Highlights
-
•
Genetic variations contribute to phenotypic individual vulnerabilities to sleep debt.
-
•
LAMP-MC is a recently developed method to characterize Single Nucleotide Polymorphism.
-
•
Detection is performed directly from whole blood or buccal cells.
-
•
LAMP-MC method produced specific melting curves for 5 sleep debt-related SNPs.
-
•
High concordance of results was observed between LAMP-MC and Taqman referent method.
Keywords: SNP detection, Loop-mediated isothermal amplification, Melting curve, Sleep debt
Abstract
Genetic variations contribute to phenotypic individual vulnerabilities to sleep debt, particularly for five single nucleotide polymorphisms (SNPs). Loop-mediated isothermal amplification and melting curve analysis (LAMP-MC) is a recently developed method to characterize SNPs. The aim of present study was to evaluate the LAMP-MC method on blood and buccal cells for detection of five SNPs of interest in healthy humans.
We first analyzed signals obtained from LAMP-MC method on 42 samples. Then we compared the results with those of referent TaqMan method.
The LAMP-MC method produced specific melting curves for the five SNPs. A high concordance of genotyping results was observed between the two methods for rs5751876_ADORA2A, rs1800629_TNF-α, rs73598374_ADA and rs228697_PER3 in blood and saliva (Cohen’s kappa coefficient >0.80). A good agreement ( = 0.61) was observed for rs4680_COMT in blood only.
LAMP-MC is a simple and reliable method to study genetic influences on health, sleep debt-related performance impairments and countermeasures.
1. Introduction
Single nucleotide polymorphisms (SNPs) are the simplest form of DNA sequence variability, contributing to phenotypic variations between individuals. They are also a relevant class of human biomarkers that may be associated to anthropometric traits, risk to specific diseases or response to environmental inputs. Sleep has a critical role in promoting health and is affected by genetic polymorphisms [1]. Two examples of conditions associated to SNP are individual sensitivity to sleep loss and to caffeine effects on sleep. In this context, Adenosine A2A receptor (ADORA2A), Adenosine deaminase (ADA), Catechol-O-methyltransferase (COMT), Tumor necrosis factor alpha (TNF-α), and Period3 (PER3) are some of the genes contributing to genotype differences [2,3]. The adenosine pathway is known to affect general sleep and its antagonism by caffeine is thought to explain its effects on sleep [4]. COMT being one of the major enzymes of the metabolic degradation of catecholamines plays an important role in cortical dopamine metabolism and consequently impacts sleep-wake regulation [5]. The pro-inflammatory TNF-α cytokine is a well-characterized sleep regulatory substance [6] and PER3 is circadian gene that regulates rhythmicity of multiple functions within the body [7]. Therefore based on previous studies we have focused on the five following SNPs, rs5751876 ADORA2A [8], rs1800629 TNF-α [9], rs73598374 ADA [10], rs4680 COMT [11] and rs228697 PER3 [7] in healthy humans participating in an experimental sleep deprivation protocol.
Methods for SNP genotyping are diverse and include array-based hybridization, PCR, and sequencing. Array-based hybridization uses probes immobilized on a solid support and, although limited-throughput arrays can be built in research laboratories, the design and manufacture of commercial arrays is an industrial undertaking, thus this method of SNP genotyping ranks low in assay flexibility. PCR and sequencing are classic methods but they require expertise and expensive lab equipment. Our objective is to validate a rapid genotyping method facilitating the detection of SNPs. The conventional TaqMan method is highly reliable for SNP genotyping applications [12,13] but time-consuming due to the necessary four steps, which are DNA extraction, DNA quality control, DNA amplification and fluorescence signal analysis. Loop mediated isothermal amplification (LAMP) is a recent isothermal nucleic acid amplification technique known for its simplicity, sensitivity and speed [14,15]. Since its invention, this technique has been one of the most extensively used molecular tools in the field of diagnostics, providing rapid, accurate, sensitive and cost-effective diagnosis of human pathogens including bacteria, parasites and viruses [16]. In the case of viruses, the implementation of reverse transcription loop-mediated isothermal amplification (RT-LAMP) has enabled a fast and efficient detection [17]. LAMP features have also resulted in development of rapid SNP detection methods from blood and buccal cells [18,19] and more recently in development of the LAMP melting curve (LAMP-MC) genotyping method [20,21]. The LAMP-MC system principally employs a set of six primers that recognize eight distinct regions on the target gene, so that the specificity and the amplification speed are extremely high [22]. It uses a strand displacing polymerase and, consequently, does not rely on thermal cycling, the target sequence is amplified at a constant temperature around 65 °C. Following the amplification procedure SNP are detected by fluorophore-labelled probe-based melting curve analysis. Furthermore, LAMP has been applied to complex biological matrices, such as whole blood and saliva, without prior DNA extraction [23]. Both characteristics of LAMP results in shorter “sample to answer” times than conventional PCR.
In this study we evaluated the precision of LAMP-MC method within a run and between runs and we compared genotyping results obtained by LAMP-MC with those obtained by the referent TaqMan method.
2. Materials and methods
2.1. Samples
Blood and saliva samples were obtained from healthy European adult volunteers participating in research protocols on the link between genetic polymorphisms, sleep and caffeine consumption [24]. Genotype determinations were carried out in two separate biology centers, the French National Center for Research in Human Genomics (CNRGH) for the referent Taqman method and the French Biomedical Research Institute of Armed Forces (IRBA) for the LAMP-MC method. The study received the agreement of the Cochin – CPP Ile de France 1 (Paris) Ethics Committee and was approved by the Agence Nationale pour la Sécurité du Médicament (ANSM) (2017-A02793-50) for the genetic collection. It was conducted according to the principles expressed in the Declaration of Helsinki of 1975, as revised in 2001 after obtaining written informed consent for all the participants.
Whole saliva samples were collected using Oragene DNA kits OG-500 (DNAgenotek, Ottawa, Canada) from volunteers (n = 1023) after rinsing the mouth with water and at least 30 min after eating or drinking. DNA from saliva collected in Oragene containers should be stable for at least 5 years at ambient temperature. These saliva samples were dedicated to TaqMan assays in the laboratories of CNRGH.
In addition among these volunteers, 42 blood and buccal cells samples were collected for the blinded methods comparison. EDTA blood were aliquoted in sterile microtubes and stored at −20 °C until LAMP-MC analysis. Sterile omniswab devices (Whatman FTA collection, GE Healthcare, UK) were used for buccal cells collection, each brush-like swab head were ejected in a microtube contained 200 μL of Tris-EDTA buffer and stored at −20 °C until LAMP-MC genotyping.
2.2. TaqMan SNP genotyping assays
After manual cell lysate preparation the genomic DNA purification was performed on an Autopure LS instrument (Qiagen, Hilden, Germany) at CNRGH. DNA quantity, integrity and ability to PCR amplification were evaluated by Quality Controls. 1023 DNA extracts were transferred on 384 wells plates. Participants were genotyped using predesigned probes and TaqMan SNP genotyping assays provided by Thermo Fisher Scientific (Whaltham, USA). PCR was performed on GeneAmp PCR System 9700 and a 7900 H T system with SDS software version 2.4 (Applied Biosystems, Foster City, USA) was used for fluorescence detection and allelic discrimination. A check by Sanger sequencing of PCR products was carried out for rs5751876 because ADORA2A gene includes many SNPs in this region.
2.3. LAMP-MC assays
LAMP-MC consists in a lysis of cells from whole blood or collected on buccal swabs followed by the amplification of the target sequence at a constant temperature around 65 °C using simultaneously three sets of primers, a polymerase with high strand displacement activity in addition to a replication activity and a fluorophore-labelled probe. Detection of homozygous wild, heterozygous and homozygous mutant genotypes is performed by melting curve analysis after amplification.
LAMP-MC assays were realized by use of the customized Human Sleep Deprivation Combo kit (Cat#LC-SDC-LP-24, LaCAR MDX, Liège, Belgium). A positive control and a negative control were supplied for each SNP. Samples, controls and reagents were thawed at room temperature and the reactions were set up with 5 μL blood or control, or one swab, in 1 mL of lysis buffer. After 10 min of incubation at room temperature, 5 μL of the lysed sample or control were added to 20 μL reaction buffer per well. Each SNP was detected by a separate reaction buffer, containing all reagents specific for the amplification and detection of one SNP. The 8 wells strip, containing 6 unknown samples and 2 controls, was briefly centrifuged and then placed in the analyzer (LC-Genie III™ v3.17). Following the 40 min amplification step at 65 °C, the mix were cooled down to 35 °C to allow fluorophore-labelled probe annealing. Then melting curves were generated in the temperature range of 35−80 °C with ramp rate of 0.2 °C/s. The increasing of temperature induces the separation of the fluorophore from the quencher and the generation of a fluorescence signal. According to the design of each SNP probe, the presence or the absence of mutation decreases the dehybridization temperature of probe.
2.4. Analytical validation and statistics
For the validation of each SNP, 30 blood samples and 12 buccal cells samples were blindly assayed performing 7 runs. The quality and specificity of the signal were checked from the observed melting profiles as well as the adequacy between the measured melting peaks and the acceptance windows mentioned by the manufacturer. Intra-assay precision was evaluated by the analysis of one set of 6 replicates from one heterozygote blood sample for each studied SNP. Inter-assay precision was calculated from melting temperatures that were obtained after testing the same heterozygote sample during 6 assays, by three different operators. At last we have compared the genotyping results of the two methods for the 42 assayed samples. To assess the concordance of genotype data between technologies, we computed the Cohen’s kappa coefficient, a commonly used index to quantify agreement between two measurements [25]. The κ statistic measures agreement between two raters that is beyond chance, with chance being a value of zero and with 1.00 being complete agreement, not by chance. κ Values were calculated and ranked as minimal agreement (κ<0.20), weak agreement (κ<0.40), moderate agreement (κ<0.60), good agreement (κ<0.80) and excellent agreement (κ≥0.80) [26].
3. Results
3.1. LAMP-MC method
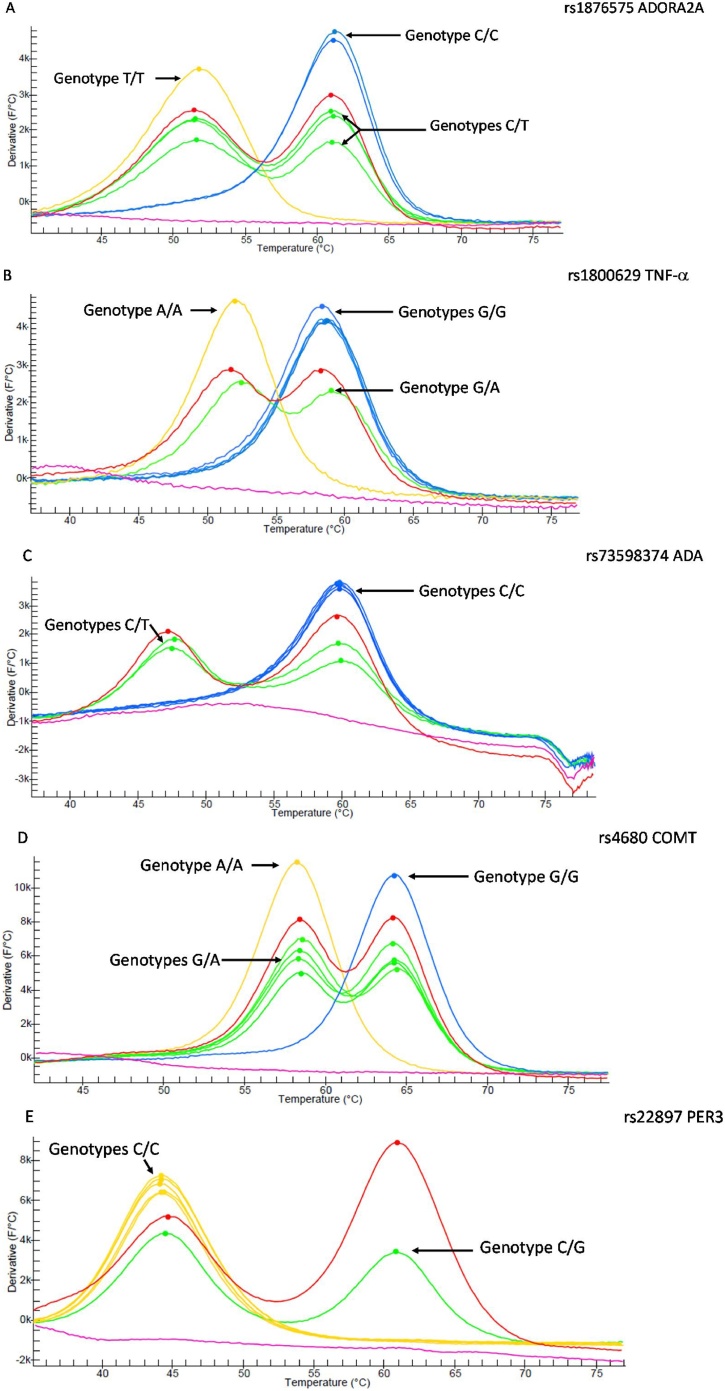
Genetic variations for the rs5751876 ADORA2A, rs1800629 TNF-α , rs73598374 ADA, rs4680 COMT and rs228697 PER3 are T > C, A > G, T > C, A > G and G > C respectively. The customized kit included the accepted range of melting temperature for each allele of the 5 SNPs. In both runs negative controls have certified that no contamination was present. The efficiency of the procedure was assessed by systematically using positive controls that consist in different plasmids containing wild type and mutated gene fragments for each target polymorphism. Observed fluorescence levels were satisfactory. Curves were generated by derivative of fluorescence signal increases. Melting curves of the different genotypes detected by LAMP-MC are shown in Fig. 1. They well illustrate the quality of the signal for each of the 5 assayed SNPs; each profile includes a negative control, a positive control and six unknown samples. Overall the analysis of the 42 samples (30 blood cells plus 12 buccal cells) by the LAMP-MC method gave specific signals for the 5 SNPs. In detail, all melting peaks were in the accepted range excepted for one sample for rs1800629 TNF-α and rs4680 COMT and two samples for rs228697 PER3. On the other hand no signal was obtained from the 12 buccal cells samples for the rs4680 COMT. Supplementary lysis tests, consisting to add a buccal swab to blood lysis, have shown that presence of saliva systematically inhibits the fluorescence signal.
Fig. 1.
LAMP melting curves of blood samples for the 5 tested SNPs. A) rs5751876 ADORA2A, B) rs1800629 TNF-alpha, C) rs73598374 ADA, D) rs4680 COMT and E) rs22897 PER3. Signals from homozygous subjects are represented by yellow and blue lines, signals from heterozygous subjects are represented by green lines. Red and pink lines correspond respectively to positive and negative controls (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Both means, minimum and maximum temperatures of accepted melting peaks obtained from the 7 runs analysis, are presented in Table 1. Intra- and inter-assay precisions are described in Table 2. The assays have good performances (CV < 1.5 % in the inter-assay precision analysis).
Table 1.
Observed melting peak temperatures from 42 samples.
| SNP | Mean | Observed range | Observed range size | LaCAR MDX range acceptance |
|---|---|---|---|---|
| rs5751876 ADORA2A | ||||
| C allele (n = 39) | 60.98 | 60.66 – 61.77 | 1.11 | 59.6 – 62.6 |
| T allele (n = 26) | 51.52 | 51.18 – 52.31 | 1.13 | 50.2 – 53.2 |
| TNF-α rs1800629 | ||||
| G allele (n = 39) | 58.65 | 58.05 – 59.47 | 1.42 | 57.1 – 60.1 |
| A allele (n = 11) | 52.24 | 51.52 – 53.46 | 1.94 | 50.8 – 53.8 |
| rs73598374 ADA | ||||
| C allele (n = 42) | 59.66 | 58.37 – 60.49 | 2.12 | 58.1 – 61.1 |
| T allele (n = 7) | 47.67 | 47.41 – 48.02 | 0.61 | 45.8 – 48.8 |
| rs4680 COMT | ||||
| G allele (n = 16) | 64.18 | 63.45 – 64.63 | 1.18 | 62.8 – 65.8 |
| A allele (n = 18) | 58.43 | 57.45 – 58.99 | 1.54 | 57.0 – 60.0 |
| rs228697 PER3 | ||||
| C allele (n = 40) | 44.80 | 43.65 – 46.06 | 2.41 | 43.1 – 46.1 |
| G allele (n = 5) | 60.97 | 60.67 – 61.29 | 0.62 | 59.0 – 62.0 |
All data are in °C.
Table 2.
Analysis of assay precision (n = 6).
| Intra-assay precision |
Inter-assay precision |
|||
|---|---|---|---|---|
| Tm mean (°C) | CV (%) | Tm mean (°C) | CV (%) | |
| rs5751876 ADORA2A | ||||
| C allele | 60.95 | 0.34 | 61.00 | 0.49 |
| T allele | 51.50 | 0.38 | 51.61 | 0.46 |
| rs1800629 TNF-α | ||||
| G allele | 58.55 | 0.39 | 58.67 | 0.71 |
| A allele | 51.98 | 0.45 | 52.12 | 0.87 |
| rs73598374 ADA | ||||
| C allele | 59.84 | 0.45 | 59.63 | 0.67 |
| T allele | 47.56 | 0.88 | 47.57 | 1.07 |
| rs4680 COMT | ||||
| G allele | 64.28 | 0.38 | 64.06 | 0.37 |
| A allele | 58.46 | 0.39 | 58.37 | 0.31 |
| rs228697 PER3 | ||||
| C allele | 45.58 | 0.44 | 44.60 | 1.24 |
| G allele | 61.36 | 0.38 | 60.79 | 0.33 |
Tm: melting peak temperature; CV: Coefficient of variation.
3.2. Comparison of genotypes frequencies with the referent TaqMan method
The TaqMan-determined genotypes frequencies of the 42 humans volunteers from our 1023 European panel and those obtained in 1000 Genomes Project are shown in Table 3. The genotypes frequencies for our 1023 panel are similar to the 1000 Genomes Project data on the GRCh38 reference assembly [24]. All alleles were represented in the 42 volunteers, all existing genotypes for each SNP were therefore tested.
Table 3.
Genotypes frequencies.
| Gene | Reference SNP | Allele (Wild/Mutated) | Frequency W/W | Frequency W/M | Frequency M/M |
|---|---|---|---|---|---|
| ADORA2A | rs5751876 | C/T | 33.3 | 59.3 | 7.4 |
| (35.9) | (45.6) | (18.5) | |||
| [37.4] | [47.1] | [15.5] | |||
| TNF-α | rs1800629 | G/A | 75.5 | 18.9 | 5.6 |
| (74.1) | (24.6) | (1.3) | |||
| [74.4] | [24.5] | [1.2] | |||
| ADA | rs73598374 | C/T | 87.2 | 12.8 | 0 |
| (89.2) | (10.4) | (0.4) | |||
| [88.1] | [11.5] | [0.4] | |||
| COMT | rs4680 | G/A | 29.2 | 45.8 | 25 |
| (30.4) | (49) | (20.6) | |||
| [26.4] | [47.1] | [26.4] | |||
| PER3 | rs228697 | C/G | 89.8 | 10.2 | 0 |
| (82.7) | (16.4) | (0.9) | |||
| [81.7] | [17.3] | [1] |
Data in percent (%) of the n = 42 subjects, % of the 1023 European volunteers in parentheses and % of the 1000 Genomes project in brackets. W is the wild allele, M is the mutated allele and W/M correspond to heterozygous subject.
The comparison was only performed for the sample showing melting peak temperature in the manufacturer accepted range (see above for excluded samples). Moreover, as no signal was obtained for rs4680 COMT on the 12 buccal cells samples, this comparison was not possible. Thus number of interpreted melting curves was comprised between 42 and 29 after running 30 blood samples and 12 buccal cells samples for each SNP (Table 4). The concordance analysis between the two genotyping methods is summarized in Table 4. A majority of genotyping results was valid except for a few heterozygous profiles. In these profiles, one of the two allelic peaks was too weak and 1 false homozygous result was displayed.
Table 4.
Concordance between genotyping results of LAMP-MC method with the referent method.
| Gene /SNP | Sample type | Sample number | Interpreted melting curves | Valid result | False resulta | Concordance (%) | Cohen’s κ Coefficientb |
|---|---|---|---|---|---|---|---|
| ADORA2A / | Blood | 30 | 30 | 29 | 1 | 96.7 | 0.936 |
| rs5751876 | Bu Cells | 12 | 12 | 12 | 0 | 100 | 1 |
| TNF-α / | Blood | 30 | 29 | 28 | 1 | 96.6 | 0.910 |
| rs1800629 | Bu Cells | 12 | 12 | 11 | 1 | 91.7 | 0.824 |
| ADA / | Blood | 30 | 30 | 30 | 0 | 100 | 1 |
| rs73598374 | Bu Cells | 12 | 12 | 12 | 0 | 100 | 1 |
| COMT / | Blood | 30 | 29 | 25 | 4 | 86.2 | 0.610 |
| rs4680 | Bu Cells | 12 | 0 | – | – | – | – |
| PER3 / | Blood | 30 | 28 | 27 | 1 | 96.4 | 0.870 |
| rs228697 | Bu Cells | 12 | 12 | 12 | 0 | 100 | 1 |
Bu Cells means Buccal cells.
Heterozygous not detected (one of the two peaks too weak).
Cohen’s kappa values are ranked as minimal agreement (κ<0.20), weak agreement (κ<0.40), moderate agreement (κ<0.60), good agreement (κ<0.80) and excellent agreement (κ≥0.80).
The results showed an excellent concordance with the referent TaqMan method for 4 studied SNPs (i.e. rs5751876 ADORA2A, rs1800629 TNF-α, rs73598374 ADA and rs228697 PER3 in blood and buccal cells). Cohen’s Kappa coefficients higher than 0.80 in blood and buccal cells attest to an excellent agreement between the two genotyping methods (Table 4) [26]. Concerning the rs4680 COMT, Cohen’s Kappa coefficient 0.61 in blood signs a good agreement.
In addition it should be noted that in the case of an invalid melting peak temperature or invalid result, samples were tried again and a half was detected successfully (Data not shown).
4. Discussion
Initially described by Notomi et al. [14], the LAMP approach allows to rapidly amplify a target DNA sequence in high specific and sensitive manner despite the use of untreated biological fluids. These properties have been used by LaCAR MDX to develop kits for genetic polymorphisms detections. The present study shows that the LAMP technique and subsequent melting curve analysis may be chosen for simple and rapid detection of number of single nucleotide variants. The observed temperature ranges were lower than 2 °C for 8 alleles out of 10 which attests the LAMP-MC robustness (Table 1).
A reliable genotype identification of 4 SNPs of interest was achieved in more than 90 % of samples (i.e. rs5751876 ADORA2A, rs1800629 TNF-α, rs73598374 ADA and rs228697 PER3 in blood and buccal cells) and the success rate is 86 % for rs4680 COMT with blood samples. Moreover for each invalid signal (melting peak temperature out of range or one of the two peaks too weak) a second assay of sample has allowed to obtain a correct result in 50 % of the cases. Thus the set of primers and labelled fluorophore probe designed by LaCAR MDX has permitted to reliably detect the five SNPs in blood even in the presence of several mutations in the targeted DNA region especially for rs5751876 ADORA2A and rs4680 COMT. A LAMP-MC assay has been previously developed for a rapidly diagnosis of genetic predisposition to lactose intolerance in human [21] and our results showed its promising interest in future research protocols to rapidly identify sensitivity to extreme environmental conditions such as sleep debt.
In addition we are the first to detect SNP in buccal cells of healthy human using LAMP-MC method without prior DNA extraction. Except for rs4680 COMT, this method is good or excellent. Indeed, a previous work has been conducted by Carlos et al. [19] on buccal cells but with a prior DNA extraction before LAMP analysis.
In our study, both blood and buccal cells have given good results, except for the rs4680 COMT. In the case of this SNP, the lack of signal is closely related to the use of the salivary matrix. Previous studies have shown that saliva-derived genomic DNA contains significant bacterial contamination, which may reduce the amplification and genotyping success rates compared to the use of blood-derived genomic DNA [[27], [28], [29]]. Thus Hansen et al. [27] and Rogers et al. [28] compared the amplification and genotyping performances obtained after extraction of DNA from blood and from different saliva collection devices. They particularly reported more important bacterial DNA and protein contaminations and lower genotyping call rates when buccal swab rather than whole saliva Oragen devices are used. The loss of efficiency between blood and different salivary samples is likely related to the quantity of protein and bacterial DNA in saliva samples. In our case of a specific lack of signal for rs4680 COMT assay, we can put forward two explanations: (i) a common bacterial nuclease cleaves human genomic DNA near the rs4680 target or (ii) some contaminating bacterial DNA may competitively interfere with the binding of the labelled probe to the rs4680 target and adjacent sequences. On this assumption, purification of the salivary sample will therefore not necessarily bring a better result. Furthermore, Abildgaard et al. [20] have observed more consistent melting curve profiles when whole blood was used as raw material compared to the DNA extract. To overcome problems encountered with salivary samples, future assays will be conducted using non-absorbent materials. Small scarpers cells will be tested in place of buccal swabs to maximize rapid recovery of epithelial cells and minimize salivary proteins binding. For the genotyping of saliva samples, we therefore recommend that preliminary tests be performed for each newly developed kit to verify the quality of the amplification signal. It should be noted that all biological fluids can be tested [23] and that in any case only 5 μL of blood is required.
Compared to conventional genotyping methods (array-based hybridization, PCR or sequencing), LAMP-MC exhibits several interesting advantages: (i) no prior DNA extraction is required, (ii) the assays are simple and easy to perform once the appropriate primers are prepared, (iii) the maximum amplification signal is achieved within 40 min, (iv) the equipment required is neither expensive nor cumbersome. These characteristics of LAMP-MC results in shorter response times between sample and reliable genotyping than conventional methods. This allows us to quickly and individually advise each of our healthy subjects during sleep and caffeine awareness training. Furthermore, due to the absence of genomic DNA extraction, an ancestral DNA library is not built up, and this guarantee facilitates the participation of volunteers in health follow-ups.
In conclusion, the LAMP-MC method is fast and does not request expensive equipment which makes it useful for low throughput research studies. A major advantage of the genotyping LAMP-MC method is its ability to amplify DNA directly from whole blood or buccal cells by the easy use of custom-designed kits.
Our results show that this genotyping method is adapted to help understanding individual vulnerability to sleep debt or individual adaptability to countermeasures.
Authors statment
All authors agree the revised manuscript BTRE_2020_94 and state it has not been submitted elsewhere.
Ethical approval
The study received the agreement of the Cochin – CPP Ile de France 1 (Paris) Ethics Committee and was approved by the Agence Nationale pour la Sécurité du Médicament (ANSM) (2017-A02793-50).
Author contributions
CD: Conceptualization, Methodology, Validation, Formal analysis and Writing - original draft. FS: Investigation and Funding acquisition. ME: Validation. LD: Methodology. CeD: Methodology. MCE: Methodology. AB and JFD: Data curation and Supervision. DGM: Writing - review & editing. MC: Project administration and Supervision.
Declaration of Competing Interest
L.D. is a full-time employee of LaCAR MDX. Other authors have no competing interests with this study to declare. This study has been funded by the Armaments Procurement Agency (Direction Générale de l'Armement, DGA, Ministry of Armed Forces, contract PDH-1-SMO-2-509).
Acknowledgements
The sponsor of this study was the French armed forces heath service (Service de santé des armées). We thank Dr Robert Olaso, Ms Marie-Laure Moutet and Mr Bertrand Fin from the CNRGH (Centre National de Recherche en Génomique Humaine, Université Paris-Saclay, CEA, Evry, France) for their technical support and expertise. We thank Mederic de la Bourdonnaye for his technical participating.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2020.e00468.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Goel N. Genetics of sleep timing, duration and homeostasis in humans. Sleep Med. Clin. 2011;6:171–182. doi: 10.1016/j.jsmc.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goel N. Neurobehavioral effects and biomarkers of sleep loss in healthy adults. Curr. Neurol. Neurosci. Rep. 2017;17 doi: 10.1007/s11910-017-0799-x. [DOI] [PubMed] [Google Scholar]
- 3.Landolt H.P. Genetic determination of sleep EEG profiles in healthy humans. Prog. Brain Res. 2011;193:51–61. doi: 10.1016/B978-0-444-53839-0.00004-1. [DOI] [PubMed] [Google Scholar]
- 4.Rétey J.V., Adam M., Khatami R., Luhmann U.F.O., Jung H.H., Berger W., Landolt H.P. A genetic variation in the adenosine A2A receptor gene (ADORA2A) contributes to individual sensitivity to caffeine effects on sleep. Clin. Pharmacol. Ther. 2007;81:692–698. doi: 10.1038/sj.clpt.6100102. [DOI] [PubMed] [Google Scholar]
- 5.Dauvilliers Y., Tafti M., Landolt H.P. Catechol-O-methyltransferase, dopamine, and sleep-wake regulation. Sleep Med. Rev. 2015;22:47–53. doi: 10.1016/j.smrv.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Krueger J.M., Rector D.M., Roy S., Van Dongen H.P.A., Belenky G., Panksepp J. Sleep as a fundamental property of neuronal assemblies. Nat. Rev. Neurosci. 2008;9:910–919. doi: 10.1038/nrn2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hida A., Kitamura S., Katayose Y., Kato M., Ono H., Kadotani H., Uchiyama M., Ebisawa T., Inoue Y., Kamei Y., Okawa M., Takahashi K., Mishima K. Screening of clock gene polymorphisms demonstrates association of a PER3 polymorphism with morningness-eveningness preference and circadian rhythm sleep disorder. Sci. Rep. 2014;4:1–6. doi: 10.1038/srep06309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodenmann S., Hohoff C., Freitag C., Deckert J., Rétey J.V., Bachmann V., Landolt H.P. Polymorphisms of ADORA2A modulate psychomotor vigilance and the effects of caffeine on neurobehavioural performance and sleep EEG after sleep deprivation. Br. J. Pharmacol. 2012;165:1904–1913. doi: 10.1111/j.1476-5381.2011.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satterfield B.C., Wisor J.P., Field S.A., Schmidt M.A., Van Dongen H.P. TNFα G308A polymorphism is associated with resilience to sleep deprivation-induced psychomotor vigilance performance impairment in healthy young adults. Brain Behav. Immun. 2015;47:66–74. doi: 10.1016/j.bbi.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazzotti D.R., Guindalini C., Pellegrino R., Barrueco K.F., Santos-Silva R., Bittencourt L.R.A., Tufik S. Effects of the adenosine deaminase polymorphism and caffeine intake on sleep parameters in a large population sample. Sleep. 2011;34:399–402. doi: 10.1093/sleep/34.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valomon A., Holst S.C., Bachmann V., Viola A.U., Schmidt C., Zürcher J., Berger W., Cajochen C., Landolt H.P. Genetic polymorphisms of DAT1 and COMT differentially associate with actigraphy-derived sleep-wake cycles in young adults. Chronobiol. Int. 2014;31:705–714. doi: 10.3109/07420528.2014.896376. [DOI] [PubMed] [Google Scholar]
- 12.Livak K.J. Allelic discrimination using fluorogenic probes and the 5% nuclease assay. Genet. Anal. Biomol. Eng. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 13.Shen G.Q., Abdullah K.G., Wang Q.K. The TaqMan method for SNP genotyping. Methods Mol. Biol. 2009;578:293–306. doi: 10.1007/978-1-60327-411-1_19. [DOI] [PubMed] [Google Scholar]
- 14.Notomi T., Okayama H., Masubuchi H. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 16.Chakraborty G., Karunagasar I., Chakraborty A. Loop-mediated isothermal amplification (LAMP): a rapid molecular diagnosis technique for detection of human pathogens. NUJSH. 2017;7:42–48. [Google Scholar]
- 17.Kim J.-G., Baek S.H., Kim S., Kim H.I., Lee S.W., Phan L.M.T., Kailasa S.K., Park T.J. Rapid discriminative detection of dengue viruses via loop mediated isothermal amplification. Talanta. 2018;190:391–396. doi: 10.1016/j.talanta.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C., Yao Y., Zhu J.L., Zhang S.N., Zhang S.S., Wei H., Hui W.L., Cui Y.L. Establishment and application of a real-time loop-mediated isothermal amplification system for the detection of CYP2C19 polymorphisms. Sci. Rep. 2016;6:1–7. doi: 10.1038/srep26533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlos F.F., Veigas B., Matias A.S., Doria G., Flores O., Baptista P.V. Allele specific LAMP- gold nanoparticle for characterization of single nucleotide polymorphisms. Biotechnol. Rep. 2017;16:21–25. doi: 10.1016/j.btre.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abildgaard A., Tovbjerg S.K., Giltay A., Detemmerman L., Nissen P.H. Lactase persistence genotyping on whole blood by loop-mediated isothermal amplification and melting curve analysis. Clin. Chim. Acta. 2018;482:50–56. doi: 10.1016/j.cca.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 21.Delacour H., Leduc A., Louçano-Perdriat A., Plantamura J., Ceppa F. Diagnosis of genetic predisposition for lactose intolerance by high resolution melting analysis. Ann. Biol. Clin. 2017;75:67–74. doi: 10.1684/abc.2016.1210. [DOI] [PubMed] [Google Scholar]
- 22.Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 23.Francois P., Tangomo M., Hibbs J., Bonetti E.J., Boehme C.C., Notomi T., Perkins M.D., Schrenzel J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011;62:41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- 24.Erblang M., Drogou C., Gomez-Merino D., Metlaine A., Boland A., Deleuze J.F., Thomas C., Sauvet F., Chennaoui M. The impact of genetic variations in ADORA2A in the association between caffeine consumption and sleep. Genes. 2019;10 doi: 10.3390/genes10121021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen J.A. Coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960;20:37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 26.Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. https://159 [DOI] [PubMed] [Google Scholar]
- 27.Hansen Tv.O., Simonsen M.K., Nielsen F.C., Andersen Hundrup Y. Collection of blood, saliva, and buccal cell samples in a pilot study on the danish nurse cohort: comparison of the response rate and quality of genomic DNA. Cancer Epidemiol. Biomarkers Prev. 2007;16:2072–2076. doi: 10.1158/1055-9965.EPI-07-0611. [DOI] [PubMed] [Google Scholar]
- 28.Rogers N.L., Cole S.A., Lan H.-C., Crossa A., Demerath E.W. New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am. J. Hum. Biol. 2007;19:319–326. doi: 10.1002/ajhb.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y., Ehli E.A., Nelson K., Bohlen K., Lynch C., Huizenga P., Kittlelsrud J., Soundy T.J., Davies G.E. Genotyping performance between saliva and blood-derived genomic DNAs on the DMET array: a comparison. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.