Abstract
Spermatogenesis depends on precise epigenetic and genetic regulation of spermatogonial stem cells (SSCs). However, it remains largely unknown about the roles and mechanisms of small noncoding RNA in regulating the self-renewal and apoptosis of human SSCs. Notably, we have found that Homo sapiens-microRNA (hsa-miR)-1908-3p is expressed at a higher level in human spermatogonia than pachytene spermatocytes. MiR-1908-3p stimulated cell proliferation and DNA synthesis of the human SSC line. Allophycocyanin (APC) Annexin V and propidium iodide staining, determined by flow cytometric analysis and TUNEL assays, showed that miR-1908-3p inhibited early and late apoptosis of the human SSC line. Furthermore, Kruppel-like factor 2 (KLF2) was predicted and verified as the target of miR-1908-3p, and, significantly, KLF2 silencing resulted in the increase of proliferation and DNA synthesis, as well as reduction of apoptosis of the human SSC line. Moreover, KLF2 silencing ameliorated the decrease in the proliferation and DNA synthesis and the enhancement in the apoptosis of the human SSC line caused by miR-1908-3p inhibition. Collectively, these results implicate that miR-1908-3p stimulates the self-renewal and suppresses the apoptosis of human SSCs by targeting KLF2. This study thus provides a novel epigenetic regulatory mechanism underlying the fate determinations of human SSCs, and it offers new endogenous targets for treating male infertility.
Keywords: hsa-miR-1908-3p, human spermatogonial stem cells, self-renewal, apoptosis, KLF2
Graphical Abstract
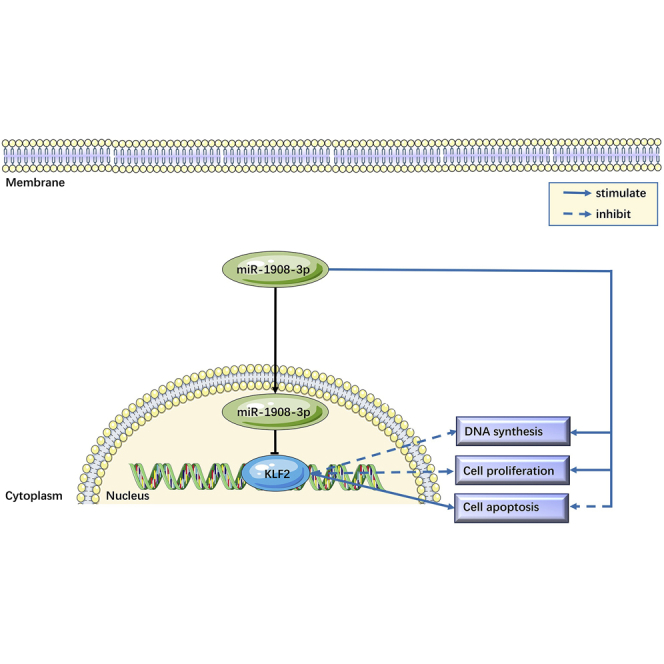
This study has demonstrated that miR-1908-3p stimulates the self-renewal and suppresses the apoptosis of human spermatogonial stem cells (SSCs) by targeting KLF2, which provides a novel epigenetic regulatory mechanism underlying the fate determinations of human SSCs and offers new targets for the diagnosis and treatment of male infertility.
Introduction
Abnormality in spermatogenesis results in male infertility, which seriously affects human reproduction and health. It has been estimated that infertility affects about 15% of the couples worldwide,1 and the male factor accounts for around one-half of this disease. Infertility rates are higher in Africa and Central/Eastern Europe. Additionally, rates of male infertility in North America, Australia, and Central and Eastern Europe vary from 5% to 6%, 9%, and 8% to 12%, respectively.2 A comprehensive meta-regression analysis indicates the significant decline in sperm counts between 1973 and 2011 from North America, Europe, Australia, and New Zealand.3 Therefore, male infertility has become a major disease that seriously affects the human population. Moreover, this problem is gradually increasing, due to environmental pollution, unhealthy lifestyle, late marriage, and late childbearing.4,5
Spermatogenesis refers to the process by which spermatogonial stem cells (SSCs) self-renew and differentiate into mature sperm. Abnormal spermatogenesis may cause azoospermia, which eventually leads to male infertility.6 Human SSCs transmit genetic information to the next generations,7,8 which are the basis of male fertility, and their self-renewal and apoptosis are essential for maintaining human spermatogenesis.9 Increasing evidence has indicated the importance of small noncoding RNA (ncRNA) in male infertility.10,11 MicroRNAs (miRNAs) are endogenous ncRNAs of 18 to 25 nt in length, and they regulate mRNA stability at the post-transcriptional level by binding to the 3′ UTR (3′ untranslated region) of the target genes.12 Drosha and Dicer1 are key enzymes in miRNA biogenesis and maturation, and their mutation may lead to abnormal spermatogenesis and male infertility.13,14 Notably, human SSCs can obtain pluripotency to become embryonic stem (ES)-like cells that differentiate into all cell types of three germ layers, and they are able to transdifferentiate to cells of other lineages,15, 16, 17, 18, 19 reflecting that human SSCs have significant implication in the fields of regenerative medicine.
It has been shown that miRNAs have important regulatory effect on cell proliferation, differentiation, and apoptosis,20, 21, 22, 23 and they mediate rodent spermatogenesis.24 It has been reported that miRNA (miR)-21 affects mouse spermatogonial stem cell self-renewal by regulating Ets variant gene 5 (Ev5) to maintain the pool of stem cells.25 MiR-544 and miR-204 negatively regulate the self-renewal of goat male germline stem cells by inhibiting promyelocytic leukemia zinc finger (PLZF) and Sirt1.26,27 It has been implied that the fate determinations of mouse and human SSCs are subject to the regulation of multiple miRNAs.
Nevertheless, the roles and mechanisms of miRNAs in regulating human SSCs remain largely unknown. We have recently compared large-scale differential expression profiles of miRNAs in human spermatogonia and pachytene spermatocytes, and we found that 110 miRNAs were significantly differentially expressed between the above two types of cells, indicating that these miRNAs may regulate the self-renewal and differentiation of human spermatogonia. Among them, the expression level of Homo sapiens (hsa)-miR-1908-3p in human spermatogonia was significantly higher than pachytene spermatocytes, which suggests that hsa-miR-1908-3p may play an important role in fate decisions of human SSCs. Therefore, this study was designed to explore the function and target of hsa-miR-1908-3p in regulating self-renewal and apoptosis of human SSCs, which will provide a new epigenetic regulation mechanism underlying human spermatogenesis.
Results
hsa-miR-1908-3p Is Abundantly Expressed in the Human SSC Line and Human Primary SSCs
To find the interesting miRNAs that are involved in the fate determinations of human SSCs, we performed an miRNA microarray showing that hsa-miR-1908-3p was expressed at a higher level in human spermatogonia than pachytene spermatocytes (Table S1). Real-time PCR illustrated that the relative level of miR-1908-3p in the human SSC line was abundant compared to other miRNAs, including miR-1908-5p, miR-1469, and miR-1224-5p (Figure S1A). We isolated human THY1-positive spermatogonia, i.e., human SSCs, from testicular tissues of obstructive azoospermia (OA) patients using a two-step enzymatic digestion and magnetic-activated cell sorting (MACS), and we found that hsa-miR-1908-3p was expressed at a high level in these cells (Figure S1B). Together, these data reflect that hsa-miR-1908-3p may play a critical role in regulating the fate determinations of human SSCs.
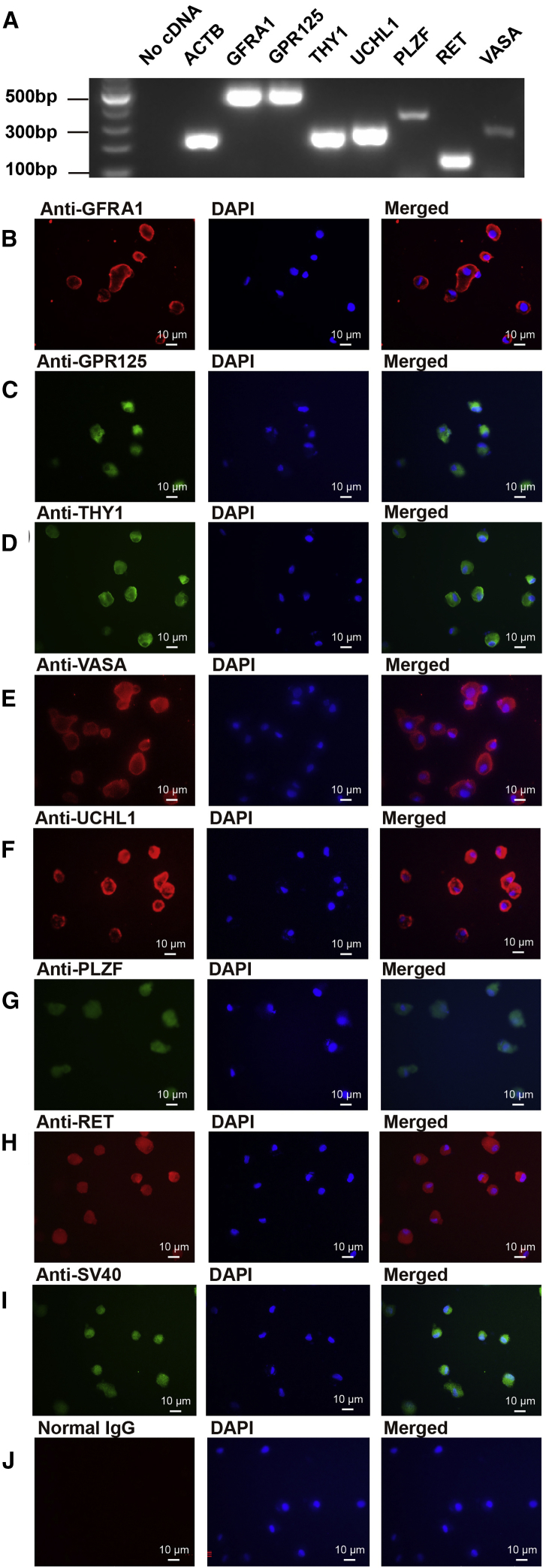
In order to explore the biological function of hsa-miR-1908-3p in human SSCs, we used the human SSC line as working cells, since this cell line possesses the features of human primary SSCs in vivo and in vitro and can proliferate in culture to obtain sufficient cells for our research.28 For the identification of the human SSC line, reverse transcription (RT)-PCR showed that a number of hallmarks for human SSCs and germ cells, including GFRA1, GPR125, THY1, UCHL1, PLZF, RET, and VASA, were detected in this cell line (Figure 1A). Immunocytochemistry further revealed that GFRA1 (Figure 1B), GPR125 (Figure 1C), THY1 (Figure 1D), VASA (Figure 1E), UCHL1 (Figure 1F), PLZF (Figure 1G), and RET (Figure 1H) proteins were present in these cells. Additionally, SV40 protein was found in this cell line (Figure 1I), whereas no immunostaining was observed when primary antibodies were replaced with isotype immunoglobulin G (IgG) (Figure 1J), thus verifying the specific immunostaining of the antibodies mentioned above. Collectively, these data implicate that the human SSC line is human SSCs phenotypically.
Figure 1.
Identification of the Human SSC Line
(A) RT-PCR revealed gene expression of GFRA1, GPR125, THY1, UCHL1, PLZF, RET, and VASA in the human SSC line. RNA, without RT but with PCR, was used as a negative control, and ACTB served as the loading control of total RNA. (B–J) Immunocytochemistry showed the expression of GFRA1 (B), GPR125 (C), THY1 (D), VASA (E), UCHL1 (F), PLZF (G), RET (H), SV40 (I), and isotype IgGs (J) in the human SSC line. (B–J) Scale bars, 10 μm.
hsa-miR-1908-3p Stimulates the Proliferation of the Human SSC Line
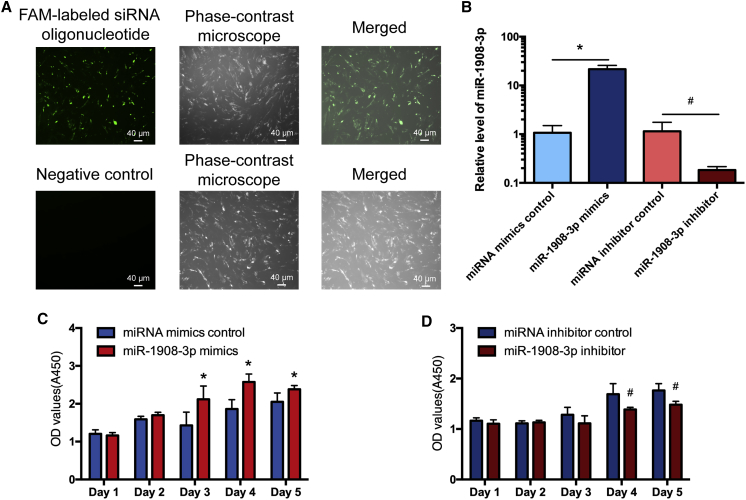
We next explored whether miR-1908-3p contributes to the proliferation and DNA synthesis of the human SSC line. Fluorescein amidite (FAM)-labeled small interfering RNA (siRNA) oligonucleotides showed that the transfection efficiency of miRNAs was over 80% (Figure 2A). Real-time PCR demonstrated that the level of miR-1908-3p was enhanced by miR-1908-3p mimics in the human SSC line compared to miRNA mimics control (Figure 2B), and conversely, its level was decreased by the miR-1908-3p inhibitor in the human SSC line in comparison to miRNA inhibitor control (Figure 2B). The Cell Counting Kit 8 (CCK-8) assay indicated that, compared to miRNA mimics control, miR-1908-3p mimics stimulated the proliferation of the human SSC line at day 3 to day 5 of culture (Figure 2C). In comparison to the miRNA inhibit control, the miR-1908-3p inhibitor suppressed the proliferation of the human SSC line at day 4 to day 5 of cell culture (Figure 2D). Together, these results imply that miR-1908-3p promotes the proliferation of the human SSC line.
Figure 2.
Effect of miR-1903-3p on the Proliferation of the Human SSC Line
(A) Fluorescence microscope and phase-contrast microscope displayed transfection efficiency of miR-1908-3p mimics and inhibitor using the FAM-labeled siRNA oligonucleotides. Scale bars, 40 μm. (B) Real-time PCR demonstrated the relative levels of miR-1908-3p in the human SSC line after transfection of miR-1908-3p mimics, miRNA mimics control, miR-1908-3p inhibitor, and miRNA inhibitor control for 24 h. The level of miR-1908-3p was quantified with U6 as a loading control. (C and D) CCK-8 assays displayed the growth curve of the human SSC line treated with miR-1908-3p mimics and miRNA mimics control (C), as well as the miR-1908-3p inhibitor and miRNA inhibitor control (D) for 5 days. ∗p < 0.05, statistically significant differences compared to miRNA mimics control; #p < 0.05, statistically significant differences compared to miRNA inhibitor control.
hsa-miR-1908-3p Accelerates the DNA Synthesis of the Human SSC Line
PCNA (proliferating cell nuclear antigen) has been regarded as a marker for DNA synthesis of cells. Western blots demonstrated that the relative level of PCNA protein was increased by miR-1908-3p mimics in the human SSC line when compared with miRNA mimics control (Figures 3A and 3B), whereas its expression level was reduced in the human SSC line by the miR-1908-3p inhibitor compared to miRNA inhibitor control (Figures 3A and 3B). Moreover, 5-ethynyl-2′-deoxyuridine (EDU) assays showed the percentages of EDU-positive cells and DNA synthesis in the human SSC line after the transfection for 48 h. Compared to the miRNA mimics control (6.33% ± 1.47% of EDU-positive cells), 11.5% ± 4.9% of EDU-positive cells were observed in the human SSC line treated with miR-1908-3p mimics (Figures 3C and 3D). Compared to the miRNA inhibit control (7.16% ± 1.33% of EDU-positive cells), 4.08% ± 2.20% of EDU-positive cells were seen in human SSC treated with the miR-1908-3p inhibitor (Figures 3E and 3F). Considered together, these data suggest that miR-1908-3p accelerates DNA synthesis of the human SSC line.
Figure 3.
Influence of miR-1903-3p on the DNA Synthesis of the Human SSC Line
(A) Western blots demonstrated PCNA expression in the human SSC line at 48 h after transfection of miRNA mimic control, miR-1908-3p mimics, miRNA inhibitor control, and miR-1908-3p inhibitor. ACTB served as the loading control of protein. (B) The relative expression of PCNA in the human SSC line at 48 h after transfection of miR-1908-3p mimics to miRNA mimics control and miR-1908-3p inhibitor to miRNA inhibitor control through normalization to the signals of their loading control.(C–F) EDU incorporation assays showed the percentages of EDU-positive cells in the human SSC line treated with miRNA mimics control compared to the miR-1908-3p mimics for 48 h (C and D), as well as miR-1908-3p inhibitor compared to the miRNA inhibitor for 48 h (E and F). (C and E) Scale bars, 40 μm. ∗p < 0.05, statistically significant differences compared to miRNA mimics control; #p < 0.05, statistically significant differences compared to miRNA inhibitor control.
hsa-miR-1908-3p Suppresses the Apoptosis of the Human SSC Line
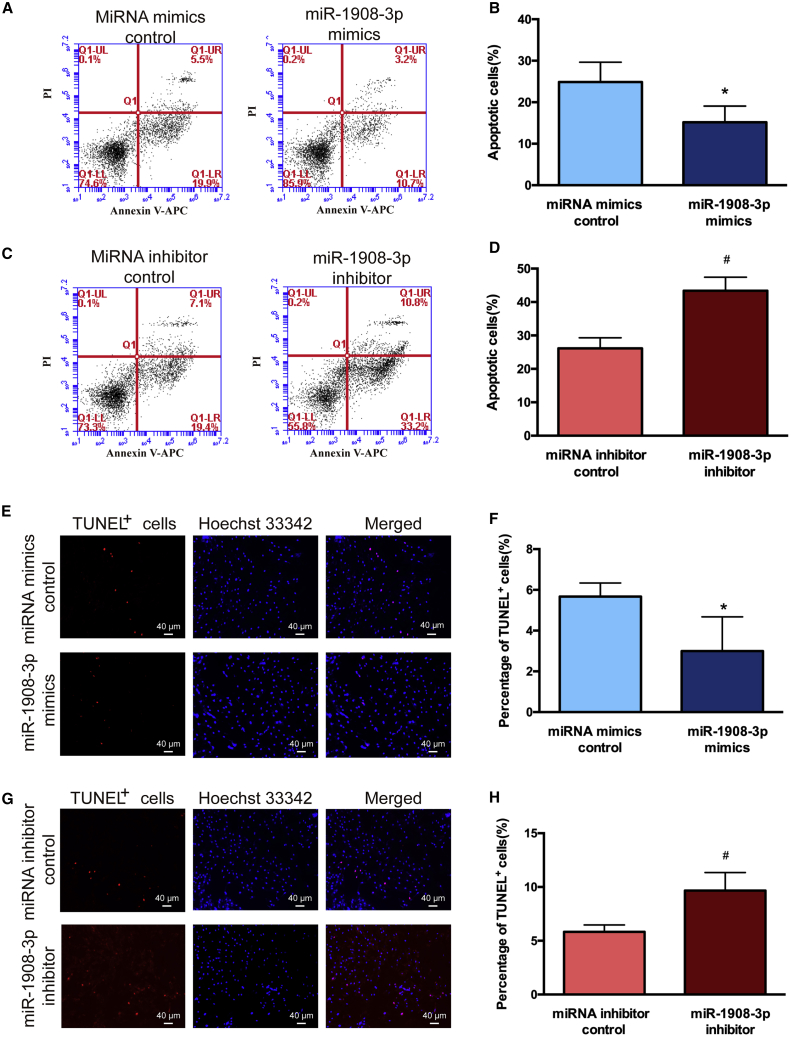
To determine the role of miR-1908-3p in controlling the apoptosis in human SSCs, the human SSC line was transfected by miRNA mimics control, miR-1908-3p mimics, miRNA inhibitor control, and miR-1908-3p inhibitor for 48 h, and the cells were stained with allophycocyanin (APC) Annexin V and propidium iodide (PI) and analyzed by flow cytometric analysis. The percentages of both early and late apoptosis were lower by miR-1908-3p mimics than miRNA mimics control in the human SSC line (Figures 4A and 4B). In contrast, the percentages of early and late apoptosis were higher by the miR-1908-3p inhibitor than miRNA inhibitor control (Figures 4C and 4D). Furthermore, TUNEL assays displayed that the percentages of TUNEL-positive cells in the human SSC line were less in miR-1908-3p mimics-treated cells than miRNA mimics control (Figures 4E and 4F), and there were more TUNEL-positive cells in the human SSC line by the miR-1908-3p inhibitor than miRNA inhibitor control (Figures 4G and 4H). The data mentioned above indicate that miR-1908-3p suppresses the apoptosis of the human SSC line.
Figure 4.
Impact of miR-1908-3p on the Apoptosis of the Human SSC Line
(A–D) APC Annexin V and flow cytometric analysis showed the percentages of early and late apoptosis in the human SSC line affected by miRNA mimics control and miR-1908-3p mimics (A and B) and miRNA inhibitor control and miR-1908-3p inhibitor (C and D) for 48 h. (E–H) TUNEL assays displayed the percentages of TUNEL-positive cells in the human SSC line treated with miRNA mimics control and miR-1908-3p mimics (E and F) and miR-1908-3p inhibitor and miRNA inhibitor (G and H) for 48 h. (E and G) Scale bars, 40 μm. ∗p < 0.05, statistically significant differences compared to miRNA mimics control; #p < 0.05, statistically significant differences compared to miRNA inhibitor control.
Kruppel-like Factor 2 (KLF2) Is a Direct Downstream Target of miR-1908-3p in the Human SSC Line
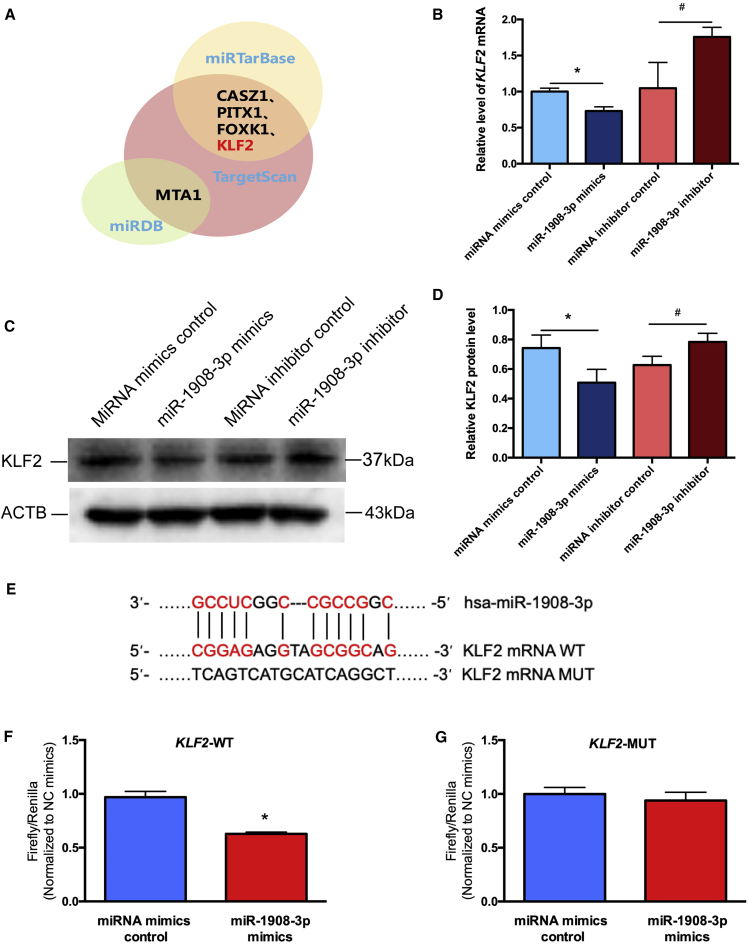
We predicted the targets of miR-1908-3p with 3 bioinformatics tools, namely, TargetScan, miRTarBase, and miRDB, and the targets of the database comprehensive predictions were KLF2, CASZ1, FOXK1, PITX1, and MTA1 (Figure 5A). Real-time PCR demonstrated the relative levels of KLF2, CASZ1, PITX1, FOXK1, and MTA1 mRNA in the human SSC line after transfection of miR-1908-3p mimics compared to the miRNA mimics control, as well as the miR-1903-3p inhibitor compared with the miRNA inhibitor control (Figures 5B and S2). Only KLF2 mRNA (Figure 5B) and protein (Figures 5C and 5D) were decreased in the human SSC line after transfection of miR-1908-3p mimics compared to the miRNA mimics control, and its transcript and translation were increased by the miR-1908-3p inhibitor in the human SSC line compared to the miRNA inhibitor control in the human SSC line for 24 h, reflecting that KLF2 is a potential target of miR-1908-3p in the human SSC line. The 3-dimensional homology model of the KLF2 zinc finger domain was illustrated in Figure S3A.
Figure 5.
KLF2 Is a Direct Downstream Target of miR-1908-3p in the Human SSC Line
(A) Target genes of miR-1908-3p were predicted with three bioinformatics tools (TargetScan, miRTarBase, and miRDB). (B) Real-time PCR demonstrated the relative level of KLF2 mRNA in the human SSC line after transfection of miR-1908-3p mimics, miRNA mimics control, miR-1908-3p inhibitor, and miRNA inhibitor control for 24 h. The level of KLF2 mRNA was quantified with ACTB. (C) Western blots demonstrated KLF2 expression in the human SSC line at 48 h after transfection of miRNA mimic control, miR-1908-3p mimics, miRNA inhibitor control, and miR-1908-3p inhibitor. ACTB served as the loading control of protein. (D) The relative expression of KLF2 in the human SSC line at 48 h after transfection of miR-1908-3p mimics to miRNA mimics control and miR-1908-3p inhibitor to miRNA inhibitor control through normalization to the signals of their loading control.(E) The binding site of miR-1908-3p to the wild type and a mutated type of KLF2 mRNA. (F and G) Dual luciferase reporter assays validated the targeting of miR-1908-3p to wild-type KLF2 (F) or mutated KLF2 (G) after transfection of miR-1908-3p mimics compared to the miRNA mimics control. ∗p < 0.05, statistically significant differences compared to miRNA mimics control; #p < 0.05, statistically significant differences compared to miRNA inhibitor control.
To demonstrate further that KLF2 is the direct target of miR-1908-3p in human SSCs, we used dual luciferase reporter assays. The human SSC line was transfected with KLF2 or mutated KLF2 after transfection of miR-1908-3p mimics or the miRNA mimics control. The predicted sequence in 3′ UTR of KLF2 mRNA luciferase activity of fusion genes (Figure 5E) responded to miR-1908-3p mimics in KLF2 wild type (Figure 5F) rather than mutated KLF2 (Figure 5G). Collectively, these findings illustrate that KLF2 is a direct downstream target of miR-1908-3p in the human SSC line.
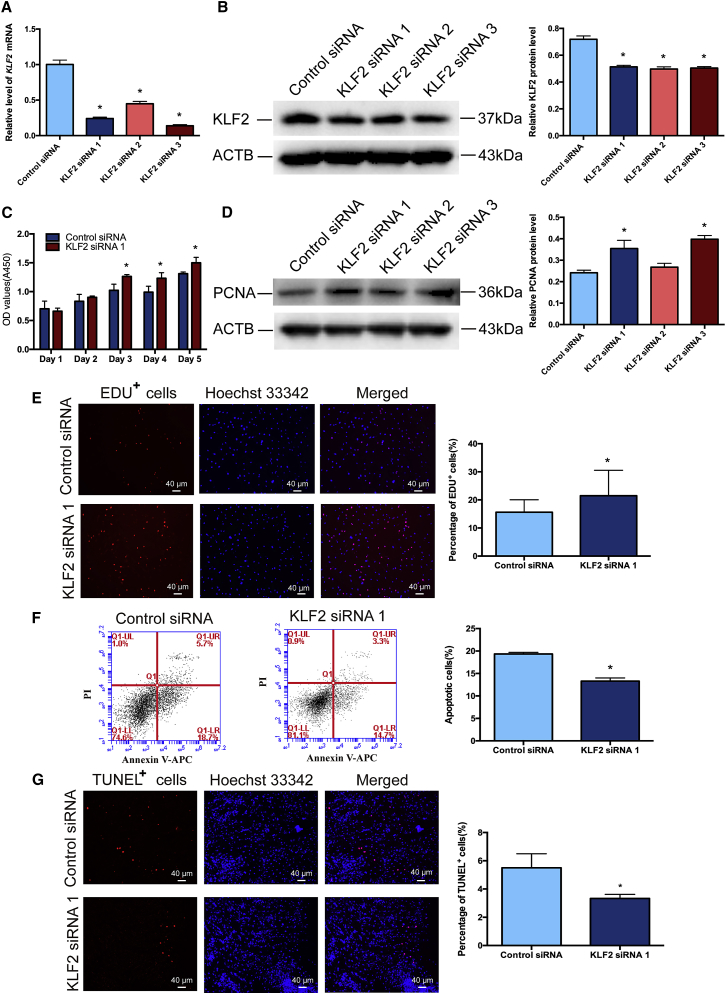
KLF2 Inhibits the Proliferation and DNA Synthesis and Stimulates the Apoptosis of the Human SSC Line
To further explore the influence of KLF2 silencing on the proliferation, DNA synthesis, and apoptosis of the human SSC line, we transfected KLF2 siRNA 1, KLF2 siRNA 2, and KLF2 siRNA 3 to these cells. Real-time PCR showed that the relative level of KLF2 mRNA was significantly decreased after treatment of KLF2 siRNAs for 24 h (Figure 6A). Western blots demonstrated that KLF2 siRNA 1–3 reduced the protein level of KLF2 in the human SSC line (Figure 6B). CCK-8 assays revealed that KLF2 siRNA 1 stimulated the proliferation of the human SSC line (Figure 6C). PCNA protein (Figure 6D) and the percentages of EDU-positive cells (Figure 6E) were increased after the transfection of KLF2 siRNA 1 for 48 h. On the other hand, as determined by flow cytometric analysis (Figure 6F) and TUNEL assays (Figure 6G), the apoptosis was decreased by KLF2 siRNA 1 in the human SSC line. In summary, these results suggest that KLF2 suppresses the proliferation and DNA synthesis and enhances the apoptosis of the human SSC line.
Figure 6.
Influence of KLF2 Silencing on the Proliferation, DNA Synthesis, and Apoptosis of the Human SSC Line
(A) Real-time PCR demonstrated the relative level of KLF2 mRNA in the human SSC line after transfection of KLF2 siRNA 1, KLF2 siRNA 2, and KLF2 siRNA 3 compared to the control siRNA for 24 h. The level of KLF2 mRNA was quantified with ACTB as a loading control. (B) Western blots showed the relative protein level of KLF2 in the human SSC line affected by KLF2 siRNA 1, KLF2 siRNA 2, and KLF2 siRNA 3 compared to the control siRNA for 48 h. The band densities of KLF2 protein were quantified with ACTB as a loading control. (C) CCK-8 assays showed the proliferation of the human SSC line treated with KLF2 siRNA 1 compared with the control siRNA for 5 days. (D) Western blots revealed the relative protein level of PCNA in the human SSC line affected by KLF2 siRNA 1, KLF2 siRNA 2, and KLF2 siRNA 3 compared to the control siRNA for 48 h. The band densities of PCNA protein were quantified with ACTB as a loading control. (E) EDU assays showed the DNA synthesis of human SSC line treated with KLF2 siRNA 1 compared to the control siRNA for 48 h. Scale bars, 40 μm. (F and G) The percentages of apoptosis in the human SSC line affected by KLF2 siRNA 1 or control siRNA for 48 h, as determined by flow cytometric analysis (F) and TUNEL assays (G). (G) Scale bars, 40 μm. ∗p < 0.05, statistically significant differences compared to control siRNA.
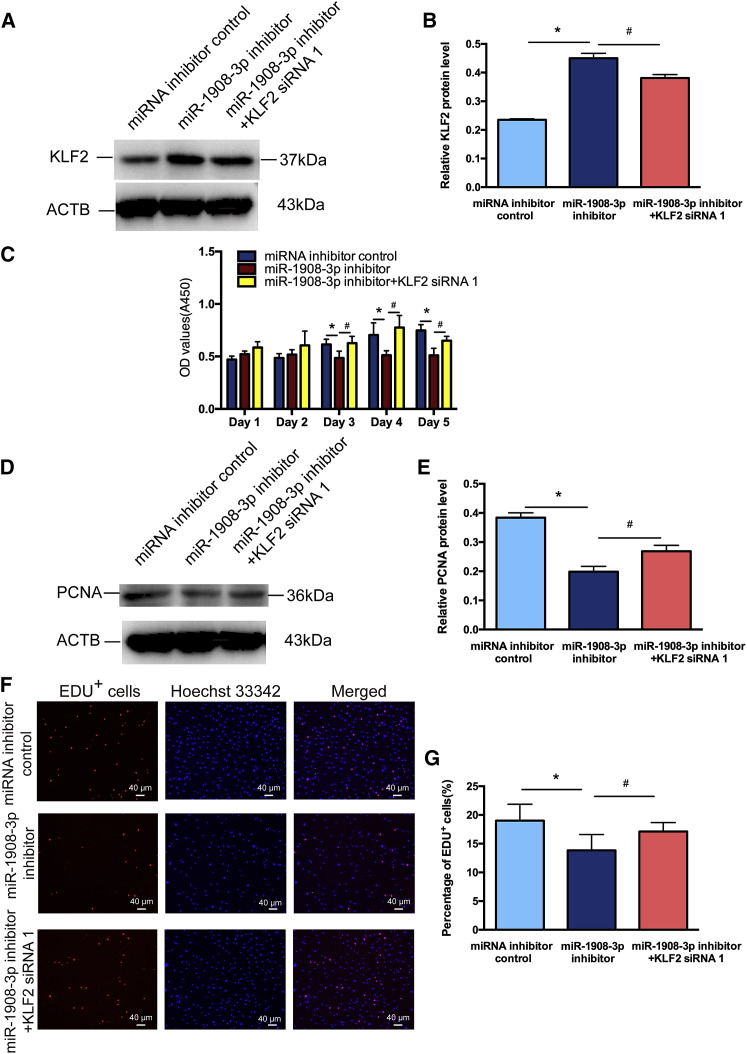
KLF2 Silencing Ameliorates the Decrease in Proliferation and DNA Synthesis as well as the Increase in Apoptosis of the Human SSC Line Caused by the miR-1908-3p Inhibitor
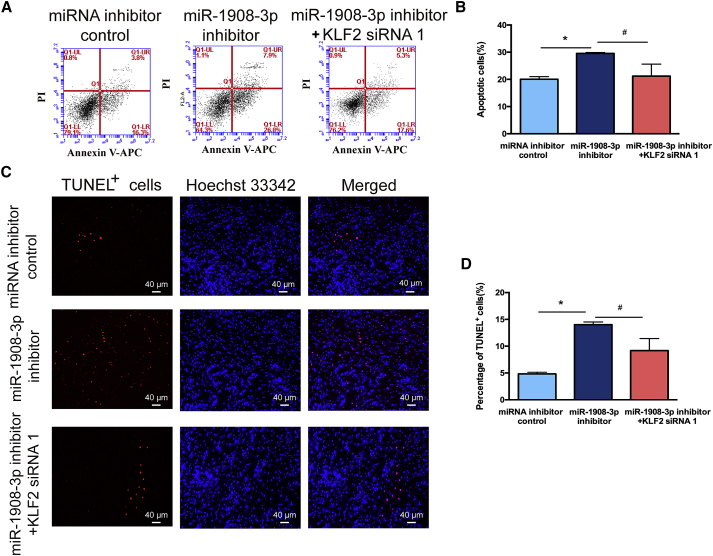
We predicted functional partners of KLF2, including NANOG, NOS3, MYC, SOX2, POU5F1 (OCT4), and FOX1 (Figure S3B). Since we have demonstrated that KLF2 is a direct downstream target of miR-1908-3p and that it is involved in the fate decisions of the human SSC line, we hypothesized that KLF2 silencing could ameliorate the adverse effect caused by inhibition of miR-1908-3p. As shown in Figures 7A and 7B, KLF2 siRNA 1 reduced the increase of the KLF2 protein level induced by the miR-1908-3p inhibitor. CCK-8 assays, Western blots, and EDU assays further revealed that KLF2 siRNA 1 abolished the decrease of cell proliferation (Figure 7C), PCNA level (Figures 7D and 7E), and EDU-positive cells (Figures 7F and 7G) of the human SSC line induced by the miR-1908-3p inhibitor. Moreover, the increase in percentages of early and late apoptosis (Figures 8A and 8B), as well as the TUNEL-positive cells (Figures 8C and 8D) caused by miR-1908-3p suppression, was significantly reduced by KLF2 siRNA 1. Taken together, these results demonstrate that KLF2 silencing ameliorates the decreased proliferation and DNA synthesis and the increased apoptosis of the human SSC line induced by miR-1908-3p inhibition.
Figure 7.
The Function of the miR-1908-3p Inhibitor in the Effect of KLF2 Silencing on the Proliferation and DNA Synthesis of the Human SSC Line
(A) Western blots demonstrated KLF2 expression in the human SSC line at 48 h after transfection of miRNA inhibitor control, miR-1908-3p inhibitor, and miR-1908-3p inhibitor and KLF2 siRNA 1. ACTB served as the loading control of protein. (B) The relative expression of KLF2 in the human SSC line at 48h after transfection of miR-1908-3p inhibitor to miRNA inhibitor control, and miR-1908-3p inhibitor and KLF2 siRNA 1 to miR-1908-3p inhibitor through normalization to the signals of their loading control. (C) CCK-8 assays demonstrated the proliferation of the human SSC line treated with miRNA inhibitor control, miR-1908-3p inhibitor, and miR-1908-3p inhibitor and KLF2 siRNA 1 of 5 days. (D) Western blots demonstrated PCNA expression in the human SSC line at 48 h after transfection of miRNA inhibitor control, miR-1908-3p inhibitor, and miR-1908-3p inhibitor and KLF2 siRNA 1. ACTB served as the loading control of protein. (E) The relative expression of PCNA in the human SSC line at 48 h after transfection of miR-1908-3p inhibitor to miRNA inhibitor control, and miR-1908-3p inhibitor and KLF2 siRNA 1 to miR-1908-3p inhibitor through normalization to the signals of their loading control. (F) EDU assays showed the DNA synthesis of human SSC line treated with miRNA inhibitor control, miR-1908-3p inhibitor, and miR-1908-3p inhibitor and KLF2 siRNA 1 for 48 h. (G) Qualification of EDU-positive cells in the human SSC line affected by miRNA inhibitor control, miR-1908-3p inhibitors, and miR-1908-3p inhibitors and KLF2 siRNA 1 in the human SSC line. (F) Scale bars, 40 μm. ∗p < 0.05, statistically significant differences compared to miRNA inhibitor control; #p < 0.05, statistically significant differences compared to miR-1908-3p inhibitor.
Figure 8.
The Role of the miR-1908-3p Inhibitor in the Influence of KLF2 Silencing on the Apoptosis of the Human SSC Line
(A–D) The percentage of apoptosis in the human SSC line affected by miRNA inhibitor control, miR-1908-3p inhibitor, and miR-1908-3p inhibitor and KLF2 siRNA 1 for 48 h, as determined by flow cytometric analysis (A and B) and TUNEL assays (C and D). (C) Scale bars, 40 μm. ∗p < 0.05, statistically significant differences compared to miRNA inhibitor control; #p < 0.05, statistically significant differences compared to miR-1908-3p inhibitor.
Discussion
Human SSCs can be induced to produce mature and functional spermatids, and they acquire pluripotency to become ES-like cells and directly transdifferentiate to functional cells and tissues.15, 16, 17, 18, 19 We have recently reported the differentiation of human SSCs from cryptorchidism patients into spermatids with fertilization and developmental potential.29 These studies highlight that human SSCs have significant application in both reproductive and regenerative medicine. Therefore, it is of great significance to uncover the molecular mechanisms that determine the fate decisions of human SSCs. Nevertheless, epigenetic regulation by key miRNAs that control the self-renewal and apoptosis of human SSCs remained largely elusive. Here, we have demonstrated for the first time that miR-1908-3p stimulates the proliferation and DNA synthesis and suppresses the apoptosis of the human SSC line by targeting KLF2.
In order to identify the key miRNA, we compared large-scale differential expression profiles of human spermatogonia, pachytene spermatocytes, and round spermatids, and we found that miR-1908-3p was significantly upregulated in human spermatogonia compared to pachytene spermatocytes. We have established the human SSC line for the first time in the world. Notably, this cell line can proliferate for a long time without tumorigenicity, and it has the biochemical characteristics of primary human SSCs in vivo and in vitro.28 In this study, we verified the identity of the human SSC line, since numerous markers for human SSCs and male germ cells, including GFRA1, GPR125, THY1, UCHL1, PLZF, RET, and VASA, were expressed in our human SSC line. We also found that the miR-1908-3p level was the highest compared to miR-1908-5p, miR-1469, and miR-1224-5p in the human SSC line, and there was a high level of miR-1908-3p in primary human SSCs, reflecting a potential role of the hsa-miR-1908 in maintaining the stemness of human SSCs.
It has been reported that miR-1908 can inhibit the differentiation of human pluripotent adipose-derived stem cells and promotes the proliferation of these cells, suggesting that miR-1908 is involved in the regulation of adipocyte differentiation and metabolism.30 MiR-1908 has been reported to be associated with the development of glioma by promoting glioma cell proliferation, invasion, anti-apoptosis, and regulation of the SPRF4/RAF1 axis.31 However, it remains unknown about the function and mechanism of hsa-miR-1908-3p in mediating the fate determinations of human SSCs. Based on the effective transfection of miR-1908-3p mimics and the miR-1908-3p inhibitor in the human SSC line, we have demonstrated that miR-1908-3p stimulates the proliferation, accelerates DNA synthesis, and suppresses the apoptosis of the human SSC line. These results support the conclusion that miR-1908-3p is clearly an important mediator of fate decisions of human SSCs.
In order to unveil the downstream regulatory factors and biological function of hsa-miR-1908 in human SSC self-renewal and apoptosis, we predicted the targeting transcriptional factors of miR-1908-3p with 3 bioinformatics tools (TargetScan, miRTarBase, and miRDB). With the use of real-time PCR and Western blots and verification by dual luciferase reporter assays, we have revealed that KLF2 is a direct downstream target of miR-1908-3p in the human SSC line. KLF2 is the major mechanosensitive transcription factor that regulates vascular homeostasis, and thus, it represents a promising therapeutic target to prevent atherosclerosis by pharmacological intervention.32,33 KLF2 also controls homeostatic natural killer (NK) cell proliferation and survival,34 and overexpression of KLF2 impairs hepatocellular carcinoma (HCC) cell and hepatocyte proliferation and induces cell apoptosis.35,36 In the current study, we found that KLF2 silencing in the human SSC line stimulated the proliferation and DNA synthesis and suppressed the apoptosis of the human SSC line, which is consistent with the effect of miR-1908-3p overexpression. Notably, the impairment of DNA synthesis, proliferation, and apoptosis of the human SSC line caused by the miR-1908-3p inhibitor was abolished by KLF2 silencing. Together, these data support the hypothesis that KLF2 has been identified as a direct target of miR-1908-3p in the human SSC line.
In summary, we have demonstrated for the first time that hsa-miR-1908-3p regulates the self-renewal and apoptosis of human SSCs by targeting KLF2. This study offers a novel epigenetic regulatory mechanism underlying the fate determinations of human SSCs and human spermatogenesis, and it provides an important basis and target for the diagnosis and treatment of male infertility.
Materials and Methods
Human SSC Line Culture and miRNA and siRNA Transfection
The human SSC line was established, as we described previously,28 and these cells were cultured in DMEM/F-12 containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco, USA) at 37°C in 5% CO2. The human SSC line was transfected with four different miRNAs: miR-1908-3p mimics, miRNA mimics control, miR-1908-3p inhibitor, and miRNA inhibitor control (GenePharma, China). The transfection efficiency of miRNAs was assessed using the FAM-labeled siRNA oligonucleotides. For siRNA assays, the human SSC line was transfected with four different siRNAs: control siRNA, KLF2 siRNA 1, KLF2 siRNA 2, and KLF2 siRNA 3. These miRNAs or siRNAs were delivered by transfection agent Lipofectamine 3000 pursuant to the instruction of manufacture (Life Technologies, USA). After transfection for 48 h, the cells were harvested to detect the expression of various genes and proteins.
Isolation of Human SSCs from OA Patients
Testicular tissues from OA patients were washed three times in DMEM with 1% penicillin and streptomycin. This study was approved by the Institutional Ethical Committee of Hunan Normal University. Human THY1-positive spermatogonia were separated using two-step enzymatic digestion and MACS. In brief, seminiferous tubules were isolated from human testis biopsies using 2 mg/mL collagenase IV (Sigma, USA) and 1 μg/μL DNase I (Gibco). Human testicular cells were obtained using a second enzymatic digestion with 4 mg/mL collagenase IV, 2.5 mg/mL hyaluronidase (Sigma), 2 mg/mL trypsin (Sigma), and 1 μg/μL DNase I. For differential plating, cells were seeded into culture plates in DMEM/F-12 (Gibco), supplemented with 10% FBS (Biological Industries, Israel) and incubated at 34°C in 5% CO2 for 12 h. After incubation, Sertoli cells attached to the culture plates, whereas male germ cells remained in suspension, and they were collected by centrifugation at 1,000 rpm for 5 min. THY1-positive spermatogonia were separated by MACS using an antibody to THY1 (Miltenyi Biotec, Germany), pursuant to the procedures in accordance with MACS instruction (Miltenyi Biotec).
RNA Extraction, RT-PCR, and Real-Time PCR
Total RNA was extracted from the cells using the RNAiso Plus reagent (Takara, Kusatsu, Japan), according to the manufacturer’s instruction. After the determination of RNA quality, the RNA was used for RT-PCR and real-time PCR. RT was performed using the First Strand cDNA Synthesis Kit (Thermo Scientific, USA) and TransScript miRNA First-Strand cDNA Synthesis SuperMix (TransGen Biotech, China).
RT-PCR was conducted by the protocol, as described previously.37 The PCR reactions started at 94°C for 2 min, and they were performed in terms of the following conditions: denaturation at 94°C for 30 s; annealing at 55°C–60°C for 45 s, as listed in Table S1; and elongation at 72°C for 45 s for 35 cycles. The PCR samples were incubated for an additional 5 min at 72°C. RNA, without RT but with PCR, served as a negative control. PCR products were separated by electrophoresis with 2% agarose gel, and they were visualized with ethidium bromide.
Quantitative real-time PCR reactions were conducted using Power SYBR Green PCR Master Mix (Applied Biosystems, USA) and real-time PCR system (Applied Biosystems). The comparative CT (threshold cycle) method was used to quantify the PCR products. The CT value of gene was normalized against the U6 or the housekeeping gene ACTB. The primer sequences of the chosen genes for RT-PCR and real-time PCR were designed and listed in Table S2.
Immunocytochemistry
For immunocytochemistry, the human SSC line was washed three times with cold PBS (HyClone, UT, USA) and fixed with 4% paraformaldehyde (PFA) for 15 min, and these cells were washed three times with cold PBS and permeabilized with 0.4% Triton X-100 (Sigma, USA) for 10 min. After extensive washes with PBS, the cells were blocked in 5% BSA for 1 h at room temperature and followed by incubation with primary antibodies, including GFRA1 (Abcam, Cambridge, UK; ab8026, 1:200), GPR125 (Abcam; ab51705, 1:200), THY1 (Abcam; ab133350, 1:200), UCHL1 (Bio-Rad; MCA4750, 1:200), PLZF (Abcam; ab104854, 1:200), RET (Abcam; ab134100, 1:200), and VASA (Santa Cruz Biotechnology, USA; sc-517247, 1:200), overnight at 4°C. After extensive washes with PBS, the cells were incubated with IgGs conjugated with fluorescein isothiocyanate (FITC) (Sigma) or rhodamine-conjugated IgG (Sigma) at a 1:200 dilution for 1 h at room temperature. Replacement of primary antibodies with isotype IgGs served as negative controls. DAPI (4′,6-diamidino-2-phenylindole) was employed to label cell nuclei, and the images were captured with a microscope (Nikon, Tokyo, Japan).
CCK-8 Assays
The human SSC line was transfected with miRNAs and/or siRNAs. Cell culture medium was replaced with 10% CCK-8 solution (Dojin Laboratories, Kumamoto, Japan) for incubating 3 h, and the absorbance was measured at 450 nm by a microplate reader during the following 5 days of culture.
Western Blots
After transfection with miRNAs and/or siRNAs, the human SSC line was lysed with radioimmunoprecipitation assay (RIPA) buffer (Dingguo, Beijing, China) on ice for about 30 min. The proteins of cells were obtained by centrifugation at 12,000 × g for 15 min, and the concentration of proteins was measured by a bicinchoninic acid (BCA) kit (Dingguo). Total protein extracts (30 μg) were separated on 8%–12% Bis-Tris gels and followed by transferring onto polyvinylidene fluoride membranes. After blocking with 5% skimmed milk for 2 h at room temperature, the blots were incubated with a primary antibody overnight at 4°C and then with horseradish peroxidase-conjugated secondary antibody for 1 h. The primary antibodies included PCNA (Cell Signaling Technology, MA, USA; 2586, 1:2,000), KLF2 (Abcam; ab203591, 1:1,000), and ACTB (Proteintech, Chicago, IL, USA; 60008, 1:2,000). ACTB was used as a loading control of protein. The intensities of protein bands were visualized using chemiluminescence (Bio-Rad, GE Healthcare, CA, USA).
EDU Incorporation Assay
The human SSC line was seeded at a density of 5,000 cells/well in a 96-well plate containing DMEM/F-12 medium with 50 mM EDU (RiboBio, Guangzhou, China), and it was treated with miRNAs or siRNAs. After 12 h of culture, the cells were washed with DMEM and fixed with 4% PFA. Cells were neutralized with 2 mg/mL glycine and permeabilized in 0.5% Triton X-100 for 10 min at room temperature. EDU immunostaining was performed with Apollo staining reaction buffer. The nuclei of cells were stained with Hoechst 33342, and the EDU-positive cells were counted from at least 500 cells under fluorescence microscopy (Nikon, Tokyo, Japan).
Flow Cytometric Analysis
To quantify the apoptotic percentages of the human SSC line after transfection with miRNAs and/or siRNAs for 48 h, cells were digested and collected, and they were washed twice with cold PBS. In total, 106 cells were resuspended with Annexin V Binding Buffer (BioLegend, London, UK), and 5 μL of APC Annexin V and 10 μL of PI solution were added to the cells. After incubation for 15 min at room temperature in the dark, the cells were analyzed by flow cytometry with a C6 instrument (BD Biosciences, NJ, USA).
TUNEL Assay
The apoptotic percentages of the human SSC line were detected using the TUNEL Apoptosis Detection Kit (Yeasen, Shanghai, China). After transfection with miRNA or siRNAs, cells were treated with 20 μg/mL proteinase K for 20 min at room temperature and incubated with FITC-12-deoxyuridine 5'-triphosphate (dUTP) labeling/terminal deoxynucleotidyl transferase (TdT) enzyme buffer for 1 h in the dark. PBS without TdT enzyme was used as the negative control. Hoechst 33342 was used to label cell nuclei, and images were observed under a Nikon microscope. The percentages of TUNEL-positive cells were counted from at least 500 cells, and three independent experiments were performed.
Dual Luciferase Reporter Assays
After transfection miRNA mimics and 500 ng plasmids with the binding sequence in 3′ UTRs of KLF2 (pmirGLO; Genecreate, Wuhan, China) for 48 h, the culture medium was discarded, and the cells were washed with PBS. Fifty microliters of the diluted 1× Passive lysate buffer (PLB) was added to each well of cells and followed by shaking at room temperature for 15 min. In total, 100 μL of premixed LAR II and 100 μL of premixed Stop & Glo Reagent were utilized, and luciferase activities were measured using the tube luminometer (Berthold, Germany), according to the manufacturer’s protocol. Data were normalized to miRNA mimic control-transfected cells.
Statistical Analysis
Data were expressed as mean ± SEM. Comparisons between two groups were performed using unpaired t test, and p values <0.05 were considered statistically significant. When individual studies were demonstrated, there was a representative of at least three independent experiments.
Author Contributions
W.C. performed the experiments, wrote the manuscript, and helped with data analysis. Y.C., C.L., L.D., L.Q., Y.J., and J.L. assisted with the experiments. B.L. and R.T. assisted with the experiments and ordered reagents. X.Y. and Q.H. assisted with the structure and functional partners’ analysis of KLF2. Z.H. was responsible for the conception and design, supervision of all aspects of the laboratory experiments, data analysis, writing of the manuscript, and final approval of the manuscript. All authors approved the manuscript.
Conflicts of Interest
The authors declare no competing interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31671550 and 31872845); National Key R&D Project (2016YFC1000606); High Level Talent Gathering Project in Hunan Province (2018RS3066); Major Scientific and Technological Projects for Collaborative Prevention and Control of Birth Defect in Hunan Province (2019SK1012); Open Fund of the NHC Key Laboratory of Male Reproduction and Genetics (KF201802); and Shanghai Hospital Development Center (SHDC12015122).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.04.016.
Supplemental Information
References
- 1.Winters B.R., Walsh T.J. The epidemiology of male infertility. Urol. Clin. North Am. 2014;41:195–204. doi: 10.1016/j.ucl.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal A., Mulgund A., Hamada A., Chyatte M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine H., Jørgensen N., Martino-Andrade A., Mendiola J., Weksler-Derri D., Mindlis I., Pinotti R., Swan S.H. Temporal trends in sperm count: a systematic review and meta-regression analysis. Hum. Reprod. Update. 2017;23:646–659. doi: 10.1093/humupd/dmx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonde J.P., Flachs E.M., Rimborg S., Glazer C.H., Giwercman A., Ramlau-Hansen C.H., Hougaard K.S., Høyer B.B., Hærvig K.K., Petersen S.B. The epidemiologic evidence linking prenatal and postnatal exposure to endocrine disrupting chemicals with male reproductive disorders: a systematic review and meta-analysis. Hum. Reprod. Update. 2016;23:104–125. doi: 10.1093/humupd/dmw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carré J., Gatimel N., Moreau J., Parinaud J., Léandri R. Does air pollution play a role in infertility?: a systematic review. Environ. Health. 2017;16:82. doi: 10.1186/s12940-017-0291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Punab M., Poolamets O., Paju P., Vihljajev V., Pomm K., Ladva R., Korrovits P., Laan M. Causes of male infertility: a 9-year prospective monocentre study on 1737 patients with reduced total sperm counts. Hum. Reprod. 2017;32:18–31. doi: 10.1093/humrep/dew284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagano M., Brinster C.J., Orwig K.E., Ryu B.Y., Avarbock M.R., Brinster R.L. Transgenic mice produced by retroviral transduction of male germ-line stem cells. Proc. Natl. Acad. Sci. USA. 2001;98:13090–13095. doi: 10.1073/pnas.231473498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dym M. Spermatogonial stem cells of the testis. Proc. Natl. Acad. Sci. USA. 1994;91:11287–11289. doi: 10.1073/pnas.91.24.11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia T.X., Hofmann M.C. Regulation of germ line stem cell homeostasis. Anim. Reprod. 2015;12:35–45. [PMC free article] [PubMed] [Google Scholar]
- 10.Gunes S., Arslan M.A., Hekim G.N.T., Asci R. The role of epigenetics in idiopathic male infertility. J. Assist. Reprod. Genet. 2016;33:553–569. doi: 10.1007/s10815-016-0682-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao C., Liu Y., Sun M., Niu M., Yuan Q., Hai Y., Guo Y., Chen Z., Hou J., Liu Y., He Z. MicroRNAs and DNA methylation as epigenetic regulators of mitosis, meiosis and spermiogenesis. Reproduction. 2015;150:R25–R34. doi: 10.1530/REP-14-0643. [DOI] [PubMed] [Google Scholar]
- 12.Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tahmasbpour E., Balasubramanian D., Agarwal A. A multi-faceted approach to understanding male infertility: gene mutations, molecular defects and assisted reproductive techniques (ART) J. Assist. Reprod. Genet. 2014;31:1115–1137. doi: 10.1007/s10815-014-0280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Procópio M.S., de Avelar G.F., Costa G.M.J., Lacerda S.M.S.N., Resende R.R., de França L.R. MicroRNAs in Sertoli cells: implications for spermatogenesis and fertility. Cell Tissue Res. 2017;370:335–346. doi: 10.1007/s00441-017-2667-z. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z., Niu M., Sun M., Yuan Q., Yao C., Hou J., Wang H., Wen L., Fu H., Zhou F. Transdifferentiation of human male germline stem cells to hepatocytes in vivo via the transplantation under renal capsules. Oncotarget. 2017;8:14576–14592. doi: 10.18632/oncotarget.14713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., Sun M., Yuan Q., Niu M., Yao C., Hou J., Wang H., Wen L., Liu Y., Li Z., He Z. Generation of functional hepatocytes from human spermatogonial stem cells. Oncotarget. 2016;7:8879–8895. doi: 10.18632/oncotarget.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y., Liu L., Sun M., Hai Y., Li Z., He Z. Expansion and long-term culture of human spermatogonial stem cells via the activation of SMAD3 and AKT pathways. Exp. Biol. Med. (Maywood) 2015;240:1112–1122. doi: 10.1177/1535370215590822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon L., Ekman G.C., Kostereva N., Zhang Z., Hess R.A., Hofmann M.C., Cooke P.S. Direct transdifferentiation of stem/progenitor spermatogonia into reproductive and nonreproductive tissues of all germ layers. Stem Cells. 2009;27:1666–1675. doi: 10.1002/stem.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kossack N., Meneses J., Shefi S., Nguyen H.N., Chavez S., Nicholas C., Gromoll J., Turek P.J., Reijo-Pera R.A. Isolation and characterization of pluripotent human spermatogonial stem cell-derived cells. Stem Cells. 2009;27:138–149. doi: 10.1634/stemcells.2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 21.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 22.Yi R., Poy M.N., Stoffel M., Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 24.Kotaja N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014;101:1552–1562. doi: 10.1016/j.fertnstert.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 25.Jamsai D., Clark B.J., Smith S.J., Whittle B., Goodnow C.C., Ormandy C.J., O’Bryan M.K. A missense mutation in the transcription factor ETV5 leads to sterility, increased embryonic and perinatal death, postnatal growth restriction, renal asymmetry and polydactyly in the mouse. PLoS ONE. 2013;8:e77311. doi: 10.1371/journal.pone.0077311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song W., Mu H., Wu J., Liao M., Zhu H., Zheng L., He X., Niu B., Zhai Y., Bai C. miR-544 Regulates Dairy Goat Male Germline Stem Cell Self-Renewal via Targeting PLZF. J. Cell. Biochem. 2015;116:2155–2165. doi: 10.1002/jcb.25172. [DOI] [PubMed] [Google Scholar]
- 27.Niu B., Wu J., Mu H., Li B., Wu C., He X., Bai C., Li G., Hua J. miR-204 Regulates the Proliferation of Dairy Goat Spermatogonial Stem Cells via Targeting to Sirt1. Rejuvenation Res. 2016;19:120–130. doi: 10.1089/rej.2015.1719. [DOI] [PubMed] [Google Scholar]
- 28.Hou J., Niu M., Liu L., Zhu Z., Wang X., Sun M., Yuan Q., Yang S., Zeng W., Liu Y. Establishment and Characterization of Human Germline Stem Cell Line with Unlimited Proliferation Potentials and no Tumor Formation. Sci. Rep. 2015;5:16922. doi: 10.1038/srep16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S., Ping P., Ma M., Li P., Tian R., Yang H., Liu Y., Gong Y., Zhang Z., Li Z., He Z. Generation of haploid spermatids with fertilization and development capacity from human spermatogonial stem cells of cryptorchid patients. Stem Cell Reports. 2014;3:663–675. doi: 10.1016/j.stemcr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L., Shi C.M., Chen L., Pang L.X., Xu G.F., Gu N., Zhu L.J., Guo X.R., Ni Y.H., Ji C.B. The biological effects of hsa-miR-1908 in human adipocytes. Mol. Biol. Rep. 2015;42:927–935. doi: 10.1007/s11033-014-3830-1. [DOI] [PubMed] [Google Scholar]
- 31.Chai Z., Fan H., Li Y., Song L., Jin X., Yu J., Li Y., Ma C., Zhou R. miR-1908 as a novel prognosis marker of glioma via promoting malignant phenotype and modulating SPRY4/RAF1 axis. Oncol. Rep. 2017;38:2717–2726. doi: 10.3892/or.2017.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu N., Xu S., Xu Y., Little P.J., Jin Z.G. Targeting Mechanosensitive Transcription Factors in Atherosclerosis. Trends Pharmacol. Sci. 2019;40:253–266. doi: 10.1016/j.tips.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pi J., Tao T., Zhuang T., Sun H., Chen X., Liu J., Cheng Y., Yu Z., Zhu H.H., Gao W.Q. A MicroRNA302-367-Erk1/2-Klf2-S1pr1 Pathway Prevents Tumor Growth via Restricting Angiogenesis and Improving Vascular Stability. Circ. Res. 2017;120:85–98. doi: 10.1161/CIRCRESAHA.116.309757. [DOI] [PubMed] [Google Scholar]
- 34.Rabacal W., Pabbisetty S.K., Hoek K.L., Cendron D., Guo Y., Maseda D., Sebzda E. Transcription factor KLF2 regulates homeostatic NK cell proliferation and survival. Proc. Natl. Acad. Sci. USA. 2016;113:5370–5375. doi: 10.1073/pnas.1521491113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang M.D., Chen W.M., Qi F.Z., Xia R., Sun M., Xu T.P., Yin L., Zhang E.B., De W., Shu Y.Q. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J. Hematol. Oncol. 2015;8:50. doi: 10.1186/s13045-015-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manavski Y., Abel T., Hu J., Kleinlützum D., Buchholz C.J., Belz C., Augustin H.G., Boon R.A., Dimmeler S. Endothelial transcription factor KLF2 negatively regulates liver regeneration via induction of activin A. Proc. Natl. Acad. Sci. USA. 2017;114:3993–3998. doi: 10.1073/pnas.1613392114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., Yuan Q., Niu M., Zhang W., Wen L., Fu H., Zhou F., He Z. Transcriptional regulation of P63 on the apoptosis of male germ cells and three stages of spermatogenesis in mice. Cell Death Dis. 2018;9:76. doi: 10.1038/s41419-017-0046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.