Abstract
Introduction
We sought to determine if proteomic profiles could predict risk for incident mild cognitive impairment (MCI) and Alzheimer's disease (AD) among adults with Down syndrome (DS).
Methods
In a cohort of 398 adults with DS, a total of n = 186 participants were determined to be non‐demented and without MCI or AD at baseline and throughout follow‐up; n = 103 had incident MCI and n = 81 had incident AD. Proteomics were conducted on banked plasma samples from a previously generated algorithm.
Results
The proteomic profile was highly accurate in predicting incident MCI (area under the curve [AUC] = 0.92) and incident AD (AUC = 0.88). For MCI risk, the support vector machine (SVM)‐based high/low cut‐point yielded an adjusted hazard ratio (HR) = 6.46 (P < .001). For AD risk, the SVM‐based high/low cut‐point score yielded an adjusted HR = 8.4 (P < .001).
Discussion
The current results provide support for our blood‐based proteomic profile for predicting risk for MCI and AD among adults with DS.
Keywords: dementia, Down syndrome, mild cognitive impairment, plasma, proteomic
1. INTRODUCTION
Down syndrome (DS) is the most common genetic cause of intellectual disability. 1 Virtually all individuals with DS show the neuropathological changes associated with Alzheimer's disease (AD) by age 40 and most will have developed dementia by age 60. 2 This association has become a major focus of interest due to the increasing prevalence of AD that has accompanied extended life expectancy within this population. 3 , 4 The neuropathological manifestations of AD in adults with DS—including deposition of extracellular amyloid beta (Aβ) in neuritic plaques, neurofibrillary tangles, neuroinflammation, and neurodegeneration—have been attributed to triplication and overexpression of the gene for amyloid precursor protein (APP) on chromosome 21, leading to elevated levels of Aβ peptides. 5 However, there remains a wide variation in age of onset and clinical progression suggesting that additional biological and environmental factors may be important contributors to disease vulnerability. 2
All studies of AD are dependent upon valid determination of clinical status. For adults with DS, this is complicated by the presence of impairments beginning early in development that vary considerably among individuals. 6 The presence of pre‐morbid cognitive impairment, especially together with variation in type and severity, makes diagnostic procedures developed for use with “neurotypical” populations uninformative in the majority of cases. 7 , 8 Identification of biomarkers related to stage and predictive of onset of mild cognitive impairment (MCI) and dementia would, therefore, be of tremendous value for studies of AD in DS. While cerebrospinal fluid (CSF) and positron emission tomography (PET) scan biomarkers are accurate in detecting AD pathology, 9 , 10 , 11 they are invasive and not cost‐effective. As has been proposed for the neurotypical AD population 12 there is a need for a multi‐tiered assessment process predicting AD risk among adults with DS. However, little work has been conducted on the identification of blood‐based biomarkers that are predictive of incident MCI and AD in adults with DS.
Schupf et al. found that among non‐demented individuals, high initial levels of Aβ42 and then declining levels of Aβ42 as well as a decrease in the Aβ42/Aβ40 ratio along with an increase in Aβ40 was associated with increased risk for incident dementia. 13 , 14 Additionally, being in the highest tertile of Aβ42 was associated with increased risk for mortality. 13 , 14 Coppus et al. 15 found that those adults with DS with the highest concentrations of Aβ40 and Aβ42 were at increased risk for incident dementia. Fortea et al. 16 found that high plasma neurofilament light protein (NfL) concentrations distinguished between healthy non‐demented adults, those with prodromal symptoms, and demented individuals. Strydom et al. found that NfL concentrations increased steeply after age 40 and were predictive of prevalent and incident dementia status. 17 Other blood‐based biomarkers have been examined by Head et al. 18 , 19 , 20 , 21 , 22
RESEARCH IN CONTEXT
Systematic review: Literature was identified and reviewed using PubMed. Several articles described growing efforts to identify blood‐based biomarkers associated with Alzheimer's disease (AD) among adults with Down syndrome (DS). Recent work has generated and cross‐validated a blood‐based proteomic profile in detecting AD and mild cognitive impairment (MCI) in the general population. No such work has sought to apply this same proteomic profile in predicting risk for incident MCI and AD among adults with DS.
Interpretation: Our findings show that blood‐based proteomic profiles can be highly accurate for predicting risk for MCI and AD. Additionally, inflammatory markers were heavily weighted in the risk scores suggesting the presence of a proinflammatory endophenotype in DS. Together, this work supports the potential utility of proteomic markers to facilitate a precision medicine approach to AD in DS.
Future directions: This article provides evidence that our blood‐based methods can be highly accurate in predicting risk for incident MCI and AD among adults with DS. Further studies are needed to cross‐validate these findings and then to prospectively test them as screening tools for novel clinical trials.
Although a majority of the work examining proteomic markers of neurodegeneration among adults with DS has focused on Aβ given the strong biological link, more recent work has expanded to explore the impact of inflammation. Among adults with DS who also have AD, a number of inflammatory proteins including tumor necrosis factor alpha (TNF‐α), interleukin 6 (IL‐6), IL‐10, IL‐8, and interferon gamma (IFN‐y) were found to be elevated compared to healthy controls without DS. 23 A meta‐analysis conducted by Zhang et al. found similar elevations across inflammatory proteins (TNF‐α, IL‐1β, INF‐y) among children with DS compared to neurotypical children reflecting that increases may be occurring over the span of the lifetime. 24 This corresponds with recent work that found similar elevations across a number of inflammatory proteins including C‐reactive protein (CRP), IL‐10, IL‐18, and TNF‐α among adults with DS with prevalent MCI and dementia. 25
In the neurotypical AD population, considerable strides have been made toward identification and refinement of blood‐based diagnostic screening modalities. 26 , 27 , 28 , 29 , 30 Our group has generated a proteomic profile for both MCI and AD within the neurotypical AD population 26 , 27 that has been validated across multiple cohorts, 26 , 27 , 31 , 32 , 33 assay technologies, 26 , 30 species, 30 and tissue. 30 We have found that our approach can also discriminate AD from other neurodegenerative diseases. 30 , 34 , 35 The aim of this study was to extend the application of this proteomic profile in an effort to predict MCI and AD risk among a cohort of adults with DS.
2. METHODS
2.1. Participants
The initial study sample included 398 members of a community‐based cohort of adults with DS. 13 , 36 , 37 Assessments included evaluations of cognition and functional abilities, behavioral and/or psychiatric conditions, and health status. Blood samples were drawn at each visit. Assessments were repeated at 14‐ to 20‐month intervals. Cognitive function was evaluated with a test battery designed for use with individuals varying widely in their initial levels of intellectual functioning. 38 Structured interviews were conducted with caregivers to collect information on adaptive behavior and medical history. Past and current medical records were reviewed for all participants. Recruitment, informed consent, and study procedures were approved by the Institutional Review Boards of the New York State Institute for Basic Research in Developmental Disabilities, The New York State Psychiatric Institute, Columbia University Medical Center, and The Johns Hopkins University School of Medicine.
2.2. Classification of dementia
To determine the occurrence of dementia and dementia subtypes in participants, data from all available sources were reviewed during a consensus conference. Following recommendations of the AAMR‐IASSID Working Group for the Establishment of Criteria for the Diagnosis of Dementia in Individuals with Developmental Disability, 39 participants were classified into three groups: (1) dementia, if there was a history of progressive memory loss, disorientation, and functional decline over a period of at least 1 year and if there were no other medical or psychiatric conditions that might result in or mimic dementia present (eg, untreated hypothyroidism, stroke); and (2) MCI‐DS, if they exhibited less substantial cognitive and few functional declines and did not meet criteria for dementia and (3) non‐demented, if they were without cognitive or functional decline.
2.3. Proteomic assays
Plasma samples were assayed via a multi‐plex biomarker assay platform using electrochemiluminescence (ECL) per our previously published methods. 29 , 30 The ECL platform has been used extensively to assay biomarkers associated with a range of human diseases including AD. 40 , 41 , 42 , 43 ECL technology uses labels that emit light when electronically stimulated, which improves the sensitivity of detection of many analytes even at very low concentrations. ECL measures have well established properties of being more sensitive and requiring less volume than conventional enzyme‐linked immunosorbent assays (ELISAs), 42 the gold standard for most assays. We recently reported the analytic performance of each of these markers for n > 1300 samples across multiple cohorts and diagnoses (normal cognition, MCI, AD). 29 The assays are reliable and our experience with these assays shows excellent spiked recovery, dilution linearity, coefficients of variation, as well as detection limits. Inter‐assay and intra‐assay variability has been excellent. Internal quality control (QC) protocols are implemented in addition to manufacturing protocols including assaying consistent controls across batches and assay of pooled standards across lots. A total of 500μl of plasma was used to assay the following markers (including coefficient of variation [CV] and lowest level of detection [LLOD]): fatty acid binding protein (CV = 4.2, LLOD = 206.8pg/mL), beta 2 microglobulin (CV = 5.5, LLOD = 96.3 pg/mL), pancreatic polypeptide (CV = 5.5, LLOD = 3436.8pg/mL), CRP (CV = 2.5; LLOD = 19.7pg/mL), ICAM‐1 (CV = 3.9; LLOD = 5.7pg/mL), thrombopoeitin (CV = 3.2; LLOD = 45.3pg/mL), α2 macroglobulin (CV = 1.7; LLOD = 4284pg/mL), exotaxin 3 (CV = 6.5; LLOD = 1.4pg/mL), tumor necrosis factor α (CV = 2.9; LLOD = 0.04pg/mL), tenascin C (CV = 3.5; LLOD = 20.8pg/mL), interleukin (IL)‐5 (CV = 4.3; LLOD = 0.05pg/mL), IL6 (CV = 4.6; LLOD = 0.07pg/mL), IL7 (CV = 5.8; LLOD = 0.1pg/mL), IL10 (CV = 2.7; LLOD = 0.02pg/mL), IL18 (CV = 5.0; LLOD = 1.7pg/mL), I309 (CV = 8.3; LLOD = 2.6pg/mL), factor VII (CV = 2.1; LLOD = 14.7pg/mL), VCAM 1 (CV = 2.5; LLOD = 9.1pg/mL), TARC (CV = 3.2; LLOD = 45.3pg/mL) and SAA (CV = 3.6; LLOD = 21.3pg/mL).
2.4. Apolipoprotein E genotypes
Apolipoprotein E (APOE) genotyping was carried out by polymerase chain reaction/restriction fragment length polymorphism (PCR/RFLP) analysis using HhaI (CfoI) digestion of an APOE genomic PCR product spanning the polymorphic (cys/arg) sites at codons 112 and 158, followed by acrylamide gel electrophoresis to document the restriction fragment sizes. 44 Participants were classified according to the presence or absence of at least one APOE e4 allele.
2.5. Statistical analysis
Statistical analyses were conducted using the R (V 3.3.3) statistical software, 45 SPSS 25 (IBM), and SAS. Support vector machine (SVM) analyses were conducted using a five‐fold internal cross‐validation to create proteomic profiles specifically for incident MCI‐DS and incident AD. SVM is based on the concept of decision planes that define decision boundaries and is primarily a classifier method that performs classification tasks by constructing hyperplanes in a multidimensional space that separates cases of different class labels. Diagnostic accuracy was calculated via receiver operating characteristic (ROC) curves with the following proteomic panel: FABP, B2M, PPY, CRP, ICAM‐1, thrombopoeitin, A2M, exotaxin 3, TNF‐α, tenascin C, IL5, IL6, IL7, IL10, IL18, I309, factor VII, VCAM 1, TARC, and SAA. Longitudinal analyses were conducted using data up to 10 years of follow‐up from the blood draw to determine if the proteomic profile could (a) predict incident MCI‐DS and (b) predict incident AD. The risk score from SVM was classified as a low/high cut point‐score for analyses examining the relation of the risk score to MCI‐DS and AD incidence. We used Cox proportional hazards modeling to determine the association of the proteomic profile risk score to cumulative incidence of MCI‐DS or AD, adjusting for age at blood draw, sex, ethnicity, level of intellectual disability, and the presence of an APOE e4 allele. All covariates were selected a priori due to their known link with AD disease progression in both the neurotypical population as well as among the DS population. For these analyses, level of intellectual ability was classified as mild/moderate and severe/profound and ethnicity was classified as non‐Hispanic white and other. The time to event variable was time from blood draw to onset in affected individuals or time from blood draw to last visit in those remaining unaffected.
3. RESULTS
Of the 398 participants, 186 participants were determined to be non‐demented, and without MCI at baseline and throughout follow‐up; n = 103 had incident MCI and n = 81 had incident AD. Participants classified as having prevalent MCI (n = 54) or AD (n = 42) at baseline were excluded from these analyses focused on risk for incident MCI or AD. There were n = 193 females and n = 96 males included in the analysis of risk for MCI. There were n = 190 females and n = 77 males included in the analysis of risk for AD. Mean age at blood draw for all participants was 48.4 (SD = 6.6) for all participants. Four participants did not have APOE genotypes. Table 1 presents the demographic characteristics of the participants by cognitive status. Those who developed MCI were older at baseline (P‐value <.001), less likely to be female (P‐value <.001), and more likely to have severe/profound intellectual disability (P‐value = .008) than those who remained unaffected over the follow‐up period. Those who developed AD were also older at baseline (P‐value <.001), less likely to be female (P‐value = .005) than those who remained unaffected, but did not differ by level of function (Table 1).
TABLE 1.
Demographic characteristics
| Characteristic | Non‐demented | Incident MCI | Incident AD |
|---|---|---|---|
| N | 186 | 103 | 81 |
| Age | 48.6 ± 6.7 | 52.9 ± 6.0b | 54.1 ± 5.6b |
| Sex | |||
| Male | 44 (23.7) | 52 (50.5) | 33(40.7) |
| Female | 142 (76.3) | 51 (49.5)b | 48 (59.3)a |
| Level of function | |||
| Mild/moderate | 127 (68.3) | 54 (52.4) | 46 (56.8) |
| Severe/profound | 59 (31.7) | 49 (47.6)a | 35 (43.2) |
| Ethnicity | |||
| White | 171 (91.9) | 91 (88.3) | 75 (92.6) |
| Non‐white | 15 (8.1) | 12 (11.7) | 6 (7.4) |
| APOE e4 allele | 32 (17.6) | 25 (24.3) | 21 (25.9)c |
| Risk score cut point | 30 (16.1) | 84(81.6)b | 66 (81.5)b |
Note: Significance P‐value <0.05a, <0.001b.
Missing data on n = 4 with incident ADc.
Abbreviations: AD, Alzheimer's disease; MCI, mild cognitive impairment
SVM was used to determine if proteomic profiles at the first blood draw would be accurate in predicting incident MCI and AD among those adults with DS who were cognitively normal at blood draw. The proteomic profile was highly accurate in predicting incident MCI among adults with DS with the optimized SVM‐based risk‐score of –0.895 resulting in an area under the curve (AUC) = 0.92, sensitivity (SN) = 0.82, and specificity (SP) = 0.84. Positive predictive value (PPV) based on the optimized model was 73.68% while the negative predictive value (NPV) was 89.14%. Figure 1 shows the variable importance plots and ROC curve for incident MCI. The top 10 proteins associated with increased risk of MCI were IL6, CRP, sICAM1, I309, PPY, SAA, TPO, IL10, TARC, and IL5. Our proteomic profile was also highly accurate in predicting incident AD with the optimized SVM‐based risk‐score of –0.978 resulting in an AUC = 0.88, SN = 0.81, and SP = 0.89. PPV was 75.86% and NPV was 91.66%. Figure 2 shows the variable importance plots and ROC curve for incident AD. The top 10 proteins associated with increased risk of AD included IL10, FABP3, IL6, CRP, TPO, IL5, SAA, Eotaxin3, TARC, and IL7.
FIGURE 1.
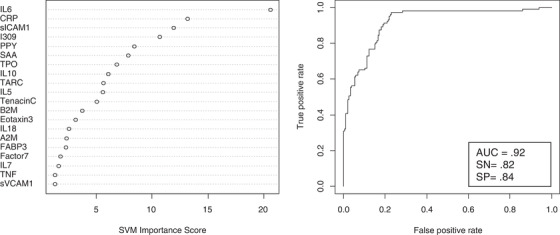
Variable importance plot and receiver operating characteristic curve for incident mild cognitive impairment
FIGURE 2.
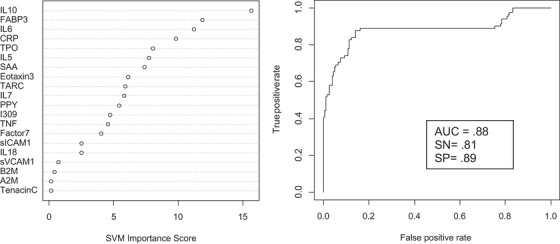
Variable importance plot and receiver operating characteristic curve for incident Alzheimer's disease
Next, the SVM‐based proteomic risk scores were dichotomized into high/low cut point scores (–0.895 for incident MCI and –0.978 for incident AD) and entered into Cox proportional hazard models for predicting incident MCI and AD. Of note, the cut point was derived from the risk score obtained through the SVM models. All models were adjusted for age at blood draw, sex, level of intellectual disability, ethnicity, and presence of the APOE e4 allele. For predicting onset of MCI, the SVM‐based proteomic high/low cut point score yielded a hazard ratio (HR) = 6.46 (95% confidence interval [CI] = 3.9–10.7), adjusted for covariates (Figure 3). For predicting onset of AD, the SVM‐based proteomic high/low cut point score yielded a HR = 8.4 (95% CI = 4.7–15.1) adjusted for covariates (P<.001; Figure 4).
FIGURE 3.

Cox proportional hazard model for predicting incident mild cognitive impairment
FIGURE 4.
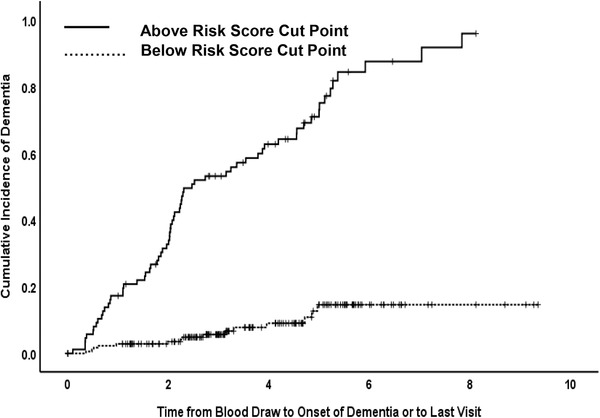
Cox proportional hazard model for predicting incident Alzheimer's disease
4. DISCUSSION
The availability of a blood‐based screening tool for predicting risk of MCI or AD among adults with DS would be a major advancement for the development of novel prevention trials. To date, however, little work has been conducted on blood‐based biomarkers for predicting risk for MCI and AD among adults with DS. The current study investigated whether our previously validated proteomic profile for detecting MCI and AD in the neurotypical population would also predict incident MCI and AD among adults with DS. The current results strongly support the possibility that proteomic profiles have the utility to predict onset of MCI (AUC = 0.92) and AD (AUC = 0.88) among adults with DS.
It is noteworthy that the proteomic profiles were heavily weighted toward inflammatory markers for both incident MCI and AD, which is consistent with our prior work in the neurotypical AD population. 26 , 29 , 30 The top 10 proteins associated with increased risk of MCI included IL6, CRP, sICAM1, I309, PPY, SAA, TPO, IL10, TARC, and IL5 and the top markers associated with incident AD included IL10, FABP3, IL6, CRP, TPO, IL5, SAA, Eotaxin3, TARC, and IL7. Therefore, inflammation plays a key role in our proteomic approach to predicting incident MCI and AD in DS. In the neurotypical AD population, we have identified a pro‐inflammatory endophenotype that identifies a specific subset of AD patients who benefitted from a “failed” nonsteroidal anti‐inflammatory drug (NSAID) trial. 28 It appears that this pro‐inflammatory endophenotype is also present within DS adults with MCI and AD, which is consistent with the increased role of inflammation among individuals with DS. 46 , 47 Prior work has shown that neuroinflammation is upregulated in fetal development in DS, which may accelerate the development of AD pathology. 46 A meta‐analysis 24 that analyzed data across 19 studies (n = 957 individuals with DS and 541 controls without DS) found alterations in multiple inflammatory markers in adults with DS. The presence of the pro‐inflammatory endophenotype among adults with DS opens the possibility of precision medicine based therapies. Specifically, the pro‐inflammatory endophenotype may identify a specific subset of adults with DS where anti‐inflammatory interventions, as a part of multi‐modal therapy (ie, anti‐inflammatory + anti‐amyloid), may be of particular use. This precision medicine approach has resulted in substantial strides in patient outcomes in cancer. 48
The availability of a blood test that can be used to determine which adults with DS should undergo more comprehensive neurodiagnostic procedures can be of tremendous value to patients, caregivers, providers, and the medical system. As is the case with all initial screening tests, the primary goal of the test is to separate low‐risk individuals (for whom no further testing is necessary) from those determined to be high risk (who should undergo additional confirmatory diagnostic testing). 29 Therefore, a blood test that can be implemented annually starting at age 40 in adults with DS can inform clinicians as to when a particular individual requires additional follow‐up or more comprehensive screening. Informing patients and their family members that the individual does not appear to be suffering from dementia at this time greatly reduces family and caregiver stress, as well as medical costs. When a positive finding arises on a blood screening test, the patient undergoes higher cost and more invasive testing. We have proposed this same multi‐tiered approach for detecting AD in primary care settings, which can have a substantial impact on cost containment. 12 When therapeutic agents become available, the availability of a sensitive and specific blood screening test will provide an opportunity to treat the individual before substantial cognitive loss occurs. This method can also increase access to novel clinical trials for underserved populations (eg, ethnically and racially diverse populations, rural populations) by providing a means for screening potential patients within primary care settings where these patients are receiving the majority of their care.
There are limitations to the current study. First, while a large‐scale retrospective study, this work needs to be cross‐validated in an independent cohort. Second, it is possible that additional proteomic markers not included in these analyses will increase the overall accuracy of the proteomic profiles. Third, the current analyses do not take into account recent work examining novel ultra‐sensitive markers related to AD pathology (amyloid, tau) and NfL that have received a great deal of attention in the recent literature. However, each of these limitations are currently being examined in the Alzheimer's Biomarker Consortium—Down Syndrome (ABC‐DS) study. Future plans through the ABC‐DS includes extending the current findings to evaluate if this specific proteomic panel is also able to detect conversion from MCI to AD among adults with DS. It will also allow for a closer examination concerning the impact of APOE e4 on risk for MCI and AD in this specific population. Taken together, the current results strongly support the utility of blood‐based biomarkers in predicting MCI and AD risk among adults with DS.
AUTHOR CONTRIBUTIONS
Conception and design of study: Sid E. O'Bryant, Sharon J. Krinsky‐McHale, Wayne Silverman, Joseph H. Lee, Nicole Schupf. Acquisition, and analysis of data: Sid E. O'Bryant, Fan Zhang, Sharon J. Krinsky‐McHale, Wayne Silverman, Deborah Pang, James Hall, Nicole Schupf. Drafting manuscript or figures: Sid E. O'Bryant, Fan Zhang, Sharon J. Krinsky‐McHale, Wayne Silverman, Joseph H. Lee, James Hall, Nicole Schupf.
CONFLICTS OF INTEREST
Sid E. O'Bryant has patents and patents pending regarding blood‐biomarkers for precision medicine in neurodegenerative diseases and served on an Advisory Board to Roche Diagnostics. No other authors have conflicts of interest to disclose.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grants: P01 HD035897, U54 HD079123, U01 AG051412 and R01 AG014673, and by grant IIRG‐08‐90655 from the Alzheimer's Association as well as by funds from the New York State Office for People with Developmental Disabilities. This work was also supported by the National Institute on Aging grant R01 AG058537.
We thank the study participants and participating agencies from the tri‐state area that made these studies possible. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Institute on Aging.
O'Bryant SE, Zhang F, Silverman W, et al. Proteomic profiles of incident mild cognitive impairment and Alzheimer's disease among adults with Down syndrome. Alzheimer's Dement. 2020;12:e12033 10.1002/dad2.12033
REFERENCES
- 1. Presson AP, Partyka G, Jensen KM, et al. Current estimate of Down Syndrome population prevalence in the United States. J Pediatr. 2013;163:1163‐1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lott IT, Head E. Alzheimer disease and down syndrome: factors in pathogenesis. Neurobiol Aging. 2005;26(3):383‐389. [DOI] [PubMed] [Google Scholar]
- 3. Zigman WB. Atypical aging in Down syndrome. Dev Disabil Res Rev. 2013;18(1):51‐67. [DOI] [PubMed] [Google Scholar]
- 4. Zigman WB, Lott IT. Alzheimer's disease in Down syndrome: neurobiology and risk. Ment Retard Dev Disabil Res Rev. 2007;13:237‐246. [DOI] [PubMed] [Google Scholar]
- 5. Rumble B, Retallack R, Hilbich C, et al. Amyloid a4 protein and its precursor in Down's syndrome and Alzheimer's disease. N Engl J Med. 1989;320(22):1446‐1452. [DOI] [PubMed] [Google Scholar]
- 6. Sabbagh M, Edgin J. Clinical assessment of cognitive decline in adults with Down syndrome. Curr Alzheimer Res. 2015;13(1):30‐34. [DOI] [PubMed] [Google Scholar]
- 7. Firth NC, Startin CM, Hithersay R, et al. Aging related cognitive changes associated with Alzheimer's disease in Down syndrome. Ann Clin Transl Neurol. 2018;5(6):741‐751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krinsky‐McHale SJ, Silverman W. Dementia and mild cognitive impairment in adults with intellectual disability: issues of diagnosis. Dev Disabil Res Rev. 2013;18:31‐42. [DOI] [PubMed] [Google Scholar]
- 9. Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. 2010;6(3):131‐144. [DOI] [PubMed] [Google Scholar]
- 10. Sabri O, Sabbagh MN, Seibyl J, et al. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer's disease: phase 3 study. Alzheimer's Dement. 2015;11(8):964‐974. [DOI] [PubMed] [Google Scholar]
- 11. Vlassenko AG, Benzinger TLS, Morris JC. PET amyloid‐beta imaging in preclinical Alzheimer's disease. Biochim Biophys Acta Mol Basis Dis. 2012;1822(3):370‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Bryant SE, Mielke MM, Rissman RA, et al. Blood‐based biomarkers in Alzheimer disease: current state of the science and a novel collaborative paradigm for advancing from discovery to clinic. Alzheimers Dement. 2017;13:45‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schupf N, Patel B, Pang D, et al. Elevated plasma beta‐amyloid peptide Abeta(42) levels, incident dementia, and mortality in Down syndrome. Arch Neurol. 2007;64:1007‐1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schupf N, Zigman WB, Tang MX, et al. Change in plasma Aβ peptides and onset of dementia in adults with Down syndrome. Neurology. 2010;75(18):1639‐1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coppus AM, Schuur M, Vergeer J, et al. Plasma beta amyloid and the risk of Alzheimer's disease in Down syndrome. Neurobiol Aging. 2012;33:1988‐1994. [DOI] [PubMed] [Google Scholar]
- 16. Fortea J, Carmona‐Iragui M, Benejam B, et al. Plasma and CSF biomarkers for the diagnosis of Alzheimer's disease in adults with Down syndrome: a cross‐sectional study. Lancet Neurol. 2018;17(10):860‐869. [DOI] [PubMed] [Google Scholar]
- 17. Strydom A, Heslegrave A, Startin CM, et al. Neurofilament light as a blood biomarker for neurodegeneration in Down syndrome. Alzheimers Res Ther. 2018;10:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perluigi M, Di Domenico F, Buttterfield DA. Unraveling the complexity of neurodegeneration in brains of subjects with Down syndrome: insights from proteomics. Proteomics Clin Appl. 2014;8:73‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Butterfield DA, Di Domenico F, Swomley AM, Head E, Perluigi M. Redox proteomics analysis to decipher the neurobiology of Alzheimer‐like neurodegeneration: overlaps in Down's syndrome and Alzheimer's disease brain. Biochem J. 2014;463:177‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cenini G, Fiorini A, Sultana R, et al. An investigation of the molecular mechanisms engaged before and after the development of Alzheimer disease neuropathology in Down syndrome: a proteomics approach. Free Radic Biol Med. 2014;76:89‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barone E, Head E, Butterfield DA, Perluigi M. HNE‐modified proteins in Down syndrome: involvement in development of Alzheimer disease neuropathology. Free Radic Biol Med. 2017;111:262‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tramutola A, Di Domenico F, Barone E, et al. Polyubiquitinylation profile in down syndrome brain before and after the development of Alzheimer neuropathology. Antioxid Redox Signal. 2017;26:280‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iulita MF, Ower A, Barone C, et al. An inflammatory and trophic disconnect biomarker profile revealed in Down syndrome plasma: relation to cognitive decline and longitudinal evaluation. Alzheimer's Dement. 2016;12(11):1132‐1148. [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y, Che M, Yuan J, et al. Aberrations in circulating inflammatory cytokine levels in patients with Down syndrome: a meta‐analysis. Oncotarget. 2017;8(48):84489‐8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petersen M, Zhang F, Krinsky‐McHale SJ, et al. Proteomic profiles of prevalent mild cognitive impairment and Alzheimer's disease among adults with Down syndrome. Alzheimer's & Dement Diagnosis, Assess Dis Monit. 2020;12: 1: 10.1002/dad2.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Bryant SE, Xiao G, Barber R, et al. A serum protein‐based algorithm for the detection of Alzheimer disease. Arch Neurol. 2010;67:1077‐1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O'Bryant SE, Xiao G, Barber R, et al. A blood‐based screening tool for Alzheimer's disease that spans serum and plasma: findings from TARC and ADNI. PLoS One. 2011;6(12):e28092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Bryant SE, Zhang F, Johnson LA, et al. A precision medicine model for targeted nsaid therapy in Alzheimer's disease. J Alzheimers Dis. 2018;66:97‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Bryant SE, Edwards M, Johnson L, et al. A blood screening test for Alzheimer's disease. Alzheimer's Dement Diagnosis. Assess Dis Monit. 2016;3:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Bryant SE, Xiao G, Zhang F, et al. Validation of a serum screen for alzheimer's disease across assay platforms, species, and tissues. J Alzheimer's Dis. 2014;42(4):1325‐1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Villarreal AE, O'Bryant SE, Edwards M, Grajales S, Britton GB. Serum‐based protein profiles of Alzheimer's disease and mild cognitive impairment in elderly Hispanics. Neurodegener Dis Manag. 2016;6:203‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edwards M, Hall J, Williams B, Johnson L, O'Bryant S. Molecular markers of amnestic mild cognitive impairment among Mexican Americans. J Alzheimers Dis. 2016;49:221‐228. [DOI] [PubMed] [Google Scholar]
- 33. O'Bryant SE, Xiao G, Edwards M, et al. Biomarkers of Alzheimer's disease among Mexican Americans. J Alzheimers Dis. 2013;34:841‐849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Bryant SE, Edwards M, Zhang F, et al. Potential two‐step proteomic signature for Parkinson's disease: Pilot analysis in the Harvard Biomarkers Study. Alzheimer's Dement Diagnosis, Assess Dis Monit. 2019;11:374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Bryant SE, Ferman TJ, Zhang F, et al. A proteomic signature for dementia with Lewy bodies. Alzheimer's Dement Diagnosis. Assess Dis Monit. 2019;11:270‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zigman WB, Devenny DA, Krinsky‐McHale SJ, et al. Chapter 4 Alzheimer's disease in adults with Down syndrome. Int Rev Res Ment Retard. 2008;36:103–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee JH, Lee AJ, Dang LH, et al. Candidate gene analysis for Alzheimer's disease in adults with Down syndrome. Neurobiol Aging. 2017;56:150‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Silverman W, Schupf N, Zigman W, et al. Dementia in adults with mental retardation: assessment at a single point in time. Am J Ment Retard. 2004;109:111‐125. [DOI] [PubMed] [Google Scholar]
- 39. Janicki MP, Heller T, Seltzer GB, Hogg J. Practice guidelines for the clinical assessment and care management of Alzheimer's disease and other dementias among adults with intellectual disability. J Intellect Disabil Res. 1996;40 (Pt 4):374‐382. [PubMed] [Google Scholar]
- 40. Alves G, Brønnick K, Aarsland D, et al. CSF amyloid‐β and tau proteins, and cognitive performance, in early and untreated Parkinson's disease: the Norwegian Park West study. J Neurol Neurosurg Psychiatry. 2010;81(10):1080‐1086. [DOI] [PubMed] [Google Scholar]
- 41. Bjerke M, Portelius E, Minthon L, et al. Confounding Factors Influencing Amyloid Beta Concentration in Cerebrospinal Fluid. Int J Alzheimer's Disease. 2010;2010: 1–11. 10.4061/2010/986310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kuhle J, Regeniter A, Leppert D, et al. A highly sensitive electrochemiluminescence immunoassay for the neurofilament heavy chain protein. J Neuroimmunol. 2010;220(1‐2):114‐119. [DOI] [PubMed] [Google Scholar]
- 43. Oh ES, Mielke MM, Rosenberg PB, et al. Comparison of conventional ELISA with electrochemiluminescence technology for detection of amyloid‐β in plasma. J Alzheimer's Dis. 2010;21(3):769‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545‐548. [PubMed] [Google Scholar]
- 45. R_Development_Core_Team . R: A language and environment for statistical computing 2009. http://www.r-project.org.
- 46. Wilcock DM, Griffin WS. Down's syndrome, neuroinflammation, and Alzheimer neuropathogenesis. J Neuroinflammation. 2013;10:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Head E, Powell DK, Schmitt FA. Metabolic and vascular imaging biomarkers in Down syndrome provide unique insights into brain aging and Alzheimer disease pathogenesis. Front Aging Neurosci. 2018;10:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jorgensen JT. Clinical application of companion diagnostics. Trends Mol Med. 2015;21:405‐407. [DOI] [PubMed] [Google Scholar]


