Abstract
Circular RNAs (circRNAs), a novel family of non-coding RNAs, play crucial roles in cancer progression. While the existing research focuses on nuclear genome-derived (nu)-circRNAs, the biological and clinical characteristics of mitochondrial genome-derived (mt)-circRNAs remain largely unknown, especially in chronic lymphocytic leukemia (CLL). In this study, we attempted to identify the novel characteristics of mc-COX2 (mitochondrial genome-derived circRNAs [mc]), one of the mt-circRNAs that can be involved in CLL progression. mt-circRNAs were found to be highly expressed in the plasma exosomes of CLL patients. The endogenous reduction of mc-COX2 can affect mitochondrial functions, suppress cell proliferation, and induce cell apoptosis. The upregulation of mc-COX2 was positively associated with leukemogenesis and worsening survival of CLL patients. Notably, functional analysis revealed that mc-COX2, as differing from conventional nu-circRNAs, was less stable and may function through novel mechanisms other than acting as the competing endogenous RNA. We also screened and tested several chemical compounds and small-molecule inhibitors that can decrease the generation of mc-COX2. It was found that the silencing of mc-COX2 in CLL cells strengthened the anti-tumor effects of drugs used in coordination. Our findings prove that mc-COX2, a critical mt-circRNA highly expressed in plasma, derived from CLL cells and delivered by exosomes, is associated with the progression and prognosis of CLL.
Keywords: circular RNA, mitochondria, exosome, chronic lymphocytic leukemia, small-molecule inhibitors
Graphical Abstract

Li et al. explored the biological and pathological functions of the mitochondrial genome-derived circRNA mc-COX2. mc-COX2, highly expressed in plasma and delivered by exosomes, is not only associated with the progression and prognosis of CLL patients, but it is also a potentially important therapeutic target for CLL treatment.
Introduction
Chronic lymphocytic leukemia (CLL), characterized by highly variable clinical outcomes, is the most prevalent incurable B cell neoplasm in Western countries.1 In China, the incidence rate of CLL is increasing along with the aging population, Westernization of lifestyle, and the improvement of clinical diagnosis capacity. A number of patients can survive without progression, but other patients with high-risk factors suffer from poor prognosis.2
Mitochondria are viewed as important fuels to maintain the bioenergetic, biosynthetic, and metabolic demands of cancer cells and are indicated to be the prime target for cancer therapy. Mitochondrial metabolism contributes to oxidative stress in CLL, and the mitochondrial mass and number are aberrant in CLL cells.3,4 Inspiringly, we have accidentally discovered a distinct expression pattern of mitochondrial genome-derived circular RNAs (mt-circRNAs) in the plasma of CLL patients. mt-circRNAs seemingly exhibit novel biological characteristics different from nuclear genome-derived (nu)-circRNAs. Moreover, Shan et al.5 reported the roles of circRNAs encoded by the mitochondrial genome in the communication between mitochondria and nucleus. circRNAs, a special family of endogenous non-coding RNAs characterized by covalently closed continuous loops without 5′ to 3′ polarity, are reportedly widespread and stable in various tissues and body fluids with high exonuclease tolerance.6 In addition, mounting evidence indicates that circRNAs are enriched and stable in exosomes,7 which are small membrane vesicles with a typical diameter of 30–100 nm.8 Exosomes may play critical roles in regulating signaling transduction by delivering mRNAs, non-coding RNAs (e.g., microRNAs [miRNAs], long non-coding RNAs, circRNAs), and proteins between neighboring and distant cells.9,10 Since CLL patients show specific expression of exosomal circRNAs in plasma, the function and clinical significance of circRNAs are important to explore. However, circRNAs as potential therapeutic targets have rarely been studied in CLL, let alone mt-circRNAs.
Ibrutinib, a small-molecule BTK inhibitor, can induce apoptosis of CLL cells and has shown remarkable efficacy for CLL patients.11 However, the main problems of ibrutinib are the reliability of patients and the occurrence of resistance.12 Venetoclax, a B cell lymphoma 2 (Bcl-2) inhibitor, is another new therapeutic option for relapsed or refractory CLL patients with 17p deletion. The protein BCL-2 family plays vital roles in the “programmed” cell death process, protecting cells from apoptosis via the mitochondria-mediated pathway, and it is associated with drug resistance.13, 14, 15 Although these small-molecule drugs have significantly improved the management of CLL patients, it is still urgent to screen out convenient and effective biomarkers and potential therapeutic targets related to the occurrence and development of CLL.
Herein, we report a novel circRNA, mc-COX2, which is generated by back-splicing from the COX2 gene on the mitochondrial genome. mc-COX2 highly expressed in the plasma exosomes of CLL patients is closely related to prognosis. Moreover, mc-COX2 may exhibit some biological characteristics different from nuclear genome-derived (nu)-circRNAs. mc-COX2 is found to protect cells from apoptosis and promotes cell proliferation. Notably, we suggest that several chemical compounds and inhibitors targeting mitochondria have efficacy on CLL cells and have a synergistic anti-tumor effect with mc-COX2 RNA interference. This study demonstrates for the first time that mc-COX2 from mitochondria is highly expressed in CLL patients’ plasma and exosomes and may be involved in disease progression.
Results
Identification and Validation of mt-circRNAs in CLL Plasma and Cells
circRNAs are reportedly involved in various diseases and play important roles in tumorigenesis.16,17 However, little is known about the functions of circRNAs in hematological diseases, especially CLL. To unveil the expression profiles and potential biomarkers of circRNAs in CLL, we collected plasma samples from five treatment-naive CLL patients and five age- and sex-matched healthy donors (HDs) for circRNA microarray analysis. Results showed that 51 circRNAs were remarkably and abnormally expressed (fold change ≥ 2, p < 0.05) in CLL plasma (Figure 1A). Surprisingly, among 28 upregulated circRNAs, the top four circRNAs were all mt-circRNAs (Figure 1B). The details of the top 10 upregulated circRNAs are listed in Table 1. To verify the existence of mt-circRNAs, we then focused on hsa_circ_0089762 (mc-COX2), one of the four mt-circRNAs mentioned above. RNA fluorescence in situ hybridization (FISH) was performed to confirm the enrichment of mc-COX2 (Figure 1C). Moreover, northern blot based on the head-to-tail probe of mc-COX2 showed that mc-COX2 was detectable within the splice sites. ciRS-7, the first reported nu-circRNA with regulatory function,18 was used as a control (Figure 1D). The results together showed that mc-COX2 was indeed circularly arranged and derived from mitochondria in CLL cells.
Figure 1.
Identification and Validation of mt-circRNAs in CLL Plasma and Cells
(A) Differentially expressed circRNAs on a cluster heatmap (fold change of ≥2). Red, high expression; blue, low expression. (B) circRNAs with four mt-circRNAs identified by a volcano plot with different colors and sizes. (C) Distribution of mc-COX2 revealed by RNA FISH. Scale bar, 10 μm. Representative images from one of three independent experiments. (D) Circular isoform of circ-RPL15 verified by northern blot. CiRS-7 was used as a quality control.
Table 1.
Top 10 Upregulated circRNAs in CLL Plasma
| circBase ID | p Value | Fold Change | Chromosome | Genomic Length (nt) | Spliced Length (nt) |
|---|---|---|---|---|---|
| hsa_circ_0089763 | 0.002 | 23.21 | chrM | 5,783 | 5,783 |
| hsa_circ_0008882 | 0.005 | 13.52 | chrM | 81 | 81 |
| hsa_circ_0002363 | 0.003 | 13.40 | chrM | 63 | 63 |
| hsa_circ_0089762 | 0.008 | 9.81 | chrM | 396 | 396 |
| hsa_circ_0013745 | 0.011 | 3.66 | chr1 | 32,774 | 334 |
| hsa_circ_0134501 | 0.021 | 3.32 | chr7 | 420 | 420 |
| hsa_circ_0109320 | 0.011 | 3.26 | chr19 | 672 | 672 |
| hsa_circ_0051886 | 0.048 | 3.25 | chr19 | 3,492 | 424 |
| hsa_circ_0109322 | 0.006 | 3.24 | chr19 | 1,680 | 1,680 |
| hsa_circ_0039943 | 0.025 | 3.18 | chr16 | 129 | 129 |
The four circRNAs with the highest expression were all from mitochondria.
Abundant mc-COX2 Enrichment in Exosomes Predicts Prognosis of CLL Patients
Regarding the remarkable expression of mc-COX2 in CLL plasma, we planned to further investigate its biological and clinical characteristics. First, plasma samples were collected from 54 untreated CLL patients and 40 HDs (age- and sex-matched) for quantitative polymerase chain reaction (qPCR) to evaluate the clinical values of mc-COX2. As expected, mc-COX2 was obviously more expressed in CLL patients compared with HDs (Figure 2A). Then, the CLL patients were divided into two groups according to the plasma mc-COX2 expression. Kaplan-Meier analysis showed that mc-COX2high CLL patients had a significantly worse overall survival (OS) compared with the mc-COX2low group (Figure 2B). The clinical characteristics of CLL patients are shown in Table S2. As reported, mitochondrial metabolism and function play important roles in the occurrence and progression of CLL.3 TP53, which is regarded as a crucial prognostic indicator in CLL, is closely correlated with mitochondria respiration.19 Thus, we tried to determine the interrelationship between mc-COX2 and TP53. Results showed that the CLL patients with TP53 deletion rather than mutation displayed higher expression of mc-COX2 (Figures 2C and S1A).
Figure 2.
mc-COX2 Enriched in Exosomes from CLL Plasma Predicts Prognosis of CLL Patients
(A) Expression of mc-COX2 in plasma samples from 54 untreated CLL patients and 40 HDs. p = 0.0011. (B) Kaplan-Meier survival curves of CLL patients with high (mc-COX2high, n = 27) or low (mc-COX2low, n = 27) mc-COX2 expression. p = 0.0073. (C) mc-COX2 expression associated with TP53 deletion. p = 0.001. (D) Plasma exosomes isolated and characterized by electron microscopy. (E and F) Flow cytometry (E) and western blotting (F) of exosomal markers CD63, CD81, and TSG101. (G) Size ranges of exosomes from particle size analysis. (H) High-expression of mc-COX2 in exosomes of CLL plasma via qRT-PCR. Data are shown as mean ± SD.
Evidence demonstrates that circRNAs are enriched and stable in exosomes, by which circRNAs can enter the circulation and be readily detected in the serum.7 Based on the result that mc-COX2 was highly expressed in CLL plasma, we speculated that mc-COX2 was delivered by exosomes into plasma from CLL cells. Thus, we collected plasma samples and isolated plasma exosomes from 20 CLL patients and 20 HDs. These vesicles were then characterized by electron microscopy (Figure 2D). In addition, the presence of exosomal markers CD63, CD81, and TSG101 were detected by flow cytometry and western blot, which further confirmed the purity of the isolated exosomes from plasma (Figures 2E and 2F). Measurement of exosome particle size is shown in Figure 2G. As expected, the results of qRT-PCR showed that mc-COX2 was highly enriched in exosomes from CLL plasma compared with normal controls (Figure 2H). The results imply that mc-COX2 is abundant in exosomes and can be delivered into plasma from CLL cells.
Characterization of mc-COX2
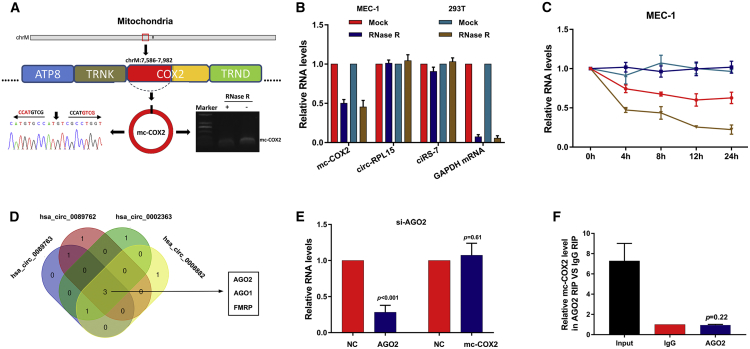
hsa_circ_0089762 was generated from COX2 on the mitochondrial genome and thus was termed mc-COX2 here. The presence of mc-COX2 in MEC-1 (a CLL cell line) and HEK293T (a human embryonal kidney cell line) was validated by sequencing the RT-PCR products amplified with specific spanning junction primers (Figure 3A; Table S1). RNase R exonuclease and actinomycin D were used to validate the stability of RNA isoforms. Unexpectedly, compared with ciRS-7 and circ-RPL15 (another nu-circRNA identified and confirmed to be highly expressed in CLL plasma in our previous study20), mc-COX2 can be degraded by RNase R to a certain extent. However, mc-COX2 was more tolerant against RNase R compared with linear mRNA GAPDH (Figure 3B). Consistent results were shown after actinomycin D treatment (Figures 3C and S2A). Furthermore, the results of qRT-PCR using random hexamer primers and oligo(dT)18 primers showed that mc-COX2 was a circRNA isoform without a poly(A) tail (Figure S2B). The abundance of ciRS-7, circ-RPL15, and GAPDH was detected at the same time both for comparisons and as controls. The above results suggest that mc-COX2 is less stable than ciRS-7 and circRPL15, but is much more stable than linear RNAs.
Figure 3.
Characterization of mc-COX2
(A) Genomic loci and basic characteristics of mc-COX2. Amplified products from MEC-1 and HEK293T cells with spanning junction primers with or without RNase R treatment verified by Sanger sequencing. (B and C) qRT-PCR for the abundance of mt-circRNA, nu-circRNAs, and liner mRNA treated with (B) RNase R and (C) actinomycin D. (D) Venn diagram of overlapping RBPs with binding sites of mt-circRNAs predicted by CircInteractome. (E) Expression change of mc-COX2 after knockdown of AGO2. (F) RIP assay of AGO2 compared with immunoglobulin G (IgG) antibody. Abundance of mc-COX2 was detected by qRT-PCR. Data are shown as mean ± SD.
RNA-binding proteins (RBPs) are involved in the biosynthetic process of circRNAs.21 Given that plentiful binding sites of RBPs are present on the mitochondrial genome, which can generate mt-circRNAs, we wondered whether RBPs can modulate the biogenesis of mt-circRNAs. Of the top five RBPs, which were predicted to bind with the four mt-circRNAs via CircInteractome (https://circinteractome.nia.nih.gov), the Venn diagram screened out three potential RBPs (AGO1, AGO2, and FMRP) (Figure 3D). The binding sites of AGO2 are shown in Figure S2C. However, the results of qRT-PCR found that after the effective knockdown of AGO2, AGO1, and FMRP with small interfering RNAs (siRNAs), the mc-COX2 expression did not significantly change in MEC-1 (Figures 3E, S2D, and S2E).
Since circRNA has been widely demonstrated to function as competitive endogenous RNA (ceRNA),22,23 we then evaluated the potential of mt-circRNAs to sponge miRNAs. An RNA immunoprecipitation (RIP) assay with an AGO2 antibody was performed to examine the binding ability of circRNAs to AGO2 protein, which is regarded as the precondition for circRNAs functioning as ceRNAs. Unlike ciRS-7, mc-COX2 cannot be enriched with the AGO2 antibody (Figures 3F and S2F). Thus, we speculated that mc-COX2 and even other mt-circRNAs may function through special mechanisms that are different from nu-circRNAs.
mc-COX2 Impacts Mitochondrial Functions and Regulates CLL Cell Growth
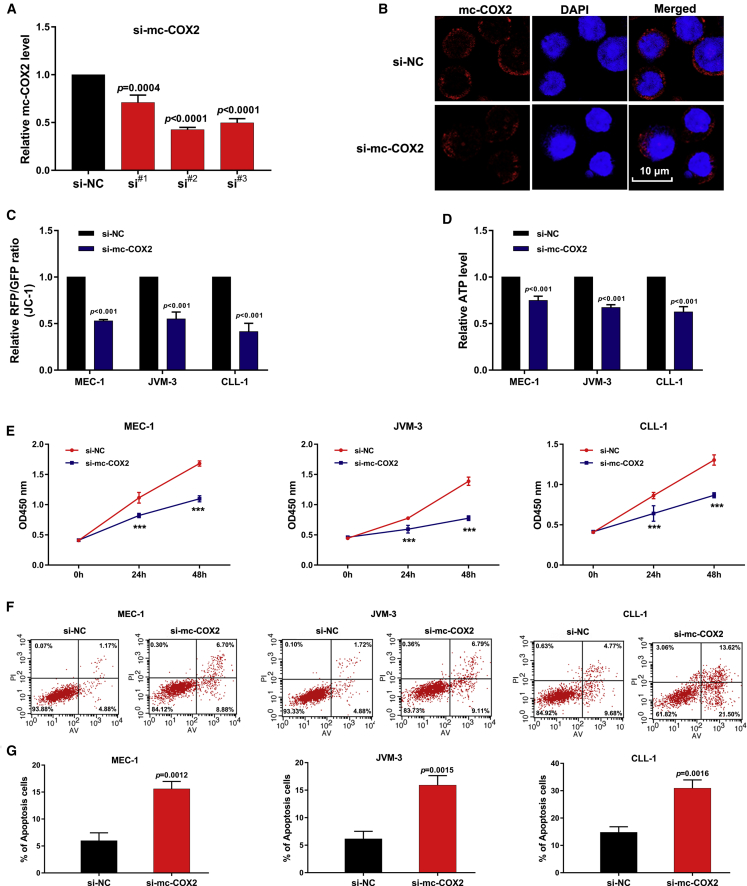
circRNAs are reportedly associated with the occurrence and development of various human diseases.24,25 To investigate the functional effects of mc-COX2 in CLL, we performed a loss-of-function analysis. Three siRNAs that targeted mc-COX2 were designed and transfected in CLL cell lines. The second one with the highest transfection effect was finally selected (Figure 4A). RNA FISH further verified that si-mc-COX2#2 can reduce the abundance of mc-COX2 (Figure 4B).
Figure 4.
Knockdown of mc-COX2 Affects Mitochondrial Functions and Promotes Apoptosis of CLL Cells
(A and B) Efficiency of siRNAs was detected by (A) qRT-PCR and (B) RNA FISH. Scale bar, 10 μm. (C and D) MMP analyzed by JC-1 fluorescent probe (C) and ATP levels in CLL cells after knockdown of mc-COX2 (D). (E–G) Cell proliferation (E and F) and apoptosis analysis (G) of CLL cell lines and CLL primary cells. The apoptosis ratio was determined. CLL cell lines: MEC-1 and JVM3; primary cells: CLL-1. Data are shown as mean ± SD.
Given that mc-COX2 is derived from mitochondria genome, we wondered whether mc-COX2 expression can regulate mitochondrial functions. We analyzed mitochondrial membrane potential using a JC-1 fluorescent probe and found that the knockdown of mc-COX2 reduced the red fluorescent protein (RFP)/GFP ratio (Figure 4C). Moreover, regulation of the ATP level by mc-COX2 was examined via an ATP assay. ATP production significantly decreased after suppressing mc-COX2 (Figure 4D). Then, to explore the roles of mc-COX2 in cell growth, we tested CLL cell lines (MEC-1, JVM-3) and the primary cells from CLL patients (CLL-1) via a Cell Counting Kit-8 (CCK-8) assay. The results indicated that cell viability was significantly higher in cells transfected with negative controls than those with si-mc-COX2 (Figure 4E). Flow cytometry also revealed that the decrease of mc-COX2 levels evidently induced cell apoptosis (Figure 4F). The same result was verified in the primary cells from one CLL patient, and the percentage of apoptotic cells was quantified (Figure 4G). These results indicate that mc-COX2 not only modulates mitochondrial function, but it also markedly regulates cell proliferation and apoptosis.
Mitochondria-Related Compounds and Inhibitors Can Reduce mc-COX2 and Suppress Proliferation of CLL Cells
Reportedly, mitochondrial biogenesis and the number of mitochondria are increased in CLL cells compared to normal B lymphocytes.3 We then wondered whether the mc-COX2 expression was positively associated with the mass and number of mitochondria. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP), an uncoupler of the electron transport chain (ETC), can lead to mitochondrial failure and cell apoptosis.26 After the CLL cell lines were treated with CCCP even at a low dose, mc-COX2 was dramatically downregulated at different time points (Figure 5A). The treatment with 5 μM or a higher dose of CCCP clearly inhibited cell proliferation. However, silencing of mc-COX2 changed the results at low concentrations and enhanced the inhibition compared with the single treatment of 1 μM CCCP or mc-COX2 (Figure 5B). After incubation with doxycycline, an antibiotic that can disrupt mitochondrial biogenesis and reduce cancer stem cells,27,28 mc-COX2 was predominantly reduced in the CLL cells (Figure 5C). Moreover, cell growth was inhibited by a certain dose of doxycycline, and this effect was potentiated by the knockdown of mc-COX2 (Figure 5D). Furthermore, after treatment with metformin, an effective inhibitor of the mitochondrial respiratory chain that can decrease the incidence, progression, and mortality of different human cancers,29,30 mc-COX2 was significantly downregulated in CLL cells (Figure 5E), and cell proliferation was inhibited in a dose- and time-dependent manner, which are consistent with a previous study.31 Moreover, the anti-leukemic activity was enhanced in combination with si-mc-COX2 (Figure 5F).
Figure 5.
Effects of Chemical Compounds on CLL Cells
(A, C, and E) Expression levels of mc-COX2 in CLL cells treated with a range dose of three chemical compounds (A, CCCP; C, doxycycline; E, metformin) at different time points. (B, D, and F) CCK-8 analysis of cell growth with the three chemical compounds (B, CCP; D, doxycycline; F, metformin) and/or with si-mc-COX2. Data are shown as mean ± SD.
To further investigate the effect of mitochondria on mc-COX2, we selected four mitochondria-related small-molecule inhibitors (daporinad, PI-103, OSI-027, ABT-199). Daporinad,32 PI-103,33,34 and ABT-19935,36 are potent inducers of CLL cell apoptosis, even in patients with TP53 dysfunction or other high-risk features. All four inhibitors can inhibit mc-COX2 expression and cell proliferation in CLL cells (Figures 6A–6F). Moreover, OSI-027, a potent and selective mTOR inhibitor, has the potential for anti-pancreatic-cancer activity,37,38 but its function in CLL is still unknown. Results showed that the 3 μM OSI-027 treatment exhibited a dose-dependent anti-leukemic activity and completely inhibited cell growth (Figure 6E). These data suggest that the damage and number of mitochondria both affect the biogenesis of mc-COX2. More importantly, mc-COX2 reduction was already evident at 6 h after treatment, indicating it is an early event in biological generation. Additionally, CLL cells were significantly more responsive to these compounds with the cooperation of mc-COX2 interference. Thus, mc-COX2 may be a complement target to enhance their efficacy and/or allow dose reduction for improved drug sensitivity.
Figure 6.
Effects of Small Molecular Inhibitors on CLL Cells
(A and B) Expression levels of mc-COX2 in CLL cells treated with a range dose of inhibitors in (A) MEC-1 and (B) JVM-3 at different time points are shown. (C–F) CCK-8 analysis of cell growth with (C) daporinad, (D) PI-103, (E) OSI-027, and (F) ABT-199. Data are shown as mean ± SD.
Discussion
circRNAs have drawn extensive attention from the cancer research field recently.39,40 The role of circRNAs in CLL has also been emphasized, including their effects on modulating disease progression. Additionally, circRNAs are potential diagnostic biomarkers and therapeutic targets in CLL.20,22,41 With the rapid development of CLL diagnosis and therapy strategies during the last few decades, several high-risk features, such as IGHV mutation status, TP53 mutation, and 17p deletion, have been regarded as useful assistants for Rai and Binet staging systems, which were proved to be robust prognostic tools for CLL patients.42, 43, 44, 45 However, the lack of convenient and reliable biomarkers is a hindrance in monitoring CLL progression. Thus, it is urgent to screen effective new biomarkers and explore potential therapeutic targets associated with CLL initiation and progression.
As reported, circRNAs as proto-oncogene or tumor suppressor factors play crucial roles in tumor occurrence and development. circRNAs in exosomes have been found in certain cancers.7,46 However, the functions and clinical significance of circRNAs in CLL have been rarely studied. In our research, first, we demonstrated the existence of mt-circRNAs and the evident upregulation of mc-COX2 in CLL plasma. mc-COX2 was positively correlated with TP53 deletion, which is crucial to the evaluation of prognosis. Also, the results highlight its applicability as a promising prognostic indicator for CLL. Remarkably, the high mc-COX2 expression can be examined and stabilized in plasma exosomes of CLL patients, which has not been reported before. Next, we unraveled the biological characteristic of mc-COX2. Compared with nu-circRNAs, mt-COX2 was less stable, but it was still significantly more stable than linear RNAs. Meanwhile, mc-COX2 cannot bind to AGO2 protein, suggesting it probably functions via other mechanisms instead of acting as ceRNA. Mitochondria play an important role in cellular metabolism and are involved in cell fate. Targeting mitochondrial bioenergetics and metabolic alterations has become an interest in the realm of cancer therapy.47 We noticed the modulating effect of mitochondria on mc-COX2 expression level and explored the effects of mitochondria-related compounds on CLL cell growth. Low-dose or transient treatment with chemical compounds was sufficient to damage mitochondria and interrupt the mc-COX2 expression. Additionally, small-molecule inhibitors also abrogated cell proliferation and this effect was enhanced in combination with mc-COX2 silencing. The inference of these results is two-sided: they indicate that the status of mitochondria modulates the expression of mt-circRNAs, but they also suggest a strategy for dual inhibition treatment.
In summary, (1) mt-circRNAs are highly expressed in the plasma of CLL patients. (2) mc-COX2 is stably expressed in exosomes, and it may not only serve as a new “liquid biopsy” biomarker for CLL prognosis, but it may also provide potential diagnostic implications for patients. (3) mc-COX2 exhibits some biological and functional characteristics that differ from nu-circRNAs. (4) mc-COX2 can dampen mitochondrial function and modulate cell fate. Moreover, mitochondria can be severely compromised following the treatment with mitochondria-related compounds, including novel inhibitors that have not been studied in CLL, such as OSI-027. Shan et al.5 first reported the existence of circRNAs from human and mouse mitochondria. They revealed the physiological role of mt-circRNAs in regulating protein importation, which has broadened our view of understanding circRNAs and mitochondria. In the present study, we proposed the existence and unique biological function of mt-circRNAs in CLL. Notably, it is inspiring to find the presence of mt-circRNAs in plasma exosomes. Nevertheless, much remains unknown about mt-circRNAs, including their origin and degradation. More importantly, it is necessary to explore how mt-circRNAs regulate mitochondria and affect disease progression.
Materials and Methods
Patient Samples and Isolation of Exosomes from Plasma
Plasma samples were collected from 54 untreated CLL patients and 40 HDs at Jiangsu Province Hospital between 2013 and 2018. After cells and other debris were removed from plasma by centrifugation at 300 and 3,000 × g, the supernatants were centrifuged at 10,000 × g for 30 min to remove the shedding vesicles and other larger-sized vesicles. Finally, plasma exosomes were isolated using an ExoQuick-TC kit (Systems Biosciences, Mountain View, CA, USA) following the manufacturer’s protocol. All patients were followed up on a regular basis, and OS time was determined from diagnosis to the date of death due to any cause or to the time of the last follow-up. This study was approved by the Ethics Committee of Jiangsu Province Hospital. Written informed consent was obtained from each participant.
circRNA Microarray
The circRNA expression profiles in plasma of five treatment-naive CLL patients and age- and sex-matched HDs were analyzed using a human circRNA array v2 (CapitalBio Technology, Beijing, China). circRNAs with fold changes ≥2.0 and p ≤0.05 were identified as significantly and differentially expressed.
Cell Culture and Transfection
The CLL cell lines (MEC-1 [TP53-mutant], JVM-3 [TP53-wild-type]) and HEK293T cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium and Dulbecco’s modified Eagle’s medium (DMEM) (Biological Industries), respectively, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin and maintained at 37°C with 5% CO2. MEC-1 cells and HEK293T cells were transfected with a Lipofectamine stem reagent and a Lipofectamine 2000 reagent (both Invitrogen, Carlsbad, CA, USA), respectively, according to the manufacturer’s instructions. The siRNAs used in our research were designed and synthesized by RiboBio (Guangzhou, China).
Reagents
Daporinad (catalog no. S2799), venetoclax (catalog no. S8048), OSI-027 (catalog no. S2624), PI-103 (catalog no. S1038), and doxycycline (catalog no. S5159) were obtained from Selleck Chemicals (Houston, TX, USA). CCCP (catalog no. HY-100941) and metformin hydrochloride (catalog no. HY-17471A) were purchased from MedChemExpress (Greenville, SC, USA). Anti-CD63 (ab59479), anti-CD81 (ab79559), and anti-TSG101 (ab125011) antibodies for immunoblotting and flow cytometry were purchased from Abcam (Cambridge, UK).
ATP Assay
ATP concentrations were tested with an enhanced ATP assay kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. Cells were lysed with an ATP lysis buffer and centrifuged at 12,000 × g and 4°C for 5 min. The supernatants were collected and added with an ATP working solution. After incubation for 3–5 min at room temperature, the intracellular ATP contents were measured with a luminometer.
Mitochondrial Membrane Potential (MMP)
MMP was detected using an MMP assay kit with JC-1 (Beyotime). Cells were incubated with 500 μL of a JC-1 working solution at 37°C for 20 min. The fluorescence-labeled cells were then washed twice and analyzed by flow cytometry.
CCK-8 Assay
Cells were seeded in 96-well plates at a density of 1 × 104 cells/well in triplicate and incubated at 37°C. Cell viability was determined by a CCK-8 kit (APExBIO, Houston, TX, USA) according to the manufacturer’s instructions. The solution was measured spectrophotometrically at 450 nm.
Cell Apoptosis Assay
Cells after different treatments were collected and washed in cold phosphate-buffered saline (PBS). After incubation with propidium iodine (PI)/annexin V-fluorescein isothiocyanate (FITC) staining (annexin V-Alexa Fluor 647/PI apoptosis detection kit) at room temperature and away from light for 15 min, cell apoptosis was detected by flow cytometry.
RNA Preparation and qRT-PCR
Total RNA was extracted from cells using TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. Plasma RNA was extracted using a TIANamp virus RNA kit (Tiangen, Beijing, China). For the detection of RNA stability, total RNA (2 μg) was incubated with 1 μL of RNase R (Epicenter Technologies, Madison, WI, USA) at 37°C for 20 min. Actinomycin D (2 μg/mL) was added to the culture medium, and cells were collected at the appointed time. RIP experiments were performed with a Magna RIP RBP immunoprecipitation kit (Millipore, Bedford, MA, USA) to obtain RNAs, which were used to detect the abundance of circRNAs. RNAs were reverse transcribed using Goldenstar RT6 cDNA synthesis kits (Tsingke, Beijing, China) with random primers or oligo(dT)18 primers. Also, qPCR was performed with Hieff qPCR SYBR Green master mix (Yeasen, Shanghai, China). GAPDH mRNA was used as a control. Primers are listed in Table S1.
Northern Blotting
RNA (10–20 mg) for detection of endogenous circRNAs was denatured and loaded onto a 1% agarose gel. Electrophoresis was carried out overnight at 25 V and 4°C. RNA was transferred onto Hybond-N+ membranes (Amersham, USA) by capillarity for 20 h after being washed in 20× saline sodium citrate (SSC). Prehybridization and hybridization were then performed in 10.0 mL of DIG Easy Hyb with denatured probes (designed by Zoonbio Biotechnology, Nanjing, China) at 68°C. The membranes were then washed, blocked, and exposed on phosphorimager screens for analysis.
RNA FISH
An RNA FISH assay was performed using a FISH kit (RiboBio, Guangzhou, China) according to the manufacturer’s guidelines. Cy3-labeled mc-COX2 probes (RiboBio) and mitochondrion-selective probes (MitoTracker Green FM, Invitrogen) were measured by visualization under confocal microscopy.
Statistical Analysis
Statistical analyses were performed using Student’s t test in GraphPad Prism 7.0. OS was assessed with the Kaplan-Meier method and compared by the log rank test. Correlations were analyzed by Pearson’s correlation test. All data generated or analyzed during this study are included in the published article and accompanying Supplemental Information.
Author Contributions
J.L. and H.J. were involved in the study design and supervised the study. Z.W. and H.S. performed the experiments and drafted the manuscript. Z.W., C.W., M.L., and W.X. analyzed the data. C.W., W.L., and Y.Z. collected and provided the clinical samples. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81700155 and 81720108002), the Jiangsu Provincial Special Program of Medical Science (BE2017751), and by the National Science and Technology Major Project (2018ZX09734-007).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.04.017.
Contributor Information
Hui Jin, Email: jinzi817@live.cn.
Jianyong Li, Email: lijianyonglm@126.com.
Supplemental Information
References
- 1.Hallek M. Chronic lymphocytic leukemia: 2017 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2017;92:946–965. doi: 10.1002/ajh.24826. [DOI] [PubMed] [Google Scholar]
- 2.Raponi S., Del Giudice I., Marinelli M., Wang J., Cafforio L., Ilari C., Piciocchi A., Messina M., Bonina S., Tavolaro S. Genetic landscape of ultra-stable chronic lymphocytic leukemia patients. Ann. Oncol. 2018;29:966–972. doi: 10.1093/annonc/mdy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jitschin R., Hofmann A.D., Bruns H., Giessl A., Bricks J., Berger J., Saul D., Eckart M.J., Mackensen A., Mougiakakos D. Mitochondrial metabolism contributes to oxidative stress and reveals therapeutic targets in chronic lymphocytic leukemia. Blood. 2014;123:2663–2672. doi: 10.1182/blood-2013-10-532200. [DOI] [PubMed] [Google Scholar]
- 4.Carew J.S., Nawrocki S.T., Xu R.H., Dunner K., McConkey D.J., Wierda W.G., Keating M.J., Huang P. Increased mitochondrial biogenesis in primary leukemia cells: the role of endogenous nitric oxide and impact on sensitivity to fludarabine. Leukemia. 2004;18:1934–1940. doi: 10.1038/sj.leu.2403545. [DOI] [PubMed] [Google Scholar]
- 5.Liu X., Wang X., Li J., Hu S., Deng Y., Yin H., Bao X., Zhang Q.C., Wang G., Wang B. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 2020 doi: 10.1007/s11427-020-1631-9. Published online January 21, 2020. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan B.T., Teng K., Wu C., Adam M., Johnstone R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Théry C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 10.Becker A., Thakur B.K., Weiss J.M., Kim H.S., Peinado H., Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kater A.P., Levin M.D., Niemann C.U. Ibrutinib and venetoclax for first-line treatment of CLL. N. Engl. J. Med. 2019;381:788–789. doi: 10.1056/NEJMc1908754. [DOI] [PubMed] [Google Scholar]
- 12.Woyach J.A., Ruppert A.S., Guinn D., Lehman A., Blachly J.S., Lozanski A., Heerema N.A., Zhao W., Coleman J., Jones D. BTKC481S-mediated resistance to ibrutinib in chronic lymphocytic leukemia. J. Clin. Oncol. 2017;35:1437–1443. doi: 10.1200/JCO.2016.70.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giménez-Cassina A., Danial N.N. Regulation of mitochondrial nutrient and energy metabolism by BCL-2 family proteins. Trends Endocrinol. Metab. 2015;26:165–175. doi: 10.1016/j.tem.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song T., Chai G., Liu Y., Yu X., Wang Z., Zhang Z. Bcl-2 phosphorylation confers resistance on chronic lymphocytic leukaemia cells to the BH3 mimetics ABT-737, ABT-263 and ABT-199 by impeding direct binding. Br. J. Pharmacol. 2016;173:471–483. doi: 10.1111/bph.13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan R., Hogdal L.J., Benito J.M., Bucci D., Han L., Borthakur G., Cortes J., DeAngelo D.J., Debose L., Mu H. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4:362–375. doi: 10.1158/2159-8290.CD-13-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L., Zhang S., Wu J., Cui J., Zhong L., Zeng L., Ge S. Retraction. Oncogene. 2019;38:5750. doi: 10.1038/s41388-019-0828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 18.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Wang S., Peng Z., Wang S., Yang L., Chen Y., Kong X., Song S., Pei P., Tian C., Yan H. KRAB-type zinc-finger proteins PITA and PISA specifically regulate p53-dependent glycolysis and mitochondrial respiration. Cell Res. 2018;28:572–592. doi: 10.1038/s41422-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z., Sun H., Liu W., Zhu H., Fu J., Yang C., Fan L., Wang L., Liu Y., Xu W. circ-RPL15: a plasma circular RNA as novel oncogenic driver to promote progression of chronic lymphocytic leukemia. Leukemia. 2020;34:919–923. doi: 10.1038/s41375-019-0594-6. [DOI] [PubMed] [Google Scholar]
- 21.Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., Rosa A., De Santis R., Scarfò R., Peruzzi G. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu W., Wu Z., Xia Y., Qin S., Li Y., Wu J., Liang J., Wang L., Zhu H., Fan L. Downregulation of circ_0132266 in chronic lymphocytic leukemia promoted cell viability through miR-337-3p/PML axis. Aging (Albany NY) 2019;11:3561–3573. doi: 10.18632/aging.101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X., Chen R.X., Wei W.S., Li Y.H., Feng Z.H., Tan L., Chen J.W., Yuan G.J., Chen S.L., Guo S.J. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin. Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 24.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Liu H., Hou L., Wang G., Zhang R., Huang Y., Chen X., Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández-Mosquera L., Diogo C.V., Yambire K.F., Santos G.L., Luna Sánchez M., Bénit P., Rustin P., Lopez L.C., Milosevic I., Raimundo N. Acute and chronic mitochondrial respiratory chain deficiency differentially regulate lysosomal biogenesis. Sci. Rep. 2017;7:45076. doi: 10.1038/srep45076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scatena C., Roncella M., Di Paolo A., Aretini P., Menicagli M., Fanelli G., Marini C., Mazzanti C.M., Ghilli M., Sotgia F. Doxycycline, an inhibitor of mitochondrial biogenesis, effectively reduces cancer stem cells (CSCs) in early breast cancer patients: a clinical pilot study. Front. Oncol. 2018;8:452. doi: 10.3389/fonc.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newby A.C. Matrix metalloproteinase inhibition therapy for vascular diseases. Vascul. Pharmacol. 2012;56:232–244. doi: 10.1016/j.vph.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Owen M.R., Doran E., Halestrap A.P. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J. 2000;348:607–614. [PMC free article] [PubMed] [Google Scholar]
- 30.Morales D.R., Morris A.D. Metformin in cancer treatment and prevention. Annu. Rev. Med. 2015;66:17–29. doi: 10.1146/annurev-med-062613-093128. [DOI] [PubMed] [Google Scholar]
- 31.Voltan R., Rimondi E., Melloni E., Gilli P., Bertolasi V., Casciano F., Rigolin G.M., Zauli G., Secchiero P. Metformin combined with sodium dichloroacetate promotes B leukemic cell death by suppressing anti-apoptotic protein Mcl-1. Oncotarget. 2016;7:18965–18977. doi: 10.18632/oncotarget.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gehrke I., Bouchard E.D., Beiggi S., Poeppl A.G., Johnston J.B., Gibson S.B., Banerji V. On-target effect of FK866, a nicotinamide phosphoribosyl transferase inhibitor, by apoptosis-mediated death in chronic lymphocytic leukemia cells. Clin. Cancer Res. 2014;20:4861–4872. doi: 10.1158/1078-0432.CCR-14-0624. [DOI] [PubMed] [Google Scholar]
- 33.Niedermeier M., Hennessy B.T., Knight Z.A., Henneberg M., Hu J., Kurtova A.V., Wierda W.G., Keating M.J., Shokat K.M., Burger J.A. Isoform-selective phosphoinositide 3′-kinase inhibitors inhibit CXCR4 signaling and overcome stromal cell-mediated drug resistance in chronic lymphocytic leukemia: a novel therapeutic approach. Blood. 2009;113:5549–5557. doi: 10.1182/blood-2008-06-165068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shehata M., Schnabl S., Demirtas D., Hilgarth M., Hubmann R., Ponath E., Badrnya S., Lehner C., Hoelbl A., Duechler M. Reconstitution of PTEN activity by CK2 inhibitors and interference with the PI3-K/Akt cascade counteract the antiapoptotic effect of human stromal cells in chronic lymphocytic leukemia. Blood. 2010;116:2513–2521. doi: 10.1182/blood-2009-10-248054. [DOI] [PubMed] [Google Scholar]
- 35.2014). ABT-199 shows effectiveness in CLL. Cancer Discov. 4, OF7. [DOI] [PubMed]
- 36.Souers A.J., Leverson J.D., Boghaert E.R., Ackler S.L., Catron N.D., Chen J., Dayton B.D., Ding H., Enschede S.H., Fairbrother W.J. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 37.Chen B., Xu M., Zhang H., Xu M.Z., Wang X.J., Tang Q.H., Tang J.Y. The antipancreatic cancer activity of OSI-027, a potent and selective inhibitor of mTORC1 and mTORC2. DNA Cell Biol. 2015;34:610–617. doi: 10.1089/dna.2015.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhi X., Chen W., Xue F., Liang C., Chen B.W., Zhou Y., Wen L., Hu L., Shen J., Bai X., Liang T. OSI-027 inhibits pancreatic ductal adenocarcinoma cell proliferation and enhances the therapeutic effect of gemcitabine both in vitro and in vivo. Oncotarget. 2015;6:26230–26241. doi: 10.18632/oncotarget.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang S., Zeng X., Ding T., Guo L., Li Y., Ou S., Yuan H. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci. Rep. 2018;8:2878. doi: 10.1038/s41598-018-21300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsiao K.Y., Lin Y.C., Gupta S.K., Chang N., Yen L., Sun H.S., Tsai S.J. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia L., Wu L., Bao J., Li Q., Chen X., Xia H., Xia R. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018;503:385–390. doi: 10.1016/j.bbrc.2018.06.045. [DOI] [PubMed] [Google Scholar]
- 42.Delgado J., Doubek M., Baumann T., Kotaskova J., Molica S., Mozas P., Rivas-Delgado A., Morabito F., Pospisilova S., Montserrat E. Chronic lymphocytic leukemia: a prognostic model comprising only two biomarkers (IGHV mutational status and FISH cytogenetics) separates patients with different outcome and simplifies the CLL-IPI. Am. J. Hematol. 2017;92:375–380. doi: 10.1002/ajh.24660. [DOI] [PubMed] [Google Scholar]
- 43.Parikh S.A., Shanafelt T.D. Prognostic factors and risk stratification in chronic lymphocytic leukemia. Semin. Oncol. 2016;43:233–240. doi: 10.1053/j.seminoncol.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Collado R., Puiggros A., López-Guerrero J.A., Calasanz M.J., Larráyoz M.J., Ivars D., García-Casado Z., Abella E., Orero M.T., Talavera E. Chronic lymphocytic leukemia with isochromosome 17q: an aggressive subgroup associated with TP53 mutations and complex karyotypes. Cancer Lett. 2017;409:42–48. doi: 10.1016/j.canlet.2017.08.041. [DOI] [PubMed] [Google Scholar]
- 45.Rai K.R., Jain P. Chronic lymphocytic leukemia (CLL)—then and now. Am. J. Hematol. 2016;91:330–340. doi: 10.1002/ajh.24282. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H., Zhu L., Bai M., Liu Y., Zhan Y., Deng T., Yang H., Sun W., Wang X., Zhu K. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int. J. Cancer. 2019;144:2501–2515. doi: 10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]
- 47.Simula L., Nazio F., Campello S. The mitochondrial dynamics in cancer and immune-surveillance. Semin. Cancer Biol. 2017;47:29–42. doi: 10.1016/j.semcancer.2017.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.