Abstract
Perfluorinated alkyl acids (PFAAs) are persistent in marine biota and are toxic to many species, including marine mammals. We measured the concentrations of 15 PFAAs in liver and kidney samples of 16 species of stranded cetaceans from Hawai‘i and other tropical North Pacific regions utilizing high performance liquid chromatographytandem mass spectrometry (LC-MS/MS). Eleven PFAAs in liver and nine PFAAs in kidney were detected, including substantial perfluorooctanesulfonate (PFOS) and perfluoroundecanoic acid (PFUnA). Regression models indicated that phylogenetic family and age class significantly influenced concentrations of certain PFAAs. PFAAs can activate transcription factor peroxisome proliferator-activated receptor alpha (PPARα), which induces transcription of cytochrome P450 4A (CYP4A). Relative expression of PPARα and CYP4A mRNA was quantified using real-time PCR (qPCR) and CYP4A protein expression, using Western blot and then compared to PFAA concentrations in liver and kidney. Concentrations of four PFAA congeners, summation of perfluoroalkyl carboxylic acids (ΣPFCAs), and ΣPFAAs correlated significantly with PPARα mRNA expression and CYP4A protein expression in kidney, suggesting either may be biomarkers of PFAA exposure in cetaceans. This is the first study to quantify PFAAs in marine mammals from this region and the first observation of a direct relationship between PFAA exposure and PPARα and CYP4A expression in cetaceans.
Graphical Abstract
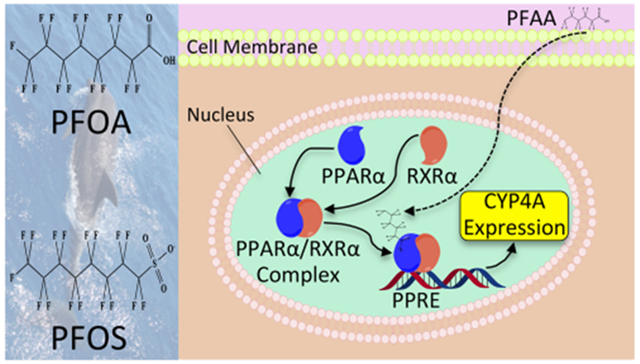
INTRODUCTION
Perfluorinated alkyl acids (PFAAs) are contaminants of interest because of their widespread distribution, high abundance, and the negative health effects attributed to their persistence in animals and the environment. Extensive industrial production, consumer usage, and the inability to be broken down naturally contribute to their high global abundances. These compounds were also readily utilized in popular household products, most notably in carpets, nonstick cookware, and food packaging.1,2
PFAAs persist in all parts of the world, especially in the marine biota. The chemical stability of PFAAs allows them to be persistent in the environment and in tissues of wildlife, particularly in the liver, blood serum, and kidney.3 High concentrations have been detected in tissues of invertebrates, fish, reptiles, birds, and marine mammals.4 The persistence of several types of PFAAs has been extensively examined in marine mammals. However, there are several geographical areas with marine mammals that have not been adequately examined, particularly in the southern hemisphere, Russian Arctic, and tropical North Pacific.
Several factors can potentially influence PFAA bioaccumulation in marine mammals, including type of PFAA (i.e., carbon chain length and functional group), species, age, and sex. PFAAs with longer carbon chains (i.e., 8 carbons or more) and compounds with odd numbered carbon chains tend to bioaccumulate more in organisms compared to shorter chain PFAAs5–8 and even numbered PFAAs,9 respectively. Also, perfluoroalkyl sulfonic acids (PFSAs) typically bioaccumulate more than perfluoroalkyl carboxylic acids (PFCAs) of the same carbon chain length, likely because sulfonates tend to bind more strongly to proteins than carboxylates.6 Bioaccumulation of PFAAs may also be influenced by a species’ trophic level, as these contaminants biomagnify in food webs,8,10–15 and a population’s geographical proximity to sources and atmospheric or oceanic pathways of PFAAs.4,10,16 Many researchers speculate that placental and/or lactational offloading of PFAAs, as evidenced in multiple studies measuring concentrations in marine mammal mother/offspring pairs,13,16–19 is the reason why concentrations in marine mammals likely decrease with age,10,18,20 and males tend to have markedly higher concentrations than females.16,21,22 A growing amount of evidence indicates a global increase in PFAA concentrations in tissues of marine mammals over the last few decades. However, concentrations of some PFAA congeners appear to have leveled off or even decreased in recent years, possibly because of a voluntary switch from perfluorooctanoic acid (PFOA) perfluorooctanesulfonate (PFOS) and PFOS-related products to less bioaccumulative alternatives with shorter carbon chains.23,24
PFAA exposure in mammals has been shown to activate peroxisome proliferator-activated receptors (PPARs), and these proteins are theorized to be a biomarker of this class of contaminants.25 PPARs are proteins involved in lipid and glucose regulation.26 There are three isoforms of PPARs (PPARα, PPAR5δ, and PPARγ) that are similar in structure but can vary in their respective location in the body and physiological roles.27 A key component of PPAR-dependent transcriptional activity is the binding of a ligand. Endogenous ligands that bind to PPARs include monounsaturated and polyunsaturated fatty acids.26 Several environmental contaminants and industrial byproducts can also act as PPAR agonists, including several types of PFAAs as agonists for PPARα.28 PPARα activation by natural and synthetic ligands typically results in downstream transcription of target genes responsible for the regulation of lipids and glucose, in particular the metabolic enzyme cytochrome P450 4A (CYP4A).29,30
CYP4A, a fatty acid and prostaglandin hydroxylase,31,32 is a family of enzymes within the cytochrome P450 group of enzymes that are integral for several metabolic functions, including detoxifying xenobiotic compounds.33 Metabolites of the CYP4A substrates are speculated to have a major role in many hepatic and renal functions.34 Peroxisome proliferators regulate CYP4A expression, which in turn acts as a modulator with other PPARα target genes to maintain lipid homeostasis.29,30,35,36 The introduction of peroxisome proliferator fatty acids leads to PPARα-mediated expression of CYP4A proteins.35 Elevated activation of the PPARα–CYP4A pathway coincident with exposure to harmful PFAAs in various species of animals could result in negative health effects in mammals, including decreased liver function, developmental toxicity, immunotoxicity, and feeding disorders.25,28,37–41
The characterization of the PPARα–CYP4A pathway has not yet been investigated in most marine mammals. Ishibashi et al.42 concluded that this pathway was conserved in the livers of Baikal seals, in that expression of PPARα was correlated with perfluorononanoic acid (PFNA) concentrations and CYP4A expression was correlated with both PFNA and perfluorodecanoic acid (PFDA) concentrations. The same study also reported a significant dose–response relationship between both PFOA and PFOS and PPARα activation, and the authors inferred that this correlation might hold true with elevated levels of these compounds in wild Baikal seal livers. Another study examined organochlorine contaminants (OCs), including PCBs, DDEs, and chlordanes, in minke whale livers, but the researchers did not ascertain a significant correlation with any of the CYPs investigated, including CYP4A.43 Despite these findings, there is insufficient information to evaluate the importance of the PPARα–CYP4A pathway in cetaceans, particularly in response to PFAA exposure.
Predicting health risk of contaminants in protected species generally requires a “weight of evidence” approach. Measuring PFAA contaminant loads provides crucial insight into the distribution and range of the toxicants from initial sources to eventual tissue sequestration in animals. Cetaceans and other marine mammals act as key sentinels for contaminant-related human health risks because of their long life spans, close proximity to coastal regions, and high trophic level status (see refs 44 and 45 for a review). Despite substantial evidence on the persistence and toxicity of PFAAs in cetaceans and other marine mammals in various parts of the world,4,12 there has been no examination of PFAA exposure in cetaceans in the tropical North Pacific. One purpose of this study was to quantify PFAAs and examine bioaccumulation trends in an opportunistic sample set of 16 different cetacean species that were stranded throughout the Hawaiian Islands and other parts of the tropical North Pacific. Increased PPARα activation and CYP4A expression in liver and kidney cells are well-established biomarkers of susceptibility and effect, respectively, of PFAA exposure in several species, but there has been no research to date examining PFAA induction of the PPARα–CYP4A pathway in cetaceans. Another purpose of this study is to quantify PPARα and CYP4A expression in liver and kidney of stranded cetaceans. Published evidence on PFAA effects in cetaceans should be made a priority for future contaminant biomarker research. By correlating these responses with PFAA concentrations in liver and kidney, we will assess whether these physiological responses (1) are related to PFAA exposure and (2) serve as potential biomarkers of PFAA exposure and effect. This is the first study to profile expression of both PPARα and CYP4A in cetaceans, and it lays a foundation for future PFAA biomarker research in these marine sentinels.
MATERIALS AND METHODS
Sample Collection and Storage.
Liver and kidney samples (n = 53) from cetaceans stranded on the main Hawaiian Islands, Micronesia, Saipan, and Guam were collected from 1997 to 2013 during necropsies performed by the Hawai‘i Pacific University Marine Mammal Stranding and Response Team (Table S1). The tissue samples selected for analysis were limited to those from stranded cetaceans considered either code 1 (alive then euthanized) or code 2 (freshly dead). Samples were stored at temperatures between −20 and −80 °C at Hawai‘i Pacific University in Kaneohe, HI and shipped to the National Institute of Standards and Technology (NIST) Gaithersburg, MD under liquid nitrogen for analysis.
Chemicals.
Calibration solutions were created by combining two solutions produced by the NIST Reference Materials (RMs) 8446 Perfluorinated Carboxylic Acids and Perfluorooctane Sulfonamide in Methanol and RM 8447 Perfluorinated Sulfonic Acids in Methanol. Together, the solution contained 15 PFAAs as follows: perfluorobutyric acid (PFBA), perfluoropentanoic acid (PFPeA), perfluorohexanoic acid (PFHxA), perfluoroheptanoic acid (PFHpA), PFOA, PFNA, PFDA, perfluoroundecanoic acid (PFUnA), perfluorododecanoic acid (PFDoA), perfluorotridecanoic acid (PFTriA), perfluorotetradecanoic acid (PFTA), perfluorobutanesulfonic acid (PFBS), perfluorohexanesulfonic acid (PFHxS), PFOS, and perfluorooctanesulfonamide (PFOSA).
Internal standards (IS) were purchased from Cambridge Isotope Laboratories (Andover, MA), RTI International (Research Triangle Park, NC) and Wellington Laboratories (Guelph, Ontario) to create an IS mixture comprised of 11 isotopically labeled PFAAs, and they were as follows: 13C4-PFBA, 13C5-PFPeA, 13C5-PFHxA, 13C4-PFHpA, 13C8-PFOA, 13C9-PFNA, 13C6-PFDA, 13C7-PFUnA, 13C2-PFDoA (IS for PFDoA, PFTriA, and PFTA), 13C3-PFBS, 13C3-PFHxS, 13C8-PFOS, and 13C8-PFOSA.
Chemical Analysis.
Tissue samples, calibrants, blanks, and Standard Reference Material (SRM) 1947 Lake Michigan Fish Tissue were extracted using a method previously described in detail by Reiner et al.46 Briefly, samples were extracted twice using 2.5 mL of 0.01 mol/L KOH in methanol after being spiked with approximately 200 μL of the IS mixture. All samples, blanks, SRMs, and calibrants were filtered using a Whatman UniPrep 0.2 μm filter (Stanford, ME), further purified in methanol using an Envi-carb cartridge (Supelco, Bellefonte, PA), and analyzed using an Agilent 1200 high performance liquid chromatography system (HPLC; Santa Clara, CA) coupled to an Applied Biosystems API 5000 triple quadrupole mass spectrometer (Applied Biosystems, Foster City, CA). The PFAA levels of SRM 1947 processed during our extraction met established values reported on the Certificate of Analysis. Measured compounds were considered above the reporting limit (RL) if the mass of an analyte in the sample was greater than the mean plus three standard deviations of all blanks. Quantitative analysis of the PFAAs are described in detail by Reiner et al.46 and in the Supporting Information.
mRNA Quantification.
The total RNA of each sample was extracted and purified using the SV Total RNA Isolation System from Promega (Madison, WI). Primers for PPARα and CYP4A were designed using MacVector with Assembler version 12.5.1 (Cary, NC) and Primer3web version 4.0.0.47,48 A consensus sequence of four mammal species (PPARα: human, cow, mouse, rat; CYP4A: human, rat, dog, pig) for each gene was created to design primers for a segment of the cetacean gene. A segment of the cetacean gene sequence was used to design real-time PCR (qPCR) primers for the samples. The primer sequences for the reference gene tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ) were obtained from Spinsanti et al.49 Primer pairs were validated with cetacean samples using traditional PCR and sequencing of the PCR product.
qPCR was performed on the Eppendorf Mastercycler ep realplex. All samples were run in triplicate. Relative concentrations were calculated using the ΔΔCq method with nomenclature from Bustin et al.50 The sample with the least detected mRNA expression for each gene was designated as the baseline expression (i.e., PPARα ΔCq baseline = 14.98 and CYP4A ΔCq baseline = 14.56 in animal 15063-001). Statistical analyses were performed using the −ΔΔCq to represent a positive relationship.
Consistent melt curves and Cq values with a standard deviation of <2.00 for at least two of the three triplicates must occur for a sample to be considered satisfactory for analysis. Prior to quantifying target genes, candidates for reference genes were thoroughly investigated (see the Supporting Information).
Protein Quantification.
Tissue preparation, protein extraction, total protein concentration quantification, and gel electrophoresis were performed according to Bachman et al.51 Only CYP4A proteins in kidney samples were quantified. The CYP4A positive control was Sprague–Dawley rat liver microsomes induced with clofibric acid (XenoTech). Band densities with sizes of ~55, ~65, and ~85 kDa were visible in the positive control. These bands likely correspond to CYP4A isoforms CYP4A2, CYP4A1, and CYP4A3, respectively.52 Antibodies used for the analysis are described in detail in the Supporting Information. All bands of these approximate sizes for the unknown samples were included in the density quantification. Protein quantification was performed using a standard curve with dilutions of the positive control. The manufacturer’s measurement of the amount of CYP4A protein in the positive control is 1.11 nmol CYP4A protein/mg total protein, and the standard curve set the limit of detection (LOD) at 22.2 pmol CYP4A protein/mg total protein.
In Western blot analyses, each blot was stripped with ReBlot Plus Strong Stripping Solution 10× (Millipore) and probed for alpha tubulin in order to discern between low expression and substantial protein degradation. Three internal standards were included in each blot (50 μg of positive control, 5 μg of positive control, and 50 μg of NIST SRM QC03LH3 pygmy sperm whale liver homogenate). These standards were used to determine the efficiency of each blot relative to the standard curve blot, and sample concentrations were corrected accordingly.
Statistical Methods.
Descriptive statistics, comparisons, and correlations were performed using IBM SPSS Statistics Version 21. Parametric statistics were utilized if the distribution could be normalized by a logarithmic transformation. Nonparametric statistics were utilized for distributions that could not be normalized. PFAA concentrations below the LOD were set as half the calculated LOD for statistical tests. Sample concentrations for mRNA and protein quantification were excluded from analysis if the output was not satisfactory or if expression was below the LOD. Backward stepwise multiple linear regressions for PFAA quantification were performed using JMP 11 (SAS Institute). The parameters of the model included phylogenetic family (Balaenopteridae, Delphinidae, Kogiidae, Ziphiidae), age class (adults, juveniles, calves), sex (females, males), and day of stranding (where day 1 = 01/01/1997, day 2 = 01/02/1997, etc.). Phylogenetic family, age class, and sex were considered categorical variables, and day of stranding was considered a continuous variable. All parameters were initially entered into the model, and the p-value threshold to eliminate a parameter from the model was set at 0.200. The parameters considered for analyses are those that maintained a p-value <0.200 in the final model. Categorical parameters were grouped by the program to describe significant influence of each individual variable on the model. All p-values stated in the regression results are the parameter p-values and represent each parameter’s influence on the model (α < 0.10 denoting significant influence).
RESULTS AND DISCUSSION
Liver and Kidney PFAA Concentrations.
Eleven individual PFAA congeners were detected in liver and kidney from 53 stranded cetaceans from the tropical North Pacific (Figure 1). PFOS and PFUnA were the most commonly detected PFAAs found in the liver and kidney samples (Table 1). PFOS concentrations in liver samples ranged from below the LOD to 795 ng/g wet weight (ww) and in kidney samples from below the LOD to 34.3 ng/g ww. PFUnA concentrations measured in the liver samples ranged from below the LOD to 175 ng/g ww, and the concentrations measured in the kidney samples ranged from below the LOD to 149 ng/g ww. Eleven PFAAs were detected in the liver samples, and nine PFAAs were detected in the kidney samples. Four PFAAs (i.e., PFBA, PFPeA, PFHpA, and PFBS) were not detected in any of the samples and were excluded from further analyses. In addition, PFHxA and PFHxS were also excluded because they were not detected in kidney; PFHxA was only detected in two liver samples and PFHxS in one liver sample. There was a strong positive correlation between PFAA concentrations measured in matching liver and kidney samples with a p-value <0.0001 for all analyzed samples (Table 1).
Figure 1.
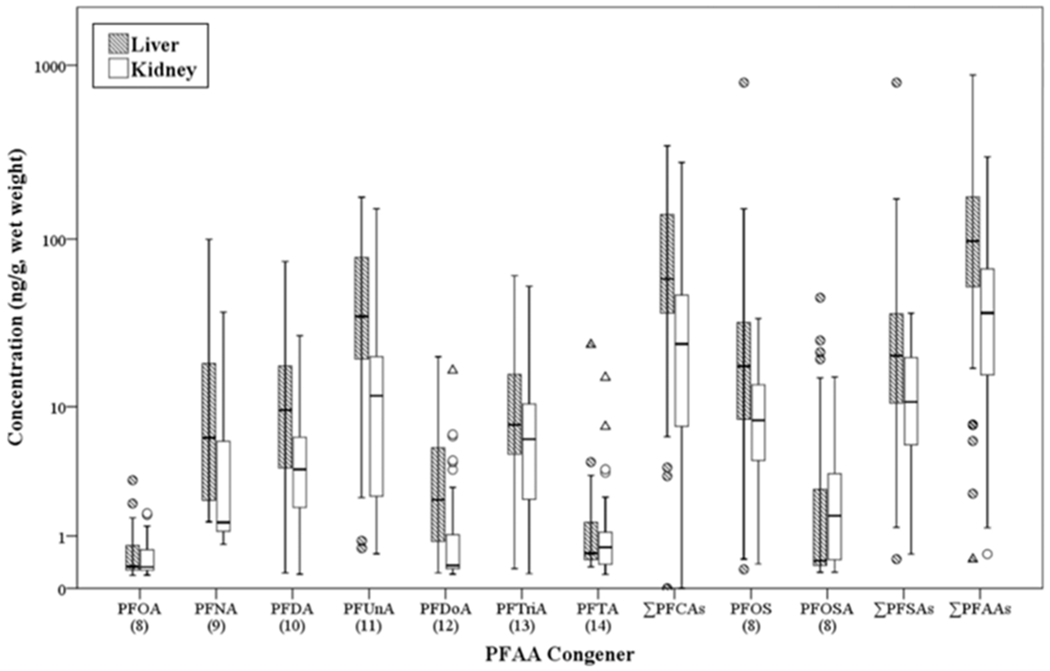
Summary of liver and kidney PFAA concentrations. The PFAAs are ordered by carbon chain length and class. The y-axis is the concentration of the PFAAs (ng/g, ww), and it is in a logarithmetic scale. The circles and triangles represent extreme values or concentrations that extend past the interquartile range >1.5 times and >3 times the value of the interquartile range, respectively.
Table 1.
Descriptive Statistics for PFAA Concentrations in Liver and Kidneya
| PFAA | tiss. | #-% samp. det. | mean (ng/g ww) | med. (ng/g ww) | comp. stat. p-value | corr. stat. p-value |
|---|---|---|---|---|---|---|
| PFOA | liv. | 16-31.4 | 0.579 | <LOD | z = 1.26 | rs = 0.629 |
| kid. | 15-31.9 | 0.503 | <LOD | p = 0.210 | p < 0.0001 | |
| PFNA | liv. | 30-58.8 | 13.2 | 6.30 | z = 5.46 | rs = 0.725 |
| kid. | 18-38.3 | 5.73 | <LOD | p < 0.0001 | p < 0.0001 | |
| PFDA | liv. | 45-88.2 | 13.9 | 9.52 | t = 7.18 | r = 0.872 |
| kid. | 44-93.6 | 5.45 | 3.82 | p < 0.0001 | p < 0.0001 | |
| PFUnA | liv. | 48-94.1 | 51.9 | 35.3 | z = 5.84 | rs = 0.801 |
| kid. | 36-76.6 | 17.5 | 11.7 | p < 0.0001 | p < 0.0001 | |
| PFDoA | liv. | 40-78.4 | 4.14 | 2.23 | z = 5.22 | rs = 0.566 |
| kid. | 15-31.9 | 1.37 | <LOD | p < 0.0001 | p < 0.0001 | |
| PFTriA | liv. | 48-94.1 | 12.8 | 7.66 | t = 5.60 | r = 0.858 |
| kid. | 41-87.2 | 8.90 | 6.16 | p < 0.0001 | p < 0.0001 | |
| PFTA | liv. | 16-31.4 | 1.42 | <LOD | z = 0.085 | rs = 0.689 |
| kid. | 32-68.1 | 1.32 | 0.719 | p = 0.933 | p < 0.0001 | |
| ΣPFCAs | liv. | 50-98.0 | 96.3 | 60.9 | t = 11.2 | r = 0.849 |
| kid. | 44-93.6 | 39.3 | 25.9 | p < 0.0001 | p < 0.0001 | |
| PFOS | liv. | 50-98.0 | 40.7 | 17.8 | t = 7.72 | r = 0.786 |
| kid. | 46-97.9 | 10.0 | 8.18 | p < 0.0001 | p < 0.0001 | |
| PFOSA | liv. | 19-37.3 | 9.90 | <LOD | z = 2.01 | rs = 0.537 |
| kid. | 34-72.3 | 3.74 | 1.61 | p = 0.044 | p < 0.0001 | |
| ΣPFSAs | liv. | 51-100 | 43.4 | 21.0 | t = 6.66 | r = 0.802 |
| kid. | 47-100 | 12.5 | 10.7 | p < 0.0001 | p < 0.0001 | |
| ΣPFAAs | liv. | 51-100 | 140 | 97.4 | z = 5.71 | rs = 0.764 |
| kid. | 47-100 | 51.8 | 37 | p < 0.0001 | p < 0.0001 |
The mean and medians represent all of the samples measured for each tissue (tiss.). Summations of concentrations for ΣPFCAs, ΣPFSAs, and ΣPFAAs do not include any concentrations below the LOD. The detection rate (% samp. det.) was calculated using the number of samples detected (# samp. det.) and the total number of samples measured (51 liver and 47 kidney). Comparisons (comp.) between liver (liv.) and kidney (kid.) used paired sample t tests (t), and correlations (corr.) used Pearson correlation (r) when data were normalized using log10 transformation. Comparisons for non-parametric untransformed data used Wilcoxon signed ranks tests (z), and correlations used Spearman’s rank tests (rs). PFBA, PFPeA, PFHpA, and PFBS were not detected in any the samples, and PFHxA and PFHxS were only detected in two samples and one sample in liver, respectively, and are not displayed. For comparisons and correlations, eight animals were excluded because either liver or kidney samples were not analyzed for the animal (i.e., 15028-001, 15121-001, 15377-001, KW2007003, KW2008003, KW2008005, KW2008006, and KW2011021).
Liver PFAA concentrations were significantly higher than kidney for all PFAA congeners except PFOA and PFTA, for which the concentration means for both were higher in liver but the differences were not significant (Table 1). This trend is supported by several studies where higher PFAA concentrations are reported in liver compared to kidney, particularly in marine mammal studies.3,13,53 Because of this, liver is the most commonly examined tissue when measuring PFAA concentrations in marine mammals. However, a study by Van de Vijver et al.54 on harbor seals in the Dutch Wadden Sea measured higher PFAA concentrations in kidney than liver. Even though our finding of significantly higher PFAA concentrations in liver compared to kidney agrees with most of the literature, conflicting evidence in marine mammals demonstrates the need for more research on PFAA toxicodynamics.
The overall highest concentrations of PFAAs in liver and kidney were PFUnA (median 35.3 ng/g ww and 11.7 ng/g ww, respectively), followed by PFOS (median 17.8 ng/g ww and 8.18 ng/g ww, respectively). PFUnA concentrations have also been highest among PFCAs measured in previous marine mammal studies.8,17,55,56 Abundant PFUnA is consistent with the observation that longer chain, odd numbered PFCAs tend to bioaccumulate more than shorter chain and even-numbered PFCAs.5–9 Martin et al.14 also postulated that PFCAs with carbon chains >11 carbons may be too large to bioaccumulate effectively. This could explain why we observed lower concentrations of PFCAs with >11 carbons (i.e., PFDoA, PFTriA, and PFTA) compared to PFUnA in both liver and kidney samples, even though they have longer carbon chains. PFOS is the most commonly examined PFAA in marine mammals and also typically has the highest concentration of PFSAs (see ref 4 for a review), with some exceptions.8,16,17,56,57 PFSAs tend to bioaccumulate more than PFCAs of the same carbon chain length in marine mammals (e.g., PFOS concentrations higher than PFOA16,18,20,53,58), likely because sulfonates bind more strongly to proteins than carboxylates.6 The results of this study provided evidence of that trend, with PFOS levels being substantially higher than PFOA in every sample where both contaminants were detected (Tables S4 and S5).
Phylogenetic Family Trends.
In general, PFCA concentrations in liver and kidney are higher in Ziphiidae and Delphinidae than Kogiidae and Balaenopteridae. PFSA concentrations in Delphinidae liver and kidney are also very high comparatively. However, PFOSA was not detected in Ziphiidae liver, and only low concentrations were detected in kidney (m = 0.707 ng/g ww) compared to Delphinidae (m = 2.67 ng/g ww) and Kogiidae (m = 2.51 ng/g ww). PFOS concentrations in Delphinidae and Ziphiidae liver and kidney are generally higher than Kogiidae and Balaenopteridae (Table S6).
To explore patterns in this diverse data set, we used a backward-stepwise multiple linear regression model approach to examine life history influences on PFAA concentrations (Table 2). It is important to note that the purpose of the model is to identify the most influential parameters affecting PFAA concentrations, not to characterize differences within those parameters.
Table 2.
Backwards-Stepwise Multiple Linear Regression for Liver and Kidneya
| liver |
kidney |
||||||
|---|---|---|---|---|---|---|---|
| PFAA | parameter | comparison | p-value | model R2 | comparison | p-value | model R2 |
| PFOA | family | Del. > Kog., Zip., Bal. | 0.005 | Del. > Bal., Kog., Zip. | 0.002 | ||
| sex | female > males | 0.094 | 0.333 | 0.247 | |||
| age class | calves, juv. > adults | 0.012 | calves, juv. > adults | 0.009 | |||
| PFNA | family | Zip., Del. > Kog., Bal. | 0.008 | 0.229 | Zip., Del. > Bal., Kog. | 0.015 | 0.213 |
| age class | juv., calves > adults | 0.007 | juv., calves > adults | 0.006 | |||
| PFDA | family | Zip., Del. > Kog., Bal. | 0.004 | Zip. > Del., Bal., Kog. | 0.013 | ||
| age class | juv. > calves, adults | 0.018 | 0.285 | juv. > calves, adults | 0.077 | 0.207 | |
| calves > adults | 0.076 | calves > adults | 0.093 | ||||
| PFUnA | family | Zip. > Del., Kog., Bal. | 0.002 | Zip. > Del., Bal., Kog. | 0.005 | ||
| Del. > Kog., Bal. | 0.007 | 0.406 | 0.318 | ||||
| age class | juv. > adults, calves | 0.015 | juv. > calves, adults | 0.054 | |||
| PFDoA | family | Zip. > Del., Kog., Bal. | <0.0001 | Zip. > Del., Bal., Kog. | 0.0002 | ||
| Del. > Kog., Bal. | 0.033 | 0.428 | 0.388 | ||||
| sex | males > females | 0.054 | |||||
| age class | juv. > calves, adults | 0.042 | |||||
| PFTriA | family | Zip. > Del., Kog., Bal. | <0.0001 | Zip. > Del., Kog., Bal. | <0.0001 | ||
| Del., Kog. > Bal. | 0.038 | 0.479 | Del. > Kog., Bal. | 0.047 | 0.606 | ||
| sex | males > females | 0.045 | |||||
| age class | juv. > adults, calves | 0.044 | |||||
| PFTA | family | N/A | Zip. > Del., Kog., Bal. | 0.030 | 0.249 | ||
| age class | calves > juv., adults | 0.052 | |||||
| ΣPFCAs | family | Zip., Del. > Kog., Bal. | 0.003 | Zip. > Del., Bal., Kog. | 0.0004 | ||
| Zip. > Del. | 0.069 | 0.345 | Del. > Bal., Kog. | 0.030 | 0.408 | ||
| age class | juv. > calves, adults | 0.064 | juv. > calves, adults | 0.082 | |||
| PFOS | family | Del. > Zip., Kog., Bal. | 0.095 | Del., Zip. > Bal., Kog. | 0.0005 | ||
| age class | juv. > calves, adults | 0.008 | 0.167 | juv. > calves, adults | 0.059 | 0.302 | |
| calves > adults | 0.052 | ||||||
| PFOSA | family | 0.099 | Del., Kog. > Zip., Bal. | 0.064 | 0.246 | ||
| age class | adults > juv., calves | 0.026 | adults > juv., calves | 0.022 | |||
| ΣPFSAs | family | Del. > Zip., Kog., Bal. | 0.076 | 0.169 | Del., Zip. > Bal., Kog. | 0.0004 | 0.254 |
| age class | juv. > calves, adults | 0.009 | |||||
| ΣPFAAs | family | Zip., Del. > Kog., Bal. | 0.013 | Zip. > Del., Bal., Kog. | 0.002 | ||
| 0.316 | Del. > Bal., Kog. | 0.012 | 0.395 | ||||
| age class | juv. > calves, adults | 0.0009 | juv. > calves, adults | 0.081 | |||
The regression R2 and p-value describe the influence that each measured life history parameter had on the model to predict PFAA bioaccumulation in liver and kidney. The parameters considered were phylogenetic family, age class, and sex. Only parameters that significantly influenced the model are shown (α < 0.10 denoting significant influence). Commas between variables in the comparison column denote variables that are grouped together by the algorithm, and “>” indicates where the influence significantly diverges between each group within the parameter (e.g., A > B,C means that group A has higher concentrations than both groups B and C, and this difference is what significantly influences the model). Lower parameter p-values indicate stronger influence on the model. Sperm whale KW2011008 was excluded from the analysis because it was the only animal representing the family Physeteridae. The following are abbreviations listed in the table: Delphinidae = Del.; Kogiidae = Kog.; Ziphiidae = Zip.; Balaenopteridae = Bal.; Juvenile = Juv.
Phylogenetic family had significant influence on the model for all PFAAs except for PFTA in liver. In general, Ziphiidae and Delphinidae were higher than Kogiidae and Balaenopteridae for most of the PFAAs. It is widely accepted that animals in the families Ziphiidae and Delphinidae occupy a higher trophic level than those in the family Balaenopteridae.59 Since PFAAs are thought to biomagnify in marine food webs,12–15 it follows that these animals will have significant differences in PFAA concentrations. However, Pauly et al.59 also found that Kogiidae (i.e., pygmy sperm whales and dwarf sperm whales) have a very similar trophic level to many species of Ziphiidae and Delphinidae. If trophic level has a significant impact on PFAA concentrations, then these three families should have similar concentrations and markedly higher concentrations than Balaenopteridae. However, our results do not indicate this, in that Kogiidae have statistically similar concentrations to Balaenopteridae. It is possible that there are other family specific characteristics that affect PFAA bioaccumulation besides trophic level, such as differences in PFAA metabolism, elimination rates, and the spatial distribution of prey. Since this data set has a relatively low number of both Ziphiidae (n = 4) and Kogiidae (n = 4), uncertainty is high. Despite this, the difference between Delphinidae/Ziphiidae and Balaenopteridae suggests that the phylogeny and trophic level of cetaceans influence bioaccumulation of these contaminants.
Age Class Trends.
Age class influenced PFAA concentrations in both liver and kidney. For this study, we designated adults as animals that have reached sexual maturity, juveniles as animals that have not reached sexual maturity, but were no longer nursing, and calves as animals that were still nursing. Juveniles have the highest median concentrations of PFAAs above the LOD in both livers and kidneys with the exception of PFOSA in kidney, where adults have the highest median concentration (Table S7).
The regression model indicates that age class is an important factor driving PFAA concentrations (Table 2). Age class has strong influence over the model for all PFAAs except for PFDoA, PFTriA, and PFTA in liver samples and ΣPFSAs in kidney. High concentrations in juveniles significantly influenced the model for PFDA, PFUnA, ΣPFCAs, PFOS, ΣPFSAs, and ΣPFAAs in liver samples and PFDA, PFUnA, PFDoA, PFTriA, ΣPFCAs, PFOS, and ΣPFAAs in kidney. Adults have the highest PFOSA concentrations in both liver and kidney. High concentrations of PFOA in calf livers (significantly higher than adults but not juveniles) and PFOA and PFTA in calf kidneys significantly influenced the model.
In summary, age class has a strong influence on PFAA concentration in these cetaceans, and juveniles typically have the highest concentrations. The literature demonstrates a clear trend of PFAA concentrations decreasing with age in cetaceans10,18,20 and pinnipeds,42,56,60 likely because of placental and/or lactational offloading of the contaminants from mother to offspring.13,16–19 Most studies that characterize age class split animals into two classes, adults and calves, with juveniles being divided between the two. In our study, juveniles have significantly higher concentrations of most PFAAs than calves and adults. A study on Pacific harbor seals in San Francisco bay observed significantly higher PFOS concentrations in weaned pups compared to yearlings and juveniles.61 This seal study, along with our cetacean results, suggests that individuals that are no longer nursing, but have not yet reached offloading potential at sexual maturity, could merit one or more distinct age class when investigating PFAAs in future studies.
Sex and Temporal Trends.
In general, the data indicate that PFAA concentrations are higher in male kidney samples than female kidney samples; however, no relationship is established in liver samples (Table S8). This study is somewhat consistent with the literature, where the trend is ultimately inconclusive, but most evidence suggests male marine mammals typically have higher PFAA concentrations than females16,21,22 likely because of female placental and/or lactational offloading.13,16–19 In this study, PFCA compounds with shorter carbon chain length appear to have higher concentrations in females, and PFCAs with longer carbon chain length and PFSAs have higher concentrations in males. The regression model provided only minor support for both trends (Table 2). PFOA was significantly influenced by sex, with female livers having higher levels than male livers, but detection of PFOA was inconsistent, with most samples <LOD. High concentrations in male kidneys significantly influenced the model for longer carbon chain PFDoA and PFTriA. In summary, there is no clear trend of PFAA concentrations in regards to sex; however, the data indicate that male kidneys may have higher long carbon chain PFAAs than female kidneys.
Although several studies suggest an increase in PFAAs in marine mammal tissue over the last few decades, there were no clear temporal trends in the data.
PPARα and CYP4A mRNA Expression.
PPARα mRNA was expressed in 33 livers (13 species, 64.7% of samples) and 18 kidneys (7 species, 38.3%), and CYP4A mRNA was expressed in 37 livers (14 species, 72.5%) and 31 kidneys (11 species, 66.0%). The highest PPARα expression was in a bottlenose dolphin liver (15121-001, 21 600-fold change from the baseline), and the lowest detected expression was in a humpback whale kidney (15063-001, baseline expression). The highest CYP4A expression was in a striped dolphin liver (KW2009011, 231 000-fold change from the baseline), and the lowest detected expression was in a humpback whale liver (15063-001, baseline expression) (Table S9).
In general, mRNA expression was greater in liver than in kidney, especially with regard to CYP4A expression (Table 3). This was reflected in the means and medians of both PPARα and CYP4A fold change over the baseline sample, but statistical tests were only significant for CYP4A and not PPARα. It is apparent that liver expresses more CYP4A mRNA than kidney, but a tissue-specific trend for PPARα expression is not established. Also, there was a significant positive correlation between PPARα and CYP4A expression in both liver (r = 0.703, p < 0.001, df = 27) and kidney (r = 0.527, p = 0.044, df = 13), thus suggesting that the PPARα–CYP4A pathway is conserved in cetaceans (Table 3). This relationship was expected because PPARα is a transcription factor for CYP4A in many other species,29,30 but this is the first study to examine the pathway in cetaceans.
Table 3.
Comparisons and Correlations of Liver vs. Kidney mRNA Expression and Correlations Between PPARα and CYP4A mRNA Expressiona
| PPARα |
CYP4A |
|||
|---|---|---|---|---|
| liver vs kidney | ti = 1.52 | p = 0.134 | ti = 5.20 | p < 0.001 |
| df = 49 | df = 54.6b | |||
| r = 0.495 | p = 0.086 | r = 0.179 | p = 0.381 | |
| df = 11 | df = 24 | |||
| liver |
kidney |
|||
| PPARα vs CYP4A | r = 0.703 | p < 0.001 | r = 0.527 | p = 0.044 |
| df = 27 | df= 13 | |||
Comparisons and correlations were performed using the untransformed −ΔΔCq values of PPARα and CYP4A. The test utilized is indicated by the test statistic, where ti = independent-samples t-test and r = Pearson’s correlation coefficient. The degrees of freedom (df) are listed with equal variances assumed unless otherwise specified.
Adjusted degrees of freedom, equal variances not assumed.
CYP4A Protein Expression.
CYP4A proteins were detected in 43 kidneys (91.5% of samples) representing 15 cetacean species (mean = 231 pmol/mg; median = 195 pmol/mg; st. dev. = 180). The highest kidney CYP4A protein expression was in a Longman’s beaked whale (KW2010005, 985 pmol/mg), and the lowest detectable concentration was in a humpback whale (KW2013010, 40.7 pmol/mg). Individuals with CYP4A concentrations below the LOD (based on detectable alpha tubulin) included a humpback whale (15063-001), a spinner dolphin (KW2011018), and two melon-headed whales (KW2011002 and KW2011009). A significant correlation between CYP4A mRNA and CYP4A protein expression in kidney (r = 0.493, p = 0.007, df = 28) indicates that either may serve as a biomarker.
PPARα and CYP4A as Biomarkers for PFAA Exposure.
PPARα and CYP4A are potential contaminant biomarkers for PFAA exposure in stranded cetaceans. PPARα mRNA expression and CYP4A protein expression in kidney positively correlated with PFDA, PFUnA, PFTriA, PFTA, ΣPFCAs, and ΣPFAAs (Figure 2). PPARα and CYP4A mRNA expression in liver and CYP4A mRNA expression in kidney did not significantly correlate with any PFAAs. These results suggest that certain congeners of this class of contaminants interact with the PPARα–CYP4A pathway in cetacean kidney. The relationship between PFAAs and the PPARα–CYP4A pathway was not established in liver.
Figure 2.
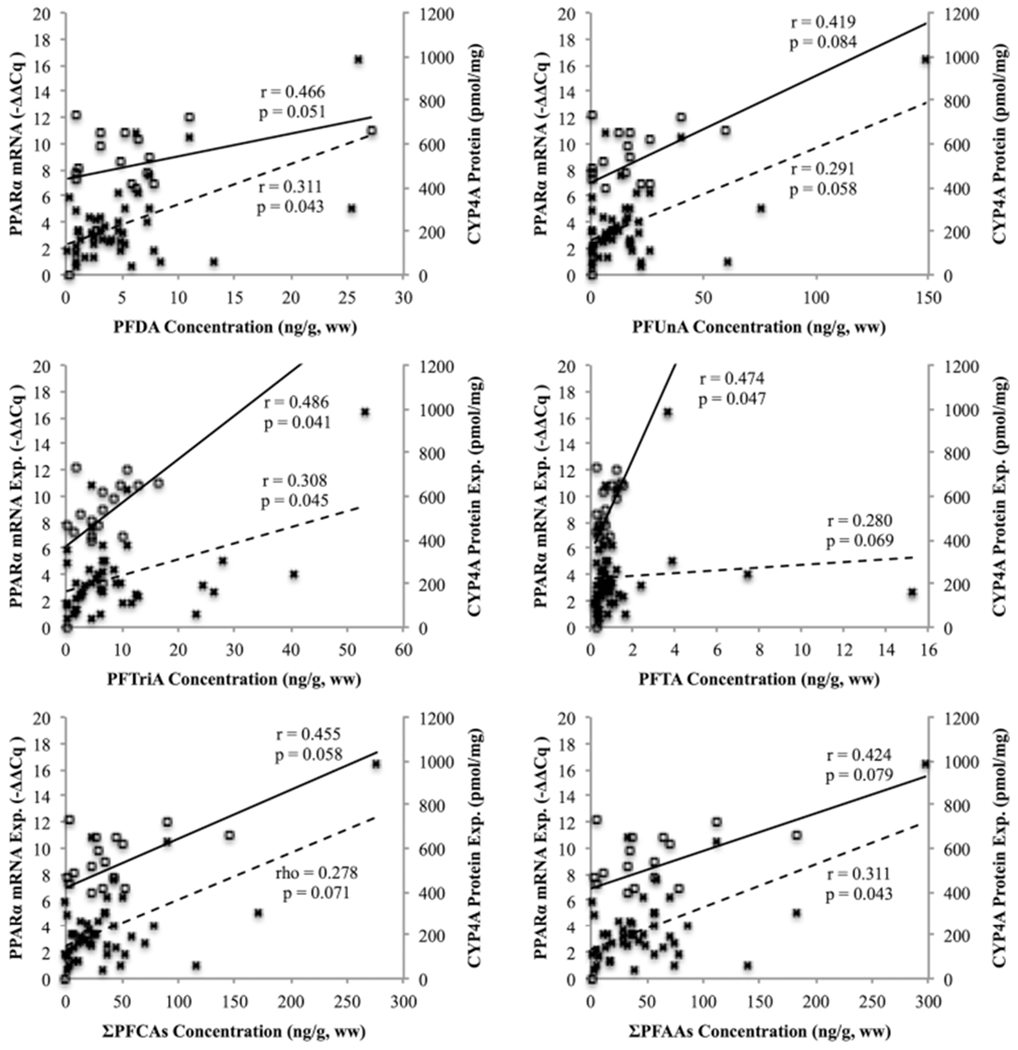
Relationship between PPARα mRNA expression (open squares with solid line) and CYP4A protein expression (×s with dashed line) and PFAA congeners PFDA, PFUnA, PFTriA, PFTA, ΣPFCAs, and ΣPFAAs in kidney. Pearson’s correlation coefficient was used for PPARα mRNA expression with all PFAA congeners; PFDA and ΣPFCAs used log10 transformed data (df = 16). Pearson’s correlation coefficient was used with log10 transformed data for CYP4A protein expression with all PFAA congeners, excluding ΣPFCAs (Spearman’s rank test) (df = 31) (α < 0.10 denoting significance).
PFAA-induced activation of the PPARα–CYP4A pathway has been studied in several different species,42,62–65 but very little research is available on marine mammals. A study that examined PFAA concentrations and activation of the PPARα–CYP4A pathway in Baikal seal liver concluded that this relationship was conserved for PPARα expression with PFNA concentrations and CYP4A expression with PFNA and PFDA concentrations.42 Although they claim that the high abundance of PFNA and PFDA in the analyzed livers may have masked the effects of the other PFAAs examined, there was also a strong correlation between ΣPFAA and both PPARα mRNA and CYP4A protein expression. Our results corroborate this trend for PFAAs in cetacean kidney but not liver.
PFCA concentrations, not PFSA concentrations, appear to be influencing the activation of the PPARα–CYP4A pathway in cetaceans. Past PFAA research and subsequent PFAA production regulation have focused more on the negative health effects of PFSAs, particularly PFOS more than PFCAs.4 It was only recently that focus has shifted to PFCA production, where major PFAA producers in the U.S. made a consent agreement to stop or phase out the manufacturing of PFAAs with chains of eight or more carbons by 2015.66 Despite this decrease in production of longer-chained PFAAs, their inability to naturally degrade67 and long-distance transport and dispersal characteristics2 will continue to make them prevalent in the marine biota. Longer-chained PFCA concentrations may induce activation of the PPARα–CYP4A pathway in cetaceans apart from normal biological processes, a response that has been implicated with several deleterious effects in other mammal species.25,39,41,64
The PPARα–CYP4A pathway plays a pivotal role in lipid and glucose regulation, so anthropogenic inducers, such as PFAAs, are not the only activators of this response. These genes have some baseline level of expression independent of contaminant exposure, and many other biological factors regulate this pathway. Despite these potential confounding influences, PPARα mRNA expression and CYP4A protein expression in cetacean kidney significantly correlated with PFDA, PFUnA, PFTriA, and PFTA.
This initial survey of PFAAs in cetaceans that were stranded on Hawai‘i and the tropical North Pacific revealed that many factors influence concentrations of PFAAs in cetaceans, including tissue type, phylogenetic family, age class, and to a lesser extent sex. Although our analysis indicated clear trends, it is important to consider that measurements were performed on stranded cetaceans representing 16 different cetacean species with different life histories. The results confirm the PPARα–CYP4A pathway in cetaceans and suggest strongly that both PPARα and CYP4A expression are affected by PFAA exposure (particularly PFCAs), with kidney as a potential target organ. Overall, both PPARα and CYP4A remain good biomarker candidates for PFAA exposure and effect in cetaceans. This study can be used as a foundation for future contaminant research and the physiological responses and health risks of PFAAs in cetaceans.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Yongli Chen for providing comments and Melannie Bachman, Erin Urekew, Angela Hanson, Jessica Jacob, Lila Jones, and Lauren Shinego for laboratory assistance. Laboratory analyses and sample collection were funded by National Institute of Standards and Technology (PI B.A.J. 70NANB10H246, 60NANB12D224, 60NANB15D026) and the National Oceanic and Atmospheric Administration John H. Prescott Grant (PI K.L.W. NA12NMF4390125, NA11NMF4390087, N A 1 0 N M F 4 3 9 0 2 6 7 , N A 0 9 N M F 4 3 9 0 2 0 9 , NA08NMF4390556, NA07NMF4390256).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.8b05619.
Additional materials and methods for quantification of PFAAs, mRNA, and protein, description of experimental quality controls, and supporting tables (S1, animal information for PFAA measurement samples; S2, concentrations of PFAAs measured in SRM samples; S3, PCR primers; S4 and S5, concentrations of all PFAAs in liver and kidney; S6-S8, descriptive statistics for PFAA concentrations by phylogenetic family, age class, and sex; S9, descriptive statistics for mRNA expression) (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Rumsby PC; McLaughlin CL; Hall T Perfluorooctane sulfonate and perfluorooctanoic acid in drinking and environmental waters. Philos. Trans. R. Soc., A 2009, 367, 4119–4136. [DOI] [PubMed] [Google Scholar]
- (2).Prevedouros K; Cousins I; Buck R; Korzeniowski S Sources, fate and transport of perfluorocarboxylates. Environ. Sci. Technol 2006, 40 (1), 32–44. [DOI] [PubMed] [Google Scholar]
- (3).Lau C; Anitole K; Hodes C; Lai D; Pfahles-Hutchens A; Seed J Perfluoroalkyl Acids: A review of monitoring and toxicological findings. Toxicol. Sci 2007, 99 (2), 366–394. [DOI] [PubMed] [Google Scholar]
- (4).Houde M; De Silva AO; Muir DCG; Letcher RJ Monitoring of perfluorinated compounds in aquatic biota: An updated review. Environ. Sci. Technol 2011, 45, 7962–7973. [DOI] [PubMed] [Google Scholar]
- (5).Goecke-Flora CM; Reo NV Influence of carbon chain length on the hepatic effects of perfluorinated fatty acids. A 19F-and 31P-NMR investigation. Chem. Res. Toxicol 1996, 9 (4), 689–695. [DOI] [PubMed] [Google Scholar]
- (6).Jones PD; Hu W; De Coen W; Newsted JL; Giesy JP Binding of perfluorinated fatty acids to serum proteins. Environ. Toxicol. Chem 2003, 22 (11), 2639–2649. [DOI] [PubMed] [Google Scholar]
- (7).Conder JM; Hoke RA; Wolf WD; Russell MH; Buck RC Are PFCAs bioaccumulative? A critical review and comparison with regulatory criteria and persistent lipophilic compounds. Environ. Sci. Technol 2008, 42 (4), 995–1003. [DOI] [PubMed] [Google Scholar]
- (8).Kelly BC; Ikonomou MG; Blair JD; Surridge B; Hoover D ; Grace R; Gobas FA Perfluoroalkyl contaminants in an arctic marine food web: Trophic magnification and wildlife exposure. Environ. Sci. Technol 2009, 43 (11), 4037–4043. [DOI] [PubMed] [Google Scholar]
- (9).Ellis DA; Martin JW; De Silva AO; Mabury SA; Hurley MD; Sulbaek Andersen MP; Wallington TJ Degradation of fluorotelomer alcohols: A likely atmospheric source of perfluorinated carboxylic acids. Environ. Sci. Technol 2004, 38, 3316–3321. [DOI] [PubMed] [Google Scholar]
- (10).Van de Vijver KI; Hoff PT; Das K; Van Dongen W; Esmans EL; Jauniaux T; Bouquegneau JM; Blust R; De Coen W Perfluorinated chemicals infiltrate ocean waters: link between exposure levels and stable isotope ratios in marine mammals. Environ. Sci. Technol 2003, 37 (24), 5545–5550. [DOI] [PubMed] [Google Scholar]
- (11).DeNiro MJ; Epstein S Influence of diet on the distribution of nitrogen isotopes in animals. Geochim. Cosmochim. Acta 1981, 45 (3), 341–351. [Google Scholar]
- (12).Butt CM; Berger U; Bossi R; Tomy GT Levels and trends of poly- and perfluorinated compounds in the arctic environment. Sci. Total Environ 2010, 408, 2936–2965. [DOI] [PubMed] [Google Scholar]
- (13).Houde M; Bujas TAD; Small J; Wells RS; Fair PA; Bossart GD; Solomon KR; Muir DCG Biomagnification of perfluoroalkyl compounds in the bottlenose dolphin (Tursiops truncatus) food web. Environ. Sci. Technol 2006, 40, 4138–4144. [DOI] [PubMed] [Google Scholar]
- (14).Martin JW; Whittle DM; Muir DC; Mabury SA Perfluoroalkyl contaminants in a food web from Lake Ontario. Environ. Sci. Technol 2004, 38 (20), 5379–5385. [DOI] [PubMed] [Google Scholar]
- (15).Tomy GT; Budakowski W; Halldorson T; Helm PA; Stern GA; Friesen K; Pepper K; Tittlemier SA; Fisk AT Fluorinated organic compounds in an eastern Arctic marine food web. Environ. Sci. Technol 2004, 38 (24), 6475–6481. [DOI] [PubMed] [Google Scholar]
- (16).Reiner JL; O’Connell SG; Moors AJ; Kucklick JR; Becker PR; Keller JM Spatial and temporal trends of perfluorinated compounds in beluga whales (Delphinapterus leucas) from Alaska. Environ. Sci. Technol 2011, 45, 8129–8136. [DOI] [PubMed] [Google Scholar]
- (17).Hart K; Kannan K; Isobe T; Takahashi S; Yamada TK; Miyazaki N; Tanabe S Time trends and transplacental transfer of perfluorinated compounds in melon-headed whales stranded along the Japanese coast in 1982, 2001/2002, and 2006. Environ. Sci. Technol 2008, 42 (19), 7132–7137. [DOI] [PubMed] [Google Scholar]
- (18).Houde M; Wells RS; Fair PA; Bossart GD; Hohn AA; Rowles TK; Sweeney JC; Solomon KR.; Muir DCG. Polyfluoroalkyl compounds in free-ranging bottlenose dolphins (Tursiops truncatus) from the Gulf of Mexico and the Atlantic Ocean. Environ. Sci. Technol 2005, 39 (17), 6591–6598. [DOI] [PubMed] [Google Scholar]
- (19).Van de Vijver KI; Holsbeek L; Das K; Blust R; Joiris C; De Coen W Occurrence of perfluorooctane sulfonate and other perfluorinated alkylated substances in harbor porpoises from the Black Sea. Environ. Sci. Technol 2007, 41 (1), 315–320. [DOI] [PubMed] [Google Scholar]
- (20).Fair PA; Adams J; Mitchum G; Hulsey TC; Reif JS; Houde M; Muir D; Wirth E; Wetzel D; Zolman E; McFee W; Bossart GD Contaminant blubber burdens in Atlantic bottlenose dolphins (Tursiops truncatus) from two southeastern US estuarine areas: Concentrations and patterns of PCBs, pesticides, PBDEs, PFCs, and PAHs. Sci. Total Environ 2010, 408 (7), 1577–1597. [DOI] [PubMed] [Google Scholar]
- (21).Kannan K; Koistinen J; Beckmen K; Evans T; Gorzelany JF; Hansen KJ; Jones PD; Helle E; Nyman M; Giesy JP Accumulation of perfluorooctane sulfonate in marine mammals. Environ. Sci. Technol 2001, 35 (8), 1593–1598. [DOI] [PubMed] [Google Scholar]
- (22).Kannan K; Perrotta E; Thomas NJ Association between perfluorinated compounds and pathological conditions in southern sea otters. Environ. Sci. Technol 2006, 40 (16), 4943–4948. [DOI] [PubMed] [Google Scholar]
- (23).Renner R The long and the short of perfluorinated replacements. Environ. Sci. Technol 2006, 40 (1), 12–13. [DOI] [PubMed] [Google Scholar]
- (24).Betts KS Perfluoroalkyl acids: what is the evidence telling us? Environ. Health Perspect 2007, 115 (5), A250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Abbott BD; Wolf CJ; Das KP; Zehr RD; Schmid JE; Lindstrom AB; Strynar MJ; Lau C Developmental toxicity of perfluorooctane sulfonate (PFOS) is not dependent on expression of peroxisome proliferator activated receptor-alpha (PPAR alpha) in the mouse. Reprod. Toxicol 2009, 27 (3–4), 258–265. [DOI] [PubMed] [Google Scholar]
- (26).Desvergne BWW Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev 1999, 20 (5), 649–688. [DOI] [PubMed] [Google Scholar]
- (27).Francis G; Fayard E; Picard F; Auwerx J Nuclear receptors and the control of metabolism. Annu. Rev. Physiol 2003, 65, 261–311. [DOI] [PubMed] [Google Scholar]
- (28).Abbott B Review of the expression of peroxisome proliferator-activated receptors alpha (PPARγ), beta (PPARβ), and gamma (PPARγ) in rodent and human development. Reprod. Toxicol 2009, 27, 246–257. [DOI] [PubMed] [Google Scholar]
- (29).Aldridge TC; Tugwood JD; Green S Identification and characterization of DNA elements implicated in the regulation of CYP4A1 transcription. Biochem. J 1995, 306 (2), 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Johnson EF; Palmer CN; Griffin KJ; Hsu MH Role of the peroxisome proliferator-activated receptor in cytochrome P450 4A gene regulation. FASEB J. 1996, 10 (11), 1241–1248. [DOI] [PubMed] [Google Scholar]
- (31).Simpson AECM The cytochrome P450 4 (CYP4) family. Gen. Pharmacol 1997, 28 (3), 351–359. [DOI] [PubMed] [Google Scholar]
- (32).Aoyama T; Hardwick JP; Imaoka S; Funae Y; Gelboin HV; Gonzalez FJ Clofibrate-inducible rat hepatic P450s IVA1 and IVA3 catalyze the omega-and (omega-1)-hydroxylation of fatty acids and the omega-hydroxylation of prostaglandins E1 and F2 alpha. J. Lipid Res 1990, 31 (8), 1477–1482. [PubMed] [Google Scholar]
- (33).Guengerich FP Human cytochrome P450 enzymes In Cytochrome P450 (473–535); Springer: New York, 1995. [Google Scholar]
- (34).Bell DR; Bars RG; Gibson GG; Elcombe CR Localization and differential induction of cytochrome P450IVA and acyl-CoA oxidase in rat liver. Biochem. J 1991, 275 (1), 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hardwick JP; Song BJ; Huberman E; Gonzalez FJ Isolation, complementary DNA sequence, and regulation of rat hepatic lauric acid omega-hydroxylase (cytochrome P-450LA omega). Identification of a new cytochrome P-450 gene family. J. Biol. Chem 1987, 262 (2), 801–810. [PubMed] [Google Scholar]
- (36).Rakhshandehroo M; Knoch B; Muller M; Kersten S Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010, 2010, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Klaunig J; Babich M; Baetcke K; Cook J; Corton J; David R; DeLuca J; Lai D; McKee R; Peters J; Roberts R; Fenner-Crisp P PPARα Agonist-Induced Rodent Tumors: Modes of Action and Human Relevance. Crit. Rev. Toxicol 2003, 33 (6), 655–780. [DOI] [PubMed] [Google Scholar]
- (38).Abbott BD; Wolf CJ; Schmid JE; Das KP; Zehr RD; Helfant L; Nakayama S; Lindstrom AB; Strynar MJ; Lau C Perfluorooctanoic acid-induced developmental toxicity in the mouse is dependent on expression of perioxisome proliferator-activated receptor-alpha. Toxicol. Sci 2007, 98 (2), 571–581. [DOI] [PubMed] [Google Scholar]
- (39).Yang Q; Xie Y; Alexson S; Nelson B; De Pierre J Involvement of the peroxisome proliferator-activated receptor alpha in the immunomodulation caused by peroxisome proliferators in mice. Biochem. Pharmacol 2002, 63, 1893–1900. [DOI] [PubMed] [Google Scholar]
- (40).Corsini E; Sangiovanni E; Avogadro A; Galbiati V; Viviani B; Marinovich M; Galli CL; Dell’Agli M; Germolec DR In vitro characterization of the immunotoxic potential of several perfluorinated compounds (PFCs). Toxicol. Appl. Pharmacol 2012, 258, 248–255. [DOI] [PubMed] [Google Scholar]
- (41).Asakawa A; Toyoshima M; Harada K; Fujimiya M; Inoue K; Koizumi A The ubiquitous environmental pollutant perfluoroctanoic acid inhibits feeding behavior via peroxisome proliferator-activated receptor-alpha. Int. J. Mol. Med 2008, 21 (4), 439–445. [PubMed] [Google Scholar]
- (42).Ishibashi H; Iwata H; Kim EY; Tao L; Kannan K; Amano M; Miyazaki N; Tanabe S; Batoev VB; Petrov EA Contamination and effects of perfluorochemicals in Baikal seal (Pusa sibirica). 1. Residue level, tissue distribution, and temporal trend; 2. Molecular characterization, expression level, and transcriptional activation of peroxisome proliferator-activated receptor a. Environ. Sci. Technol 2008, 42, 2295–2308. [DOI] [PubMed] [Google Scholar]
- (43).Niimi S; Kim EY; Iwata H; Watanabe MX; Yasunaga G; Fujise Y; Tanabe S Identification and hepatic expression profiles of cytochrome P450 1–4 isozymes in common minke whales (Balaenoptera acutorostrata). Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol 2007, 147 (4), 667–681. [DOI] [PubMed] [Google Scholar]
- (44).Ross PS Marine mammals as sentinels in ecological risk assessment. Hum. Ecol. Risk Assess 2000, 6 (1), 29–46. [Google Scholar]
- (45).Bossart GD Marine mammals as sentinel species for oceans and human health. Oceanography 2006, 19 (2), 134–137. [DOI] [PubMed] [Google Scholar]
- (46).Reiner JL; O’Connell SG; Butt CM; Mabury SA; Small JM; De Silva AO; Muir DCG; Delinsky AD; Strynar MJ; Lindstrom AB; Reagen WK; Malinsky M; Schafer S; Kwadijk CJAF; Schantz MM; Keller JM Determination of perfluorinated alkyl acid concentrations in biological standard reference materials. Anal. Bioanal Chem 2012, 404 (9), 2683–2692. [DOI] [PubMed] [Google Scholar]
- (47).Untergasser A; Cutcutache I; Koressaar T; Ye J; Faircloth BC; Remm M; Rozen SG Primer3 - new capabilities and interfaces. Nucleic Acids Res. 2012, 40 (15), e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Koressaar T; Remm M Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23 (10), 1289–1291. [DOI] [PubMed] [Google Scholar]
- (49).Spinsanti G; Panti C; Lazzeri E; Marsili L; Casini S; Frati F; Fossi CM Selection of reference genes for quantitative RT-PCR studies in striped dolphin (Stenella coeruleoalba) skin biopsies. BMC Mol. Biol 2006, 7 (1), 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Bustin SA; Benes V; Garson JA; Hellemans J; Huggett J; Kubista M; Mueller R; Nolan T; Pfaffl M; Shipley G; Vandesompele J; Wittwer CT The MIQE guidelines: minimum information for publication of quantitative real-time PCR experi-ments. Clin. Chem 2009, 55 (4), 611–622. [DOI] [PubMed] [Google Scholar]
- (51).Bachman MJ; Foltz KM; Lynch JM; West KL; Jensen BA Using cytochrome P4501A1 expression in liver and blubber to understand effects of persistent organic pollutant exposure in stranded Pacific Island cetaceans. Environ. Toxicol. Chem 2015, 34 (9), 1989–1995. [DOI] [PubMed] [Google Scholar]
- (52).Xu Y; Knipp GT; Cook TJ Expression of CYP4A isoforms in developing rat placental tissue and rat trophoblastic cell models. Placenta 2005, 26 (2), 218–225. [DOI] [PubMed] [Google Scholar]
- (53).Ahrens L; Siebert U; Ebinghaus R Total body burden and tissue distribution of polyfluorinated compounds in harbor seals (Phoca vitulina) from the German Bight. Mar. Pollut. Bull 2009, 58 (4), 520–525. [DOI] [PubMed] [Google Scholar]
- (54).Van de Vijver KI; Hoff P; Das K; Brasseur S; Van Dongen W; Esmans E; Reijnders P; Blust R; De Coen W Tissue distribution of perfluorinated chemicals in harbor seals (Phoca vitulina) from the Dutch Wadden Sea. Environ. Sci. Technol 2005, 39 (18), 6978–6984. [DOI] [PubMed] [Google Scholar]
- (55).Bossi R; Riget FF; Dietz R Temporal and spatial trends of perfluorinated compounds in ringed seal (Phoca hispida) from Greenland. Environ. Sci. Technol 2005, 39 (19), 7416–7422. [DOI] [PubMed] [Google Scholar]
- (56).Butt CM; Muir DC; Stirling I; Kwan M; Mabury SA Rapid response of Arctic ringed seals to changes in perfluoroalkyl production. Environ. Sci. Technol 2007, 41 (1), 42–49. [DOI] [PubMed] [Google Scholar]
- (57).Bossi R; Riget FF; Dietz R; Sonne C; Fauser P; Dam M; Vorkamp K Preliminary screening of perfluorooctane sulfonate (PFOS) and other fluorochemicals in fish, birds and marine mammals from Greenland and the Faroe Islands. Environ. Pollut 2005, 136 (2), 323–329. [DOI] [PubMed] [Google Scholar]
- (58).Kannan K; Corsolini S; Falandysz J; Oehme G; Focardi S; Giesy JP Perfluorooctanesulfonate and related fluorinated hydrocarbons in marine mammals, fishes, and birds from coasts of the Baltic and the Mediterranean Seas. Environ. Sci. Technol 2002, 36 (15), 3210–3216. [DOI] [PubMed] [Google Scholar]
- (59).Pauly D; Trites AW; Capuli E; Christensen V Diet composition and trophic levels of marine mammals. ICES J. Mar. Sci 1998, 55 (3), 467–481. [Google Scholar]
- (60).Shaw S; Berger ML; Brenner D; Tao L; Wu Q; Kannan K Specific accumulation of perfluorochemicals in harbor seals (Phoca vitulina concolor) from the northwest Atlantic. Chemosphere 2009, 74 (8), 1037–1043. [DOI] [PubMed] [Google Scholar]
- (61).Sedlak MD; Greig DJ Perfluoroalkyl compounds (PFCs) in wildlife from an urban estuary. J. Environ. Monit 2012, 14 (1), 146–154. [DOI] [PubMed] [Google Scholar]
- (62).Sohlenius AK; Eriksson AM; Hogstrom C; Kimland M; DePierre JW Perfluorooctane sulfonic acid is a potent inducer of peroxisomal fatty acid β-oxidation and other activities known to be affected by peroxisome proliferators in mouse liver. Pharmacol. Toxicol 1993, 72 (2), 90–93. [DOI] [PubMed] [Google Scholar]
- (63).Maloney EK; Waxman DJ trans-Activation of PPARα and PPARγ by structurally diverse environmental chemicals. Toxicol. Appl. Pharmacol 1999, 161, 209–218. [DOI] [PubMed] [Google Scholar]
- (64).Rosen MB; Lee JS; Ren H; Vallanat B; Liu J; Waalkes MP; Abbott BD; Lau C; Corton JC Toxicogenomic dissection of the perfluorooctanoic acid transcript profile in mouse liver: Evidence for the involvement of nuclear receptors PPAR-α and CAR. Toxicol. Sci 2008, 103 (1), 46–56. [DOI] [PubMed] [Google Scholar]
- (65).Cheng X; Klaassen CD Perfluorocarboxylic acids induce cytochrome P450 enzymes in mouse liver through activation of PPAR-α and CAR transcription factors. Toxicol. Sci 2008, 106 (1), 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).US Environmental Protection Agency. Fact Sheet: 2010/2015 PFOA Stewardship Program; Available at https://www.epa.gov/assessing-and-managing-chemicals-under-tsca/fact-sheet-20102015-pfoa-stewardship-program.
- (67).Kudo N; Kawashima Y Toxicity and toxicokinetics of perfluorooctanoic acid in humans and animals. J. Toxicol. Sci 2003, 28 (2), 49–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


