar]
- Battin T.J., Besemer K., Bengtsson M.M., Romani A.M., Packmann A.I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 2016;14:251–263. doi: 10.1038/nrmicro.2016.15. [DOI] [PubMed] [Google Scholar]
- Berlanga M., Guerrero R. Living together in biofilms : the microbial cell factory and its biotechnological implications. Microb. Cell Factories. 2016:1–11. doi: 10.1186/s12934-016-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks A.N., Beer K.D., Turkarslan S., Baliga N.S., Li F.Y. Adaptation of cells to new environments. Wiley Interdiscip Rev Syst Biol Med. 2011;3:544–561. doi: 10.1002/wsbm.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., He S., Liang Z., Li Q.X., Yan H., Hu J. Biodegradation of pyraclostrobin by two microbial communities from Hawaiian soils and metabolic mechanism. J. Hazard Mater. 2018;354:225–230. doi: 10.1016/j.jhazmat.2018.04.067. [DOI] [PubMed] [Google Scholar]
- Chen W., Wilkes G., Khan I.U.H., Pintar K.D.M., Thomas J.L., Lévesque C.A., Chapados J.T., Topp E., Lapen D.R. Aquatic bacterial communities associated with land use and environmental factors in agricultural landscapes using a metabarcoding approach. Front. Microbiol. 2018;9:2301. doi: 10.3389/fmicb.2018.02301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comte J., Fauteux L., Giorgio P.A. Links between metabolic plasticity and functional redundancy in freshwater bacterioplankton communities. Front. Microbiol. 2013;4:112. doi: 10.3389/fmicb.2013.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decho A.W., Gutierrez T. Microbial extracellular polymeric substances (EPSs) in ocean systems. Front. Microbiol. 2017;8:1–28. doi: 10.3389/fmicb.2017.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Deng C., Yang J., Li B., Wang E. Novel Butane-Oxidizing bacteria and diversity of bmoX genes in Puguang gas field. Front. Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.01576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P.G., Kukulies T., Forstner C. The effects of glyphosate, glufosinate, paraquat and paraquat-diquat on soil microbial activity and bacterial, archaeal and nematode diversity. Sci. Rep. 2018;8:2119. doi: 10.1038/s41598-018-20589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desta A.F., Assefa F., Leta S., Stomeo F., Wamalwa M., Njahira M. Microbial community structure and diversity in an integrated system of anaerobic-aerobic reactors and a constructed wetland for the treatment of tannery wastewater in Modjo, Ethiopia. PloS One. 2014;9:1–22. doi: 10.1371/journal.pone.0115576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzanski T., Gravina F., Steckling B., Olchanheski L.R., Sprenger R.F., Espírito Santo B.C. Bacillus megaterium strains derived from water and soil exhibit differential responses to the herbicide mesotrione. PloS One. 2018;13:1–24. doi: 10.1371/journal.pone.0196166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea T.C., Silva R., Boscolo M., Rigonato J., Monteiro D.A., Grünig D., Silva H., Wielen F., Helmus R., Parsons J.R., Gomes E. Diuron degradation by bacteria from soil of sugarcane crops. Heliyon. 2017;3(12) doi: 10.1016/j.heliyon.2017.e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017 doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- Flemming H.C., Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- Gholizadeh M.H., Melesse A.M., Reddi L. A comprehensive review on water quality parameters estimation using remote sensing techniques. Sensors (Switzerland) 2016;16 doi: 10.3390/s16081298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy-Lozano E.E., Escobar-Zepeda A., Raggi L., Merino E., Gutierrez-Rios R.M., Juarez K. Bacterial diversity and the geochemical landscape in the southwestern Gulf of Mexico. Front. Microbiol. 2018;9:1–15. doi: 10.3389/fmicb.2018.02528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl D.M., Vangay P., Garbe J., MacLean A., Hauge A., Becker A. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016;34:942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- Gutarowska B., Celikkol-Aydin S., Bonifay V., Otlewska A., Aydin E., Oldham A.L. Metabolomic and high-throughput sequencing analysis-modern approach for the assessment of biodeterioration of materials from historic buildings. Front. Microbiol. 2015;6:1–13. doi: 10.3389/fmicb.2015.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O., Hersen P., Ramanathan S., Nelson D.R. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl. Acad. Sci. 2007;104:19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschend J., Damho Z.B.V., Marquard A.M., Sven B., Sørensen S.J., Hä P. 2017. A Meta-Proteomics Approach to Study the Interspecies Interactions Affecting Microbial Biofilm Development in a Model Community; pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse S.M., Welch D.M., Morrison H.G., Sogin M.L. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imfeld G., Vuilleumier S. Measuring the effects of pesticides on bacterial communities in soil: a critical review. Eur. J. Soil Biol. 2012;49:22–30. [Google Scholar]
- Jang J., Di D.Y.W., Lee A., Unno T., Sadowsky M.J., Hur H.G. Seasonal and genotypic changes in Escherichia coli phylogenetic groups in the Yeongsan River basin of South Korea. PloS One. 2014;9:5–12. doi: 10.1371/journal.pone.0100585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., Rhoads W.J., Edwards M.A., Pruden A. Impact of water heater temperature setting and water use frequency on the building plumbing microbiome. ISME J. 2017;11:1318–1330. doi: 10.1038/ismej.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Wang M., Chen W., Li X., Balseiro-Romero M. Changes in the integrated functional stability of microbial community under chemical stresses and the impacting factors in field soils. Ecol. Indicat. 2020;110:105919. [Google Scholar]
- Komarova N.L. Spatial interactions and cooperation can change the speed of evolution of complex phenotypes. Proc. Natl. Acad. Sci. 2014;111:10789–10795. doi: 10.1073/pnas.1400828111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger M.E., Delmont T.O., Eren A.M., Meyer K.M., Guo J., Khan K. New biological insights into how deforestation in Amazonia affects soil microbial communities using metagenomics and metagenome-assembled genomes. Front. Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn-Molt I., Alawi M., Förstner K.U., Wiegandt A., Burkhardt L., Indenbirken D. Insights into Microalga and bacteria interactions of selected phycosphere biofilms using metagenomic, transcriptomic, and proteomic approaches. Front. Microbiol. 2017;8:1–14. doi: 10.3389/fmicb.2017.01941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M.G.I., Clemente J.C., Knight R., Burkepile D.E., Caporaso J.G., McDonald D. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch T.Z., Chenu C., Dignac M.F., Barriuso E., Mariotti A. Biofilm vs. Planktonic lifestyle: consequences for pesticide 2,4-D metabolism by Cupriavidus necator JMP134. Front. Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewenza S., Abboud J., Poon K., Kobryn M., Humplik I., Bell J.R. 2018. Pseudomonas aeruginosa Displays a Dormancy Phenotype during Long-Term Survival in Water; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan A., Sindhu R., Parameswaran B., Sukumaran R.K., Pandey A. Metagenome analysis: a powerful tool for enzyme bioprospecting. Appl. Biochem. Biotechnol. 2017;183:636–651. doi: 10.1007/s12010-017-2568-3. [DOI] [PubMed] [Google Scholar]
- Martins P.F., Martinez C.O., Carvalho G De, Irajara P., Carneiro B., Azevedo R.A. Selection of microorganisms degrading S-Metolachlor herbicide. 2007;50:153–159. [Google Scholar]
- Maunders E., Welch M. Matrix exopolysaccharides; the sticky side of biofilm formation. FEMS Microbiol. Lett. 2017;364:1–10. doi: 10.1093/femsle/fnx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende D.R., Bryant J.A., Aylward F.O., Eppley J.M., Nielsen T., Karl D.M. Environmental drivers of a microbial genomic transition zone in the ocean’s interior. Nat. Microbiol. 2017;2:1367–1373. doi: 10.1038/s41564-017-0008-3. [DOI] [PubMed] [Google Scholar]
- Ofaim S., Ofek-Lalzar M., Sela N., Jinag J., Kashi Y., Minz D. Analysis of microbial functions in the rhizosphere using a metabolic-network based framework for metagenomics interpretation. Front. Microbiol. 2017;8:1–14. doi: 10.3389/fmicb.2017.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olchanheski L.R., Dourado M.N., Beltrame F.L., Zielinski A.A.F., Demiate I.M., Pileggi S.A.V. Mechanisms of tolerance and high degradation capacity of the herbicide mesotrione by Escherichia coli strain DH5-α. PloS One. 2014;9:1–8. doi: 10.1371/journal.pone.0099960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson G.M., Gao H., Meyer B.M., Miles M.S., Overton E.B. Effect ofCorexit 9500A on MississippiCanyon crude oil weatheringpatterns using artificial andnatural seawater. Heliyon. 2017;3 doi: 10.1016/j.heliyon.2017.e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oueriaghli N., Castro D.J., Llamas I., Béjar V., Martínez-Checa F. Study of bacterial community composition and correlation of environmental variables in Rambla Salada, a hypersaline environment in South-Eastern Spain. Front. Microbiol. 2018;9:1–17. doi: 10.3389/fmicb.2018.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prione L.P., Olchanheski L.R., Tullio L.D., Santo B.C.E., Reche P.M., Martins P.F. GST activity and membrane lipid saturation prevents mesotrione-induced cellular damage in Pantoea ananatis. AMB Express. 2016;6:1–12. doi: 10.1186/s13568-016-0240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W.G., Peplies J. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R., Rani A., Metwally A., McGee H.S., Perkins D.L. Vol. 469. Elsevier Ltd; 2016. (Analysis of the Microbiome: Advantages of Whole Genome Shotgun versus 16S Amplicon Sequencing). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller M., Lucić V., Nagy I., Perica T., Vlahoviček K. Environmental shaping of codon usage and functional adaptation across microbial communities. Nucleic Acids Res. 2013;41:8842–8852. doi: 10.1093/nar/gkt673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva T.M., Stets M.I., Mazzetto A.M., Andrade F D., Pileggi S.A.V., Fávero P.R. Degradation of 2,4-D herbicide by microorganisms isolated from Brazilian contaminated soil. Braz. J. Microbiol. 2007;38:522–525. [Google Scholar]
- Staley C., Gould T.J., Wang P., Phillips J., Cotner J.B., Sadowsky M.J. Evaluation of water sampling methodologies for amplicon-based characterization of bacterial community structure. J. Microbiol. Methods. 2015;114:43–50. doi: 10.1016/j.mimet.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Stephens W.Z., Wiles T.J., Martinez E.S., Jemielita M., Burns A.R., Parthasarathy R. Identification of population bottlenecks and colonization factors during assembly of bacterial communities within the zebrafish intestine. mBio. 2015;6:1–11. doi: 10.1128/mBio.01163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storck V., Nikolaki S., Perruchon C., Chabanis C., Sacchi A., Pertile G. Lab to field assessment of the ecotoxicological impact of chlorpyrifos, isoproturon, or tebuconazole on the diversity and composition of the soil bacterial community. Front. Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.01412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G. Structure and function of the global. Ocean Microbiom. 2015;348:1–10. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- Tekere M., Lötter A., Olivier J., Jonker N., Venter S. Metagenomic analysis of bacterial diversity of Siloam hot water spring, Limpopo, South Africa. Afr. J. Biotechnol. 2011;10:18005–18012. [Google Scholar]
- Thompson L.R., Williams G.J., Haroon M.F., Shibl A., Larsen P., Shorenstein J. Metagenomic covariation along densely sampled environmental gradients in the Red Sea. ISME J. 2017;11:138–151. doi: 10.1038/ismej.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeriani F., Margarucci L.M., Gianfranceschi G., Ciccarelli A., Tajani F., Mucci N., Ripani M., Spica V.R. Artificial-turf surfaces for sport and recreational activities: microbiota analysis and 16S sequencing signature of synthetic vs natural soccer fields. Heliyon. 2019;5(8) doi: 10.1016/j.heliyon.2019.e02334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde J.R., Marín S., Mellado R.P. Effect of herbicide combinations on bt-maize rhizobacterial diversity. J. Microbiol. Biotechnol. 2014;24:1473–1483. doi: 10.4014/jmb.1405.05054. [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hatt J.K., Tsementzi D., Rodriguez -R.L.M., Ruiz-Pérez C.A., Weigand M.R., Kizer H., Maresca G., Krishnan R., Poretsky R., Spain J.C., Konstantinidis K.T. Quantifying the importance of the rare biosphere for microbial community response to organic pollutants in a freshwater ecosystem. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.03321-16. e03321-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . fourth ed. World Heal Organ; Geneva: 2011. Guidelines for Drinking-Water Quality; p. 340. WHO. [Google Scholar]
- Yao R., Yang Z., Zhang Z., Hu T., Chen H., Huang F., Gu X., Yang X., Lu G., Zhu L. Are the gut microbial systems of giant pandas unstable? Heliyon. 2019;5(9) doi: 10.1016/j.heliyon.2019.e02480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeglin L.H. Stream microbial diversity in response to environmental changes: review and synthesis of existing research. Front. Microbiol. 2015;6:454. doi: 10.3389/fmicb.2015.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Li Y., Yue B., Wu M. Genes as early responders regulate quorum-sensing and control bacterial cooperation in Pseudomonas aeruginosa. PloS One. 2014;9:1–8. doi: 10.1371/journal.pone.0101887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., He Z., Yang Y., Deng Y., Tringe S.G., Alvarez-cohen L. High-throughput metagenomic technologies for complex microbial community analysis: Open and closed formats. CEUR Workshop Proc. 2015;1542:33–36. doi: 10.1128/mBio.02288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łukasik P., Nazario K., Van Leuven J.T., Campbell M.A., Meyer M., Michalik A. Multiple origins of interdependent endosymbiotic complexes in a genus of cicadas. Proc. Natl. Acad. Sci. 2018;115:E226–E235. doi: 10.1073/pnas.1712321115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A1