Highlights
-
•
Coris COVID-19 Ag Respi-Strip should not be used alone for COVID-19 diagnosis.
-
•
Coris COVID-19 Ag Respi-Strip shows no benefit in reducing the use of RT-qPCR.
-
•
Highest viral load is associated with better antigen detection rates.
Keywords: COVID-19, Rapid antigen detection test, SARS-CoV-2
Abstract
Background
Ensuring accurate diagnosis is essential to limit the spread of SARS-CoV-2 and for the clinical management of COVID-19. Although real-time reverse transcription polymerase chain reaction (RT- qPCR) is the current recommended laboratory method to diagnose SARS-CoV-2 acute infection, several factors such as requirement of special equipment and skilled staff limit the use of these time-consuming molecular techniques. Recently, several easy to perform rapid antigen detection tests were developed and recommended in some countries as the first line of diagnostic.
Objectives
The aim of this study was to evaluate the performances of the Coris COVID-19 Ag Respi-Strip test, a rapid immunochromatographic test for the detection of SARS-CoV-2 antigen, in comparison to RT-qPCR.
Results
148 nasopharyngeal swabs were tested. Amongst the 106 positive RT-qPCR samples, 32 were detected by the rapid antigen test, given an overall sensitivity of 30.2%. All the samples detected positive with the antigen rapid test were also positive with RT-qPCR.
Conclusions
Higher viral loads are associated with better antigen detection rates. Unfortunately, the overall poor sensitivity of the COVID-19 Ag Respi-Strip does not allow using it alone as the frontline testing for COVID-19 diagnosis.
1. Background
A week after alerting the WHO of a cluster of pneumonia of unknown etiology in Wuhan, the Chinese authorities announced on 7 January 2020 that a novel coronavirus was identified as the cause of these pneumonia. According to phylogenetic analysis, this novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), previously named 2019-nCoV, belongs to the B lineage of Betacoronavirus genus and the Sarbecovirus subgenus and has more than 85% nucleotide sequence identity with a bat SARS-like CoV genome published previously [1,2]. Initially described in China, the coronavirus disease (COVID-19) caused by SARS-CoV-2 rapidly gained ground and evidence of human-to-human transmission rose. On January 30, WHO declared COVID-19 outbreak a public health emergency of international concern and the disease has now spread worldwide. Highly sensitive and specific tests are crucial to identify and manage COVID-19 patients and implement control measures to limit the outbreak. Real time reverse transcription polymerase chain reaction (RT-qPCR) in respiratory samples is the current recommended laboratory method to diagnose SARS-CoV-2 acute infection [3,4]. However, performing RT-qPCR requires special equipment and skilled laboratory personnel familiar with molecular techniques. Moreover, molecular tests are costly and often time consuming. COVID-19 Ag Respi-Strip (Coris BioConcept, Gembloux, Belgium) is a dipstick immunochromatographic test designed to detect SARS-CoV-2 antigen in nasopharyngeal secretions within 15 min. This rapid test was approved by the belgian federal agency for medicines and health products (AFMPS) and included in the first line of diagnostic tests for COVID-19 by the public health institute in Belgium (Sciensano).
2. Objectives
The aim of this study was to assess the performances of COVID-19 Ag Respi-Strip as a frontline testing in comparison to molecular technique.
3. Study design
3.1. Clinical specimens
Nasopharyngeal swab specimens were collected from samples received at the microbiology department of the Cliniques universitaires Saint-Luc Hospital, a tertiary hospital of more than 900 beds in Brussels, between April 6 and April 21, 2020. Samples were selected at random. If the rapid antigen test was not performed immediately, samples were stored at 4 °C until the test.
3.2. RT-qPCR
COVID-19 laboratory diagnosis relies on the genesig® Real-Time PCR assay (Primerdesign Ltd, Chandler’s Ford, UK), a RT-qPCR assay performed on RNA extracts to detect viral RNA by targeting the RNA dependant RNA polymerase (RdRp) gene. The amplification was performed on a LightCycler 480 instrument (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s recommendations. Samples with SARS-CoV-2 RT-qPCR cycle threshold value (Ct) under 40 were considered positive.
3.3. Ultracentrifugation
In a second time, some samples were ultracentrifuged at 31,510g for 2 h at 4 °C. After discarding the supernatant, the 150 μL of residual sample were vortexed and analyzed by the rapid test.
3.4. Rapid antigen detection test
COVID-19 Ag Respi-Strip (Coris Bioconcept, Gembloux, Belgium) is a ready to use test which allows rapid and qualitative detection of SARS-CoV-2 antigen in nasopharyngeal secretions. This test, based on a membrane technology with colloidal gold nanoparticules, uses monoclonal antibodies to detect highly conserved SARS-CoV and SARS-CoV-2 nucleoprotein antigen. Another monoclonal antibody is conjugate to colloidal gold nanoparticules. These antibodies are immobilized onto the nitrocellulose membrane. The test was performed according to manufacturer’s instruction by mixing 100 μL of nasopharyngeal secretions with 4 drops (approximately 100 μL) of LY-S dilution buffer in a tube and the strip was added. When the nasopharyngeal secretions come into contact with the strip, passive diffusion allows the solubilized conjugate to migrate with the sample and react with the anti-SARS-CoV-2 antibodies immobilized onto the membrane. A control line is included in the strip to assess the correct migration of the sample. Visual interpretation of the result is performed after 15 min. Two versions of the test were evaluated. On the second version, conjugate was coupled on a different way and the control line was optimized.
3.5. Statistics
The criteria used for the performance assessment of COVID-19 Ag Respi-Strip were sensitivity and specificity. RT-qPCR was considered as the gold standard for this evaluation, therefore positive and negative samples by molecular techniques were considered to be true positive and true negative samples, respectively. The overall percentage of agreement and Cohen’s kappa coefficient (κ) were used to evaluate assay agreement. Analyses were performed using GraphPad Prism 7.0 (GraphPad Prism Software Inc., San Diego, California) and MedCalc 19.2.0 (MedCalc Software Ltd, Ostend, Belgium).
4. Results
We collected 148 nasopharyngeal samples from 148 patients. The median age of the study population was 57.5 (range: 0–94) with a sex ratio of 0.8 (64 men and 84 women). According to RT-qPCR results, 42 samples were negative and 106 were positive, with a median Cycle threshold (Ct) value of 33 (mean: 31.4; range:16–38), equivalent to a median of 1.6 × 103 copies/mL (mean: 7.4 × 105; range: 84.5–3.6 × 107). Data about the symptoms were available for 131 patients (88.5%). Amongst the 45 (30.4%) patients who had no symptoms, RT-qPCR and COVID-19 Ag Respi-strip were both negative for 31 patients, 14 had a positive RT-qPCR result and only four of them had also a positive rapid test. The median Ct value of these positive RT-qPCR amongst patients with no symptoms was 32.5 (mean: 31.6; range: 18–38), equivalent to a median of 2.2 × 103 copies/mL. The median time of symptom duration before the sampling date was four days (mean: 6.6; range: 0–34). Amongst the 86 symptomatic patients, 34 (39.5%) had concordant results between RT-qPCR and rapid test with nine negative results and 25 positive results obtained with both detection methods. Discordant results with positive RT-qPCR and negative rapid test were observed for 52 samples (62.7%).
Overall, amongst the 106 positive samples, the COVID-19 Ag Respi-Strip detected 32 samples (Table 1 ). For samples with Ct < 25 (n = 10), <30 (n = 34) and <35 (n = 64), 1.8 × 105, 9.4 × 103 and 494.8 copies/mL respectively, COVID-19 Ag Respi-Strip has a sensitivity of 100%, 70.6% and 46.9%. However, in our study population, the overall sensitivity is 30.2% (95% IC: 21.7–39.9).
Table 1.
Performances of the COVID-19 Ag Respi-strip.
| RT-qPCR |
|||
|---|---|---|---|
| Detected | Not detected | ||
| COVID-19 Ag Respi-Strip | Detected | 32 | 0 |
| Not detected | 74 | 42 | |
The 32 concordant positive samples (positive results with RT-qPCR and rapid test) had a median Ct of 26 (range: 16–36), equivalent to a median of 9.9 × 104 copies/mL, whereas the median Ct of the 74 discordant (positive RT-qPCR with negative rapid test) samples was 35 (range: 25–38), corresponding to a median of 494.8 copies/mL (Fig. 1 ). The 42 samples with a negative result with RT-qPCR technique were also negative with the rapid test, giving an overall specificity of 100%. The accuracy was 50% (95% CI: 41.7–58.3) and the agreement κ index between the methods was 0.2 (95% CI: 0.1−0.3) indicating a slight agreement between the two methods. Five positive COVID-19 Ag Respi-Strip shew no control line.
Fig. 1.
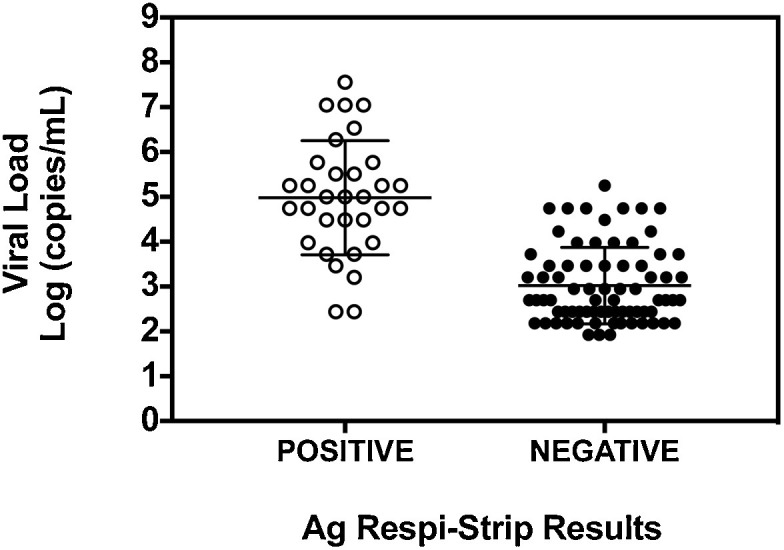
COVID-19 Ag Respi-Strip results according to viral load.
With the aim of gaining sensitivity, we ultracentrifuged 24 discordant samples with positive result in RT-qPCR but negative antigen rapid test. Only two of the 24 samples were very weak positive with the COVID-19 Ag rapid test performed on centrifuged material.
5. Discussion
In the ongoing pandemic context of COVID-19, diagnostic testing for SARS-CoV-2 is crucial in order to limit the spread of the virus as well as appropriately manage infected patients. Different diagnostic test manufacturers have developed rapid tests based on SARS-CoV-2 proteins detection in respiratory samples. However, the analytical performances of these rapid antigenic tests depend on different factors including the viral load, the quality of the specimen and how it is processed. The performances also depend on the setting of patients tested. Although the COVID-19 Ag Respi-Strip has several advantages such as the ease and fast achievement of the test, the rapid answer, the lower cost and the non-requirement of special equipment or skills compared with molecular techniques, data presented here suggested that this rapid test is suffering from poor sensitivity. The rapid antigen detection test is able to detect SARS-CoV-2 with high sensitivity in nasopharyngeal samples with high viral load equivalent at least to 1.7 × 105 copies/mL (Ct < 25), but the sensitivity declines substantially when the viral load decreases with Ct values over 30, equivalent to 9.4 × 103 copies/mL, which is often the case in patients suffering of COVID-19. Actually, in our study, while the specificity was 100%, the overall sensitivity of the COVID-19 Ag Respi-Strip was 30.2%. This lack of sensitivity of rapid diagnostic tests for virus detection was already observed during the Influenza A (H1N1) pandemic [5]. Ensuring accurate diagnosis is essential to limit the spread of the virus. The poor sensitivity of the COVID-19 Ag Respi-Strip leads to false negative results, which in these times of pandemic can be of great consequence. This rapid test was thought to be used as a first line COVID-19 diagnostic test in Belgium in order to possibility reduce the number of RT-qPCR testing in case of positive results, but requiring confirmation for negative results. However, because negative results cannot rule out SARS-CoV-2 infection, this test is of little use in a pandemic setting. Pending more evidence of their performances, our data suggest that COVID-19 Ag Respi-Strip should not be used alone for COVID-19 diagnosis and shows no benefit in reducing the use of RT-qPCR.
CRediT authorship contribution statement
Anaïs Scohy: Conceptualization, Investigation, Formal analysis, Writing - original draft, Writing - review & editing. Ahalieyah Anantharajah: Methodology, Formal analysis. Monique Bodéus: Writing - review & editing. Benoît Kabamba-Mukadi: Conceptualization, Methodology, Writing - review & editing. Alexia Verroken: Writing - review & editing. Hector Rodriguez-Villalobos: Conceptualization, Methodology, Writing - review & editing.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
COVID-19 Ag Respi-Strip tests were provided by Coris BioConcept, Gembloux, Belgium.
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.OMS . 2020. Laboratory Testing Strategy Recommendations for COVID-19. Interim Guidance. 21 march. [Google Scholar]
- 5.Centers for Disease C Prevention. Evaluation of rapid influenza diagnostic tests for detection of novel influenza A (H1N1) Virus - United States, 2009. MMWR Morb. Mortal. Wkly. Rep. 2009;58(30):826–829. [PubMed] [Google Scholar]


