Highlights
-
•
Detection of a SARS-CoV-2 genome through metatranscriptome next-generation sequencing.
-
•
Direct metatranscritptome sequencing from nasopharyngeal swab of a COVID-19 patient.
-
•
Depletion of human rRNA and in-house bioinformatics strategy for Ion torrent sequencing.
Abbreviations: RT-qPCR, real-time reverse transcription PCR; Ct, cycle threshold; NGS, next-generation sequencing; SNP, single nucleotide polymorphism
Keywords: Covid-19, SARS-CoV-2, next-generation sequencing, metatranscriptomics
Abstract
Herein, we describe the detection of a SARS-CoV-2 genome through metatranscriptome next-generation sequencing directly from the nasopharyngeal swab of a suspected case of local transmission of Covid-19, in Brazil. Depletion of human ribosomal RNA and use of an optimized in-house developed bioinformatics strategy contributed to successful detection of the virus.
COVID-19, the disease caused by the novel coronavirus SARS-CoV-2, was firstly recognized as an atypical pneumonia in China, in December 2019. By early-April 2020, the disease had already spread globally to more than 150 countries, with over 1,000,000 confirmed cases according to the World Health Organization (WHO) (Fauci et al., 2020). The first confirmed imported case in South America was in February 26, 2020, from a traveler returning from Northern Italy to the city of São Paulo, the largest city in Brazil (Candido et al., 2020). Soon after, first suspected cases of local transmission within the country were reported, including in the Bahia state, in early March 2020.
Ascertaining the roles of different diagnostic technologies during an ongoing epidemic of viral infection is challenging, and recent studies have already reported sensitivity issues when using real-time reverse transcription PCR (RT-qPCR) for diagnosing COVID-19 (Kim et al., 2020; Wu et al., 2020). Possible interfering factors in RT-qPCR may arise from high viral recombination of coronaviruses (Shen et al., 2020) and varying viral loads in the natural course of disease (Zou et al., 2020), but this still needs further studies in the COVID-19 diagnostics context (Lo and Chiu, 2020). Therefore, developing new protocols for untargeted clinical metagenomics may provide valuable diagnostic tools for detecting SARS-CoV-2 and other pathogens in a current global scenario of emerging viral infections. Other studies have recently reported on metagenome-based strategies for next-generation sequencing of SARS-CoV-2 and varying results were reached, when comparing amplicon-based sequencing or direct metatranscriptome sequencing. Also, metagenome sequencing from bronchoalveolar lavage specimens rendered better results when compared to nasal swabs (Shen et al., 2020; Moore et al., 2020).
In this study, we successfully detected a SARS-CoV-2 genome directly from a nasopharyngeal swab specimen of a suspected case of local transmission of COVID-19 in Brazil, using a metatranscriptomics approach. On March 03, 2020, a female patient presented to a public hospital, in Feira de Santana-Bahia, Brazil, complaining of sore-throat and mild-dyspnea. Due to self-reported close contact with a returning traveler from Italy who had a confirmed COVID-19 diagnosis, specific diagnostic investigation was warranted, and a nasopharyngeal swab sample was immediately collected. Viral RNA extraction and RT-qPCR amplification were carried out following a protocol validated in our group at the Institute of Health Sciences, Federal University of Bahia, Salvador-BA, Brazil, according to a Promega application report (Promega Application Report, 2020; https://www.promega.com.br/applications/virus-detection-assay-coronavirus-detection-covid-19-sars-cov-2/). In brief, 200 μL of samples stored in transport medium (Leibovitz`s L15) were submitted in triplicate to RNA extraction in the Maxwell 16 instrument, following the Maxwell® 16 Viral Total Nucleic Acid System protocol (Promega). Detection of SARS-CoV-2 viral genome by RT-qPCR was done according to the CDC 2019-Novel Coronavirus protocol (CDC, 2020), using the GoTaq® Probe 1-Step RT-qPCR System (Promega) and primers/ probe targeting the nucleocapsid (N) encoding region. A moderately high viral load was detected in the nasopharyngeal swab sample, with a cycle threshold (Ct) of 21, which is similar to the highest values reported for the first days since symptoms onset in eighteen Chinese patients (Zou et al., 2020).
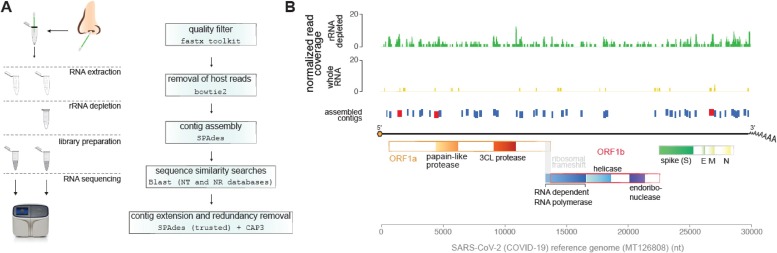
In parallel, the extracted RNA (ca. 14 ng/μL) was submitted to metatranscriptome next-generation sequencing in the Ion S5 platform (ThermoFisher) using an Ion 540™ chip and the Ion Total RNA-Seq kit v2, as per the manufacturer’s protocol (ThermoFisher). One sample was previously processed with the Low Input RiboMinus™ Eukaryote System v2 (ThermoFisher), for depletion of human ribosomal RNA from total RNA (Fig. 1 ). Sequence reads were pre-processed by quality filtering (Phred >20), presence of ambiguous bases (filtering out of reads with >1% Ns), and size (reads > = 30 nt); all reads deriving from human transcripts were also discarded: 77.29% in the rRNA depleted library and 84.49% in the whole RNA library. The remaining sequence reads (size range: 30 - 358 nt) were then used for contig assembly (see detailed pipeline in Fig. 1). Sequencing data is available at NCBI’s SRA database under accession number PRJNA613951.
Fig. 1.
Identification of SARS-Cov-2 through nasopharyngeal swab metatranscriptomics. A) Left panel: extraction of RNA from nasopharyngeal swab was performed according to a protocol validated in our laboratory (COVID-19 Extraction and Amplification with Maxwell® 16 Viral Total Nucleic Acid and GoTaq® Probe 1-Step RT-qPCR; Application Report, Promega 2020). Total RNA sequencing after rRNA depletion followed the protocol of the Ion Total RNA-Seq kit v2, in the Ion S5 platform. Depletion of human ribosomal RNA was done with the Low Input RiboMinus™ Eukaryote System v2 kit. Right panel: raw reads were submitted to quality filter where reads larger than 30 nt with Phred quality > 20 were aligned into human reference genome. Unaligned reads were used to perform contig assemblage. Assembled contigs were compared to NCBI databases using Blast software. Contigs that presented sequence similarity to SARS-Cov2-2 with e-value lower than 1e-5 were considered as from viral origin. Viral contigs were further submitted to contig extension using SPAdes and ‘trusted contigs’ option that was followed by cap3 tool to remove sequence redundancy. B) SARS-Cov-2 coverage profile of reads and assembled contigs. Reads were normalized by number of reads from each library. Contigs in red indicate high quality contigs larger than 400 nt with coverage of reads in both libraries.
Overall, depletion of human rRNA rendered significantly higher numbers of contigs (n = 51; N50 = 180 nt) confidentially aligned to the SARS-CoV-2 viral genome (Fig. 1b), yielding a genome coverage of 29.9% for contigs (as opposed to 5.4 % for the non-depleted sample) and 59.9% for total reads, which is higher than the coverage reported for nasopharyngeal swabs in recent studies (Moore et al., 2020). SNP analysis using Atlas 2 (https://sourceforge.net/p/atlas2/wiki/Atlas-SNP/) indicated that mutations accumulate mainly in 4 ORFs (ORF1a, M, ORF8 and N) in addition to 3’UTR, in a similar profile observed for strains derived from Brazilian patients present in NexStrain platform (https://nextstrain.org/).
Our results confirm that direct metatranscritptome sequencing from nasopharyngeal swabs can be used as an additional strategy in the detection of SARS-CoV-2 infection, besides providing information on possible nucleotide mutations that could interfere with sensitivity of diagnostics based on RT-qPCR (Wu et al., 2020). Noteworthy, handling of the sample and preparation of the sequencing library, particularly regarding depletion of ribosomal RNA, may significantly impact metatranscriptomic results.
CRediT authorship contribution statement
Gubio S. Campos: Investigation, Resources, Project administration, Supervision, Writing - review & editing. Silvia I. Sardi: Investigation, Resources, Formal analysis, Supervision, Writing - review & editing. Melissa B. Falcao: Investigation, Supervision. Emilia M.M.A. Belitardo: Investigation, Formal analysis. Danilo J.P.G. Rocha: Investigation, Formal analysis. Carolina A. Rolo: Investigation, Formal analysis. Aline D. Menezes: Investigation, Formal analysis. Carina S. Pinheiro: Investigation, Formal analysis. Rejane H. Carvalho: Investigation, Methodology, Formal analysis. João P.P. Almeida: Investigation, Methodology, Software. Eric R.G.R. Aguiar: Conceptualization, Methodology, Writing - original draft, Writing - review & editing, Formal analysis, Supervision. Luis G.C. Pacheco: Conceptualization, Methodology, Writing - original draft, Writing - review & editing, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that there is no conflict of interest related to this article content.
Acknowledgments
This study was partially funded by the following research grants: MCTI/FINEP/CT-INFRA -01/2013 (NGP-Saúde); MCTI/FINEP/FNDCT 01/2016 -Zika and Universal CNPq-2019. We acknowledge the technical support from the Laboratory of Immunology and Laboratory of Alergy and Acarology, both at the Institute of Health Sciences, Federal University of Bahia (UFBA), Brazil.
Contributor Information
Eric R.G.R. Aguiar, Email: ericgdp@gmail.com.
Luis G.C. Pacheco, Email: luis.pacheco@ufba.br.
References
- Candido D.D.S., Watts A., Abade L., Kraemer M.U.G., Pybus O.G., Croda J. Routes for COVID-19 importation in Brazil. Journal of Travel Medicine. 2020;18(March) doi: 10.1093/jtm/taaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel.https://www.fda.gov/media/134922/download [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci A.S., Lane H.C., Redfield R.R. N Engl J Med. 2020;382:1268–1269. doi: 10.1056/NEJMe2002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim D.-M., Lee B. 2020. Preprints. [cited 2020 Mar 22] [DOI] [Google Scholar]
- Lo Y.M.D., Chiu R.W.K. Clinical Chemistry. 2020;66:503–504. doi: 10.1093/clinchem/hvaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S.C., Penrice-Randal R., Alruwaili M., Dong X., Pullan S.T., Carter D. 2020. MedRxiv Infectious Diseases (except HIV/AIDS) Mar [cited 2020 Mar 22] [DOI] [Google Scholar]
- Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L. Clinical Infectious Diseases. 2020;9(March) doi: 10.1093/cid/ciaa203. [DOI] [Google Scholar]
- Wu X., Cai Y., Huang X., Yu X., Zhao L., Wang F. Emerg Infect Dis. 2020;26:6. doi: 10.3201/eid2606.200299. [cited 2020 Mar 22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]