Abstract
Objectives
We aimed to develop a simple algorithm to help early identification of SARS-CoV-2 infection patients with severe progression tendency.
Methods
The univariable and multivariable analysis were computed to identify the independent predictors of COVID-19 progression. The prediction model was established in a retrospective training set of 322 COVID-19 patients and was re-evaluated in a prospective validation set of 317 COVID-19 patients.
Results
The multivariable analysis identified age (OR = 1.061, p = 0.028), lactate dehydrogenase (LDH) (OR = 1.006, p = 0.037), and CD4 count (OR = 0.993, p = 0.006) as the independent predictors of COVID-19 progression. Consequently, the age-LDH-CD4 algorithm was derived as (age × LDH)/CD4 count. In the training set, the area under the ROC curve (AUROC) of age-LDH-CD4 model was significantly higher than that of single CD4 count, LDH, or age (0.92, 0.85, 0.80, and 0.75, respectively). In the prospective validation set, the AUROC of age-LDH-CD4 model was also significantly higher than that of single CD4 count, LDH, or age (0.92, 0.75, 0.81, and 0.82, respectively). The age-LDH-CD4 ≥ 82 has high sensitive (81%) and specific (93%) for the early identification of COVID-19 patients with severe progression tendency.
Conclusions
The age-LDH-CD4 model is a simple algorithm for early identifying patients with severe progression tendency following SARS-CoV-2 infection, and warrants further validation.
Keywords: Severe acute respiratory syndrome coronavirus 2, 2019 novel coronavirus disease, Severe progression, Risk factors
Introduction
Since November 2019, an outbreak of 2019 novel coronavirus disease (COVID-19) in Wuhan, China, caused by a novel coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the rapidly spread has caused a major public health issue around the world [1]. As of March 12th, 2020, there have been 80,955 confirmed cases and 3162 deaths in China, and 118,326 confirmed cases and 4292 deaths globally [2].
In previous studies, the clinical characteristics of SARS-CoV-2 infection have been described in detail [3–5]. Most patients were mild and often experienced fever, cough, and fatigue after an incubation period of median 3–7 days, and then recovered in 2–3 weeks [6]. About 17–32% of patients developed into severe cases, and might progress rapidly to complications including acute respiratory distress syndrome, shock, secondary infection, and so on [3–5]. The reported proportion of severe cases were 17% [4], 26.1% [5], and 32% [3], and the mortalities were 4.3% [5], 11.0% [4], and 14.6% [3], respectively, in Wuhan, China.
Although the first patient of SARS-CoV-2 infection in the United States responding well to remdesivir [7], there were no clinically effective antiviral drugs for SARS-CoV-2 infection [8]. Early identification of COVID-19 progression is urgently needed not only to guide appropriate supportive care for patients, but also assist medical staff in triaging patients when allocating limited healthcare resources. However, to date, there is no clinically available method to early identify patients with severe progression tendency following SARS-CoV-2 infection. In this study, we designed a simple algorithm which helps in early identification of COVID-19 progression, and aimed to provide a clinically available method to regulate the large flow of SARS-CoV-2 infection patients between primary health care and tertiary centers.
Patients and methods
Patients
To establish a prediction model, we retrospectively enrolled 322 confirmed SARS-CoV-2 infection patients, who hospitalized in Shanghai Public Health Clinical Center, Shanghai, China, a designated tertiary teaching hospital for the treatment of SARS-CoV-2 infection, between January 20th 2020 and February 23th 2020. The 322 patients were used as the training set, and grouped into severe (n = 26) and non-severe (n = 296). Of 26 severe cases, 11 were diagnosed as severe cases on admission, and 15 developed to severe cases during the hospitalization.
To re-evaluate the prediction performance of the age-LDH-CD4 model, we prospectively included 317 COVID-19 patients who hospitalized in Shanghai Public Health Clinical Center, between February 24th and May 1th 2020. Among the 317 COVID-19 patients, 3 were diagnosed as severe cases on admission, 2 developed to severe cases during the hospitalization and 312 were non-severe cases.
This study was approved by the ethics committee of Shanghai Public Health Clinical Center. The clinical diagnosis and treatment complied with the Helsinki declaration.
Diagnostic criteria
Laboratory confirmation of SARS-CoV-2 infection was achieved by the Chinese Center for Disease Prevention and Control. A confirmed case was defined as a positive result with SARS-COV-2 nucleotides by polymerase-chain-reaction (PCR) assay for nasopharyngeal or throat swab specimens [9], according to the protocol established by World Health Organization [10]. Severe cases were defined as at least one of the followings: (1) respiratory distress, respiratory rates ≥ 30/min; (2) pulse oxygen saturation ≤ 93% in a resting state; (3) oxygenation index (PaO2/FiO2) ≤ 300 mmHg; (4) require mechanical ventilation; (5) shock; (6) combined with other organ failures and needed treatment in intensive care unit (ICU).
Data collection
The age, gender, and comorbidities were extracted from the electronic medical records. The laboratory findings including complete blood count, lymphocyte subsets, C-reactive protein (CRP), procalcitonin (PCT), alanine aminotransferase (ALT), aspartate aminotransferase (AST), gamma-glutamyl-transpeptidase (GGT), lactate dehydrogenase (LDH), total bilirubin (TBIL), creatinine, creatine kinase (CK), and D-dimer were obtained with data collection forms.
Statistical analysis
The normality test was performed for continuous variables using the Kolmogorov–Smirnov test. Normal distribution variables, non-normal distribution continuous variables, and categorical variables, were shown as means and standard deviations, medians and interquartile ranges (IQR), and counts and percentage, respectively. T tests, Mann–Whitney-tests, and chi-square tests were applied to normal distribution variables, non-normal distribution continuous variables, and categorical variables, respectively. The Pearson correlation analysis was performed to show the correlation between two variables. The univariable and multiple analyze was performed to identify the independent predictors of COVID-19 progression. The areas under the receiver operating characteristic (ROC) curves (AUROCs) were used to estimate the predictive accuracy, and compared using Delong test [11]. The optimal cut-offs were obtained by maximizing Youden index. All statistical analyses were conducted with SPSS software version 15.0 (SPSS Inc. USA) and MedCalc software version 16.1 (MedCalc Software, Belgium).
Results
Baseline characteristics of patients in the training set
Baseline characteristics of patients in the training set are shown in Table 1. The median age was 51 years (IQR, 36–64), 51.8% were males, and 33.2% had comorbidities. The age (65 vs. 49 years, p < 0.001) and proportion of male (76.9% vs. 49.8%, p = 0.008) in severe cases were significantly higher than that in non-severe cases. Moreover, comorbidities were more common in severe cases as compared with non-severe cases (69.2% vs. 30.1%, p < 0.001).
Table 1.
Baseline characteristics of patients in the training set
| Total (n = 322) | Severe (n = 26) | Non-severe (n = 296) | p value | |
|---|---|---|---|---|
| Age (years) | 51 (36–64) | 65 (63–76) | 49 (36–63) | < 0.001 |
| Male, n (%) | 167 (51.8%) | 20 (76.9%) | 147 (49.8%) | 0.008 |
| Comorbidities, n (%) | 107 (33.2%) | 18 (69.2%) | 89 (30.1%) | < 0.001 |
| WBC (109/L) | 4.8 (3.9–6.0) | 5.4 (3.6–10.2) | 4.8 (4.0–5.9) | 0.295 |
| Lymphocyte (109/L) | 1.11 (0.79–1.49) | 0.65 (0.48–0.87) | 1.14 (0.84–1.52) | < 0.001 |
| Platelet (109/L) | 179 (143–224) | 157 (121–211) | 181 (144–226) | 0.188 |
| CD3 (cell/μl) | 727 (504–1027) | 323 (181–542) | 773 (550–1054) | < 0.001 |
| CD8 (cell/μl) | 250 (159–388) | 124 (62–173) | 264 (171–404) | < 0.001 |
| CD4 (cell/μl) | 428 (299–633) | 159 (110–304) | 452 (313–650) | < 0.001 |
| CD45 (cell/μl) | 1089 (750–1460) | 586 (431–799) | 1120 (800–1505) | < 0.001 |
| CRP (mg/L) | 9 (2–26) | 53 (26–87) | 8 (2–22) | < 0.001 |
| PCT (ng/mL) | 0.03 (0.02–0.06) | 0.16 (0.06–0.62) | 0.03 (0.02–0.05) | < 0.001 |
| ALT (U/L) | 22 (15–34) | 26 (19–39) | 22 (15–33) | 0.067 |
| AST (U/L) | 24 (19–33) | 45 (26–53) | 23 (19–32) | < 0.001 |
| GGT (U/L) | 25 (17–42) | 28 (21–68) | 25 (16–42) | 0.119 |
| LDH (U/L) | 229 (193–293) | 399 (336–499) | 224 (192–270) | < 0.001 |
| TBIL (μmol/L) | 8.2 (6.6–10.5) | 10.3 (8.6–13.8) | 8.0 (6.5–10.4) | 0.001 |
| BUN (mmol/L) | 4.41 (3.55–5.46) | 5.10 (4.04–9.80) | 4.37 (3.55–5.36) | 0.025 |
| Creatinine (μmol/L) | 63 (51–76) | 80 (57–117) | 63 (51–75) | 0.002 |
| CK (U/L) | 82 (57–130) | 220 (113–417) | 78 (55–118) | < 0.001 |
| D-dimer (ng/mL) | 0.43 (0.29–0.79) | 1.20 (0.74–2.23) | 0.41 (0.28–0.69) | < 0.001 |
The p values indicate differences between severe and non-severe patients. p < 0.05 was considered statistically significant
WBC white blood count, CRP C-reactive protein, PCT procalcitonin, ALT alanine aminotransferase, AST aspartate aminotransferase, GGT γ-glutamyl-transpeptidase, LDH lactate dehydrogenase, TBIL total bilirubin, BUN blood urea nitrogen, CK creatine kinase
Severe cases had significantly higher CRP (53 vs. 8 mg/L, p < 0.001), PCT (0.16 vs. 0.03 ng/mL, p < 0.001), AST (45 vs. 23 U/L, p < 0.001), LDH (399 vs. 224 U/L, p < 0.001), TBIL (10.3 vs. 8.0 μmol/L, p = 0.001), BUN (5.10 vs. 4.37 mmol/L, p = 0.025), creatinine (80 vs. 63 μmol/L, p = 0.002), CK (220 vs. 78 U/L, p < 0.001), and D-dimer (1.20 vs. 0.41 μg/mL, p < 0.001); but significantly lower lymphocyte (0.65 × 109 vs. 1.14 × 109 cells/L, p < 0.001), CD3 (323 vs. 773 cell/μl, p < 0.001), CD8 (124 vs. 264 cell/μl, p < 0.001), CD4 (159 vs. 452 cell/μl, p < 0.001), and CD45 count (586 vs. 1120 cell/μl, p < 0.001) compared with non-severe cases.
Baseline characteristics of patients in the prospective validation set
Baseline characteristics of patients in the prospective validation set are shown in Table 2. The median age was 30 years (IQR, 21–42), 59.3% were males, and 14.2% had comorbidities. The age (54 vs. 30 years, p = 0.003) and proportion of comorbidities (100% vs. 12.8%, p < 0.001) in severe cases were significantly higher than that in non-severe cases. Severe cases had significantly higher CRP (13 vs. 0.5 mg/L, p = 0.025), LDH (299 vs. 193 U/L, p = 0.015), CK (170 vs. 76 U/L, p = 0.010), and D-dimer (0.47 vs. 0.28 μg/mL, p = 0.02); but significantly lower lymphocyte (1.01 × 109 vs. 1.61 × 109 cells/L, p = 0.008), CD3 (611 vs. 1176 cell/μl, p = 0.008), CD8 (151 vs. 430 cell/μl, p = 0.002), CD4 (453 vs. 622 cell/μl, p = 0.045), and CD45 count (942 vs. 1634 cell/μl, p = 0.018) compared with non-severe cases.
Table 2.
Baseline characteristics of patients in the prospective validation set
| Total (n = 317) | Severe (n = 5) | Non-severe (n = 312) | p value | |
|---|---|---|---|---|
| Age (years) | 30 (21–42) | 54 (39–66) | 30 (21–41) | 0.003 |
| Male, n (%) | 188 (59.3%) | 4 (80%) | 184 (59%) | 0.342 |
| Comorbidities, n (%) | 45 (14.2%) | 5 (100%) | 40 (12.8%) | < 0.001 |
| WBC (109/L) | 5.7 (4.5–6.8) | 4.7 (2.4–6.9) | 5.7 (4.5–6.8) | 0.343 |
| Lymphocyte (109/L) | 1.60 (1.22–2.02) | 1.01 (0.27–1.40) | 1.61 (1.22–2.02) | 0.008 |
| Platelet (109/L) | 227 (187–270) | 199 (154–243) | 228 (187–272) | 0.297 |
| CD3 (cell/μl) | 1170 (882–1529) | 611 (114–1038) | 1176 (893–1531) | 0.008 |
| CD8 (cell/μl) | 429 (301–585) | 151 (55–319) | 430 (303–588) | 0.002 |
| CD4 (cell/μl) | 621 (461–849) | 453 (54–630) | 622 (463–852) | 0.045 |
| CD45 (cell/μl) | 1630 (1234–2095) | 942 (261–1550) | 1634 (1240–2100) | 0.018 |
| CRP (mg/L) | 0.5 (0.5–1.12) | 13 (1–29) | 0.5 (0.5–1.11) | 0.025 |
| PCT (ng/mL) | 0.02 (0.02–0.03) | 0.02 (0.01–0.08) | 0.02 (0.02–0.03) | 0.299 |
| ALT (U/L) | 20 (13–31) | 26 (18–43) | 20 (12–31) | 0.281 |
| AST (U/L) | 19 (16–24) | 24 (19–49) | 19 (16–24) | 0.079 |
| LDH (U/L) | 193 (171–217) | 299 (197–385) | 193 (170–217) | 0.015 |
| TBIL (μmol/L) | 9.1 (7.2–12.3) | 15.5 (7.7–16.2) | 9.1 (7.2–12.3) | 0.241 |
| BUN (mmol/L) | 4.21 (3.57–5.03) | 5.81 (4.09–6.24) | 4.20 (3.56–5.01) | 0.072 |
| Creatinine (μmol/L) | 67 (56–76) | 74 (45–97) | 67 (56–75) | 0.720 |
| CK (U/L) | 77 (57–104) | 170 (109–209) | 76 (57–104) | 0.010 |
| D-dimer (ng/mL) | 0.28 (0.22–0.40) | 0.47 (0.31–1.64) | 0.28 (0.22–0.40) | 0.02 |
The p values indicate differences between severe and non-severe patients. p < 0.05 was considered statistically significant
WBC white blood count, CRP C-reactive protein, PCT procalcitonin, ALT alanine aminotransferase, AST aspartate aminotransferase, GGT γ-glutamyl-transpeptidase, LDH lactate dehydrogenase, TBIL total bilirubin, BUN blood urea nitrogen, CK creatine kinase
Correlations between clinical parameters and COVID-19 progression
Variables associated with COVID-19 progression are shown in Table 3. The COVID-19 progression positively correlated with LDH (r = 0.39, p < 0.001), PCT (r = 0.39, p < 0.001), CRP (r = 0.35, p < 0.001), D-dimer (r = 0.32, p < 0.001), CK (r = 0.31, p < 0.001), age (r = 0.27, p < 0.001), comorbidities (r = 0.27, p < 0.001), AST (r = 0.25, p < 0.001), TBIL (r = 0.18, p = 0.001), creatinine (r = 0.17, p = 0.002), male (r = 0.15, p = 0.008), and BUN (r = 0.12, p = 0.026); and negatively correlated with CD4 (r = − 0.34, p < 0.001), CD3 (r = − 0.33, p < 0.001), CD8 (r = − 0.32, p < 0.001), CD45 (r = − 0.31, p < 0.001), and lymphocyte count (r = − 0.30, p < 0.001).
Table 3.
Correlations between clinical parameters and COVID-19 progression
| Variables | r value | p value |
|---|---|---|
| LDH (U/L) | 0.39 | < 0.001 |
| PCT (ng/mL) | 0.39 | < 0.001 |
| CRP (mg/L) | 0.35 | < 0.001 |
| D-dimer (ng/mL) | 0.32 | < 0.001 |
| CK (U/L) | 0.31 | < 0.001 |
| Age (years) | 0.27 | < 0.001 |
| Comorbidities | 0.27 | < 0.001 |
| ALT (IU/L) | 0.25 | < 0.001 |
| TBIL (μmol/L) | 0.18 | 0.001 |
| Creatinine (μmol/L) | 0.17 | 0.002 |
| Male | 0.15 | 0.008 |
| BUN (mmol/L) | 0.12 | 0.026 |
| CD4 count (cell/μl) | − 0.34 | < 0.001 |
| CD3 count (cell/μl) | − 0.33 | < 0.001 |
| CD8 count (cell/μl) | − 0.32 | < 0.001 |
| CD45 count (cell/μl) | − 0.31 | < 0.001 |
| Lymphocyte (109/L) | − 0.30 | < 0.001 |
LDH lactate dehydrogenase, PCT procalcitonin, CRP C-reactive protein, CK creatine kinase, ALT alanine aminotransferase, TBIL total bilirubin, BUN blood urea nitrogen; r value, correlation coefficient; the higher the absolute value of r value, the stronger the correlation
Independent predictors of COVID-19 progression
The independent predictors of COVID-19 progression are shown in Table 4. Univariate analysis showed that age, male, comorbidities, D-dimer, white blood cell, lymphocyte, CRP, CD3, CD4, CD45, AST, LDH, TBIL, and creatinine were associated with COVID-19 progression (all p < 0.05). Multivariable analysis identified age (OR = 1.061, 95% CI, 1.007–1.119, p = 0.028), CD4 count (OR = 0.993, 95% CI, 0.987–0.998, p = 0.006), and LDH (OR = 1.006, 95% CI, 1.000–1.012, p = 0.037) as the independent predictors of COVID-19 progression.
Table 4.
Independent predictors of COVID-19 progression
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | |
| Age (years) | 1.086 (1.048–1.126) | < 0.001 | 1.061 (1.007–1.119) | 0.028 |
| Male | 3.379 (1.319–8.652) | 0.011 | 1.152 (0.245–5.411) | 0.857 |
| Comorbidities | 2.628 (1.805–3.826) | < 0.001 | 1.125 (0.532–2.380) | 0.757 |
| D-dimer (ng/mL) | 1.115 (1.027–1.211) | 0.009 | 0.967 (0.865–1.082) | 0.562 |
| WBC (109/L) | 1.187 (1.041–1.353) | 0.011 | 1.154 (0.917–1.454) | 0.223 |
| Lymphocyte (109/L) | 0.132 (0.041–0.425) | 0.001 | 9.406 (0.143–619.476) | 0.294 |
| CRP (mg/L) | 1.048 (1.032–1.063) | < 0.001 | 1.019 (0.998–1.039) | 0.072 |
| CD3 (cell/μl) | 0.997 (0.996–0.999) | < 0.001 | 0.999 (0.995–1.003) | 0.656 |
| CD8 (cell/μl) | 0.999 (0.997–1.002) | 0.535 | ||
| CD4 (cell/μl) | 0.991 (0.987–0.994) | < 0.001 | 0.993 (0.987–0.998) | 0.006 |
| CD45 (cell/μl) | 0.998 (0.997–0.999) | < 0.001 | 1.000 (0.998–1.002) | 0.963 |
| Platelet (109/L) | 0.997 (0.990–1.004) | 0.360 | ||
| ALT (U/L) | 1.010 (0.993–1.027) | 0.267 | ||
| AST (U/L) | 1.017 (1.003–1.032) | 0.020 | 1.005 (0.978–1.033) | 0.716 |
| GGT (U/L) | 1.007 (0.997–1.017) | 0.145 | ||
| LDH (U/L) | 1.011 (1.007–1.015) | < 0.001 | 1.006 (1.000–1.012) | 0.037 |
| TBIL (μmol/L) | 1.119 (1.042–1.202) | 0.002 | 1.003 (0.871–1.155) | 0.966 |
| BUN (mmol/L) | 1.000 (0.996–1.003) | 0.836 | ||
| Creatinine (μmol/L) | 1.025 (1.012–1.038) | < 0.001 | 1.012 (0.986–1.039) | 0.371 |
| CK (U/L) | 1.000 (1.000–1.001) | 0.123 | ||
Multivariate analysis were fitted by including the factors associated with COVID-19 progression in the univariable analyses (p < 0.05). Multivariable analysis identified age (p = 0.028), CD4 (p = 0.006), and LDH (p = 0.037) as the independent predictors of COVID-19 progression
WBC white blood count, CRP C-reactive protein, ALT alanine aminotransferase, AST aspartate aminotransferase, GGT γ-glutamyl-transpeptidase, LDH lactate dehydrogenase, TBIL total bilirubin, BUN blood urea nitrogen, CK creatine kinase
Develop a simple algorithm early identifying patients with severe progression tendency
In Table 3, we found that age and LDH had a positive correlation with COVID-19 progression (r > 0, p < 0.001), and CD4 count was negatively correlated (r < 0, p < 0.001). In Table 4, we found that age, LDH, and CD4 count were the independent predictors of COVID-19 progression. To improve the prediction performance using age, LDH, and CD4 count, a simple algorithm was derived as: age (years) × LDH (U/L)/ CD4 (cell/μl).
AUROCs comparison of the age-LDH-CD4 model and single index
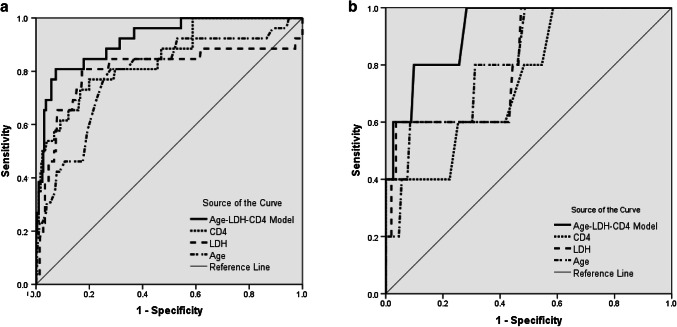
Based on the fact that only the patients developed to severe cases after admission could be counted for prediction. Therefore, ROC curve analysis was only performed in 15 patients who developed to severe cases after admission in the training set (Fig. 1a) and 2 patients who developed to severe cases after admission in the prospective validation set (Fig. 1b). Pairwise comparison of AUROCs is shown in Table 5. In the training set, the AUROC of age-LDH-CD4 model (0.92, 95%CI 0.88 to 0.95) was significantly higher than that of CD4 (0.85, 95% CI 0.81–0.89, p = 0.005), LDH (0.80, 95% CI 0.75–0.84, p = 0.025), and age (0.79, 95% CI 0.74–0.83, p < 0.001). In the prospective validation set, the AUROC of age-LDH-CD4 model (0.92, 95% CI 0.89–0.95) was also significantly higher than that of CD4 (0.75, 95% CI 0.70–0.80, p = 0.021), LDH (0.81, 95% CI 0.76–0.85, p = 0.027), and age (0.82, 95% CI 0.77–0.86, p = 0.028).
Fig. 1.
ROC curves of the age-LDH-CD4 model and single index in the training set (a) and the prospective validation set (b). The AUROC of age-LDH-CD4 model was significantly higher than that of CD4 count, LDH, and age. In the training set, the AUROC of age-LDH-CD4 model was significantly higher than that of CD4 (p = 0.005), LDH (p = 0.025), and age (p < 0.001). In the prospective validation set, the AUROC of age-LDH-CD4 model was also significantly higher than that of CD4 (p = 0.021), LDH (p = 0.027), and age (p = 0.028)
Table 5.
AUROCs comparison of the age-LDH-CD4 model and single index
| Training set | Prospective validation set | ||||
|---|---|---|---|---|---|
| AUROC | (95% CI) | AUROC | (95% CI) | ||
| Age-LDH-CD4 model | 0.92 | (0.88–0.95) | 0.92 | (0.89–0.95) | |
| CD4 (cell/μl) | 0.85 | (0.81–0.89) | 0.75 | (0.70–0.80) | |
| LDH (U/L) | 0.80 | (0.75–0.84) | 0.81 | (0.76–0.85) | |
| Age (years) | 0.79 | (0.74–0.83) | 0.82 | (0.77–0.86) | |
| Age-LDH-CD4 vs. CD4 | p = 0.005 | p = 0.021 | |||
| Age-LDH-CD4 vs. LDH | p = 0.025 | p = 0.027 | |||
| Age-LDH-CD4 vs. age | p < 0.001 | p = 0.028 | |||
Age-LDH-CD4 model = (age × LDH)/CD4 count
LDH lactate dehydrogenase
Cut-off values of the age-LDH-CD4 model and single index
The cut-off values are shown in Table 6. According to maximizing the Youden index, the optimal cut-off values were 82 for the age-LDH-CD4 model (the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) was 82%, 81%, 49%, and 98%, respectively), 295 for CD4 (the sensitivity, specificity, PPV, and NPV was 77%, 81%, 25%, and 98%, respectively), 60 for age (the sensitivity, specificity, PPV, and NPV was 81%, 72%, 20%, and 98%, respectively), and 300 for LDH (the sensitivity, specificity, PPV, and NPV was 81%, 83%, 29%, and 98%, respectively).
Table 6.
Cut-off values of the age-LDH-CD4 model and single index
| Cut-off | Se (%) | Sp (%) | PPV (%) | NPV (%) | + LR | − LR | |
|---|---|---|---|---|---|---|---|
| Age-LDH-CD4 model | 82 | 81 | 93 | 49 | 98 | 10.87 | 0.21 |
| CD4 (cell/μl) | 295 | 77 | 81 | 25 | 98 | 3.86 | 0.29 |
| Age (years) | 60 | 81 | 72 | 20 | 98 | 2.88 | 0.27 |
| LDH (U/L) | 300 | 81 | 83 | 29 | 98 | 4.69 | 0.23 |
Age-LDH-CD4 Model = (age × LDH)/CD4 count
LDH lactate dehydrogenase, Se sensitivity, Sp specificity, PPV positive predictive value, NPV negative predictive value, + LR positive likelihood ratio, − LR negative likelihood ratio
Discussion
The current SARS-CoV-2 outbreak is the third epidemic caused by coronavirus in the twenty-first century, following severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome. At present, the epidemiological and clinical characteristics of SARS-CoV-2 infection have been reported [3–5]. However, there is still a lack on clinically available method for early identifying COVID-19 patients with severe progression tendency, which is important for regulating the large flow of patients between primary health care and tertiary centers. Patients without severe progression tendency can be treated in primary health care, whereas those had severe progression tendency needed to be redirected to tertiary centers for specialized care and treatment.
This study developed a simple algorithm, named age-LDH-CD4 model, to early identify COVID-19 patients with severe progression tendency. The age-LDH-CD4 model based on three routine parameters: age, LDH, and CD4 count. In this study, the age-LDH-CD4 model ≥ 82 has high sensitive (81%) and specific (93%) for the early identification of COVID-19 progression. The NPV of the age-LDH-CD4 model is 98% for predicting COVID-19 progression, suggesting just 2% of patients with age-LDH-CD4 < 82 developed to severe cases in the training set. In the prospective validation set, 2 patients had the age-LDH-CD4 model ≥ 82 on admission, 1 patient (50%) developed to severe cases during the hospitalization. In the prospective validation set, 315 patients had the age-LDH-CD4 model < 82 on admission, and only 1 patient (0.3%) developed to severe cases. That is to say, over 99% of patients with age-LDH-CD4 model < 82 will not progress to severe cases, and can be managed at primary health care or district hospitals. About 50% of patients with age-LDH-CD4 model ≥ 82 will progress to severe cases, and can benefit from early transfer to tertiary centers.
In this study, COVID-19 patients with age > 60 years, LDH > 300 U/L, or CD4 count < 295 cell/μl had more likely to develop to severe cases. The results were consistent with previous studies. Wang et al. [5] reported that patients treated in the ICU were older (median age, 66 years vs. 51 years) compared with patients not treated in the ICU. Guan et al. [12] reported that patients in severe subgroups had higher age than patients in non-severe subgroups (mean difference, 7.0, 95% CI, 4.4–9.6). Chan et al. [13] also found older patients (aged > 60 years) had more lymphopenia and increased LDH levels.
Test of lymphocyte subsets is important for the diagnosis and treatment of virus infection. In 2003, Cui et al. [14] had found that the absolute counts of lymphocyte subsets had a clinical significance for patients with SARS, and CD4 (+) T lymphocyte levels were reduced in 100% of SARS patients. In 2004, Li et al. [15] reported that peripheral T lymphocyte subsets can facilitate an earlier and more accurate diagnosis of SARS. Therefore, at the early stages of the SARS-CoV-2 outbreak, lymphocyte subsets were routinely measured for COVID-19 patients in this study. Besides our study, Chen and colleagues found that increased lymphopenia (in CD4+ and CD8+ T cells), and decreased IFN-γ expression in CD4+ T cells are associated with severe COVID-19 [16]. In the issue of the Clinical Infectious Disease, Wang and colleagues also found that CD4+ T cells decreased in COVID-19 patients, and severe cases had a lower level than mild cases [17]. Therefore, we evaluated CD4 count when designing an algorithm that signified a severe disease course in this study.
In this study, besides patients were diagnosed as severe cases on admission (n = 11 for the training set, and n = 3 for the prospective validation set), there were only a few patients included in the study that progressed to a severe disease status after admission (n = 15 for the training set, and n = 2 for the prospective validation set). The number of patients who developed to severe cases during the hospitalization was lower in this study compared with that reported in Wuhan, China (17–32%) [3–5]. As of May 1th 2020, of 639 confirmed COVID-19 patients in training and validation sets, only 7 patients dead, a markedly lower fatality rate (1.1%) compared with that reported in Wuhan (4.3–14.6%) [3–5]. The possible reasons for the lower severe rate and fatality rate are as follows. First, the control measures (early discovery, early isolation, early diagnosis, and early management) have been undertaken, and enough health workers and medical services have been provided at the early time of the COVID-19 outbreak in Shanghai, China. Second, all imported persons were tested with PCR assay for SARS-CoV-2 before they were released from quarantine. As a result, nearly all of the COVID-19 cases were diagnosed at earlier stages of disease. Especially for the prospective validation set of 317 COVID-19 patients, 146 (46%) had no pneumonia in CT scans, and 58 (18.3%) were asymptomatic patients.
This study has some limitations. First, this study is a single-center study, because Shanghai Public Health Clinical Center is the only SARS-CoV-2 designated hospital for confirmed adult patients in Shanghai, China. Second, in this study, IL-6 had not been evaluated when designing the algorithm. Recently, a meta-analysis of 9 studies demonstrated that patients with severe COVID-19 had a significantly higher serum IL-6 levels compared to non-severe patients, and increasing IL-6 level was associated with increased mortality (Coefficient (Q) 0.01, 95% CI 0.01–0.03, p = 0.03) [18]. Another meta-analysis of 3 studies also demonstrated that severe COVID-19 patients have a higher IL-6/IFN-γ ratio than moderate patients [19]. Unfortunately, 322 patients in the training set hospitalized between January 20th 2020 and February 23th 2020, the early stages of the COVID-19 outbreak, when the role of IL-6 had not been reported in the evaluation of COVID-19 progression. As a result, IL-6 had not been routinely measured in this study. Therefore, we could not evaluate the IL-6 when designing the algorithm.
In summary, this study showed that SARS-CoV-2 infection had a low severe rate and fatality rate once the control measures (early discovery, early reporting, early quarantine and early treatment) were undertaken at the beginning of COVID-19 outbreaks. The age, LDH, and CD4 counts were the independent predictors of COVID-19 progression. The age-LDH-CD4 algorithm is a simple and accurate index for the early identification of COVID-19 patients with severe progression tendency, and warrants further validation.
Acknowledgements
We thank the doctors (Shui-Bao Xu, Yi-Xiao Lin, Feng Li, Tao Li, Zhiping Qian, Jun Chen, Bi-Jie Hu, Sheng Wang, En-Qiang Mao, Lei Zhu, Wen-Hong Zhang, Yinzhong Shen) for their efforts in the treatment of patients.
Author contributions
Study concept and design: QL, JZ, YL, LC, and HL. Data collection: QL, WL, and XZ. Analysis and interpretation of data: QL, JZ, and YL. Drafting of the manuscript: QL, JZ, and YL. Critical revision of the manuscript: LC and HL.
Funding
This study was supported by Grant no. SHDC12015129 from Shanghai Shen Kang Hospital Development Center, Grant no. 17411969700 from Shanghai Association for Science and Technology, Grant no. 19YF1441200 from Shanghai Sailing Plan Program, Grant no. IDF162005 from the First-class University and first-class discipline building project of Fudan University, and Grant no. 2020YJKY01 from Shanghai Public Health Clinical Center. Role of the Sponsor: The funding organizations are public institutions and had no role in the design and conduct of the study; collection, management, and analysis of the data; or preparation, review, and approval of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial and/or non-financial interests.
Footnotes
Qiang Li and Jianliang Zhang contributed equally in this study.
Contributor Information
Hongzhou Lu, Email: luhongzhou@fudan.edu.cn.
Liang Chen, Email: chenliang@shphc.org.cn.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Coronavirus disease 2019 Situation Report–51 (2020) https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf. Accessed 12 Mar 2020
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang LM, Wei L, Xie L, Zhu G, Dela Cruz CS, et al. Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan China. JAMA. 2020 doi: 10.1001/jama.2020.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holshue ML, Debolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen ZM, Fu JF, Shu Q, Chen YH, Hua CZ, Li FB, et al. Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World J Pediatr. 2020 doi: 10.1007/s12519-020-00345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Health Commission of the People’s Republic of China (2020) New coronavirus pneumonia prevention and control program (the fouth edition). Beijing, China. https://www.nhc.gov.cn/yzygj/s7653p/202002/3b09b894ac9b4204a79db5b8912d4440.shtml. Accessed 12 Feb 2020.
- 10.World Health Organization (2020) Laboratory diagnostics for novel coronavirus. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance. Accessed 17 Jan 2020.
- 11.Delong ER, Delong DM, Clarke DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 12.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 2020 doi: 10.1101/2020.02.06.20020974. [DOI] [Google Scholar]
- 13.Chan J, Yuan S, Kok K, To KK, Chu H, Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;2020(395):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui W, Fan Y, Wu W, et al. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin Infect Dis. 2003;37:857–859. doi: 10.1086/378587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Qiu Z, Zhang L, et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004;189:648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Nie J, Wang H, et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aziz M, Fatima R, Assaly R. Elevated interleukin-6 and severe COVID-19: a meta-analysis. J Med Virol. 2020 doi: 10.1002/jmv.25948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagunas-Rangel FA, Chavez-Valencia V. High IL-6/IFN-gamma ratio could be associated with severe disease in COVID-19 patients. J Med Virol. 2020 doi: 10.1002/jmv.25900. [DOI] [PMC free article] [PubMed] [Google Scholar]