Abstract
Objective
Statins are a group of medications that reduce cholesterol synthesis by inhibiting the activity of HMG-CoA reductase, a key enzyme in the mevalonate pathway. The clinical use of statins to lower excess cholesterol levels has revolutionized the cardiovascular field and increased the survival of millions, but some patients have adverse side effects. A growing body of data suggests that some of the beneficial and adverse effects of statins, including their anti-inflammatory, anti-tumorigenic, and myopathic activities, are cholesterol-independent. However, the underlying mechanisms for these effects of statins are not well defined.
Methods
Because Caenorhabditis elegans (C. elegans) lacks the cholesterol synthesis branch of the mevalonate pathway, this organism is a powerful system to unveil the cholesterol-independent effects of statins. We used genetic and biochemical approaches in C. elegans and cultured macrophage-derived murine cells to study the cellular response to statins.
Results
We found that statins activate a conserved p38-MAPK (p38) cascade and that the protein geranylgeranylation branch of the mevalonate pathway links the effect of statins to the activation of this p38 pathway. We propose that the blockade of geranylgeranylation impairs the function of specific small GTPases we identified as upstream regulators of the p38 pathway. Statin-mediated p38 activation in C. elegans results in the regulation of programs of innate immunity, stress, and metabolism. In agreement with this regulation, knockout of the p38 pathway results in the hypersensitivity of C. elegans to statins. Treating cultured mammalian cells with clinical doses of statins results in the activation of the same p38 pathway, which upregulates the COX-2 protein, a major regulator of innate immunity in mammals.
Conclusions
Statins activate an evolutionarily conserved p38 pathway to regulate metabolism and innate immunity. Our results highlight the cytoprotective role of p38 activation under statin treatment in vivo and propose that this activation underlies many of the critical cholesterol-independent effects of statins.
Keywords: Statins, Cholesterol-independent effect, Mevalonate pathway, Geranylgeranylation, Small GTPases, p38 pathway, COX-2, C. elegans, Innate immunity.
Highlights
-
•
Statins activate a conserved stress-related p38-MAPK in C. elegans and mammalian cells.
-
•
The blockadge of geranylgeranylation of small GTPases is a suggested mechanism for p38 activation.
-
•
p38 activation can protect C. elegans from the deleterious effects of statins.
-
•
Clinical concentrations of statins activate p38 to alter innate immunity programs in mammals.
1. Introduction
Cardiovascular diseases (CVDs) are one of the two leading causes of death worldwide and remain among the most significant medical challenges of our time [1]. A revolutionary breakthrough in the fight against CVDs occurred 40 years ago with the discovery of statins, a group of cholesterol-lowering medications [2,3] that collectively have become one of the most widely prescribed classes of drugs [4,5]. In addition to lowering cholesterol levels, statins have a plethora of benefits, including anti-inflammatory [6] and anti-cancerous [7,8] effects, as well as proposed anti-neurodegenerative properties [9,10]. However, statins can also cause adverse side effects, ranging from the development of myopathy found in 1–2% [11,12] and 9% [13] of statin-taking patients, to the onset of type II diabetes in 1–2% of these patients [11,12].
Statins inhibit HMG-CoA reductase (HMGCR), the second enzyme in the main branch of the mevalonate pathway, which converts HMG-CoA into mevalonate [14]. Statins inhibit the synthesis of cholesterol as well as other products of the mevalonate pathway, including electron carriers used in mitochondrial respiration, anchors for protein glycosylation in the ER, and lipid moieties required for the farnesylation and geranylgeranylation of proteins to intracellular membranes [15]. Thus, statin treatment potentially has a myriad of mevalonate-dependent but cholesterol-independent effects on the cells.
Intriguingly, and in keeping with this concept, an increasing number of studies suggest that key aspects of the therapeutic activity of statins may be independent of their cholesterol-reducing activity. For example, statin treatment in patients between the ages of 70 and 90 reduced mortality rates independent of their blood cholesterol levels [16]. Surprisingly, even cardioprotection by statins is at least partially or even entirely facilitated by the cholesterol-independent activity of statins on endothelial cells of the vascular system [17]. Although mevalonate pathway metabolism is required for the synthesis of many end products, a growing body of data links the cholesterol-independent effects of statins with the inhibition of geranylgeranylation. Geranylgeranylation is critical for the activity of many small GTPases from the RAS superfamily, which function as regulators of numerous signaling pathways, including pathways involved in immunity and tumorigenicity [18]. Inhibition of geranylgeranylation was proposed to account for the anti-inflammatory [6] and anti-cancerous [8,19] activities of statins but the molecular mechanism underlying these responses remains largely unknown.
Recent findings in mammalian cell cultures have suggested that the modulation of the p38 pathway, one of the conserved MAP kinase family members, is a part of the cellular response to statins. The p38 pathway plays a pivotal role in the coordination of many biological processes, including stress and immune responses, as well as tumorigenesis (review in [20,21]). The core architecture of this pathway relies on the sequential activation of three kinases: the MAPKKK, MAPKK, and MAPK proteins. Activation of the MAPK protein often results in a transcriptional response in the nucleus and the phosphorylation of target proteins in the cytoplasm [22]. In cultures of murine macrophage (RAW264.7), statin treatment results in the inactivation of the small GTPases RHO-1 and CDC-42 [23]. This inactivation triggers p38 signaling to upregulate the cyclooxygenase-2 (COX-2) enzyme. COX-2 upregulation results in the synthesis of 15-deoxy-delta-12,14-prostaglandin J2 (15d-PGJ2), which in these cells has an anti-inflammatory effect. Whether this effect is related to inhibition of the mevalonate pathway or stems from one of the many off-target effects of statins remains unclear [23]. Moreover, p38 activation and COX-2 upregulation in RAW264.7 cells were shown to take place in micromolar concentrations of statins. It is not clear if therapeutic concentrations of statins, which in many clinical settings range between 34 nM and 234 nM [23,24], can elicit the same response.
Caenorhabditis elegans lacks the cholesterol-synthesizing branch of its mevalonate pathway [25] but has all other arms, including those that are responsible for the synthesis of electron carriers and the moieties required for protein prenylation. Therefore, because C. elegans does not depend on the mevalonate pathway as its source of cholesterol, it constitutes a powerful tool to dissect and understand the cholesterol-independent effects of statins in vivo. Recently, we and others have shown that the inhibition of geranylgeranylation by statins can block the protective mitochondrial unfolded protein response (UPRmt) in C. elegans [[26], [27], [28]], thereby highlighting possible crosstalk between statins and mitochondrial homeostasis. In parallel to inhibiting UPRmt, statin treatment was shown to elicit the activation of an ire-1-dependent endoplasmic reticulum unfolded protein (UPRer) stress response [29].
Here we describe a mechanism conserved from C. elegans to mammals in which statin treatment triggers the activation of a specific p38 signaling cascade. This mechanism involves the blockade of mevalonate pathway metabolism and downstream geranylgeranylation by statins. Similar to statins, impaired geranylgeranylation and the inactivation of specific small GTPases from the RAS superfamily, including the RHO-1, RAB-10, and ARF-3 proteins, activate a p38-mediated transcriptional response. This transcriptional program confers a cytoprotective response that involves the regulation of innate immunity and stress responses. We found that treating a macrophage-derived cell line with therapeutic concentrations of statins has the same effect of activating p38. In these cells, p38 activation by statins leads to the upregulation of COX-2, a major regulator of inflammation and innate immunity.
2. Materials and methods
2.1. C. elegans strains and maintenance
Unless otherwise stated, C. elegans strains were maintained on nematode growth medium (NGM) plates at 20 °C as previously reported [30]. A list of the C. elegans strains used in this study and strain construction details are provided in Table S5.
2.2. Analysis of deletions and point mutations (dCAPS)
Deletions and point mutations were analyzed by the single-worm PCR method [31]. A list of primers is provided in Table S5. To analyze point mutations, the dCAPS method was employed [32] using dCAPS Finder 2.0 software [33].
2.3. Pharmacological C. elegans experiments
All of the pharmacological experiments were conducted using 35 mm or 55 mm diameter plates poured with either 3 ml or 10 ml autoclaved NGM, which was cooled to 55 °C. The different pharmacological agents were mixed with the NGM media before pouring it onto the plates. Mevalolactone (Sigma-Aldrich, Cat. #M4667) was dissolved in water to create a 1 M stock solution and was used at a final concentration of 10 mM in the NGM plates. Fluvastatin (Sigma-Aldrich, Cat. # PHR1620) was dissolved in water to a 50 mM stock solution that was maintained indefinitely at −20 °C. Simvastatin (Sigma-Aldrich, Cat. #S6196) was dissolved in dimethyl sulfoxide (DMSO) to a 10 mM stock solution. After adding different concentrations of fluvastatin to the NGM plates, the plates were dried at room temperature in the dark for one day. 100 μl or 350 μl of OP50 bacteria were seeded on the 35 mm or 55 mm plates, respectively, and the plates were dried at room temperature in the dark for one additional day before C. elegans were spotted on the plates. Unless stated otherwise, all of the experiments were conducted at 20 °C.
For experiments involving continuous exposure to pharmacological agents during the development of C. elegans, we spotted on 35 mm plates approximately 100 animals synchronized to larval stage one (L1) that were incubated in an S-basal medium [34] for 24 h post-bleach. In these experiments, the animals were analyzed 72 h after spotting them on the plates, a time point that corresponds to day one of adulthood. In cases of conditional exposure experiments, the animals were grown on plates seeded with OP50 or double-stranded RNA clones of interest until the larval stage four (L4) (48 h post-bleach) and then transferred to 55 mm plates with fluvastatin for 48 h of incubation before we conducted the analysis. In these experiments, we transferred 25 animals onto each of the 55 mm plates.
2.4. RNAi knockdown
All double-stranded (dsRNA) expressing clones were sequenced before use. Feeding was conducted as previously described [35].
2.5. Expression analysis by RT-qPCR
C. elegans were bleached and synchronized by incubation for 24 h in S. basal medium at 20 °C. Synchronized wild-type or dlk-1-mutant C. elegans at the L1 stage (approximately 300 animals/plate, four plates per condition) were spotted onto 55 mm NGM plates seeded with 350 μl of OP50. At the L4 stage (approximately 48 h later), the animals were washed from the plates with S. basal medium and cleaned from the OP50 bacteria by washing with S. basal medium three times for 5 min each. Next, C. elegans were added to 55 mm NGM plates with or without fluvastatin, which was seeded a day before with 350 μl of OP50 bacteria. At the time of interest, the animals were collected and washed with an S. basal medium three times and with RNase-free water two more times.
Total RNA was extracted using a Direct Zol RNA Miniprep kit (Zymo Research, Cat. #R2051) and treated with DNase according to the manufacturer's instructions. The mRNA concentration was quantified by both Nanodrop and Qubit (Thermo Fisher Scientific) and 1 μg of total RNA was used for cDNA synthesis using a qScript cDNA Synthesis Kit (QuantaBio, Cat. # 95047-025). RT-qPCR reactions were conducted using the Fast SYBR Green Master Mix (Thermo Fisher Scientific, Cat. # 4385612) in a Step One Real-Time-PCR machine (Thermo Fisher Scientific, Cat. # 4376357).
Primers were designed using the NCBI primer design tool (Primer-BLAST) [36]. All of the primers spanned at least one intron-exon junction and had a melting temperature (Tm) ranging from 58 °C to 62 °C. All of the primers were synthesized by IDT (Germany) and can be found along with their calculated TM and amplicon size in Table S5. Primer sequences for pmp-3 were used for expression normalization [37]. The ΔΔCt method was used to calculate the fold change in expression. The results are averages of three biological replicates, and error bars represent standard errors.
2.6. RNA-Seq and whole transcriptome analysis
The RNA-Seq analysis was conducted using three independent biological replicates for each condition. Synchronized wild-type or dlk-1-mutant C. elegans at the L1 stage (approximately 200 animals/plate, four plates per condition) were spotted onto 55 mm NGM plates seeded with 350 μl of OP50. At the L4 stage (approximately 48 h later), the animals were washed from the plates with S. basal medium and cleaned from the OP50 bacteria by washing the animals with S. basal medium three times for 5 min each. The animals were dropped on 55 mm NGM plates that were seeded a day before with 350 μl of OP50 with or without fluvastatin. After 9 h, the animals were collected and washed with an S. basal medium three times and with RNase-free water two more times. Total RNA was extracted using a Direct Zol RNA Miniprep kit (Zymo Research, Cat. #R2070) and treated with DNase according to the manufacturer's instructions. The mRNA concentration was quantified by both Nanodrop and Qubit (Thermo Fisher Scientific), and RNA was used for library construction and RNA sequencing.
RNA libraries were prepared using a TruSeq RNA Library Prep Kit v2 (Illumina, Cat. # RS-122-2001) according to the manufacturer's protocol. The libraries were loaded on one lane of an Illumina HiSeq 2500 instrument and ran by a 50 bp single read using V4 reagents at the Technion Genome Center, Technion Israel Institute of Technology. The reads were trimmed, collapsed, and then aligned to the WS220 gene transcripts of C. elegans (WormBase) using Bowtie [38], allowing multiple alignments to include multiple transcripts due to alternative splicing. Of the reads in all of the samples, 94–95% were mapped to the C. elegans transcriptome. The DESeq2 package [39] in R was used to identify differentially expressed genes upon the statin treatment of each strain. PCA analysis revealed a batch effect of biological replica number two, and this batch effect was included in the DESeq2 analysis. The DESeq2 package includes automatic strict filtering of low normalized gene counts to increase power and ensure reliable results. To declare significance for genes that were differentially expressed under the different conditions, we chose a p value threshold after a false discovery rate (FDR) correction of 0.05. The DAVID platform [40] was used for gene enrichment studies.
2.7. Fluorescence imaging
To determine the level of GFP expression, synchronized C. elegans harboring different GFP reporters were mounted on agar pads (2% agar in S. basal medium) with 3 μl of 10 mM levamisole (Santa Cruz Biotechnology, Cat. # 16595-80-5). Gravid adult C. elegans were visualized at a chronological age of day one of adulthood, 72 h post-release from L1 arrest, or, in cases of conditional statin exposure, at day two of adulthood. Because some genetic backgrounds and RNAi treatments result in the slow development of C. elegans, the plates were often photographed more than once to analyze the animals on both the chronological and biological ages of day 1 of adulthood. In the targeted screen, 30 C. elegans were analyzed quantitatively and positive candidates were further analyzed by the quantitative measurements of the relative fluorescence intensity.
To capture images of the C. elegans on a slide, 10 animals were collected from the plates and transferred to a 1 μl drop of 10 mM levamisole solution. Once the drop dried, the animals were oriented head to head using an eyelash pick and then covered by a coverslip. The levamisole solution was added from the margins of the slide until the entire agar pad was covered with the solution. The slides were then maintained in a humid chamber for approximately 30 min to reduce the number and size of air bubbles. The same magnification, light intensity, and exposure time were used in every experiment. The images were captured using a Nikon (DS-Ri1) camera connected to a Nikon E600 fluorescence compound microscope or using a Nikon (SMZ18) fluorescence-dissecting microscope connected to a Nikon DS-Fi3 camera. The images were processed using Adobe Photoshop software according to the ethical guidelines for the appropriate use and manipulation of scientific digital images [41].
For fluorescence intensity measurements, 20 animals were collected under each condition and mounted on an agar slide with 3 ml of 10 mM levamisole. The animals were imaged using a Nikon (SMZ18) fluorescence-dissecting microscope connected to a Nikon DS-Fi3 camera. The fluorescence intensity of three independent biological replicas of each condition was analyzed using Fiji (ImageJ) software. Each image's background signal was subtracted using the Threshold function in ImageJ. The same threshold was set for all of the images in the same experiment.
2.8. Thrashing assay
C. elegans at a chronological age of day two of adulthood were grown as previously described. Groups of five animals were placed in a drop of 20 μl of S. basal medium on a glass slide, and the number of thrashes was measured after approximately 5 min of habituation. Movies of C. elegans swimming in 30-s intervals were visualized using a Nikon (SMZ18) fluorescence-dissecting microscope and captured using Active Presenter software. The captured movies were then played in slow motion to count the number of times the head or tail crossed the animal's midline, which we defined as one thrash.
2.9. Measurement of C. elegans length
A total of 100 L1 C. elegans larvae per plate were spotted on 35 mm plates with the chemical of interest and grown for 72 h, except strains carrying the atfs-1 (et15) allele, which were analyzed after 96 h due to their slow development. Three images were captured for each plate and, because C. elegans vary in size and level of transparency, the C. elegans length was measured manually using ImageJ's “segmented” tool. Ten animals were measured in each biological replica, and the results were the sum of three independent biological replicas.
2.10. Detection of phospho-p38-PMK-3 in C. elegans
Synchronized cultures of C. elegans were grown according to standard procedures [34]. Briefly, wild-type (N2) and dlk-1(ku12) mutant C. elegans were grown in liquid cultures at 20 °C with gentle agitation (∼160 rpm) until the young-adult stage. Animals were grown in an S-complete medium at a concentration of one animal per one microliter of culture with NA22 bacteria as a food source. At the young-adult stage, the cultures were treated with 500 μM fluvastatin and C. elegans from the cultures were sampled at each time point. Time 0 corresponded to cultures before fluvastatin treatment. At each time point, the samples were cleaned from the bacterial food using four cycles of low-speed centrifugation (800 g) and S. basal washing. The animals were counted and pellets of 200 animals at the bottom of a microcentrifuge tube were frozen immediately at −80 °C. Protein sample buffer was added directly to the frozen pellets before the incubation of the tubes at 95 °C for 5 min. We loaded 100 animals per lane and used anti-actin antibody as a loading control for the SDS-PAGE analysis. Western blotting membranes were probed with rabbit polyclonal anti-phospho-p38 (Cell Signaling, Cat. #9211) at a dilution of 1:1000 and mouse monoclonal anti-actin C4 (MP Biomedicals, Cat. # 8691002) at a dilution of 1:1000. Horseradish peroxidase-conjugated goat anti-rabbit IgG or anti-mouse was used at a dilution of 1:10,000. The proteins were visualized using WesternBright ECL (Advansta, Cat. # K-12045-D20) in an Amersham Imager 600 and quantified by ImageQuant TL software.
2.11. Statin treatment in cultured mammalian cells
RAW264.7 cells were seeded at a density of 300,000 cells in six-well dishes. The media were replaced the next day, and fluvastatin, simvastatin, DMSO, and mevalonolactone were added at the indicated concentrations. At 72 h post-treatment, the cells were washed twice with ice-cold PBS and lysed in 150 μl of RIPA/SDS buffer (50 mM Tris pH 8, 150 mM NaCl, 5 mM EDTA, 1% v/v NP-40, 0.5% w/v deoxycholic acid, 0.1% w/v SDS, 10 mM NaF, 0.1 mM PMSF, and Complete Protease Inhibitor cocktail tablets). The total protein concentrations were determined using a Bradford Assay (Bio-Rad) and 40 μl of the samples were diluted in 5 x Laemmli sample buffer and resolved by SDS-PAGE. Next, Western blotting membranes were probed with antibodies against rabbit polyclonal phospho-p38 (Cell Signaling, Cat. #9211), mouse monoclonal p38 (Abcam, Cat. #AB31828), and mouse monoclonal anti-actin C4 (MP Biomedicals, Cat. # 8691002), all at 1:1000 dilution, and goat polyclonal anti-COX-2 (Santa Cruz Biotechnology, Cat. #SC-1745) at 1:400 dilution. Horseradish peroxidase-conjugated bovine anti-goat IgG, goat anti-rabbit IgG, and goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) were used at a dilution of 1:10,000. The proteins were visualized using WesternBright ECL (Advansta) in an Amersham Imager 600 and quantified by ImageQuant TL software.
3. Results
3.1. Inhibition of mevalonate metabolism by statins activates a specific stress response in C. elegans
To study the cellular response to statins, we screened a collection of reporters for various stress responses in C. elegans (Figure 1A). To phenocopy the treatment of patients with statins and circumvent the effect of a high statin concentration on the development of C. elegans [29], we exposed animals at the end of the last larval stage to fluvastatin for two days. For our screen, we used the statin fluvastatin because it is the primary statin used in many previous studies of C. elegans and mammalian cells ([23,42]). Consistent with previous reports [27,28,42], fluvastatin treatment does not activate the UPRmt reporter hsp-6p::gfp but does mildly activate the UPRer reporter hsp-4p:gfp [29] (Figure 1A). In addition, fluvastatin does not activate other well-characterized stress responses, including the hsp-16.2-mediated heat shock response, the skn-1-dependent oxidative stress response, and the daf-2/daf-16 pathway. In contrast, we found that fluvastatin triggered a strong and significant upregulation of the tbb-6p::gfp reporter (Figure 1A), which was previously shown to be a sensor of mitochondrial dysfunction independent of the canonical UPRmt response [43].
Figure 1.
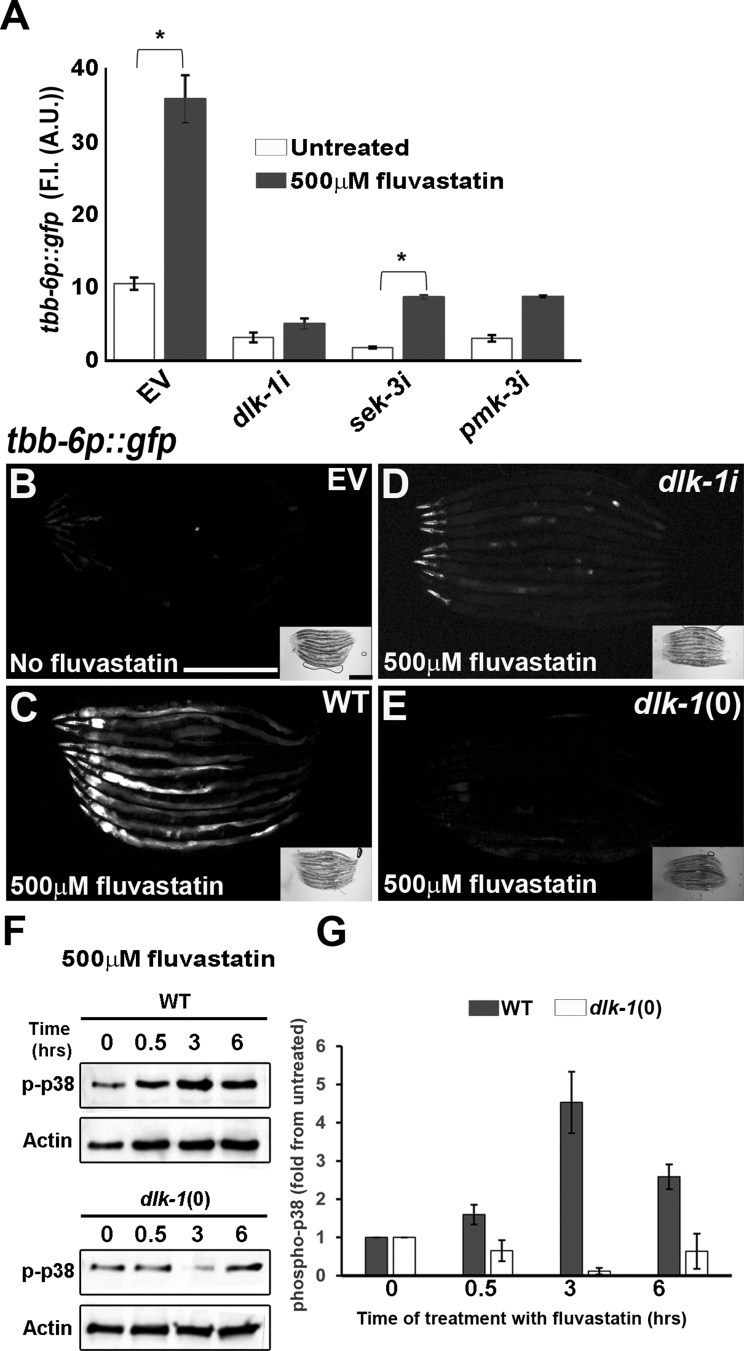
Concentration-dependent upregulation of the tbb-6p::gfp reporter by statins.A. A targeted screen for stress-related reporters identified tbb-6p::gfp as a target for statin activity in C. elegans. Hereafter, all of the fluorescence intensity measurements are the average of three independent experiments and error bars represent standard errors of the mean. ∗∗U ≤ 0.01; ∗∗∗U ≤ 0.001 using the Mann-Whitney nonparametric test. B. Fluorescence intensity measurements of the concentration-dependent upregulation of tbb-6p::gfp by statins. ∗p ≤ 0.05 using one-way ANOVA with Tukey's test for multiple comparisons. C. The upregulation of tbb-6p::gfp by statins is suppressed by 10 mM mevalonate (mevalonolactone). Hereafter, images are of 10 C. elegans oriented head to head with their anterior pole to the left. D. Fluorescence intensity measurements of the mevalonolactone rescue experiment. ∗p ≤ 0.05 using one-way ANOVA with Tukey's test for multiple comparisons. E. Inhibition of the main branch of the mevalonate pathway and the downstream geranylgeranylation sub-branch activate the tbb-6p::gfp reporter. ∗p ≤ 0.05 using one-way ANOVA with Tukey's test for multiple comparisons. F. Images of empty vector control (EV), hmgs-1, M57.2, and ggtb-1 RNAis. Scale bars represent 500 μm.
With fluvastatin treatment, tbb-6p::gfp was primarily upregulated in C. elegans’ intestine and pharynx in a concentration-dependent manner (Figures 1B and S1A). To find the time window of the transcriptional response to fluvastatin, we used RT-qPCR to measure the relative level of tbb-6 mRNA in C. elegans exposed to fluvastatin for different time intervals. Consistent with the upregulation of the tbb-6p::gfp reporter, tbb-6 mRNA was also upregulated following fluvastatin treatment (Fig. S1B). Upregulation after 6 h of exposure to fluvastatin demonstrated that a primed transcriptional response was activated upon treatment. Importantly, we found that supplementing the product of the HMGR-1 enzyme, mevalonate, in the form of mevalonolactone acid [29] suppressed the upregulation of tbb-6p::gfp by fluvastatin (Figure 1C,D). Thus, the activation of the tbb-6p::gfp reporter by fluvastatin resulted from the direct inhibition of HMG-CoA reductase and the downstream mevalonate pathway metabolism and not from the off-target effects of these drugs.
After farnesyl pyrophosphate (FPP) synthesis, the mevalonate pathway is divided into several sub-branches (Figure 1E). Pathway-level understanding of the crosstalk between mevalonate pathway metabolism and tbb-6p::gfp-related stress response required the identification of the specific sub-branch whose inactivation resulted in the upregulation of the tbb-6p::gfp reporter. To this end, we used RNA interference (RNAi) to systematically impede the metabolism of specific sub-branches of the mevalonate pathway. Consistent with the idea that statins inhibit the metabolism of the main branch, knockdown of several enzymes on the main branch of the mevalonate pathway, including HMGS-1 and HMGR-1 (Figures 1F and S1C), resulted in the upregulation of the tbb-6p::gfp reporter. Importantly, knocking down the geranylgeranyl-transferase ggtb-1 (Figure 1F) led to tbb-6p::gfp upregulation. In contrast, blocking the metabolism of the other sub-branches of the pathway, including sub-branches that are known regulators of the UPRmt [28], did not significantly alter the level of the tbb-6p::gfp reporter in comparison to the control RNAi (Figure 1E). A growing body of data suggests that statins can alter the metabolism of many species of bacteria [44], raising the possibility that the upregulation of tbb-6p::gfp in C. elegans stems from a metabolic switch in its bacterial food. However, the knockdown of mevalonate pathway enzymes using RNAi resulted in tbb-6p:gfp upregulation (Figures 1E, 1F, and S1C) pinpointing this response to the impaired metabolism of the mevalonate pathway in C. elegans. Taken together, pharmacological and genetic approaches revealed that blocking mevalonate pathway metabolism and downstream geranylgeranylation in C. elegans leads to the activation of a specific stress response.
3.2. Statin treatment results in the activation of the p38 pathway through the regulation of p38-MAPK phosphorylation
A recent genetic study demonstrated that in cases of mitochondrial stress, the upregulation of tbb-6p::gfp reporter is governed by p38 MAP kinase [43]. This pathway involves the sequential activation of DLK-1, SEK-3, and PMK-3 proteins that function as MAP3K, MAP2K, and MAPK, respectively. To test whether the DLK-1/SEK-3/PMK-3 axis was involved in the upregulation of tbb-6 following statin treatment, we exposed C. elegans with the tbb-6p::gfp reporter to fluvastatin while knocking down the genes of this axis. RNAi for dlk-1 and pmk-3 genes significantly inhibited the upregulation of tbb-6p::gfp following the treatment of C. elegans with statins (Figure 2A–D and S2A) or RNAi clones that impaired the metabolism of the main branch or downstream geranylgeranylation (Figs. S2B–D). Consistent with these results, loss of function mutations in dlk-1 (Figure 2E) or sek-3 (Fig. S2E) genes blocked the upregulation of the tbb-6p::gfp reporter upon the treatment of C. elegans with fluvastatin. Moreover, in agreement with the ability of mevalonolactone to suppresses the upregulation of tbb-6p::gfp following statin treatment (Figure 1C–D), this metabolite suppressed the upregulation of the tbb-6p::gfp reporter following the knockdown of hmgs-1 and ggtb-1 (Figs. S3A–F). The suppression of the effect of ggtb-1 RNAi by mevalonolactone probably stemmed from an increase in the level of the substrate available for the GGTB-1 enzyme that compromises the partial knockdown of the enzyme by RNAi. These results further demonstrate the role of the DLK-1/SEK-3/PMK-3 axis (the p38 pathway herein) in the activation of a specific transcriptional response in cases of impaired mevalonate pathway metabolism and downstream geranylgeranylation.
Figure 2.
Statin treatment results in the activation of the DLK-1/SEK-3/PMK-3 axis of the p38 pathway.A. Loss of the DLK-1/SEK-3/PMK-3 pathway results in the inhibition of statin-mediated upregulation of the tbb-6p::gfp reporter. ∗p ≤ 0.05 by mixed-design ANOVA with Tukey's test for multiple comparisons. B.C. elegans with the tbb-6p::gfp reporter in untreated conditions. C.tbb-6p::gfp upregulation upon exposure to statins. D and E. Knockdown or knockout of the dlk-1 gene, respectively, results in the inhibition of statin-induced upregulation of the tbb-6p::gfp reporter. F. Phosphorylation of C. elegans p-38-MAPK following fluvastatin treatment. Time 0 is the untreated culture. A total of 100 animals were used per lane and an anti-actin antibody was used as a loading control because the antibody for the detection of total p38 proteins in mammals does not recognize the C. elegans p-38 protein. G. Quantification of two independent experiments of fluvastatin-induced p-38 phosphorylation in C. elegans. Scale bars represent 500 μm.
A recent study has demonstrated the involvement of the MAP kinase phosphatase VHP-1 in the regulation of the p38 pathway [43]. To test the possible role of VHP-1 in the response to fluvastatin, we knocked down the vhp-1 gene by RNAi. Consistent with the proposed role of vhp-1 as a negative regulator of the p38 pathway, we found that its knockdown further increased the level of the tbb-6p::gfp reporter in C. elegans treated with the ggtb-1 RNAi (Figs. S3G–H). This increase demonstrated that in a wild-type background, the activation of the p38 pathway is negatively regulated by at least two mechanisms: the activity of geranylgeranylated proteins and the function of the VHP-1 phosphatase.
To directly determine the level of p38 activation, we used an antibody that detects phosphorylated p38, which is the active form of p38. In agreement with our genetic studies, this biochemical approach revealed that fluvastatin treatment led to an increase in the level of p38 phosphorylation (Figure 2F,G). Notably, the knockout of the p38-MAPKKK protein DLK-1 abolished statin-mediated phosphorylation of p38 (Figure 2F,G). Consistent with our genetic analysis (Figure 2A–E), the involvement of DLK-1 in the regulation of the level of p38 phosphorylation further demonstrated the critical function of an upstream regulatory network that controls the level of p38 activation.
Collectively, our results showed that by blocking mevalonate pathway metabolism and downstream geranylgeranylation, statins trigger the activation by phosphorylation of a primed p38 pathway in C. elegans.
3.3. Knockdown of specific small GTPases activates the p38 pathway
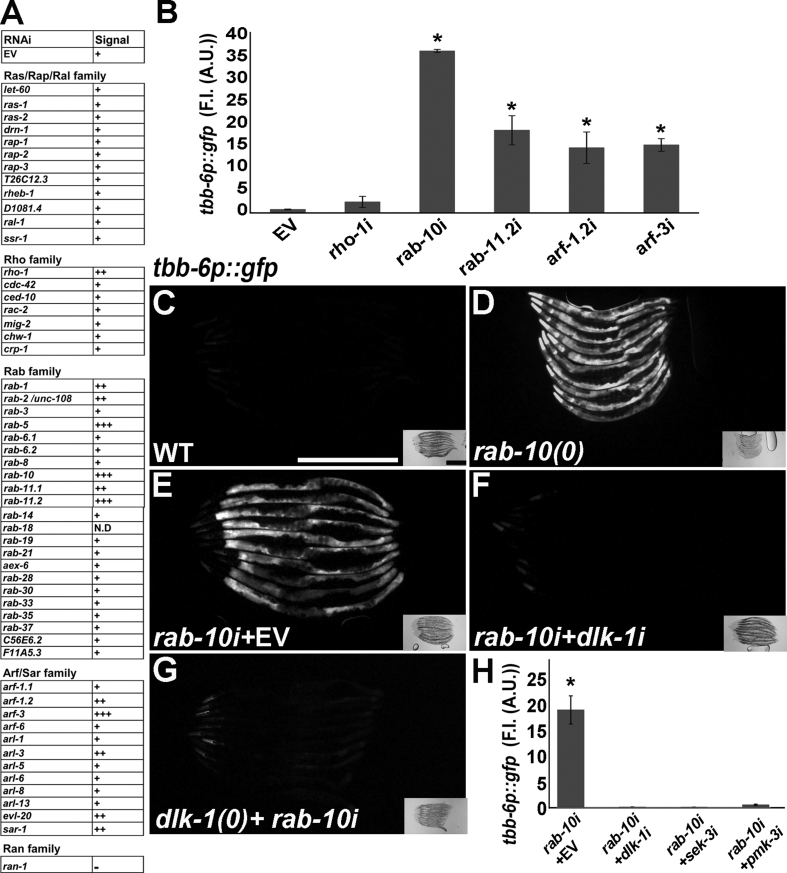
Membrane anchoring by geranylgeranylation is critical for the activation of many small GTPases, including those that regulate various stress responses [45]. Thus, it is plausible that the blocking of geranylgeranylation by statins inactivates specific small GTPases that are regulators of p38 signaling. To test this hypothesis, we used RNAi to knock down 52 out of 54 members of the small-GTPase RAS superfamily in C. elegans (a modified list from [46] and Table S1). In this targeted screen, we monitored the level of expression of the tbb-6p::gfp reporter as a readout for p38 activation. We found that the inactivation of 12 small GTPases of the RHO, RAB, and ARF/STAR families activated the p38 pathway (Figure 3A and S4A-F). The knockdown of RAB-10 resulted in the strongest upregulation of the tbb-6p::gfp reporter (Figure 3B) and only marginally affected the size or rate of development of C. elegans (Figure 3A). Therefore, we focused on the crosstalk between the small GTPase RAB-10 and the p38 pathway. Consistent with the results of our RNAi screen, we found that the loss-of-function mutation in the rab-10 gene elicited strong upregulation of the tbb-6p::gfp reporter (Figure 3C–D). The inactivation of the dlk-1 gene by RNAi (Figure 3E–F) or a loss-of-function mutation (Figure 3G) suppressed the upregulation of the tbb-6p::gfp reporter upon the loss of rab-10. Similar results obtained with RNAi clones of sek-3 and pmk-3 (Figure 3H) showed that the loss of rab-10 activated the pathway until the PMK-3 MAPK. In agreement with the notion that the mevalonate pathway is required upstream of RAB-10 function, the supplementation of mevalonate did not suppress the activation of tbb-6p::gfp in the rab-10-mutant background (Figs. S4G–J). Prediction tools for protein-protein interactions suggested that many of the small GTPases identified in the screen were clustered together in functional networks. This can explain the identification of several proteins from the RAB and ARF/STAR families as regulators of p38 activation. Thus, any perturbations in the network of small GTPases that function upstream from the DLK-1 MAPKKK may activate the p38 cascade. Taken together, the RNAi screen demonstrated the involvement of specific small GTPases in the regulation of the p38 pathway and provided a plausible mechanistic link between the blocking of geranylgeranylation and the activation of p38.
Figure 3.
Loss of specific small GTPases results in p38 pathway activation.A. The results of a targeted RNAi screen for the involvement of small GTPase proteins from the RAS superfamily in the regulation of the p38 pathway. We knocked down by RNAi 52 out of 54 members of the small-GTPase RAS superfamily in C. elegans (a modified list from [46]). B. Fluorescence intensity measurements of candidate RNAis that induced the strongest expression of the tbb-6pr::gfp reporter. ∗p ≤ 0.05 by one-way ANOVA with Tukey's test for multiple comparisons. C. The tbb-6p::gfp levels in wild-type C. elegans. D. In rab-10 knockout C. elegans, tbb-6p::gfp is upregulated, demonstrating the specificity of the screen. E. The rab-10 RNAi upregulates the tbb-6p::gfp reporter. F. The knockdown of the dlk-1 gene inhibits the upregulation of tbb-6p::gfp by the rab-10 RNAi. G. Loss of the p38 pathway completely abolishes the upregulation of tbb-6p::gfp by rab-10 RNAi. H. Fluorescence intensity measurements of the co-RNAi experiments of p38 pathway genes with rab-10 RNAi. ∗p ≤ 0.05 by one-way ANOVA with Tukey's test for multiple comparisons. Scale bars represent 500 μm.
3.4. The statin-p38 axis regulates transcriptional programs of innate immunity, stress, and metabolism
To understand the molecular details of the statin-p38 transcriptional response, we conducted an RNA-sequencing (RNA-Seq) analysis of wild-type and dlk-1-mutant C. elegans exposed to fluvastatin (Figure 4A). Importantly, in keeping with our tbb-6p::gfp and RT-qPCR results, RNA-Seq analyses indicated that the tbb-6 transcript was upregulated 8.7 fold following fluvastatin treatment (Figure 4B and Table S2). This upregulation decreased by 1.6 fold in C. elegans lacking the p38 pathway (Table S2), demonstrating the consistency of the different methods we used to assess the transcriptional response to statins. A recent study showed that following the detection of mitochondrial stress, the UPRmt pathway selectively upregulates enzymes of specific sub-branches of the mevalonate pathway [28]. This upregulation may confer the reported resistance of C. elegans in which the UPRmt is constitutively activated to statins [42]. To examine the possibility that the p38 pathway regulates the expression of mevalonate pathway genes similar to the UPRmt, we determined the expression levels of these genes under different conditions. Notably, we found that the statin-p38 axis did not significantly alter the expression levels of mevalonate pathway genes (Figure 4B). Thus, in sharp contrast to the UPRmt, the biological significance of p38 activation following statin treatment was not directly related to mevalonate pathway metabolism.
Figure 4.
The p38 pathway regulates transcriptional programs of cellular defense and metabolic processes. A. A graph of differential gene expression across different genetic backgrounds and statin concentrations. The graph shows only genes with differences in expression levels under at least one condition with a p adjust (FDR) of p ≤ 0.05. We classified genes identified by DESeq2 to significantly differ in expression between specimens grown under normal and statin-supplemented conditions as follows: genes affected by statins in both wild-type and p38 mutant C. elegans (gray triangles); genes affected by statins only in wild-type C. elegans (magenta circles); and genes affected by statins only in p38 mutant C. elegans (green squares). The dashed line represents a linear regression between fold change values of gene expression in wild-type vs p38 mutant C. elegans. Regression R2 = 0.972; the regression equation was K = 0.94N + 0.03A; plot of log2 fold change differences in expression levels of genes from statin-treated vs untreated wild-type vs p38 mutant C. elegans. B. Fluvastatin treatment does not alter the expression of mevalonate pathway genes. The genes of the pathway's main branch are in order from top to bottom and the genes of each sub-branch are grouped. We used the reference gene pmp-3 [37] to standardize the expression and the tbb-6 gene as a positive control. C. GO term enrichment analysis of differently expressed genes in wild-type C. elegans upon statin treatment. The list of genes tested for GO term enrichment contains genes that show differential expression with a p adjust (FDR) of p ≤ 0.05. D. A list of GO term enrichment analysis of p38-specific genes that were differentially expressed upon the treatment of C. elegans with statins. The list of genes tested for GO term enrichment contains genes that have a p adjust (FDR) of p ≤ 0.05 in wild-type C. elegans treated with statins but a p adjust above 0.05 in p38 mutant C. elegans treated with statins. GO term enrichment was analyzed using the DAVID platform [40]. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To elucidate the biological processes that were altered upon exposure of C. elegans to statins, we searched for gene ontology (GO) enrichment signatures in the global transcriptional response of wild-type C. elegans treated with fluvastatin. Consistent with our findings that fluvastatin upregulates both the p38 and UPRer reporters (Figure 1A), among the significant enrichments, were GO terms related to innate immunity, metabolic processes, defense from external stimuli, and the UPRer (Figure 4C and Table S3). In addition, the GO analysis suggested that fluvastatin can modify transcriptional programs dedicated to the regulation of oxidation-reduction processes and the biosynthesis of flavonoids that share structural properties with fluvastatin. Notably, the proposed perturbations in the oxidation-reduction balance after fluvastatin treatment did not upregulate the UPRmt (Figure 1A).
Next, we sought to decipher the p38-dependent portion of the global transcriptional response to statins. To this end, we searched for transcripts that are altered with fluvastatin treatment in wild-type C. elegans but not significantly altered in p38 mutant C. elegans exposed to fluvastatin. Beyond the enrichment of transcripts related to structurally similar flavonoid molecules, the GO term analysis suggested that the statin-p38 axis regulates innate immunity, metabolic processes, and the balance of cellular oxidation-reduction reactions (Figure 4D and Table S3). Importantly, the lack of enrichment of UPRer targets in the pool of transcripts regulated by the p38 pathway (Figure 4D) highlights a mechanism in which the p38 and UPRer pathways are activated independently in response to the treatment of C. elegans with fluvastatin. Collectively, our specific (Figure 1A) and global (Figure 4C–D) transcriptional analyses showed that two dedicated stress-related transcriptional responses, p38 and UPRer, were activated as part of the cellular response to statins. The activation of p38 resulted in a transcriptional response that involved the regulation of innate immunity, stress responses, and metabolism.
3.5. The p38 pathway protects C. elegans from the effects of statins
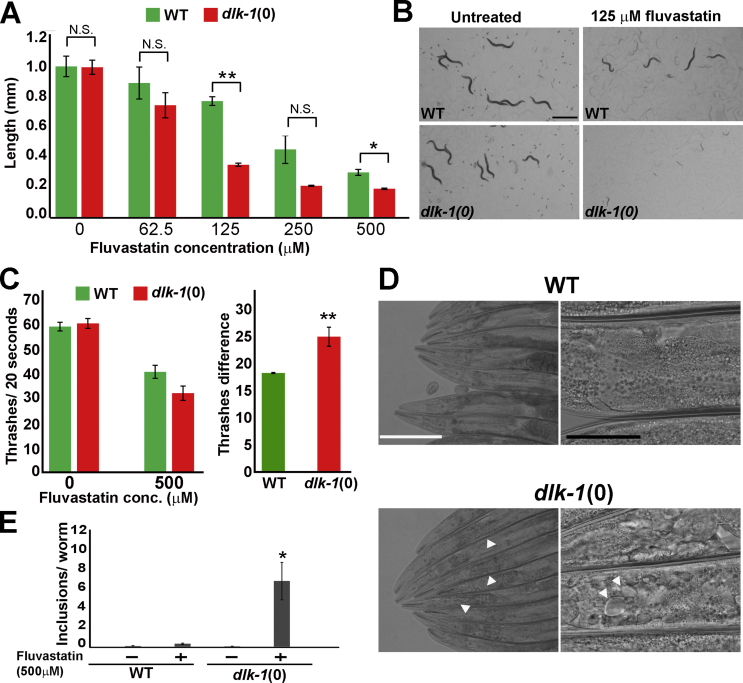
One key design principle of many stress responses, such as the UPRmt [47], is their ability to act as a compensatory mechanism to mitigate the challenge imposed by the stressor. To study the physiological significance of the statin-mediated activation of the p38 pathway, we inhibited the pathway in live animals treated with fluvastatin. To determine the effects of statins on C. elegans physiology, we measured the length of C. elegans following a 72-h exposure to different concentrations of fluvastatin. In agreement with a previous study [42], we found that fluvastatin reduced the size of wild-type C. elegans in a concentration-dependent manner (Figure 5A). To determine if the p38 pathway plays any role in the cellular response to statins, we compared the sensitivity of wild-type and p38 mutant C. elegans to increasing concentrations of fluvastatin. Importantly, in untreated conditions, the loss of the p38 pathway did not significantly alter the length of C. elegans in comparison to wild-type C. elegans. The animals mutated for the dlk-1 gene of the p38 pathway, however, were found to be hypersensitive to the effects of fluvastatin (Figure 5A–B). This hypersensitivity demonstrated the protective role the p38 pathway plays in the cellular response to statins. Next, we employed a functional assay of C. elegans thrashing to assess the effect of statins on muscle function. Consistent with the proposed protective role of p38 on the effect statins on C. elegans’ length (Figure 4A,B), the p38 pathway was required to protect the muscle-related activity of trashing from the effects of fluvastatin (Figure 4C).
Figure 5.
The p38 pathway protects C. elegans from the deleterious effect of statins.A. The length of day one adult C. elegans (chronological time) grown from the larval stage one for 72 h on plates with different concentrations of fluvastatin. Length analysis revealed that dlk-1-mutant C. elegans are hypersensitive to statins. Hereafter, the lengths of 10 C. elegans were averaged in each biological replica and the results show the average of three biological replicas. The significance of differences in C. elegans length was tested by the Mann-Whitney nonparametric test for the two strains in every concentration of fluvastatin. ∗U ≤ 0.05 and ∗∗U ≤ 0.01. B. Images of typical plates with day one adult wild-type or dlk-1-mutant C. elegans grown on plates without or with 125 μM of fluvastatin. C. dlk-1-mutant adults treated with fluvastatin for 48 h show impaired swimming in comparison to wild-type C. elegans. The significance of the difference in the number of thrashes was tested by one-tailed unpaired Student's t-tests. ∗∗p ≤ 0.01. D. The p38 pathway protects C. elegans from the formation of statin-mediated inclusions in intestinal cells. Images of the intestines of wild-type and dlk-1-mutant C. elegans treated with 500 μM of statins for 48 h from the L4 larval stage. In contrast to the intact intestines of wild-type C. elegans, dlk-1-mutant C. elegans have many inclusions in their intestines (white arrows). E. A significant increase in the number of inclusions in statin-treated dlk-1-mutant C. elegans. n (number of inclusions per animal) was calculated for 10 animals in three biological replicas. ∗p ≤ 0.05 by one-way ANOVA with Tukey's test for multiple comparisons. Scale bars represent 1 mm in panel B and 200 μm (white bar) and 50 μm (black bar) in panel D.
The expression of the reporter for p38 activation in the pharynx and intestine of C. elegans pinpoints the activity of this pathway in the digestive tract. Therefore, we investigated the structure and function of the digestive tract following the treatment of C. elegans with fluvastatin. Under untreated conditions, the dlk-1-mutant C. elegans had intact intestines typical to those of wild-type C. elegans (Fig. S5). The exposure of wild-type C. elegans from the fourth larval-stage to 50 μM of fluvastatin for 48 h did not significantly alter the morphology of the intestine. However, consistent with the protective role of the p38 pathway (Figure 5A–C), the loss of this pathway resulted in the accumulation of many inclusion vesicles in the intestines of dlk-1-mutant C. elegans (Figure 5D–E and S5A-H). The significant accumulation of these intestinal inclusions was dependent both on fluvastatin treatment and the lack of a functional p38 pathway (Figure 5E). The formation of vesicles in C. elegans lacking a functional p38 pathway underscored the physiological role of this pathway to protect intestinal cells from statin-mediated cytotoxicity.
To date, the only gene we have identified as a transcriptional target for p38 activation following fluvastatin treatment is tbb-6. (Figure 1A). TBB-6 protein showed sequence similarity to beta tubulins including the human tubulin beta 1 class VI protein that is part of the microtubule network in the hemopoietic lineage in mammals [48]. The functional significance of upregulating structural proteins such as TBB-6 in response to cytotoxic stress is unknown. To study the possible functional role of tbb-6 upregulation, we knocked down this gene using RNAi and detected a significant activation of the p38 pathway (Figs. S6A–E). This activation demonstrated the functional role of tbb-6 upregulation by the p38 pathway as part of the compensatory mechanism to the insult imposed by statins.
Taken together, our functional and transcriptional studies pinpointed the p38 pathway as a compensatory mechanism that safeguards C. elegans from the effects of statins by the regulation of immunity, stress, and metabolism.
3.6. Activation of the UPRmt stress response suppresses the p38-dependent response to statins
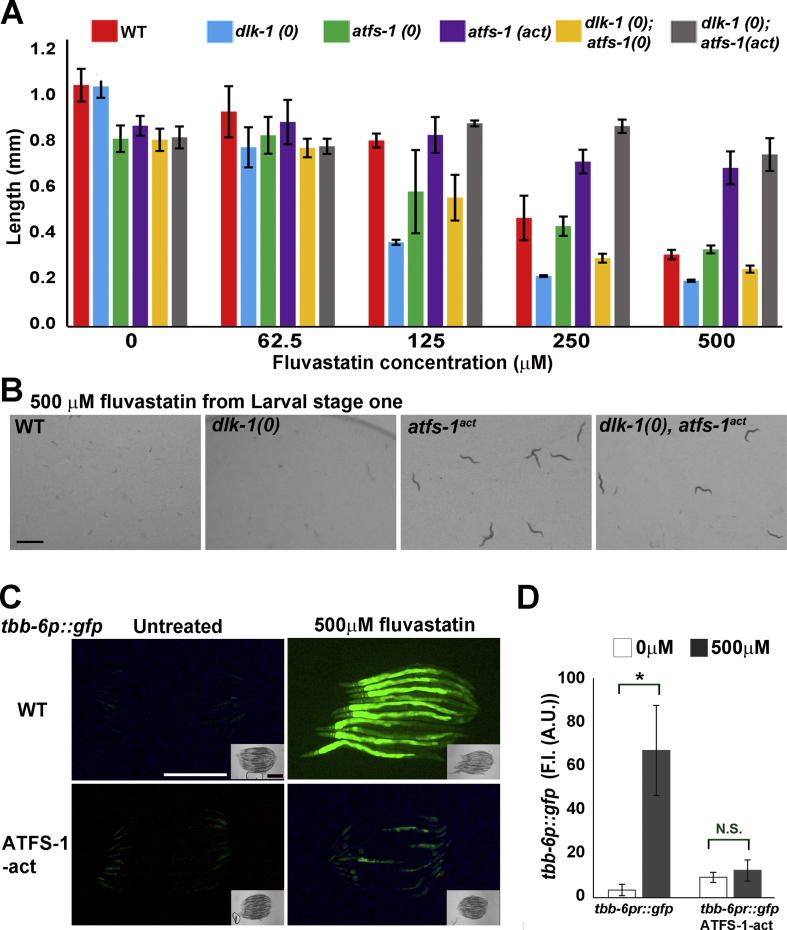
Although statin treatment does not result in the activation of the UPRmt in C. elegans, the forced activation of this stress response protects C. elegans from the adverse effects of high concentrations of statins [42]. In C. elegans, the UPRmt response is mediated by the activity of the transcription factor ATFS-1 that is translocated from the mitochondrion to the nucleus under conditions of mitochondrial dysfunction. To elucidate the possible crosstalk between the p38 and UPRmt pathways, we used combinations of double mutants and reporter strains. In agreement with a previous report [42], we found that forced activation of the UPRmt using a constitutively active ATFS-1 protein can protect C. elegans from the effects of fluvastatin (Figure 6A). The loss of the UPRmt, however, did not result in significant hypersensitivity to fluvastatin (Table S4), presumably because the UPRmt is not endogenously activated following statin treatment [27,28,42]. Blocking both the p38 and UPRmt pathways (Figure 6A) resulted in fluvastatin sensitivity comparable to the inhibition of the p38 pathway alone, further supporting the notion that in cases of impaired mevalonate pathway metabolism, p38 but not UPRmt is activated as part of a protective response. Nevertheless, the forced activation of the UPRmt by a constitutively active ATFS-1 protein rescued the sensitivity of p38 mutants to fluvastatin (Figure 6A,B), indicating a functional crosstalk between these pathways. In agreement with this model, constitutively activated UPRmt signaling suppressed the upregulation of the tbb-6p::gfp reporter following fluvastatin treatment (Figure 6C,D).
Figure 6.
Forced activation of the UPRmt suppresses the activation of the p38 pathway by statins.A. The UPRmt does not protect C. elegans from the deleterious effects of strains but induced activation of this pathway can safeguard dlk-1-mutant C. elegans from statins. Statistical significances of differences in C. elegans length were analyzed by one-way ANOVA with Tukey's test for multiple comparisons (Table S4). B. Images of typical plates with 500 μM of fluvastatin and the different strains. Wild-type and dlk-1-mutant C. elegans arrest on plates with 500 μM of fluvastatin, whereas the forced activation of the UPRmt transcription factor ATFS-1 protects wild-type and dlk-1-mutant C. elegans from the deleterious effects of statins. C. Activated ATFS-1 suppresses the statin-mediated activation of the p38 reporter tbb-6p::gfp. C. elegans were treated with statins for 48 h from the larval stage four. D. The difference in fluorescence intensity of the tbb-6p::gfp reporter in wild-type and ATFS-1-activated C. elegans. ∗p ≤ 0.05 by one-way ANOVA with Tukey's test for multiple comparisons. Scale bars represent 1 mm (B); 500 μm (C).
3.7. Treatment with therapeutic levels of statins results in the activation of the p38 pathway in a mammalian macrophage-derived cell line
Our findings in C. elegans and a report on the RAW264.7 murine macrophage-derived cell line [23] suggest an evolutionarily conserved mechanism of p38 activation following statin treatment. However, it remains unknown whether statin-mediated activation of p38 in RAW264.7 cells stems from the direct inhibition of HMG-CoA reductase or off-target effects of this medication. Moreover, little is known about the effects of statins in levels close to their therapeutic concentrations in the nanomolar range. To test the clinical relevance of our findings, we reduced the concentration of fluvastatin and extended the incubation time to mimic the therapeutic effects of statins in vivo in patients. Importantly, we were able to detect p38 activation and the upregulation of the p38 target, COX-2, following treatment for 72 h with a fluvastatin concentration as low as 200 nM (Figure 7A). Notably, similar to C. elegans, supplementing mevalonolactone suppressed p38 activation and COX-2 upregulation (Figure 7B–D), indicating that these effects stemmed from the inhibition of mevalonate pathway metabolism and not from the off-target effects of statins. COX-2 is known to regulate innate immunity by facilitating the synthesis of several known prostaglandins that control inflammation. Thus, the significant increase in the level of COX-2 following statin treatment (Figure 7D) suggested that statins can regulate innate immunity programs in RAW264.7 macrophages. Because the concentration of statins in therapeutic settings is between 34 and 234 nM [24], our analysis of p38 activation by fluvastatin is potentially clinically relevant. Finally, we tested the activation level of the p38 pathway in response to simvastatin, one of the most commonly prescribed statins that is more lipophilic than fluvastatin [49]. Simvastatin treatment resulted in the activation of the p38 reporter tbb-6p::gfp in C. elegans (Fig. S7A) and p38 phosphorylation and COX-2 upregulation in RAW264.7 cells (Fig. S7B). Thus, the regulatory circuits we characterized are activated following the inhibition of the mevalonate pathway following treatment with different types of statins. Taken together, our results demonstrated an evolutionarily conserved mechanism of statin-mediated activation of the p38 pathway that protects C. elegans from the cytotoxic effect of statins (Figure 7E).
Figure 7.
Treatment with clinical doses of statins results in the activation of the p38 pathway in mammalian cell cultures.A. Activation of the p38 pathway was measured in cultures of murine RAW264.7 macrophages following statin treatment. RAW264.7 cells were incubated with fluvastatin at the indicated concentrations for 72 h. The data indicated a marked increase in the levels of phosphorylated p38 (p-p38), with no change in the total p38 levels (T-p38), suggesting a marked activation of this pathway by fluvastatin. The levels of COX-2, a downstream target of the p38 pathway, were also elevated at 200 nM, which corresponds to clinical statin doses [24]. B. Activation of the p38-COX-2 axis stems from the inhibitory effect of statins on the metabolism of the mevalonate pathway. RAW264.7 murine macrophages were incubated for 72 h with fluvastatin in the presence or absence of mevalonate (10 mM mevalonolactone). Mevalonate supplementation suppressed statin-mediated phosphorylation of the p38 kinase and the upregulation of COX-2. C. Summary graph of the fold change in phospho-p38 levels in RAW264.7 cells treated with 500 nM fluvastatin for 72 h based on three independent experiments. Statistical significance was tested by one-way ANOVA with Tukey's test for multi-comparisons. ∗p < 0.05 vs untreated. D. Summary graph of the fold change in COX-2 levels in RAW264.7 cells treated with 200 nM fluvastatin for 72 h based on three independent experiments. Because the COX-2 data were not normally distributed, statistical significance was tested by the Kruskal-Wallis one-way analysis of variance method with Dunnett's test for multi-comparisons. ∗p < 0.05 vs untreated. E. A proposed model for the conserved activation of the p38 pathway by statins and the consequential transcriptional response. Processes shared by C. elegans and mammals are in green. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
We discovered an evolutionarily conserved mechanism in which the inhibition of mevalonate pathway metabolism and the downstream blockade of geranylgeranylation by statins activates the p38 pathway. Four independent approaches, including antibody staining and global and specific expression analyses in two model systems, C. elegans and mammalian cells in culture, show that p38 activation by statins induces specific transcriptional programs related to the regulation of innate immunity. We propose that independently of cholesterol homeostasis, a conserved p38-mediated transcriptional response has evolved to safeguard the cell in cases of mevalonate pathway dysfunction. The activity of this genetic circuit probably accounts for many of the cholesterol-independent effects of statins that are increasingly considered critical for both the beneficial and adverse effects of this widely used medication.
A growing body of clinical evidence and studies in model organisms demonstrate the pleiotropic effects of statins. These effects are proposed to stem from the versatile metabolism of the mevalonate pathway that synthesizes many different end products and the proposed off-target effects of statins on cellular targets other than HMGCR-1. The proposed mechanisms include the inhibition of HMGCR and the downstream synthesis of electron carriers [50] but also the off-target binding of statins to complex III of the respiratory chain [51]. Moreover, statins were shown to alter gene expression, for example, by regulating the activity of the CREB transcription factor [52]. These pleiotropic effects of statins are proposed to activate a network of stress responses to cope with the challenge of mevalonate pathway inhibitions. However, our findings show that the cellular response to statins is highly specific and evolutionarily conserved (Figure 1, Figure 7E). We found that the activation of the p38 pathway is dependent on blocking of geranylgeranylation; knocking down all other branches of the mevalonate pathway, including the branches that account for the production of ubiquinones, did not result in p38 activation (Figure 1E). Moreover, metabolic rescue experiments in C. elegans (Figure 1C,D) and mammalian cells (Figure 7B–D) indicated that statin-induced p38 activation and the consequential transcriptional response stem from the inhibition of mevalonate pathway metabolism and not from an off-target effect of statins.
How does statin treatment result in the activation of the p38 pathway? In mammalian cells, p38 activation was shown to originate from the knockdown of specific small GTPases, such as RHO-1 [23]. Consistent with this finding, impaired geranylgeranylation in C. elegans (Figure 1E) results in p38 activation, underscoring the possibility that similar to mammalian cells, the inactivation of specific GTPases in C. elegans triggers p38 activation. The unique amenability of C. elegans genetics enabled us to screen the entire family of small GTPases for their involvement in p38 activation. We found that specific small GTPases (Figure 3A) from different families function as regulators of p38 response. Similar to the involvement of Rho family proteins in p38 activation in murine macrophages, we identified Rho-1 as a regulator of p38 activation in C. elegans (Figure 3A and Table S1). The evolutionarily conserved role of Rho family proteins in the response to statins may stem from the misregulation of signaling cascades that are regulated by these proteins. Moreover, we identified small GTPases from the RAB and ARF families as regulators of p38 signaling. The role of proteins from the RAB and ARF families in vesicle transport [53] and cytoskeleton rearrangement [54] underscore the possibility that statin treatment impairs these processes. Notably, the protein sequence of C. elegans TBB-6 is similar to different beta-tubulins, highlighting another plausible link to the cytoskeleton network. Activation of the p38 pathway following tbb-6 knockdown (Figs. S6A–E) demonstrates that tbb-6 plays a role in the compensatory response to the insult imposed by statins (Figure 4A,B). TBB-6 and its putative mammalian orthologue TUBB2B [43] possess GTP binding domains and putative GTPases activity. Therefore, these proteins may function at the interface of the cytoskeleton network and specific small GTPases that regulate this network. Thus, our data suggest that the cholesterol-independent effect of statins may be more pleiotropic than previously appreciated, affecting not only signaling cascades but also the fine architecture of the cell.
One critical aspect of the efficacy and safety of statins is their dosage in clinical settings. The study of mammalian cells in culture offers not only the opportunity to study the effects of statins on mammalian cells but also a system to examine whether the response is within the concentration range of statins administered to patients. In our modified fluvastatin treatment protocol, this agent activates the p38 pathway at a concentration as low as 200 nM, underscoring the possibility that p38 activation is taking place in patients who are typically prescribed statins in concentrations ranging from 34 nM to 234 nM [23,24] and for a much longer time. In C. elegans, however, we found that the activation of the p38 pathway takes place at a higher fluvastatin concentration (Figures 1B and S1A). The difference in the concentration of fluvastatin required for p38 pathway activation may stem from the use of C. elegans as an in vivo model. C. elegans is protected from the environment by a cuticle that functions as a selective barrier for chemicals and drugs [55]. Thus, C. elegans cells are probably exposed to a much lower concentration of statins than their concentration in the culture medium. Moreover, previous studies revealed that long-term exposure to 20 μM fluvastatin [56] or 50 μM lovastatin [57] can extend the life span of C. elegans. To thoroughly study the dynamics of p38 pathway activation by statins, in most of our experiments we used a high dosage of fluvastatin. However, the life span assays suggest that statins can alter C. elegans physiology even at much lower concentrations, possibly even within the range of their clinical dosage in patients.
A growing body of data link many of the cholesterol-independent effects of statins, including their anti-inflammatory, anti-cancerous, and anti-degenerative activities, with the inhibition of geranylgeranylation and, consequently, the functions of small GTPases (reviewed in [17,58]). Our findings that statins activate a protective p38 response in C. elegans and the p38-COX-2 axis in RAW264.7 cells suggests that statins have an evolutionarily conserved function as regulators of innate immunity. The phosphorylation of p38 following fluvastatin treatment probably activates a transcription factor that upregulates the transcription of the cox-2 gene. The identity of this transcription factor and possible post-translational regulation of COX-2 levels will be important topics for future investigations. The significant upregulation of COX-2 in cells treated with 200 nM fluvastatin highlights the possibility that COX-2 upregulation is one of the treatment hallmarks of patients with statins. Accumulative data suggest that COX-2 activity can be either pro- or anti-inflammatory depending on the physiological context of its function [59]. By regulating inflammation, we propose that COX-2 upregulation may account for critical aspects of the cholesterol-independent effects of statins. COX-2 upregulation as a pro-inflammatory agent can be a key step in the manifestation of the adverse effects of statins, for example, the development of type 2 diabetes [11,12]. Alternatively, the anti-inflammatory function of COX-2 [60] can be a protective mechanism that is responsible for specific beneficial activities of statins [17].
Although COX-2 is not conserved in C. elegans (data not shown), a growing body of research suggests that the regulation of inflammation by statins is only partially mediated by COX-2 activity [61,62]. Thus, we posit that a conserved transcriptional response mediated by p38 activation underlies some of the critical cholesterol-independent effects of statins. Consistent with this hypothesis, we found that in C. elegans, statins elicit a global transcriptional response that involves the upregulation of protein families related to stress, immunity, and metabolism. A possible downstream target of p38 activation is the insulin pathway that is regulated by p38 activation [63] and responsible for statin-dependent life span extension in C. elegans [57]. However, our targeted screen of stress reporters (Fig. 1A) and the global expression analysis (Figure 4C–D) did not identify a signature of insulin pathway activity. Thus, we suggest that p38 activation acts independently of the insulin pathway to protect C. elegans from the cytotoxic effect of statins. Finding other targets of the statin-p38 axis in mammals and understanding how this axis is regulated in different cell types is expected to deepen our understanding of the cholesterol-independent effects of statins. Better mechanistic understanding of the effects of statins should offer more effective use of these medications prescribed to more than 200 million patients worldwide to treat cardiovascular diseases and cancer [64,65].
Data availability
The results of the screens conducted to identify the GTPases that regulate the expression levels of the tbb-6p::gfp reporter are presented in Table S1. Table S4 contains the statistical analysis of the length measurement depicted in Figure 6A. Table S5 contains data about all of the C. elegans strains, primers, and reagents used in this study. The strains and plasmids are available upon request. The RNA-seq reads were deposited into the National Center for Bioinformatic Information Gene Expression Omnibus (NCBI GEO) database under the accession #GSE132594. Further data sets supporting the RNA-seq results are included in supporting information files (Tables S2–3).
Author contributions
ILG, ST, LBH, and AS designed the study. ILG, ST, LBH, and AS conducted the experiments and analyzed the data. AS wrote the paper.
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We are grateful to Drs. Cole Haynes (UMass, Amherst, MA, USA), Paul. W. Sternberg (Caltech, Pasadena, CA, USA), Shane L. Rea (UW, Seattle, WA, USA), and Sivan Henis-Korenblit (Bar-Ilan University, Ramat Gan, Israel) for C. elegans strains. We thank Drs. Yoram Gercheman, Elah Pick, Shamsuzzama, and Mr. Benjamin Trabelsi (all from the University of Haifa, Haifa, Israel) for reagents and assistance with the analysis of the RT-qPCR results. We thank Drs. Limor Broday (Tel Aviv University, Tel Aviv, Israel) and Benjamin Podbilewicz (Technion-Israel Institute of Technology, Haifa, Israel) for providing reagents. We are grateful to Drs. Ayelet Lamm, Roni Hass, and Guy Horev (Technion-Israel Institute of Technology, Haifa, Israel) and Dr. Maya Lalzar (University of Haifa, Haifa, Israel) for assistance with the analysis of RNA-seq data. This study was supported by the Israel Science Foundation (ISF) grants 1747/16 to AS and 2240/19 to LBH.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101003.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ziaeian B., Fonarow G.C. Statins and the prevention of heart disease. JAMA Cardiology. 2017;2(4):464. doi: 10.1001/jamacardio.2016.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Endo A., Kuroda M., Tsujita Y. ML-236A, ML-236B, and ML-236C, new inhibitors of cholesterogenesis produced by Penicillium citrinium. Journal of Antibiotics. 1976;29(12):1346–1348. doi: 10.7164/antibiotics.29.1346. [DOI] [PubMed] [Google Scholar]
- 3.Endo A. A historical perspective on the discovery of statins. Proceedings of the Japan Academy Series B Physical and Biological Sciences. 2010;86(5):484–493. doi: 10.2183/pjab.86.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gazzerro P., Proto M.C., Gangemi G., Malfitano A.M., Ciaglia E., Pisanti S. Pharmacological actions of statins: a critical appraisal in the management of cancer. Pharmacological Reviews. 2012;64(1):102–146. doi: 10.1124/pr.111.004994. [DOI] [PubMed] [Google Scholar]
- 5.Ramkumar S., Raghunath A., Raghunath S. Statin therapy: review of safety and potential side effects. Acta Cardiologica Sinica. 2016;32(6):631–639. doi: 10.6515/ACS20160611A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonopoulos A.S., Margaritis M., Lee R., Channon K., Antoniades C. Statins as anti-inflammatory agents in atherogenesis: molecular mechanisms and lessons from the recent clinical trials. Current Pharmaceutical Design. 2012;18(11):1519–1530. doi: 10.2174/138161212799504803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan K.K., Oza A.M., Siu L.L. The statins as anticancer agents. Clinical Cancer Research : An Official Journal of the American Association for Cancer Research. 2003;9(1):10–19. [PubMed] [Google Scholar]
- 8.Freed-Pastor W.A., Mizuno H., Zhao X., Langerod A., Moon S.H., Rodriguez-Barrueco R. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148(1–2):244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butterfield D.A., Barone E., Mancuso C. Cholesterol-independent neuroprotective and neurotoxic activities of statins: perspectives for statin use in Alzheimer disease and other age-related neurodegenerative disorders. Pharmacological Research. 2011;64(3):180–186. doi: 10.1016/j.phrs.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McFarland A.J., Anoopkumar-Dukie S., Arora D.S., Grant G.D., McDermott C.M., Perkins A.V. Molecular mechanisms underlying the effects of statins in the central nervous system. International Journal of Molecular Sciences. 2014;15(11):20607–20637. doi: 10.3390/ijms151120607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins R., Reith C., Emberson J., Armitage J., Baigent C., Blackwell L. Interpretation of the evidence for the efficacy and safety of statin therapy. Lancet. 2016;388(10059):2532–2561. doi: 10.1016/S0140-6736(16)31357-5. [DOI] [PubMed] [Google Scholar]
- 12.Finegold J.A., Manisty C.H., Goldacre B., Barron A.J., Francis D.P. What proportion of symptomatic side effects in patients taking statins are genuinely caused by the drug? Systematic review of randomized placebo-controlled trials to aid individual patient choice. European Journal Preview Cardiology. 2014;21(4):464–474. doi: 10.1177/2047487314525531. [DOI] [PubMed] [Google Scholar]
- 13.Abramson J.D., Rosenberg H.G., Jewell N., Wright J.M. Should people at low risk of cardiovascular disease take a statin? BMJ British Medical Journal. 2013;347 doi: 10.1136/bmj.f6123. [DOI] [PubMed] [Google Scholar]
- 14.DeBose-Boyd R.A. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Research. 2008;18(6):609–621. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein J.L., Brown M.S. Regulation of the mevalonate pathway. Nature. 1990;343(6257):425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs J.M., Cohen A., Ein-Mor E., Stessman J. Cholesterol, statins, and longevity from age 70 to 90 years. Journal of the American Medical Directors Association. 2013;14(12):883–888. doi: 10.1016/j.jamda.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Keizer H.G. The "Mevalonate hypothesis": a cholesterol-independent alternative for the etiology of atherosclerosis. Lipids in Health and Disease. 2012;11:149. doi: 10.1186/1476-511X-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C.Y., Liu P.Y., Liao J.K. Pleiotropic effects of statin therapy: molecular mechanisms and clinical results. Trends in Molecular Medicine. 2008;14(1):37–44. doi: 10.1016/j.molmed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang P., Mukthavaram R., Chao Y., Nomura N., Bharati I.S., Fogal V. In vitro and in vivo anticancer effects of mevalonate pathway modulation on human cancer cells. British Journal of Cancer. 2014;111(8):1562–1571. doi: 10.1038/bjc.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zarubin T., Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Research. 2005;15(1):11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 21.Cuenda A., Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochimica et Biophysica Acta. 2007;1773(8):1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Cuadrado A., Nebreda A.R. Mechanisms and functions of p38 MAPK signalling. Biochemical Journal. 2010;429(3):403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 23.Yano M., Matsumura T., Senokuchi T., Ishii N., Murata Y., Taketa K. Statins activate peroxisome proliferator-activated receptor gamma through extracellular signal-regulated kinase 1/2 and p38 mitogen-activated protein kinase-dependent cyclooxygenase-2 expression in macrophages. Circulation Research. 2007;100(10):1442–1451. doi: 10.1161/01.RES.0000268411.49545.9c. [DOI] [PubMed] [Google Scholar]
- 24.Cho K.J., Hill M.M., Chigurupati S., Du G., Parton R.G., Hancock J.F. Therapeutic levels of the hydroxmethylglutaryl-coenzyme A reductase inhibitor lovastatin activate ras signaling via phospholipase D2. Molecular and Cellular Biology. 2011;31(6):1110–1120. doi: 10.1128/MCB.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinci G., Xia X., Veitia R.A. Preservation of genes involved in sterol metabolism in cholesterol auxotrophs: facts and hypotheses. PloS One. 2008;3(8) doi: 10.1371/journal.pone.0002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranji P., Rauthan M., Pitot C., Pilon M. Loss of HMG-CoA reductase in C. elegans causes defects in protein prenylation and muscle mitochondria. PloS One. 2014;9(2) doi: 10.1371/journal.pone.0100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., Samuel B.S., Breen P.C., Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508(7496):406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oks O., Lewin S., Goncalves I.L., Sapir A. The UPR(mt) protects Caenorhabditis elegans from mitochondrial dysfunction by upregulating specific enzymes of the mevalonate pathway. Genetics. 2018;209(2):457–473. doi: 10.1534/genetics.118.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morck C., Olsen L., Kurth C., Persson A., Storm N.J., Svensson E. Statins inhibit protein lipidation and induce the unfolded protein response in the non-sterol producing nematode Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(43):18285–18290. doi: 10.1073/pnas.0907117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams B.D., Schrank B., Huynh C., Shownkeen R., Waterston R.H. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131(3):609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neff M.M., Neff J.D., Chory J., Pepper A.E. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. The Plant Journal. 1998;14(3):387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 33.Neff M.M., Turk E., Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends in Genetics. 2002;18(12):613–615. doi: 10.1016/s0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- 34.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timmons L., Court D.L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263(1–2):103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 36.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Chen D., Smith M.A., Zhang B., Pan X. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PloS One. 2012;7(3) doi: 10.1371/journal.pone.0031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B. Aligning short sequencing reads with Bowtie. Current Protocols Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. [Chapter 11]:Unit 11 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 41.Cromey D.W. Avoiding twisted pixels: ethical guidelines for the appropriate use and manipulation of scientific digital images. Science and Engineering Ethics. 2010;16(4):639–667. doi: 10.1007/s11948-010-9201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rauthan M., Ranji P., Aguilera Pradenas N., Pitot C., Pilon M. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(15):5981–5986. doi: 10.1073/pnas.1218778110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munkacsy E., Khan M.H., Lane R.K., Borror M.B., Park J.H., Bokov A.F. DLK-1, SEK-3 and PMK-3 are required for the life extension induced by mitochondrial bioenergetic disruption in C. elegans. PLoS Genetics. 2016;12(7) doi: 10.1371/journal.pgen.1006133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hennessy E., Adams C., Reen F.J., O'Gara F. Is there potential for repurposing statins as novel antimicrobials? Antimicrobial Agents and Chemotherapy. 2016;60(9):5111–5121. doi: 10.1128/AAC.00192-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bar D.Z., Charar C., Gruenbaum Y. Small GTPases in C. elegans metabolism. Small GTPases. 2016:1–5. doi: 10.1080/21541248.2016.1247940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiner D.J., Lundquist E.A. Small GTPases. WormBook. 2016:1–99. doi: 10.1895/wormbook.1.67.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nargund A.M., Pellegrino M.W., Fiorese C.J., Baker B.M., Haynes C.M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337(6094):587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lecine P., Italiano J.E., Jr., Kim S.W., Villeval J.L., Shivdasani R.A. Hematopoietic-specific beta 1 tubulin participates in a pathway of platelet biogenesis dependent on the transcription factor NF-E2. Blood. 2000;96(4):1366–1373. [PubMed] [Google Scholar]
- 49.Sirtori C.R. The pharmacology of statins. Pharmacological Research. 2014;88:3–11. doi: 10.1016/j.phrs.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Banach M., Serban C., Ursoniu S., Rysz J., Muntner P., Toth P.P. Statin therapy and plasma coenzyme Q10 concentrations--A systematic review and meta-analysis of placebo-controlled trials. Pharmacological Research. 2015;99:329–336. doi: 10.1016/j.phrs.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 51.Schirris T.J., Renkema G.H., Ritschel T., Voermans N.C., Bilos A., van Engelen B.G. Statin-induced myopathy is associated with mitochondrial complex III inhibition. Cell Metabolism. 2015;22(3):399–407. doi: 10.1016/j.cmet.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Roy A., Jana M., Kundu M., Corbett G.T., Rangaswamy S.B., Mishra R.K. HMG-CoA reductase inhibitors bind to PPARalpha to upregulate neurotrophin expression in the brain and improve memory in mice. Cell Metabolism. 2015;22(2):253–265. doi: 10.1016/j.cmet.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wandinger-Ness A., Zerial M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harbor Perspectives in Biology. 2014;6(11):a022616. doi: 10.1101/cshperspect.a022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myers K.R., Casanova J.E. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends in Cell Biology. 2008;18(4):184–192. doi: 10.1016/j.tcb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiong H., Pears C., Woollard A. An enhanced C. elegans based platform for toxicity assessment. Scientific Reports. 2017;7(1):9839. doi: 10.1038/s41598-017-10454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreux P.A., Mouchiroud L., Wang X., Jovaisaite V., Mottis A., Bichet S. A method to identify and validate mitochondrial modulators using mammalian cells and the worm C. elegans. Scientific Reports. 2014;4:5285. doi: 10.1038/srep05285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jahn A., Scherer B., Fritz G., Honnen S. Statins induce a DAF-16/Foxo-dependent longevity phenotype via JNK-1 through mevalonate depletion in C. elegans. Aging Disease. 2020;11(1):60–72. doi: 10.14336/AD.2019.0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao J.K., Laufs U. Pleiotropic effects of statins. Annual Review of Pharmacology and Toxicology. 2005;45:89–118. doi: 10.1146/annurev.pharmtox.45.120403.095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aid S., Bosetti F. Targeting cyclooxygenases-1 and -2 in neuroinflammation: therapeutic implications. Biochimie. 2011;93(1):46–51. doi: 10.1016/j.biochi.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilroy D.W., Colville-Nash P.R., Willis D., Chivers J., Paul-Clark M.J., Willoughby D.A. Inducible cyclooxygenase may have anti-inflammatory properties. Nature Medicine. 1999;5(6):698–701. doi: 10.1038/9550. [DOI] [PubMed] [Google Scholar]
- 61.Shen Y., Wu H., Wang C., Shao H., Huang H., Jing H. Simvastatin attenuates cardiopulmonary bypass-induced myocardial inflammatory injury in rats by activating peroxisome proliferator-activated receptor gamma. European Journal of Pharmacology. 2010;649(1–3):255–262. doi: 10.1016/j.ejphar.2010.08.058. [DOI] [PubMed] [Google Scholar]
- 62.Balakumar P., Mahadevan N. Interplay between statins and PPARs in improving cardiovascular outcomes: a double-edged sword? British Journal of Pharmacology. 2012;165(2):373–379. doi: 10.1111/j.1476-5381.2011.01597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horowitz L.B., Brandt J.P., Ringstad N. Repression of an activity-dependent autocrine insulin signal is required for sensory neuron development in C. elegans. Development. 2019;146(22) doi: 10.1242/dev.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blaha M.J., Martin S.S. How do statins work? Changing paradigms with implications for statin allocation. Journal of the American College of Cardiology. 2013;62(25):2392–2394. doi: 10.1016/j.jacc.2013.08.1626. [DOI] [PubMed] [Google Scholar]
- 65.Desai C.S., Martin S.S., Blumenthal R.S. Non-cardiovascular effects associated with statins. BMJ British Medical Journal. 2014;349 doi: 10.1136/bmj.g3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results of the screens conducted to identify the GTPases that regulate the expression levels of the tbb-6p::gfp reporter are presented in Table S1. Table S4 contains the statistical analysis of the length measurement depicted in Figure 6A. Table S5 contains data about all of the C. elegans strains, primers, and reagents used in this study. The strains and plasmids are available upon request. The RNA-seq reads were deposited into the National Center for Bioinformatic Information Gene Expression Omnibus (NCBI GEO) database under the accession #GSE132594. Further data sets supporting the RNA-seq results are included in supporting information files (Tables S2–3).