Abstract
Quantitative proteomics has evolved considerably over the last decade with the advent of higher order multiplexing (HOM) techniques. With the development of methods such as—multitagging, cPILOT, hyperplexing, BONPlex, and MITNCAT, the HOM technique is rapidly taking the center stage in multiplexed quantitative proteomics. These studies combined MS1 and MS2 labels in a single experiment enabling higher sample throughput. While HOM is highly promising, the computational analysis is still a big challenge, as the available tools cannot harness its power completely. We have developed a new quantitative pipeline, HyperQuant to aid in accurately quantitating complex HOM data. The pipeline uses identification results from either MaxQuant or any other search engine and quantitation results from QuantWizIQ. The Mapper and Combiner modules of HyperQuant allow facile integration of the labeled data, along with peptide spectrum match (PSM) intensity/ratio integration for proteins, respectively, for each PSM label combination. This also includes appropriate combination of replicates/fractions before summarizing the protein intensity/ratio, leading to robust quantitation. To the best of our knowledge, this is the first tool for the quantitation of HOM data with flexibility for any combination of MS1 and MS2 labels. We demonstrate its utility in analyzing two 18-plex data sets from the hyperplexing and the BONplex studies. The tool is open source and freely available for noncommercial use. HyperQuant is a highly valuable tool that will help in advancing the field of multiplexed quantitative proteomics.
Introduction
Proteomics has enabled the high throughput study of cellular systems to uncover the mechanisms regulating cellular health and disease. Understanding the cellular signaling networks or perturbations to different types of stimuli requires robust and reproducible quantitation at a large scale. Quantitative proteomics has made it possible to identify as well as quantify proteins from multiple conditions in a single run.1 Protein quantitation in shotgun proteomics is carried out using metabolic or chemical labeling.2 Metabolic labeling of proteins with SILAC (stable isotope labeling of amino acids in cell culture) replaces essential amino acids in the cell culture with their stable isotope-labeled counterparts (such as heavy lysine or arginine).3 The separate cultures from normal (light) and labeled (heavy) samples are subsequently mixed in equal amounts, digested, and analyzed by LC–MS/MS. The peptides from the two samples are reflected in MS1 spectra as pairs separated by known mass differences between light and heavy peptides. On sequencing in MS/MS, the peptides are identified by database search followed by FDR control,4 while their MS1 intensities are a proxy for their relative quantitation. Because of mixing the cell cultures early in the workflow, it is the most accurate technique for quantitation. However, biological samples cannot always be labeled in cell culture and require chemical labeling. In chemical labeling techniques such as iTRAQ5 (isobaric tags for relative and absolute quantitation) or TMT6 (tandem mass tag), digested peptides from two to sixteen samples are labeled with different variants of isobaric tags that label the N-termini and the free amino group on lysine residues.7 The isobaric tags increase the mass of peptides from all samples equally, and the peaks in MS1 represent a sum of peptide intensities from all samples. Upon fragmentation in MS/MS, the unique mass reporter ions from iTRAQ/TMT tags are observed in the low mass region, while the sequencing peaks are used for identification. The reporter peaks help in relative quantitation between the samples.8 While these techniques have made proteome-wide quantitation possible, strategies that enhance the depth and multiplexing capacity of quantitation are desirable for systems biology studies.9
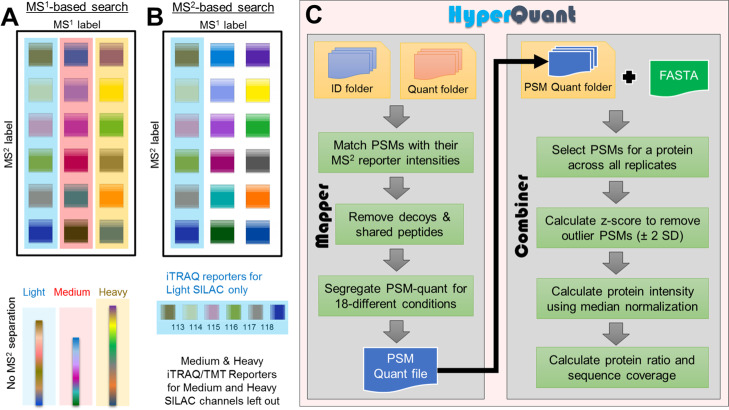
With the advancements in higher order multiplexing (HOM) technologies (combining MS1 and MS2 labels), designing a statistically robust experiment with high sample throughput is now feasible for studying proteome dynamics at systems level.10 Identifying and quantifying proteins up to 54 conditions in a single run of the mass spectrometer with the help of HOM considerably reduces the technical variability arising because of multiple runs.11 Since the first experiment performed in 2010, the technology has vastly evolved with different combinations of metabolic and chemical labeling such as multitagging,12 cPILOT,13−17 hyperplexing,18 SILAC-iTRAQ tails,19 TMT-SILAC hyperplexing,20,21 BONPlex,51 MITNCAT,22 and mPDP23 to achieve higher sample throughput in a single mass spectrometry run. Even though the technique has been around for almost a decade, the computational analysis is still lagging behind and is performed with two different searches and custom scripts for analysis. Using conventional tools, the quantitative analysis is cumbersome as the individual runs inadvertently summarize protein quantitation incorrectly. The dual search strategy is performed to identify peptides as no search engine can search for two modifications together on one amino acid. For identification of peptides labeled with both MS1 and MS2, a modified MS1 search (including mass of MS2 label) is conducted which provides a summed up MS1 quantitation (Figure 1A). For quantitation of MS2 labels, another search is performed for MS2 combination with lighter MS1, and custom scripts are written to derive MS2 quantitation of heavier MS1 combinations (Figure 1B). The downstream analysis becomes a challenge that requires custom analysis suited to each experiment type for the correct biological interpretation or designing ingenious experiments to evade the need for quantitation from one of the labels, limiting the capacity of HOM.10 For example, in the 18-plex hyperplexing study, the SILAC labels were used for biological replicates, while TMT labels were used for time points.18 Two database searches were conducted—one for each label as fixed modification and quantitation was performed using in-house program Vista.24 The use of two types of chemical labels in cPILOT allowed it to be useful for biological and clinical samples that cannot be metabolically labeled. The protein ratios in the cPILOT studies were calculated with the help of an internal standard reporter ion, thus requiring one of the reporters to be used as an internal standard.15 In TMT-SILAC hyperplexing study, the SILAC labels were used to study the kinetics of protein quantitation over time points labeled by TMT labels. Two searches were performed, one with SILAC as a fixed label, yielding SILAC ratios and one with only TMT labels. A kinetics model was calculated based on reporter ion intensities to calculate protein synthesis and degradation.20 In MITNCAT study, the protein dynamics was studied with the TMT labels at specific time points, while SILAC and AHA labels were used exclusively for protein selection in a time window.22
Figure 1.
Conceptual comparison of data analysis performed by current tools versus HyperQuant. Current tools cannot quantitate the HOM data correctly. Manipulating label definitions in search tools (SILAC-Light, SILAC-Medium, and SILAC-Heavy) can help in the identification of PSMs but not correct protein quantitation whether MS1-based search (SILAC search) or MS2-based search (iTRAQ/TMT) is used. (A) MS1 quantitative search will combine the quantitative contribution from all six time points of each SILAC into the respective MS1 peaks (L/M/H), as depicted by the colored boxes (light blue, light red, and light yellow). (B) MS2 search only allows identification and quantitation of SILAC light-based reporters. The SILAC medium and SILAC heavy reporters will not be identified or quantified. (C) HyperQuant combines data from A with reporter quantitation from QuantWizIQ to correctly carry out quantitation across each SILAC-iTRAQ combination separately.
Despite the obvious benefits of the HOM technique to provide reproducible, cost-, and time-efficient study designs, it is not yet fully exploited as a regular proteomic workflow because of challenges in computational analysis and mixing of quantitation from MS1 and MS2 labels.10 To enable easy and flexible analysis of data from HOM studies, we have designed a computational pipeline—HyperQuant, which is the first tool that collates, deconvolutes labels, summarizes, and calculates normalized protein intensities or ratios for each condition (Figure 1C). The normalized intensities can be used to calculate relative protein ratios from any combination of metabolic or chemical labels as per the experiment to facilitate uncomplicated biological interpretations. HyperQuant is flexible enough to use the results from any search engine provided in a simple predefined text format making it interoperable with many search engines. The peptide and protein quantitation is performed by combining data from multiple replicates and can output the summarized protein intensity. HyperQuant has several advantages apart from experiment-agnostic integration of HOM data. It also rescues proteins with a low number of peptide spectrum matches (PSMs) in individual replicates as combining PSMs before outlier removal or filtering saves consistently occurring replicate level one-hit wonders. Apart from rescuing protein identifications, increasing the number of peptide data points can enhance the statistical confidence in the protein quantitation. The poorer (or highly variable) quantitation values across replicates will be removed during an outlier removal. HyperQuant also provides normalized intensities for each sample used in the experiment, thus giving users the flexibility in choosing any denominator for ratio calculation. To demonstrate the utility of the pipeline; we have reanalyzed the 18-plex hyperplexing data from Dephoure and Gygi18 and compared HyperQuant results with the results from their study.
We also analyzed our previously published data of “hyperplexing and click chemistry” (later named BONPlex),51,10,25 investigating the effect of mycobacterial infection on human macrophage secretome using a combination of pSILAC, AHA, and iTRAQ. Although macrophage secretome has been previously studied using similar techniques by Eichaelbaum et al.26,27 using pSILAC with AHA and Khan et al.28 using targeted proteomics, these studies used either LPS or attenuated bacteria to create an immune response. The sample multiplexing was merely 2–3 plex and involved multiple 2-plex experiments for temporal study.27 Our study exploited AHA with the power of the HOM technique to reduce the need for multiple experiments and run-to-run variability.51 The study was also unique for using clinical and lab strains of Mtb instead of LPS, thus providing more realistic insights into the elicited immune response. The experiment focused only on newly synthesized secreted proteins and their temporal profile upon infection. Quantifying the secretome can aid in understanding the dynamics of the host–pathogen crosstalk. With the help of HyperQuant, strain specific as well as temporal changes in the host secretome were quantitated demonstrating its utility in a truly HOM experiment.
Results & Discussion
The novel design of an HOM experiment allows two types of labels for two quantitative dimensions (MS1 and MS2) in one MS run. This can be conceptually represented as a 2-D matrix to explain two types of labeling (Figure 1A,B). To ensure the complete biological understanding of the conditions labeled, the quantitation must be performed on both the dimensions of the matrix. The analysis is a challenge with the current tools, necessitating multiple searches for quantitating both labels and custom analysis scripts. This does not completely harness the power of HOM studies. To exploit the true multiplexing nature of technology and easy quantitation without any loss of information on any label, we designed the HyperQuant tool to integrate identification with quantitation along with replicate combination. There are two modules in the HyperQuant tool—Mapper and Combiner. Mapper aids in mapping of PSM identification and quantification results from MaxQuant and QuantWizIQ outputs, respectively. Apart from MaxQuant, users also have the flexibility to use any other identification engine using a simple text format that specifies PSM and label information. The Combiner module integrates the PSMs from all replicates and summarizes the protein intensity/ratio depending on the user choice. The detailed workflow of the tool is shown in Figure 1C.
Benchmarking of the HyperQuant Tool Using Hyperplexing Data
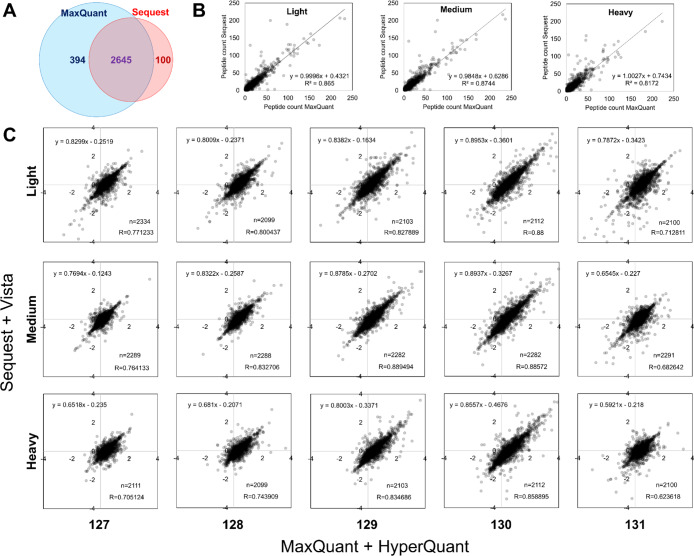
We tested the pipeline for identifying and quantifying the proteome using Dephoure and Gygi data.18 We searched the 3 × 6 experiment (18-plex) from Dephoure study using MaxQuant, performed spectra level quantitation by QuantWizIQ, and integrated the results using the HyperQuant pipeline to obtain protein ratios. For this particular data, we used the weighted average method for calculating protein ratios as per the original study. We compared our results obtained from the pipeline with their protein list. Despite using different tools (Sequest29 vs MaxQuant), we observed that ∼84% of the proteins identified were matching (Figure 2A). The number of peptides identified in light, medium, and heavy SILAC channels also showed good corroboration (Figure 2B). Our reanalysis identified 394 more proteins, while 100 proteins were missed, but we did not analyze it further as we were only interested in quantitative comparison for common proteins.
Figure 2.
Benchmarking of the HyperQuant tool using the public data. (A) Concordance of proteins identified by the two search engines MaxQuant (our search) and Sequest (Dephoure and Gygi). (B) For the common proteins, the scatter plots depict good correlation between the number of peptides identified for light, medium, and heavy SILAC channels between the two searches. (C) Overview of HyperQuant benchmarking with results from our pipeline (x-axis) compared with those from Dephoure and Gygi data (y-axis). The comparison of protein quantitation between the two pipelines depicts a high level of agreement and good correlation between quantitated values despite the use of different tools.
For the 2645 common proteins between our analysis (MaxQuant + HyperQuant) and Dephoure and Gygi results (Sequest + Vista), the protein ratios from the two pipelines were compared. Figure 2C depicts the comparison of ratios across the TMT6 labels for each SILAC channel—light, medium, and heavy separately. Because the protein ratio method used here uses peptide numbers as weights, there are some differences attributable to differences in peptide identifications (because of different search engines); thereby altering quantitation marginally. We still observed high ratio concordance across all labels. These results suggest that the HyperQuant pipeline is able to analyze HOM data and can accurately calculate the quantitative values for every label combination.
Analysis of BONPlex Data
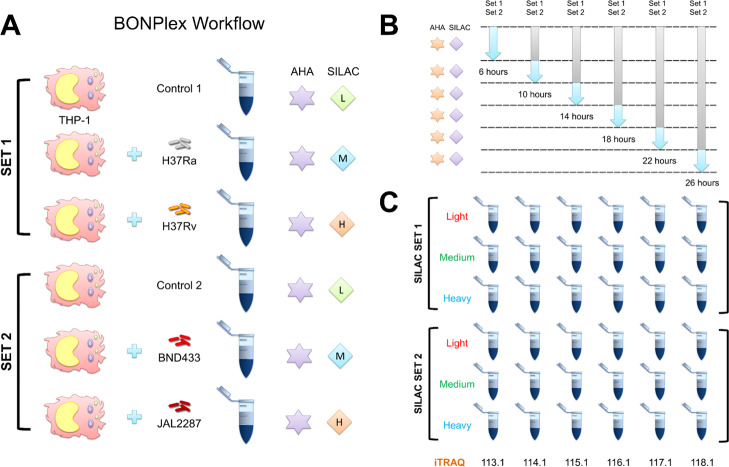
BONPlex study was designed to investigate the effect of mycobacterial infection (clinical and laboratory strains) on the human macrophage secretome. In this study, BONCAT was used to study the newly translated proteins secreted by THP-1 human macrophages after mycobacterial infection (Figure 3A). The second technique, SILAC was used as a pulsed medium and heavy labels to tag the THP-1 cells infected with different mycobacterial strains namely H37Ra (medium) and H37Rv (heavy) in one set, while BND433 (medium) and JAL2287 (heavy) in another, each with their respective light SILAC uninfected controls. The pulsed SILAC allowed selective labeling of newly synthesized proteins in a time-window postinfection, while AHA allowed their selective enrichment (Figure 3B). Although the light channel contains both pre-existing as well as newly synthesized proteins, which would be otherwise indistinguishable, the BONCAT labels allows for their selective enrichment, removing this quantitation bias. The third technique is isobaric quantitation with iTRAQ used to label six time points of each infection in the THP-1 cells (Figure 3C) from 6 to 26 h at 4 h intervals. The labeled and digested secretome from the light, medium, and heavy SILAC, each with 6-plex iTRAQ labeling for time points were mixed, separated by LC, and analyzed by tandem MS. MaxQuant was used to identify the spectra while the in-house developed tool QuantWizIQ was used to calculate iTRAQ areas for each time point. The HyperQuant tool was used to integrate the data from identification and quantitation followed by replicate combination and calculation of ratios from the areas.
Figure 3.
Overview of the BONPlex workflow and labeling. (A) Workflow comprises selective enrichment of newly synthesized proteins with BONCAT (AHA) and pulsed SILAC labeling. Two sets of SILAC were used to label THP-1 cells infected with different mycobacterial strains. (B) SILAC and AHA labels were incorporated (after 1 h of methionine, lysine, and arginine depletion) in specific time windows postinfection. (C) Cells were harvested, proteins were digested, and time points were labeled with iTRAQ reagents as shown. The supernatant was selected for LC–MS/MS, and the 18-plexed data generated from each run was analyzed using the HyperQuant pipeline.
NSS Analysis for THP-1 Cells
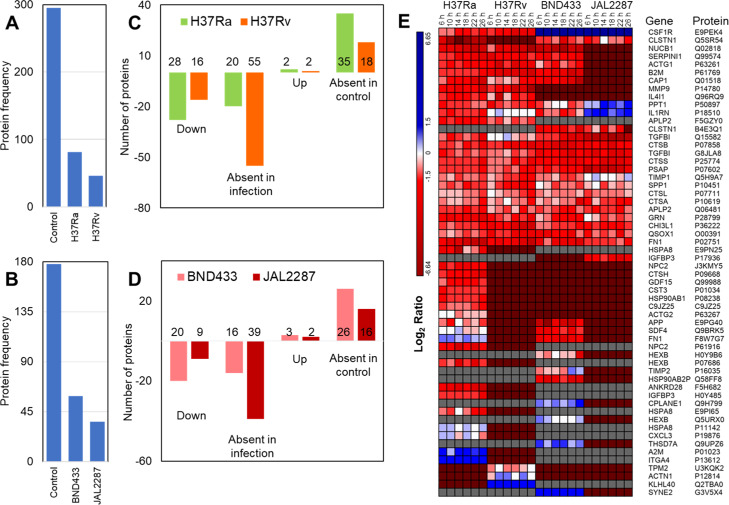
After removing contaminants and decoy proteins, we identified and quantified 454 newly synthesized secretome (NSS) proteins using BONPlex in both the experiments combined together. We observed 343 proteins in set 1 with laboratory strains and 213 proteins in set 2 with clinical strains (Table S2). We observed that the number of secreted proteins in infected cells reduces considerably as compared to uninfected cells in both the sets (Figure 4A,B). We found that 81, 46, 59, and 36 proteins were secreted in response to infection with H37Ra (avirulent), H37Rv (virulent), BND433 (clinical virulent), and JAL2287 (clinical virulent) respectively, which are hereafter referred to as Ra, Rv, BND, and JAL for the sake of brevity. We compared the proteins identified in uninfected and infected THP-1 cells in Set 1 (Figure 4A) and Set 2 (Figure 4B). We observed that majority of the proteins expressed in the control (246 in set 1 and 144 in set 2) are not expressed in infection hinting at suppression of the cellular function, as also observed in Figure S4A,B. Some proteins are expressed exclusively in infected cells (34, 17, 24, and 15 proteins in Ra, Rv, BND, and JAL, respectively, Figure S4A,B). We then combined the NSS proteins in virulent strains (Rv, BND, and JAL) for comparison against the avirulent Ra as well as the controls from both the sets (Figure S4C). We observed 27 and 48 proteins to be exclusive to avirulent and virulent strains, respectively. By observing the quantitation of proteins identified, we found that the majority of proteins secreted in different infections are downregulated (Figure 4C, down + absent in infection & Figure S5A,B). There are some proteins specifically expressed only under infection conditions (Figure 4D, up + absent in the control, Figure S6A,B), hinting at infection specific cellular response (Figure 4E). The proteins absent in any of the infection were not considered in this analysis. The macrophages secrete these proteins to probably aid in the initiation of an immune response. Failure to initiate an immune response leads to apoptosis or necrosis.30 The mycobacteria, on the other hand, tries to block the signals that can lead to immune response or tries to force production of anti-inflammatory proteins which may be a fail-safe mechanism in case some inflammatory signals leak out to the extracellular region.31 In order to identify proteins that can mount an immune response, we queried UniProt for specific immunomodulatory keywords. We found 9 chemokines, 43 cytokines, 11 laminins, 14 proteases, 6 matrix metalloproteases, and 3 caspases. We also observed that 275 proteins identified were involved in signaling mechanisms. As we were studying macrophages responses, we also observed proteins involved in bacterial response (6 proteins), immune response (55 proteins), or inflammatory response (48 proteins) in our keyword searches (Tables S1 and S3).
Figure 4.
Overview of NSS proteins identified for each strain. (A,B) represent the number of NSS proteins observed in the experiment with laboratory (A) and clinical (B) strain infections respectively. (C,D) depict the number of up- and downregulated proteins along with the unique proteins in either the infection or the control. (E) Heatmap showing the strain specific expression of proteins observed in uninfected and infected cells. The proteins expressed exclusively in control but not quantitated in any of the infections were excluded from this analysis.
Common NSS Proteins in All Four Infections
We observed sixteen proteins expressed in all four infections. Except one (CSF1R was absent in control of set 2), all of the proteins were observed in control as well as in infections. CSF1R is the macrophage colony stimulating factor-1 receptor that acts as a cell surface receptor for CSF1 and IL34 and is important for release of proinflammatory chemokines.32 It is a tyrosine kinase that activates the signaling for MAPK, STAT3, and PIP3 pathways.33 It was downregulated in Ra, Rv, and BND infections but was upregulated in initial time points of JAL infection, which later was downregulated by the last time point (Figure S7).
Among the other 15 proteins (Figure S7), all proteins were either downregulated or unchanged in all infections except for IL1RN and PPT1 in JAL. IL1RN is the receptor antagonist of IL1 that inhibits the binding of IL1 (α and β) to its receptor and reduces the inflammatory response. In unstimulated macrophages, it is expressed to prevent any unwanted inflammatory response due to nonspecific stimuli.34 During Ra infections, it gets highly downregulated in later time points as the macrophage exhibits delayed immune response and increased production of IL1β to contain Ra bacilli. In Rv infection, however, the secretion levels kept increasing in later time points, although it does not qualify as differentially expressed at our threshold criteria (±2 fold change). During BND infection, it remains downregulated in the secretome for the first few time points, while gradually increasing in the last time point. In JAL infection, it was overexpressed consistently, demonstrating its heightened capability to suppress the immune response. PPT1 is a thioesterase responsible for negative regulation of apoptotic processes in infections. It removes protein palmitoylation, thus regulating the cellular localization of substrates to cytosol.35 It was significantly downregulated only in one time point (6 h) in Ra. In Rv, the expression was initially low which then increased to normal levels by 14 h and dropped back to lower levels again by 26 h. In BND infection, the secretion behavior was similar to Ra infection, which was under expressed only in one time point. However, it was significantly upregulated in JAL infection.
Immune Regulatory Processes
We searched the immune-related pathways in UniProt and reactome to identify proteins observed in specific immune processes in our data. The common defense mechanisms during infections are cytokine production and secretion, interleukin production, inflammation, antigen processing and presentation, complement cascade, apoptosis, and autophagy. Several pathways lead to regulation of one or more defense response pathways that are observed in this experiment. The detailed integration of these pathways with the NSS proteins and how they may be regulated in the infections is shown in Figure 5, Table S3, and Figure S8. The most common pathway observed during a bacterial infection is toll-like receptor (TLR) signaling which leads to activation of the MYD88 pathway for caspase activation.36,37 CD14 is the protein that activates TLR4 for the MYD88 pathway. However, we did not observe CD14 in infection, although it was observed in uninfected cells.
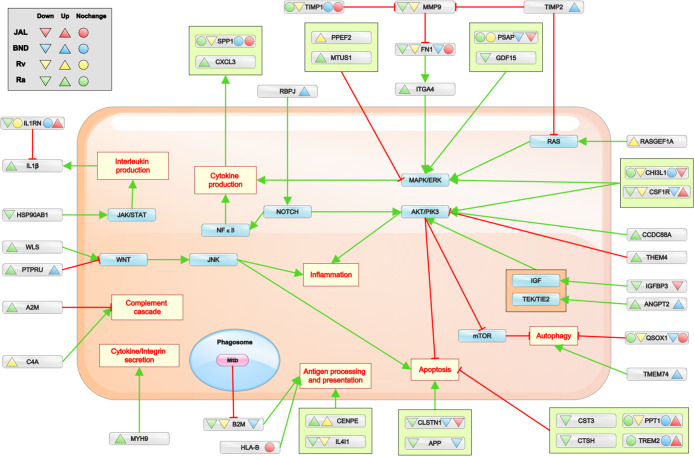
Figure 5.
Overview summary of the important NSS proteins observed in immune pathways. The common pathways regulated during immune response and the proteins observed in the secretome for the infected cells. Proteins involved in immune pathways but only observed in uninfected cells are not represented here. The green arrows represent the activation of a process by the protein, and the red arrow represents the inhibition. While the upright triangle denotes overexpression, the inverted triangle represents suppression, and the circle denotes no noticeable change in secretome. The proteins identified in only specific pathways in UniProt keyword search are represented here. The overall expression of NSS proteins for considering either up- or downregulated depends on its expression in more than three time points. The NSS proteins with no change of expression means the change is not significant enough in three or more time points to be considered as up- or downregulated.
MAPK Pathway
MAPK cascade helps in cytokine production during infection and also aids in caspase activation.38 There are several activators of MAPK cascade such as PSAP, CHI3L1, and CSF1R expressed in all four infections, but all underexpressed except CSF1R in JAL infection, indicating the bacterial control in restricting MAPK signaling. We also observed certain proteins such as GDF15 (down) and ITGA4 (up) that could activate the MAPK pathway in avirulent infection. MTUS1 was also upregulated in avirulent infection, which blocks activation of the MAPK pathway, displaying a dual control on infected cells (one by macrophages and other by infecting bacteria). PPEF2 had upregulated secretion in Rv infection which blocks MAPK, while RASGEF1A was also upregulated which activates RAS and thus MAPK indicating the dual control. In BND, RBPJ was secreted which activates the NOTCH pathway, which in turn activates MAPK and NFκB pathways. This interaction could produce SPP1 (in all infections) and CXCL3 (in Ra infection) cytokines.
AKT/PI3K Pathway
The AKT/PI3K pathway activation leads to downstream regulation of multiple immune response pathways.39 It aids in transcription of inflammation pathway proteins. It directly inhibits apoptosis processes and inhibits the mTOR pathway. The mTOR pathway inhibits autophagy processes, and in its absence, autophagy gets activated. Because the avirulent strain will not be able to block apoptosis of cells to evade infection, many proteins were observed in the secretome of avirulent infected cells to activate the AKT pathway such as CCDC88A, IGFBP3, and ANGPT2. AKT itself is observed in the secretome of the uninfected cells required to activate growth response pathways. THEM4 is observed in avirulent strain infection, which is a known inhibitor of the AKT pathway. APP and CLSTN1 (both down) were observed in avirulent and clinical infection that leads to the activation of apoptotic pathways. Also, the inhibitors of the apoptosis pathway—CST3, CTSH, TREM2, and PPT1—were observed in NSS proteins (Figure 5). QSOX1 is a known inhibitor of the autophagy process identified to be downregulated in all infections. However, in BND infection, TMEM74 is secreted out which helps in the activation of the autophagy process.
Complement Cascade, Antigen Processing, and Presentation
Complement cascade leads to cell lysis, opsonization, and inflammatory response.40 We observed A2M (up) in avirulent and C4A (up) in Rv infections that assist in regulation of the complement cascade. Interestingly, there was no known protein observed in clinical strain infections. We observed B2M (down) in Ra, Rv, and BND infections, which is known for regulating antigen processing and presentation in macrophages. B2M is a known target of the mycobacterial ESX secretion system, which aims to degrade B2M in the ER, so that it cannot be secreted out for antigen presentation.41,42 We also observed HLA-B (JAL), IL4I1, and CENPE (Ra, Rv) that are part of antigen processing and presentation.
Of the four strains, Ra is not able to block the necessary pathways of immune response. IL1β was observed in Ra infection that leads to caspase activation and thus apoptosis. In Rv infection, we observed proteins pertaining to complement cascade, MAPK pathways, antigen processing and presentation, and so forth that could lead to an immune response. In BND infection, proteins that activate NOTCH and autophagy pathways were observed. In JAL infection, which is the most aggressive virulent strain among the four, upregulated NSS proteins were observed in inhibitory pathways of apoptosis and IL1β with only one exception of CSF1R that activated the MAPK pathway. Even the antigen processing or cytokines expressed showed no significant change in secretion, indicating its competence in molding the immune response to suit its purpose.
Because the aim of this study is to demonstrate the utility of the HyperQuant pipeline in analyzing the HOM study designs, these examples highlight the advantages of using the tool for analysis of complex data. Several interesting hypothesis could be generated for further studies from this data. While we were able to observe many immune response proteins, we also observed cytoskeletal proteins, kinases, phosphatases, thiols, and proteases required for housekeeping functions. Although this analysis was aimed as the proof of concept for the tool being able to analyze a truly HOM study with 18 different conditions in one run, several immunomodulatory pathways were observed which can be a highly useful resource for the TB community.
Conclusions
Several higher order multiplexing studies have been applied to interesting biological problems over the past decade.1,10 The advances in HOM techniques, however, outpaced the associated computational developments for analysis, which prevented its widespread adoption. Recently, the number of HOM studies is increasing rapidly necessitating the need for computational tools that allow labs without computational expertise to be able to exploit this technique.
With the development of HyperQuant, we aim to fill this gap and facilitate easy analysis of data from HOM studies, allowing a true hyperplexing capacity for quantitation. We have also shown its utility by reanalyzing 18-plex data from hyperplexing study and BONPlex study. Summarizing the individual label combinations using areas or ratios, HyperQuant allows flexible analysis designs from any type of HOM data. With increased multiplexing from novel HOM studies designs, HyperQuant is already suited to analyze the data beyond 18-plex. For examples of possible multiplexing using HOM, the readers are referred to Table 1 of Aggarwal et al. 2019.10 The tool is open source, platform independent, freely available, and allows flexibility in analysis of complex HOM datasets for diverse experimental designs. By exploiting the HOM technique, including replicates and/or biological conditions in the same run, better data fidelity and robustness can be achieved that is usually lacking in most underpowered studies. To the best of our knowledge, this is the first and only tool available for analyses of HOM data. This will enable researchers to adopt HOM techniques to plan interesting studies without worrying about complicated custom data analysis pipelines.
Methods
BONPlex Dataset (PXD004281)
The data was searched against the UniProt human database (downloaded on 7th June 2014)with cRAP proteins (https://www.thegpm.org/crap) (total 65 745 sequences) and MaxQuant (1.5.0.30) added contaminants, searched using MaxQuant43 with carbamidomethylation and iTRAQ (N-term) as fixed modifications. Two new modifications were created in the MaxQuant parameter dictionary for the combined masses of iTRAQ and SILAC (medium and heavy separately) on lysine residues. SILAC pipeline search was used with SILAC light as the mass of iTRAQ, while SILAC-medium and SILAC-heavy as the new modifications created on lysine. The SILAC masses on arginine residues were 0, 6, and 10 Da, respectively, for light, medium, and heavy. BONCAT at methionine residues, deamidation of asparagine and glutamine, and methionine oxidation were used as variable modifications. The enzyme specificity was trypsin, while missed cleavage was set to 1. The data was identified at ≤1% FDR for PSMs and proteins. The precursor and fragment mass tolerances were specified as defaults for the AB Sciex 5600 instrument in MaxQuant. For isobaric quantitation, the mgf files were converted using msconvert and provided as an input to an in-house tool QuantWizIQ (https://sourceforge.net/projects/quantwiz/files/IQ) for iTRAQ quantitation (described later), using the sum intensity method, as described in Aggarwal et al. 2016.7 Spectra with the reporter ion intensity below 20 were filtered out, and the sum intensity-based method was used to calculate the iTRAQ area. The quantitation results thus obtained were merged with identifications from MaxQuant, using HyperQuant Mapper, and the normalized protein intensities were summarized using the median intensity method with the HyperQuant Combiner. The results are provided in table S2.
Hyperplexing Dataset (Dephoure and Gygi)
The 18-plex data from the hyperplexing study (kindly provided by N. Dephoure and S. Gygi) were used for benchmarking of the HyperQuant tool.18 The data in Thermo RAW format were searched using MaxQuant with search parameters followed as closely as possible to those stated in the article. The data was searched with the triplex SILAC mode in MaxQuant with carbamidomethylation and TMT at the N-term as fixed modification. The SILAC labels were redefined in the dictionary for Lysine labels as light + TMT, medium + TMT, and heavy + TMT while arginine labels were used as is. The data were filtered at ≤1% FDR. The RAW files from the 18-plex experiment were converted to mzML format using msconvert. The PSM quantitation from the mzML files was carried out using QuantWizIQ software using the sum intensity method, as described in Aggarwal et al. 2016.7 The HyperQuant mapper module combined the PSM identifications from MaxQuant with their corresponding quantitation from QuantWizIQ. The HyperQuant combiner module was used to integrate replicates and output raw intensities values from which the protein ratios were calculated using the weighted average method. The protein list and the ratios thus obtained were compared with the results from hyperplexing study.
Reporter Quantitation Using the Isobaric Quantitator (QuantWizIQ)
QuantWizIQ is an isobaric quantitation tool developed in-house for spectra level quantitation from the HUPO-PSI standard mzML data format as well the de facto standard-mascot generic format (MGF).51 It performs fast iTRAQ/TMT-labeled spectra quantitation with low memory footprints. It allows isobaric quantitation from 4/8-plex iTRAQ and 2/6/10/16-plex TMT labels. The tool also provides information of MS3 scans in case of TMT labels which HyperQuant can use later for merging identification and quantitation. It provides users with flexibility of the area method to perform quantitation. It also provides users with option of intensity normalization and purity correction. It is a platform independent, command line tool developed in Perl and freely available for noncommercial use. The details of the software can be found in the detailed documentation along with the example data at https://sourceforge.net/projects/quantwiz/files/IQ/. It is also available through Bioconda44 (https://anaconda.org/bioconda/quantwiz-iq) and Galaxy-P45 (http://galaxyp.org/) (courtesy galaxy-P team).
Protein Quantitation
For integrating the identification and quantitation results individually for each label combination and calculation of protein ratios, we developed a command-line tool HyperQuant in the Perl programming language. HyperQuant takes identification results along with MS/MS quantitation results as input and provides a list of protein areas after replicate combination. As an example, the MaxQuant text files were used to select PSM identification results. For PSMs identified and passed at ≤1% FDR threshold, quantitation values were extracted from QuantWizIQ results for corresponding spectra. If no PSM is quantified for a protein, it is reported in a separate file at this step. This module of the tool is called the Mapper, which maps the identification and quantitation results. The Mapper results for all the replicates or a single file (in case of no replicates) are subsequently provided to another module called the Combiner. The Combiner creates a dictionary (hash) of each protein group specified by MaxQuant as a key and information about all the PSMs in that group as the value. If a protein group is a subset of another protein group from other replicate(s), the two groups are combined and the PSMs are merged for these groups as one group (Note S1 and Figure S1). The quantitation values for each label combination are processed separately (Figure S2). For example, the intensity values for all the PSMs with the 113-iTRAQ reporter of SILAC light are used to calculate the standard deviation and values outside ±2 standard deviations range is discarded as outlier. This step of outlier removal ensures that during combination any extreme value will be removed (Note S2). The median intensity of the remaining values is used as the protein intensity for that sample. This enables normalization of the PSM intensities to be used for the protein-normalized intensity. The users can also select peptide ratios of isobaric labels to summarize protein ratios instead of normalized intensities. There are three methods available to the user for summarizing protein intensity/ratios that are median, average, and weighted average methods (Note S3). The results are reported with details of scan count, unique peptide count, protein length, and replicates in which the particular labels were observed along with their calculated quantitation errors. The postsearch filters on the number of peptides/replicates for each protein can be used as per the experimental design or user choice. This allows flexibility of the experimental design as the decoupling of analysis intricacies now allows innovative ways to analyze data postquantitative integration, depending on whether the labels were used as replicates. A demonstration of quantitative processing is shown in Figure S2. The tool is platform independent, open source, and freely available software for the research community, and it can be downloaded from https://sourceforge.net/projects/hyper-quant/files/ along with the example data and detailed documentation enumerating all steps for analysis.
Ratio Calculation
For BONPlex study, the ratio is calculated separately for infection by different strains, as well as temporally, for further analysis (Figure S3). The log2 fold change was calculated for labeled protein with respect to corresponding unlabeled counterparts at a given time point for studying the strain-specific variations in THP-1 responses to infection.
Some proteins were only observed in infected cells. To observe changes in these proteins, iTRAQ ratios were calculated for all time points with respect to the first one. The significance threshold chosen was ±2 fold change, that is, ±1 at log2 scale (also see Note S4 and Table S3). All absent and zero values (identified in any one channel) were converted to −6.64, (log2 of 0.01, minimum quantified value) which denoted an infinitely small value or 6.64 for highly overexpressed or only expressed in infection conditions (log2 of 100, maximum quantified value).
For hyperplexing data, the peptide ratios were used for calculating the weighted average protein ratio, as described earlier.8 The following formula was used:
where, N represents the total number of quantitated peptides for the protein.
The protein ratios were calculated from TMT labels, as the SILAC labels were used as biological replicates in this experiment. The results provided as the Supporting Information file by the authors was used for comparison with our results.
Gene Ontology/Keyword Search/Pathway Analysis
For BONPlex data, we used the peptide cut-off of ≥3 in individual conditions/biological samples and used the GeneCodis server for gene ontology46 (Table S3). The data was filtered at ≤1% Bonferroni FDR. Because the study was on immune cells, several keywords based on immune response as elucidated in the literature,26−28,47,48 were searched in UniProt49 to the fetch list of proteins. The lists were then compared to the proteins identified in this experiment. The lists were generated using the following keywords: cytokine, chemokine, interferon, interleukin, defense response, caspase, growth factor, signaling, secretory, proteases, and matrix metalloproteases26,27 (Tables S1 and S3). The proteins associated with immune response pathways were also queried in UniProt to use the resulting list to match with strain specific changes in proteins. These pathways included TLR-4, MAPK, AKT/PI3K, NOTCH, JAK/STAT, WNT, RAS, TEK/TIE2, IGF, and NF-κB pathways (Table S3). We also fetched gene ontology categories pertaining to—defense to bacterial response, inflammation, complement cascade, cytokine production, interleukin secretion, antigen processing and presentation, apoptosis, and autophagy pathways as suggested in the cited literature.26−28,47,48 The proteins were queried in the reactome50 database, and the proteins related to immune response pathways were observed (Figure S8).
Acknowledgments
S.A. is supported by the ICMR-SRF (BIC/11/(17)/2015) grant, A.K.Y. is supported by DBT-IYBA (D.O. no. BT/07/IYBA/2013), DBT-Big Data Initiative grant (BT/PR16456/BID/7/624/2016), and N.C.T. is supported by DBT Twinning NER (102/IFD/SAN/1048/2016-2017 dated 14/06/2016 serial no. 6-17). Authors would like to thank the Galaxy-P team, especially Pratik Jagtap and Praveen Kumar for making QuantWizIQ available in galaxy toolshed and Bioconda.
Glossary
Abbreviations
- BONCAT
bioorthogonal non-canonical amino acid tagging
- pSILAC
pulsed stable isotope labeling by amino acids in cell culture
- iTRAQ
isobaric tag for relative and absolute quantitation
- TMT
tandem mass tag
- Mtb
Mycobacterium tuberculosis
- NSS
newly synthesized and secreted
- MS
mass spectrometry
- LC
liquid chromatography
- HOM
higher order multiplexing
- AHA
azido homoalanine
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c00515.
Delayed protein summarization, Venn diagram, comparison of two methods for light, medium, and heavy labels, flowchart demonstrating the steps taken to calculate ratios for one protein expressed in four conditions, ratio calculation method for the BONPlex study, comparison of NSS proteins, heat maps, temporal expression of 16 NSS proteins common to all infections, overview of NSS proteins involved in various immune response pathways through Reactome analysis, NSS proteins involved in major pathways, number of proteins involved in the respective immune pathways for the infected macrophages (but not control), protein summarization for quantitation, statistical analysis BONPlex data., table of keywords representing the number of proteins found for each keyword, and details of protein names (sheet—named keywords) (PDF)
Set 1 and Set 2 protein quantitation values as obtained in the BONPlex experiment (XLSX)
Keyword search results, pathways, GO annotations of Set 1 and Set 2, and t-test results from Set 1 and Set 2 (XLSX)
Author Present Address
∥ (M.K.M.) Institute for Systems Biology, Seattle, WA 98109, United States.
Author Contributions
A.K.Y., S.J., A.K., and M.K.M. contributed to design of the study. S.J., A.K. and M.K.M. performed the experiments. S.A. and A.K.Y. wrote the code and performed computational analysis and visualization. S.A. wrote the first draft with inputs from all other authors. N.C.T., S.J., A.K., S.A., and A.K.Y. all contributed to data analysis, writing, and editing the manuscript. A.K.Y. acquired the funding, managed, and coordinated research planning and execution.
The authors declare no competing financial interest.
Supplementary Material
References
- Arul A. B.; Robinson R. A. S. Sample Multiplexing Strategies in Quantitative Proteomics. Anal. Chem. 2018, 91, 178. 10.1021/acs.analchem.8b05626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M.; Schirle M.; Sweetman G.; Rick J.; Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007, 389, 1017–1031. 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Ong S.-E.; Blagoev B.; Kratchmarova I.; Kristensen D. B.; Steen H.; Pandey A.; Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 1, 376–386. 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Aggarwal S.; Yadav A. K. False Discovery Rate Estimation in Proteomics. Methods Mol. Biol. 2016, 1362, 119–128. 10.1007/978-1-4939-3106-4_7. [DOI] [PubMed] [Google Scholar]
- Ross P. L.; Huang Y. N.; Marchese J. N.; Williamson B.; Parker K.; Hattan S.; Khainovski N.; Pillai S.; Dey S.; Daniels S.; Purkayastha S.; Juhasz P.; Martin S.; Bartlet-Jones M.; He F.; Jacobson A.; Pappin D. J. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 2004, 3, 1154–1169. 10.1074/mcp.m400129-mcp200. [DOI] [PubMed] [Google Scholar]
- Thompson A.; Schäfer J.; Kuhn K.; Kienle S.; Schwarz J.; Schmidt G.; Neumann T.; Hamon C.; Mohammed A. K.; Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003, 75, 1895–1904. 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Aggarwal S.; Yadav A. K. Dissecting the iTRAQ Data Analysis. Methods Mol. Biol. 2016, 1362, 277–291. 10.1007/978-1-4939-3106-4_18. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Jamwal S.; Midha M. K.; Hamza B.; Aggarwal S.; Yadav A. K.; Rao K. V. S. Dataset generated using hyperplexing and click chemistry to monitor temporal dynamics of newly synthesized macrophage secretome post infection by mycobacterial strains. Data in Brief 2016, 9, 349–354. 10.1016/j.dib.2016.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultin-Rosenberg L.; Forshed J.; Branca R. M. M.; Lehtiö J.; Johansson H. J. Defining, comparing, and improving iTRAQ quantification in mass spectrometry proteomics data. Mol. Cell. Proteomics 2013, 12, 2021–2031. 10.1074/mcp.m112.021592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röst H. L.; Malmström L.; Aebersold R. Reproducible quantitative proteotype data matrices for systems biology. Mol. Biol. Cell 2015, 26, 3926–3931. 10.1091/mbc.e15-07-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal S.; Talukdar N. C.; Yadav A. K. Advances in Higher Order Multiplexing Techniques in Proteomics. J. Proteome Res. 2019, 18, 2360–2369. 10.1021/acs.jproteome.9b00228. [DOI] [PubMed] [Google Scholar]
- Everley R. A.; Kunz R. C.; McAllister F. E.; Gygi S. P. Increasing throughput in targeted proteomics assays: 54-plex quantitation in a single mass spectrometry run. Anal. Chem. 2013, 85, 5340–5346. 10.1021/ac400845e. [DOI] [PubMed] [Google Scholar]
- Jayapal K. P.; Sui S.; Philp R. J.; Kok Y.-J.; Yap M. G. S.; Griffin T. J.; Hu W.-S. Multitagging proteomic strategy to estimate protein turnover rates in dynamic systems. J. Proteome Res. 2010, 9, 2087–2097. 10.1021/pr9007738. [DOI] [PubMed] [Google Scholar]
- Evans A. R.; Robinson R. A. S. Global combined precursor isotopic labeling and isobaric tagging (cPILOT) approach with selective MS(3) acquisition. Proteomics 2013, 13, 3267–3272. 10.1002/pmic.201300198. [DOI] [PubMed] [Google Scholar]
- Gu L.; Evans A. R.; Robinson R. A. S. Sample multiplexing with cysteine-selective approaches: cysDML and cPILOT. J. Am. Soc. Mass Spectrom. 2015, 26, 615–630. 10.1007/s13361-014-1059-9. [DOI] [PubMed] [Google Scholar]
- Evans A. R.; Gu L.; Guerrero R. Jr.; Robinson R. A. S. Global cPILOT analysis of the APP/PS-1 mouse liver proteome. Proteomics: Clin. Appl. 2015, 9, 872–884. 10.1002/prca.201400149. [DOI] [PubMed] [Google Scholar]
- Gu L.; Robinson R. A. S. High-throughput endogenous measurement of S-nitrosylation in Alzheimer’s disease using oxidized cysteine-selective cPILOT. Analyst 2016, 141, 3904–3915. 10.1039/c6an00417b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C. D.; Dudenhoeffer J. D.; Gu L.; Evans A. R.; Robinson R. A. S. Enhanced Sample Multiplexing of Tissues Using Combined Precursor Isotopic Labeling and Isobaric Tagging (cPILOT). J. Visualized Exp. 2017, 123, 55406. 10.3791/55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N.; Gygi S. P. Hyperplexing: a method for higher-order multiplexed quantitative proteomics provides a map of the dynamic response to rapamycin in yeast. Sci. Signaling 2012, 5, rs2. 10.1126/scisignal.2002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlage P.; Kockmann T.; Kizhakkedathu J. N.; auf dem Keller U. Monitoring matrix metalloproteinase activity at the epidermal-dermal interface by SILAC-iTRAQ-TAILS. Proteomics 2015, 15, 2491–2502. 10.1002/pmic.201400627. [DOI] [PubMed] [Google Scholar]
- Welle K. A.; Zhang T.; Hryhorenko J. R.; Shen S.; Qu J.; Ghaemmaghami S. Time-resolved Analysis of Proteome Dynamics by Tandem Mass Tags and Stable Isotope Labeling in Cell Culture (TMT-SILAC) Hyperplexing. Mol. Cell. Proteomics 2016, 15, 3551–3563. 10.1074/mcp.m116.063230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecha J.; Meng C.; Zolg D. P.; Samaras P.; Wilhelm M.; Kuster B. Peptide Level Turnover Measurements Enable the Study of Proteoform Dynamics. Mol. Cell. Proteomics 2018, 17, 974–992. 10.1074/mcp.ra118.000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg D. A.; Taliaferro J. M.; Huber S. M.; Begley T. J.; Dedon P. C.; White F. M. A Proteomics Approach to Profiling the Temporal Translational Response to Stress and Growth. iScience 2018, 9, 367–381. 10.1016/j.isci.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski M. M.; Zinn N.; Faelth-Savitski M.; Poeckel D.; Gade S.; Becher I.; Muelbaier M.; Wagner A. J.; Strohmer K.; Werner T.; Melchert S.; Petretich M.; Rutkowska A.; Vappiani J.; Franken H.; Steidel M.; Sweetman G. M.; Gilan O.; Lam E. Y. N.; Dawson M. A.; Prinjha R. K.; Grandi P.; Bergamini G.; Bantscheff M. Multiplexed Proteome Dynamics Profiling Reveals Mechanisms Controlling Protein Homeostasis. Cell 2018, 173, 260–274.e25. 10.1016/j.cell.2018.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalarski C. E.; Elias J. E.; Villén J.; Haas W.; Gerber S. A.; Everley P. A.; Gygi S. P. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J. Proteome Res. 2008, 7, 4756–4765. 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamval S.; Aggarwal S.; Yadav A. K.; Subba Rao K. V.; Kumar A.. Method of Hyperplexing in Mass Spectrometry to Elucidate Temporal Dynamics of Proteome. India Patent IN 201611029904, 2016.
- Eichelbaum K.; Winter M.; Diaz M. B.; Herzig S.; Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat. Biotechnol. 2012, 30, 984–990. 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- Eichelbaum K.; Krijgsveld J. Rapid temporal dynamics of transcription, protein synthesis, and secretion during macrophage activation. Mol. Cell. Proteomics 2014, 13, 792–810. 10.1074/mcp.m113.030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. M.; Koppenol-Raab M.; Kuriakose M.; Manes N. P.; Goodlett D. R.; Nita-Lazar A. Host-pathogen dynamics through targeted secretome analysis of stimulated macrophages. J. Proteomics 2018, 189, 34–38. 10.1016/j.jprot.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. R. 3rd; Morgan S. F.; Gatlin C. L.; Griffin P. R.; Eng J. K. Method to compare collision-induced dissociation spectra of peptides: potential for library searching and subtractive analysis. Anal. Chem. 1998, 70, 3557–3565. 10.1021/ac980122y. [DOI] [PubMed] [Google Scholar]
- Schluger N. W.; Rom W. N. The host immune response to tuberculosis. Am. J. Respir. Crit. Care Med. 1998, 157, 679–691. 10.1164/ajrccm.157.3.9708002. [DOI] [PubMed] [Google Scholar]
- Hare N. J.; Chan B.; Chan E.; Kaufman K. L.; Britton W. J.; Saunders B. M. Microparticles released from Mycobacterium tuberculosis-infected human macrophages contain increased levels of the type I interferon inducible proteins including ISG15. Proteomics 2015, 15, 3020–3029. 10.1002/pmic.201400610. [DOI] [PubMed] [Google Scholar]
- Lin H.; Lee E.; Hestir K.; Leo C.; Huang M.; Bosch E.; Halenbeck R.; Wu G.; Zhou A.; Behrens D.; Hollenbaugh D.; Linnemann T.; Qin M.; Wong J.; Chu K.; Doberstein S. K.; Williams L. T. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science 2008, 320, 807–811. 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- Chihara T.; Suzu S.; Hassan R.; Chutiwitoonchai N.; Hiyoshi M.; Motoyoshi K.; Kimura F.; Okada S. IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ. 2010, 17, 1917–1927. 10.1038/cdd.2010.60. [DOI] [PubMed] [Google Scholar]
- Witkin S. S.; Gerber S.; Ledger W. J. Influence of interleukin-1 receptor antagonist gene polymorphism on disease. Clin. Infect. Dis. 2002, 34, 204–209. 10.1086/338261. [DOI] [PubMed] [Google Scholar]
- Sobocinska J.; Roszczenko-Jasinska P.; Ciesielska A.; Kwiatkowska K. Protein Palmitoylation and Its Role in Bacterial and Viral Infections. Front. Immunol. 2017, 8, 2003. 10.3389/fimmu.2017.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A.; Inagawa H.; Kohchi C.; Nishizawa T.; Soma G. The role of toll-like receptor 2 in survival strategies of Mycobacterium tuberculosis in macrophage phagosomes. Anticancer Res. 2009, 29, 907–910. [PubMed] [Google Scholar]
- Noss E. H.; Pai R. K.; Sellati T. J.; Radolf J. D.; Belisle J.; Golenbock D. T.; Boom W. H.; Harding C. V. Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19-kDa lipoprotein of Mycobacterium tuberculosis. J. Immunol. 2001, 167, 910–918. 10.4049/jimmunol.167.2.910. [DOI] [PubMed] [Google Scholar]
- Galán J. E.; Bliska J. B. Cross-talk between bacterial pathogens and their host cells. Annu. Rev. Cell Dev. Biol. 1996, 12, 221–255. 10.1146/annurev.cellbio.12.1.221. [DOI] [PubMed] [Google Scholar]
- Ambesi A.; McKeown-Longo P. J. Anastellin, the angiostatic fibronectin peptide, is a selective inhibitor of lysophospholipid signaling. Mol. Cancer Res. 2009, 7, 255–265. 10.1158/1541-7786.mcr-08-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Crevel R.; Ottenhoff T. H. M.; van der Meer J. W. M. Innate immunity to Mycobacterium tuberculosis. Adv. Exp. Med. Biol. 2003, 531, 241–247. 10.1007/978-1-4615-0059-9_20. [DOI] [PubMed] [Google Scholar]
- Sreejit G.; Ahmed A.; Parveen N.; Jha V.; Valluri V. L.; Ghosh S.; Mukhopadhyay S. The ESAT-6 Protein of Mycobacterium tuberculosis Interacts with Beta-2-Microglobulin (β2M) Affecting Antigen Presentation Function of Macrophage. PLoS Pathog. 2014, 10, e1004446 10.1371/journal.ppat.1004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghatoleslam A.; Hemmati M.; Ebadat S.; Movahedi B.; Mostafavi-Pour Z. Macrophage Immune Response Suppression by Recombinant Mycobacterium tuberculosis Antigens, the ESAT-6, CFP-10, and ESAT-6/CFP-10 Fusion Proteins. Iran. J. Med. Sci. 2016, 41, 296–304. [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Grüning B.; Lingua::EN::Titlecase R.; Dale R.; Sjödin A.; Chapman B. A.; Rowe J.; Tomkins-Tinch C. H.; Valieris R.; Köster J. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods 2018, 15, 475–476. 10.1038/s41592-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap P. D.; Johnson J. E.; Onsongo G.; Sadler F. W.; Murray K.; Wang Y.; Shenykman G. M.; Bandhakavi S.; Smith L. M.; Griffin T. J. Flexible and accessible workflows for improved proteogenomic analysis using the Galaxy framework. J. Proteome Res. 2014, 13, 5898–5908. 10.1021/pr500812t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas-Madrid D.; Nogales-Cadenas R.; Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012, 40, W478–W483. 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann J. C.; Geiger R.; Hornburg D.; Wolf T.; Kveler K.; Jarrossay D.; Sallusto F.; Shen-Orr S. S.; Lanzavecchia A.; Mann M.; Meissner F. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 2017, 18, 583–593. 10.1038/ni.3693. [DOI] [PubMed] [Google Scholar]
- Meissner F.; Scheltema R. A.; Mollenkopf H.-J.; Mann M. Direct proteomic quantification of the secretome of activated immune cells. Science 2013, 340, 475–478. 10.1126/science.1232578. [DOI] [PubMed] [Google Scholar]
- Apweiler R.; Bairoch A.; Wu C. H.; Barker W. C.; Boeckmann B.; Ferro S.; Gasteiger E.; Huang H.; Lopez R.; Magrane M.; Martin M. J.; Natale D. A.; O’Donovan C.; Redaschi N.; Yeh L. S. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Tope G.; Gillespie M.; Vastrik I.; D’Eustachio P.; Schmidt E.; de Bono B.; Jassal B.; Gopinath G. R.; Wu G. R.; Matthews L.; Lewis S.; Birney E.; Stein L. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005, 33, D428–D432. 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.