Abstract
Abstract
This paper describes the predicted structure for the cps loci involved in capsule biosynthesis for Streptococcus parauberis serotypes III, IV, and V. Based on the specific serotype regions I, II, and III, a multiplex PCR protocol (mPCR) was designed to differentiate the main serotypes causing fish diseases. A real-time PCR method (qPCR) is also described to identify S. parauberis of serotype III in bacterial cultures and fish tissues. In silico and in vitro analyses revealed that both methods have a 100% specificity. The mPCR assay was optimized for the detection of S. parauberis strains of subtypes Ia (amplicon size 213 bp), subtypes Ib and Ic (both amplicon size 303 bp), serotype II (amplicon size 403 bp), and serotype III (amplicon size 130 bp) from bacterial cultures. The qPCR assay was optimized for the identification and quantification of S. parauberis serotype III strains in bacterial cultures and fish tissues. This assay achieved a sensitivity of 2.67 × 102 gene copies (equivalent to 3.8 × 10−9 ng/μl) using pure bacterial cultures of S. parauberis serotype III and 1.76 × 102 gene copies in fish tissues experimentally and naturally infected with S. parauberis of the serotype III. The specificity and sensitivity of the protocols described in this study suggest that these methods could be used for diagnostic and/or epidemiological purposes in clinical diagnostic laboratories.
Key Points
• Structure of loci cps for S. parauberis of serotypes III, IV and V was described.
• mPCR to differentiate S. parauberis serotypes causing disease in fish was optimized.
• qPCR assay to quantify strains of S. parauberis serotype III in fish tissues.
Electronic supplementary material
The online version of this article (10.1007/s00253-020-10683-z) contains supplementary material, which is available to authorized users.
Keywords: Real-time PCR, Streptococcus parauberis, Serotypes III, IV and V, Genomic analyses, Typing
Introduction
Streptococcus parauberis is a Gram-positive bacterium responsible for streptococcosis, a disease that causes major economic losses in the aquaculture sector. This pathology affects the central nervous system of fish, resulting in septicaemia and meningoencephalitis. S. parauberis was initially described as an aetiological agent of bovine mastitis (Williams and Collins 1990; Khan et al. 2003). Since then, S. parauberis has become an emerging pathogen in the aquaculture industry responsible for streptococcosis epizootics mainly in turbot (Scophthalmus maximus) farmed in Spain (Domenech et al. 1996), in flounder (Paralichthys olivaceus and Platichthys stellatus) farmed in Asian countries (Aoki et al. 1990; Baeck et al. 2006; Cho et al. 2008), and sea bass (Sebastes ventricosus) farmed in Japan (Oguro et al. 2014). Recently, S. parauberis was also isolated from wild striped sea bass (Morone saxatilis) in North America (USA) (Haines et al. 2013). Diversity among S. parauberis isolates was reported at genotypic and serological levels (Austin and Austin 2016).
Polysaccharide capsules are structures found on the cell surface of many bacterial species. These structures play an important role in pathogenicity, immune system escape, serum resistance, inflammation, adhesion, biofilm formation, and antigenicity, which may be used for bacterial serotyping (Okura et al. 2013; Rochat et al. 2017). The diversity of capsular polysaccharide (CPS) structure of S. parauberis, due to a variety of sugars and glycosidic linkages, led to the description of different serotypes designated as types I, II, and III (Kanai et al. 2009; Kanai et al. 2015; Tu et al. 2015a; Torres Corral et al. 2019). Serotype I was further subdivided into three subtypes, designated Ia, Ib, and Ic (Kanai et al. 2015; Tu et al. 2015a). S. parauberis isolates agglutinating with antibodies of serotypes I and II were also described and designated as non-typable strains (Kanai et al. 2015; Tu et al. 2015a).
The genes responsible for the synthesis of CPS are typically clustered in a single locus (locus cps) on the chromosome (Okura et al. 2013; Tu et al. 2015a). The genetic structure of the loci cps in S. parauberis has been determined in serotypes I and II (Tu et al. 2015a). In these serotypes a cassette-like structure was observed, in which the conserved regions flanked serotype-specific genes. At the conserved 5′ end of the cps regions, a group of five regulatory genes (lysR, cpsA. cpsB, cpsC, and cpsD) and one processing gene (cpsE) were found, while at the conserved 3′ end, one regulatory gene (cpsQ) and one hypothetical gene were found. The middle region (variable region) of the locus cps comprises genes encoding enzymes such as glycosyltransferases, acetyltransferases, aminotransferases, and modifying enzymes (Tu et al. 2015a).
This serological diversity might have important consequences for the selection of suitable candidate strains for vaccine development, in monitoring studies, epidemiological surveillance, disease control, as well as for a better understanding of host virulence and resistance traits. Serotyping is, therefore, a necessary tool for the diagnosis and epidemiological surveillance of streptococcosis caused by S. parauberis. However, serotyping has important limitations such as sensitivity, specificity, and the need to obtain antisera from animals. These limitations added to the recent determination of complete genomic sequences of a wide variety of bacteria have allowed the development of new molecular methods based on polymerase chain reaction (PCR) for bacterial typing. In recent years, different PCR bacterial typing protocols have emerged as an alternative for the use of antisera in pathogenic fish species, such as Streptococcus agalactiae (Demczuk et al. 2017; Kannika et al. 2017; Shoemaker et al. 2017), Flavobacterium psychrophylum (Rochat et al. 2017), S. parauberis (Tu et al. 2015b), and Lactococcus garvieae (Ohbayashi et al. 2017).
In the present study, genomic sequences of S. parauberis of different serotypes, hosts, and geographic regions were compared to determine the genetic structure of the locus cps of the recently described serotype III of S. parauberis (Torres Corral et al. 2019). The serotype-specific region of S. parauberis serotype III was used as a target to develop real-time PCR (qPCR) protocol to identify, detect, and quantify strains of S. parauberis serotype III. The specificity and efficacy of this assay was evaluated in silico and in vitro using target and non-target bacterial cultures and fish tissue samples. In addition, a serotyping scheme based on multiplex PCR (mPCR) capable of distinguishing subserotypes Ia, Ib/Ic, II, and III was also developed.
Material and methods
Bacterial strains and taxonomic characterization
A total of 73 bacterial strains of the genera Streptococcus, Lactococcus, Vagococcus, Carnobacterium, Aeromonas, Renibacterium, Tenacibaculum, Listeria, Flavobacterium, Yersinia, Pseudomonas, Vibrio, Listonella, and Edwarsiella were used in the present study (Table 1). This bacterial collection included reference strain representatives of the different serotypes described for S. parauberis (serotypes I, II, III. and non-typeable strains). These reference strains were used for the optimization of the qPCR and mPCR protocols and as positive control in all reactions. The bacteria were grown on Tryptic Soy Agar with 1% NaCl (w/v) (TSA-1) at 25 °C for 24 h. Tenacibaculum and Flavobacterium strains were grown at the appropriate temperatures on Flexibacter maritimus medium (FMM) (Pazos et al. 1996) and FLP medium (Cepeda et al. 2004), respectively. Stock bacterial cultures were frozen at − 80 °C in Microbank™ commercial medium (Pro-Lab Diagnostics, ON, Canada). Before the analysis, the taxonomic position of the strains used in this study was confirmed by using standard microbiological analysis and/or specie-specific PCR as described (Buller 2014; Austin and Austin 2016; Torres Corral et al. 2019). Serological typing of S. parauberis strains was carried out by Dot-Blot and agglutination tests as described by Torres-Corral et al. (2019), using formalin-killed cells (FKC) and rabbit whole cell antisera against the strains NCDO 2020 and SK451/04 (serotype III) obtained in our laboratory and antisera against the strains of subserotypes Ia, Ib, Ic, and serotype II, kindly provided by Dr. Kanai.
Table 1.
Bacterial strains used in the present study and results of serological analysis and PCR typing using the primers SP3-130F and SP3-130R
| Bacterial strains | Serotype identified | mPCR/qPCR |
|---|---|---|
| Reference strains | ||
| Streptococcus parauberis NCIMB 703043 | Serotype III | +/+ |
| S. parauberis NCDO 2020 | Serotype IV | −/− |
| Streptococcus iniae CECT 7363 | NA | −/− |
| Streptococcus agalactiae DSM 28863 | NA | −/− |
| S. agalactiae CECT 183 | NA | −/− |
| Streptococcus suis CECT 958 | NA | −/− |
| Streptococcus mutans KCCM 40105 | NA | −/− |
| Streptococcus uberis CECT 994 | NA | −/− |
| Vagococcus salmoninarum CECT 5810 | NA | −/− |
| Lactococcus garvieae NCDO 2155 | NA | −/− |
| L. garvieae CECT 5274 | NA | −/− |
| Lactococcus piscium CECT 4493 | NA | −/− |
| Lactococcus lactis ssp. lactis KCTC 3769 | NA | −/− |
| Clinical isolates | ||
| S. parauberis, diseased turbot (Spain, n = 10) | Serotype III | +/+ |
| S. parauberis, diseased flounder (Japan, n = 3) | Serotype II | +/− |
| S. parauberis, diseased flounder (Japan, n = 3) | Serotype I | +/− |
| S. parauberis, diseased flounder (Japan, n = 2) | Non-typeable | +/− |
| S. iniae, diseased marine fish (Spain, n = 5) | NA | −/− |
| S. agalactiae, unknown (Spain, n = 3) | NA | −/− |
| V. salmoninarum, diseased trout (Spain, n = 5) | NA | −/− |
| L. garvieae, diseased trout (Spain, n = 5) | NA | −/− |
| L. lactis, diseased trout (Spain, n = 2) | NA | −/− |
| Non-related strains | ||
| Carnobacterium maltaromaticum ATCC 35586 | NA | −/− |
| Carnobacterium divergens CECT 4016 | NA | −/− |
| Listeria monocytogenes CECT 934 | NA | −/− |
| Renibacterium salmoninarum ATCC 33209 | NA | −/− |
| Aeromonas salmonicida ATCC 33658 | NA | −/− |
| A. salmonicida NCIMB 2261 | NA | −/− |
| A. salmonicida subsp. achromogenes CECT 895 | NA | −/− |
| A. salmonicida subsp. masoucida CECT 896 | NA | −/− |
| Aeromonas hydrophila CECT 4330 | NA | −/− |
| Aeromonas piscicola CECT 7443 | NA | −/− |
| Vibrio anguillarum ATCC 43306 | NA | −/− |
| Listonella pelagia NCIMB 1900 | NA | |
| Tenacibaculum maritimum NCIMB 2154 | NA | −/− |
| Tenacibaculum dicentrarchi NCIMB 14598 | NA | −/− |
| Tenacibaculum gallaicum DSM 18841 | NA | −/− |
| Tenacibaculum soleae CECT 7292 | NA | −/− |
| Flavobacterium psychrophilum NCIMB 13384 | NA | −/− |
| Flavobacterium branchiophilum ATCC 35035 | NA | −/− |
| Flavobacterium plurextorum CECT 7844 | NA | −/− |
| Yersinia ruckeri CECT 955 | NA | −/− |
| Pseudomonas aeruginosa CECT 108 | NA | −/− |
| Escherichia coli CECT 99 | NA | −/− |
ATCC American Type Culture Collection (Maryland, USA), NCDO National Collection of Dairy Organism (Washington DC, USA), CECT Spanish Type Culture Collection (Valencia, Spain), DSM Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany), NCIMB National Collection of Industrial and Marine Bacteria (Aberdeen, UK), + typeable strains, − non-typeable strains, NA non-applicable
Identification of the locus cps based on genomic comparisons
Genomic comparison was performed using fifteen whole-genomes of S. parauberis and six nucleotide sequences of loci cps of S. parauberis (Supplemental Table S1). The nucleotide sequences used in this study were retrieved from the National Center for Biotechnology Information (NCBI) genome database. For comparative purposes, genomic sequences were annotated using the Rapid Annotations using Subsystems Technology (RAST) automated prokaryotic annotation server (Aziz et al. 2008). The accuracy of the RAST functional annotation of the draft genomes was compared with manual searches of homologous genes in the NCBI database and with the RAST annotation of the S. parauberis genomes classified as “complete or chromosome” by the NCBI genome database, corresponding to the reference strains KCTC 11537 and NCDFD 2020 (Supplemental Table S1), from the Korean Collection for Type Cultures (KCTC) and from National Cohort of Dairy Farms (NCDF). The capsule biosynthesis (cps) locus of the S. parauberis was identified on the basis of multiple nucleotide sequence alignments using Mauve Genome Alignment 2.4.0 (Darling et al. 2004) and ClustalW (Thompson et al. 1994). The genes of the cps loci were designated following the scheme proposed by Tu et al. (2015a).
Sequencing of cps3K gene
The nucleotide sequence of the loci cps of the strains KRS02083 (subserotype Ia), NUF1003 (subserotype Ib), NUF1071(subserotype Ic), NUF1032 (serotype II), 2007-1 and NUF1095 (non-typeable strains), strains isolated from turbot (AZ70.1 and T1), strains isolated from wild striped bass (RP17, PL23, PL9, RP15, RP25, N198_2, and N11), strains isolated from Japanese flounder (KCTC 11537, KRS02083, KCTC11980 and KRS02109), and strain isolated from Japanese black seaperch (SK-417) of S. parauberis were retrieved from the GenBank database (https://www.ncbi.nlm.nih.gov/), aligned and compared using the ClustalW (Thompson et al. 1994) and Nucleotide Basic Local Alignment Search Tool (BLASTn) programs to identify serotype-specific regions.
The cps3K gene of the variable region of the cps loci of S. parauberis, detected after in silico analysis of the genome of strains isolated from turbot and striped bass, was sequenced using the primers cps3K-F (5′-ACCCATGAGCTTTTTCTTCCAT-3′) and cps3K-R (5′-ATGTTATGTGCATGGCTCGT-3′) designed using Pick primer and Primer-BLAST tools (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The primers amplify a region of 1034 bp of the cps3K gene. Genomic DNA from three strains isolated from turbot and belonging to serotype III (NCIMB 703043, SK451/04, and SK537/10) was extracted using InstaGene matrix (BioRad, Madrid, Spain) and submitted to the Spanish Collection of Type Cultures (CECT, Valencia, Spain) for sequencing analysis using an ABI 3730xl sequencer (Applied Biosystems). The obtained sequences were cleaned and assembled using CodonCode Aligner v. 9.0.1 (CodonCode Co., USA). The partial sequences of the cps3K gene were then aligned and compared with the sequences of the loci cps of S. parauberis strains deposited in the NCBI database (Supplemental Table S1) using the ClustalW software and BLASTn to search for putative serotype III specific regions.
Typing of S. parauberis using multiplex and real-time PCR
Primer design
Pick primer and Primer-BLAST tools were used to design primers specific to identify strains of S. parauberis of serotype III, based on a sequence of 1086 bp of the S. parauberis pyruvyl transferase polysaccharide (cps3K) gene (GenBank accession no. NSGS0100000014.1). To evaluate primer properties and to optimize real-time PCR conditions, IDT SciTools Web (Integrated DNA Technologies, Coralville, IA, USA) was used. The forward primer SP3-130F (5′- GACCAACACCAGCACCAATA −3′, positions 738 to 757 in the S. parauberis cps3K gene) and the reverse primer SP3-130R (5′- TTTGGACAACCTGGAAGAGC -3′, positions 848 to 867 in the S. parauberis cps3K gene) were designed to amplify a 130-bp internal fragment of cps3K gene of S. parauberis. Primers were synthesized by IDT (Integrated DNA Technologies, Coralville, Iowa, USA). The specificity of the SP3-130F and SP3-130R primers was assessed in silico using the BLASTn tool against other sequences present in the GenBank database.
Multiplex PCR
Sequences of primer sets used to identify strains of S. parauberis of the serotypes I (subtypes Ia, Ib, and Ic), II and III using the optimized mPCR assay are shown in Table 2. Amplification was performed in 50 μl of reaction using Phire Hot Start II DNA polymerase (Thermo Scientific, Madrid, Spain), 2.4 μM of each primer and 2 μl of DNA template. The conditions of PCR amplifications were initial denaturation step of 98 °C for 10 min, followed by 30 cycles of denaturation at 98 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 90 s, and a final extension at 72 °C for 5 min. All multiplex products were checked by agarose gel electrophoresis (3% w/v in TAE 1X buffer) and stained with Redsafe nucleic acid staining solution (20,000×) (iNtRON Biotechnology, Seongnam-Si, Korea). GeneRuler 1-kb DNA ladder (Fermentas, Madrid, Spain) was used as molecular weight marker.
Table 2.
Primers used in this study
| Primers used for S. parauberis typing | Sequence (5′→3′) | Amplicon size | |
|---|---|---|---|
| Serotype I | For-Ia | ATTGTTAGTCATTCAGTTGT | 213 bp |
| Rev-Ia | AATTATAGTCAACAGTCCAG | ||
| For-Ib/Ic | ATTTCTACCAGGTTACTTTG | 303 bp | |
| Rev-Ib/Ic | ACATCTCGAAACTTCATATT | ||
| Serotype II | For-II | GAACTACTTAGGTTTAGCAT | 413 bp |
| Rev-II | ACTTGTAAATAGGATTGCT | ||
| Serotype III | SP3-130F | GACCAACACCAGCACCAATA | 130 bp |
| SP3-130R | TTTGGACAACCTGGAAGAGC | ||
Real-time PCR
A qPCR method was also optimized for specific detection of strains of S. parauberis of the serotype III in pure cultures and infected fish tissues. For this assay, bacterial suspensions containing 1 × 108 cell/ml (absorbance at 620 nm 0.131) and DNA extracted from bacterial cultures and fish tissues were used as template. Amplification was performed using Maxima SYBR Green qPCR Master Mix (2X), no ROX (Thermo Scientific, Waltham, MA, USA), 2.4 μM of each primer and 1 μl of template (bacterial suspensions or DNA from bacterial cultures or tissues of fish). The thermal cycling conditions comprised an initial denaturation of 5 min at 95 °C, followed by 30 cycles of denaturation at 95 °C for 30 s and annealing at 60 °C for 10–30 s. For bacterial suspensions, the initial denaturation step was increased to 10 min. A melt curve analysis was performed after the last cycle using a temperature gradient from 65 to 95 °C and a ramp speed of 0.5 °C s−1 (for 10 s) with continuous fluorescence data measurement. DNA isolated from pure cultures of the reference strain S. parauberis NCIMB 703043, which belongs to serotype III, was used as a positive control in qPCR reactions. Reactions lacking DNA template (no template control) and reactions containing DNA from non-infected tissues were used as negative controls. All qPCR assays were performed in triplicate. The expected size of the amplicons was confirmed by gel electrophoresis as described above.
The specificity in vitro of SP3-130F and SP3-130R primers was tested using multiplex and real-time PCR assays. In PCR assays, the existence of cross-amplifications was evaluated using as template bacterial cells and DNA from strains of serotype III of S. parauberis (n = 11 strains) and non-target bacteria (Table 1).
The sensitivity of the SP3-130F and SP3-130R primers was evaluated using amplification products of the reference strain NCIMB 703043 and a clinical isolate of S. parauberis serotype III. The amplification products were purified using the PureLink® PCR (Invitrogen, Barcelona, Spain) purification kit and quantified using Qubit® 2.0 and the Qubit® DNA HS Assay Kit (Invitrogen, Barcelona, Spain). Dilutions of the purified amplification product were used to generate a linear standard curve by plotting the Cq values obtained for each decimal amplicon dilution against the logarithm of the amount (ng) of the amplicon. Each amplicon dilution was tested in triplicate. The copy number of the amplification products was quantified as indicated by the IDT SciTools Web Tools (Integrated DNA Technologies, Coralville, IA, USA) using the formula:
where X is the amount of amplicon (ng), N is the length of dsDNA amplicon, and 660 g/mol is the average mass of 1 bp of dsDNA.
The linear standard curve was used to convert the Cq values obtained from the analysis of a sample into the copy number of the amplified product. Each decimal dilution of amplification products used to generate the linear standard curve by qPCR, were also analyzed by gel electrophoresis (2% w/v in TAE 1× buffer) to determine the analytical sensitivity of the gel electrophoresis correlated with the qPCR. The efficiency of the qPCR assay was calculated from the slope (m) of the line according to the eq. E = 10(−1/m) − 1.
Repeatability and reproducibility were evaluated by calculating the coefficient of variation (% CV) of replicates performed in the same sensitivity test (intra-assay) and of replicates performed in three independent sensitivity tests (inter-assay), respectively. For each test, three replicates were performed and only the results from concentrations that tested positive in all the three replicates were analyzed.
qPCR detection of S. parauberis serotype III in fish sample
The usefulness of primers (SP3-130F and SP3-130R) and qPCR protocol designed in this study for the detection and quantification of S. parauberis serotype III in tissues of fish inoculated with the bacterium was evaluated. Fish samples of kidney, spleen, and blood were homogenized at a concentration of 25% in saline (0.9% NaCl), seeded and incubated for 1 h with known amounts of bacterial suspension of NCIMB 703043 strain, resulting in final concentrations 5 × 107 to 5 × 100 cells/ml in the extract. To confirm the specificity of the qPCR protocol for detection of S. parauberis serotype III in fish samples, tissues samples were also artificially infected with strains representative of the other serotypes described in S. parauberis (Ia, strain KRS02083; Ib, strain NUF1003; Ic, strain NUF1071; and II, strain NUF1032). DNA from control (non-inoculated) and artificially infected fish tissues were obtained using Dynabeads DNA Direct™ (Thermo Fisher Scientific, Madrid, Spain) following manufacturer’s instructions.
The sensitivity and specificity of the qPCR was also evaluated using tissue samples (available in the laboratory), obtained from turbot experimentally infected or suffering from streptococcosis caused by S. parauberis serotype III. To exclude the possibility of non-specific amplifications, tissue samples from fish experimentally or naturally infected with other S. parauberis serotypes and other pathogens (Streptococcus iniae, Vagococcus salmoninarum, Yersinia ruckerii, L. garvieae, F. psychrophylum, and Aeromonas salmonicida) were tested in parallel. In these trials, tissue samples from healthy fish were used as negative controls. To control the quality of DNA obtained from the tissues, all fish samples were also analyzed using the primers β-actin-Fw (5′-CTGAAGTACCCCATTGAGCAT-3′) and β-actin-Rv (5′-CATCTTCTCCGTGCTT-3′) which amplify a 125 bp fragment of the β-actin gene from turbot (Fernandez-Álvarez et al. 2019). The standard curve previously performed and the Cq value obtained in those assays were used to calculate the amount of S. parauberis serotype III DNA in fish samples and thereby calculate the number of copies of the cps3K single copy gene in that sample.
Nucleotide sequence accession number
The partial sequences of the cps3K gene sequenced in this study have been deposited in the NCBI (Genbank) database under the accession numbers of MT040616 (strain NCIMB 703043), MT040617 (strain SK451/04), and MT040618 (strain SK537/10).
Results
General features of the cps loci of strains isolated from marine fish and mastitis
The genomes available at NCBI of strains isolated from marine fish (AZ70.1, T1, RP17, PL23, PL9, RP15, RP25, N198_2, N11, KCTC 11537 SK-417, KRS02083, KCTC11980, and KRS02109) and from mastitis (NCDFD 2020) were compared with the sequences of the loci cps of strains of the serotypes Ia (strain KRS02083, sequence accession no. LC060252), Ib (strain NUF1003, LC060253), Ic (strain NUF1071, LC060254), and II (strain NUF1032, LC060255) of S. parauberis previously described by Tu et al. (2015a). The locus cps detected in these genomes has the cassette-like structure previously described for serotypes I and II (Tu et al. 2015a), in which serotype-specific genes are flanked by conserved regions. The upper conserved region of the loci cps involves five regulatory genes, lysR (homology between serotypes > 99%) and cpsABCD (homology between serotypes > 95%), and a processing gene, cpsE (homology between serotypes > 98%) (Supplemental Table S2, Fig. 1). The lower conserved region involves two genes, the cpsQ gene, which encodes for a dehydrogenase responsible for capsule processing (homology between serotypes > 80%), and the cpsR gene, which encodes for a hypothetical protein (homology between serotypes > 99%) (Supplemental Table S2, Fig. 1). Based on the comparative genomic analysis, the variable regions of the cps loci previously associated (Tu et al. 2015a) with the serotypes Ia (strain KRS02083), Ib (NUF1003), Ic (NUF1071), and II (NUF1032) were clearly evidenced. The serotype-specific genes described for subserotype Ia (Tu et al. 2015a) were also identified in the whole genomes available in the NCBI database of strains isolated from Japanese flounder (KRS02083 and KCTC11980), suggesting that these strains were subserotype Ia. The serotype-specific region associated to serotype II was detected only in the whole genome of one strain isolated from Japanese flounder (KRS02109), suggesting that this strain was serotype II. Three variable regions (named as III, IV, and V), different to those described by Tu et al. (2015a) for subserotypes Ia, Ib, Ic, and serotype II, were also identified and represented in Fig. 1. The variable region III was detected in the genome of strains AZ70.1 (Supplemental Table S1 for NCBI accession no.) and T1 (Supplemental Table S1) isolated from turbot in Spain and in the genome of strains RP17, PL23, PL9, RP15, RP25, N198_2, and N11 (Supplemental Table S1) isolated from wild striped bass in USA. In this region, genes that encode transferases and epimerases were mostly detected (Supplemental Table S2 and Fig. 1). The cps3F gene of the region III, coding for a glucosyltransferase, showed homology of 99, 44, and 25% with the genes cps1aF (serotype Ia), cps1bG (serotype Ib), and cps4G (strain NCDFD2020), respectively. The genes cps3G and cps3P, both epimerases encoders, showed a homology of 75% with cps1aG (serotype Ia) and of 74% with cps1aP (serotype Ia) and cps1bP (serotype Ib), respectively (Fig. 1). The genes cps3H, cps3I, cps3J, cps3K, and cps3L were only detected in the variable region III. The variable region IV that contains mainly genes encoding for transferases and dehydrogenases (Supplemental Table S2 and Fig. 1) was identified in the genome sequence (Supplemental Table S1) of the reference strain NCDO 2020 isolated from mastitis sample milk The cps4G gene, coding for transferase, showed a homology > 25% with the genes cps1aF (serotype Ia), cps1bG (serotype Ib), and cps3F (strains AZ70.1, T1, RP17, PL23, PL9, RP15, RP25, N198_2, and N11) (Fig. 1). The genes cps4F, cps4H, cps4I, cps4J, cps4k, cps4L, cps4M, cps4N, and cps4O did not show homology with the genes of the other variable regions. The variable region V was identified in the genome of the strain KCTC 11537 (Supplemental Table S1) isolated from Japanese flounder and in the genome of strain SK-417 (Supplemental Table S1) isolated from Japanese black seaperch. No homology with the genes of the other variable regions was observed. The variable regions IV and V identified in the present study could represent serotype-specific regions of the new serotypes IV (strain representative NCDFD2020) and V (strains representatives KCTC 11537 and SK-417).
Fig. 1.
Structure of loci cps of S. parauberis detected in this study. In black are represented conserved regions of the cps locus of S. parauberis, in gray are represented genes that are present in two or more serotypes, in white are represented serotype-specific regions
To investigate if the gene cps3K of the variable region III, encoding the polysaccharide pyruvyl transferase family protein, was related with the serotype III, partial sequencing and BLAST analysis was carried out using DNA from the strains of S. parauberis serotype III isolated from turbot NCIMB 703043 (NCBI accession number MT040616), SK451/04 (NCBI accession number MT040617), and SK537/10 (NCBI accession number MT040618).The analysis of the partial sequences obtained revealed similarities greater than 99% with the gene cps3K of the variable region III of the loci cps of the strains AZ70.1, T1, RP17, PL23, PL9, RP15, RP25, N198_2, and N11 deposited in the NCBI database. This result suggests that strains AZ70.1, T1, RP17, PL23, PL9, RP15, RP25, N198_2, and N11 could belong to serotype III of S. parauberis (Fig. 1).
Typing of S. parauberis using serological tests and multiplex and real time PCR
Serological test allowed to assign the S. parauberis strains tested to the serotypes I (15% of strains), serotype II (15%) and serotype III (55% of strains). The strain NCDO 2020 only reacted with the homologous antiserum and was assigned to the new proposed serotype IV. Two strains did not react with the antisera tested and were designated as non-typeable (Table 1).
For primer design, the gene cps3K present in the variable region III of the locus cps detected in genomes of strains isolated from turbot (AZ70.1 and T1) and wild striped bass (strains RP17, PL23, PL9, RP15, RP25, N198_2, and N11) as well as in the genome of the strains NCIMB 703043, SK451/04, and SK537/10 was selected as target. In silico and in vitro analysis revealed that the primer pair designed in this study (SP3-130F and SP3-130R) for identifying strains of the serotype III of S. parauberis was specific (Table 1). Eleven strains of S. parauberis serotype III tested in this study, including the reference strain NCIMB 703043 and clinical strains isolated from diseased turbot, were positively identified by PCR at the optimal annealing temperature of 60 °C. No amplifications were found when bacterial suspensions or extracted DNA from strains of other serotypes of S. parauberis (n = 9) or other no-target bacteria (n = 53) were used (Table 1).
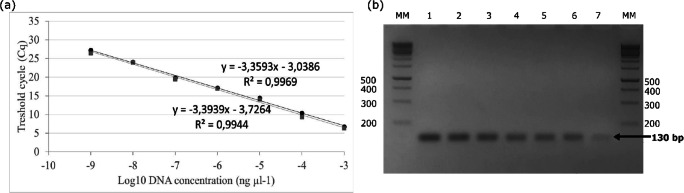
Multiplex PCR allowed the typing of S. parauberis strains belonging to subserotype Ia (specific band of 213 bp), serotype II (413 bp), and serotype III (130 bp) (Fig. 2). Non-typeable strains were also detected. The mPCR assay do not allow to differentiate subserotypes Ib and Ic, which showed a characteristic band of 303 bp (Fig. 2). No amplification products were observed using water as a negative control, DNA extracted from reference strain of S. parauberis NCDO 2020, or DNA extracted from other unrelated bacteria. Therefore, the results of molecular serotyping using the mPCR assay were comparable with the results obtained by serological tests (Table 1).
Fig. 2.
Multiplex PCR products of S. parauberis strains. Lanes: MM, Generuler DNA ladder 1 kb (Fermentas); 1, (subserotype Ia); 2, subserotype Ib; 3, subserotype Ic; 4, serotype II; 5, serotype III; 6 and 7, non-typeable strains; 8, NCDO2020 (serotype IV)
Typing of S. parauberis serotype III was also performed by qPCR using bacterial suspensions or DNA extracted from bacterial cultures or tissues from fish infected with strains of S. parauberis serotype III as template. A specific peak with a melting temperature (Tm) of 73 ± 0.33 °C (Fig. 3) was obtained with DNA samples or bacterial suspensions from strains of S. parauberis of serotype III. No amplifications were observed using bacterial suspensions or DNA extracted from other serotypes of S. parauberis or unrelated bacteria (Fig. 3, Table 1). On the standard curve, regression coefficients R2 = 0.997 (m = − 3.3593; efficiency 98.5%) were obtained using the DNA of the reference strain NCIMB 703043 (Fig. 4a) and R2 = 0.994 (m = − 3.3939; efficiency 97.08%) using the DNA of a clinical isolate (strain SK 451/04) of the serotype III of S. parauberis (Fig. 4a). The limit of detection of the assay was 2.67 × 102 amplicon copies per assay (equivalent to 3.8 × 10−9 ng μl−1) (Table 3, Fig. 4a) using as template the purified amplicon of both strains. The amplification products obtained after the qPCR sensitivity test were electrophoresed at 2% agarose gel (Fig. 4b). The intra-assay % CV values ranged from 0.17 to 7.53 (Table 3) and inter-assay % CV values ranged from 0.50 to 6.33 (Table 3).
Fig. 3.
Melting curve analysis using qPCR and SP3-130F and SP3-130R primers, showing a specific melting peak at Tm 73 ± 0.40 °C using bacterial DNA from strains of Streptococcus parauberis serotype III (n = 11 strains) and the lack of amplification obtained using DNA from strains of other serotypes of S. parauberis or unrelated bacteria and negative control
Fig. 4.
Standard curves obtained by qPCR from the amplification of 10-fold dilutions of purified amplicon obtained using SP3-130F and SP3-130R primers and DNA from the reference strain S. parauberis NCIMB 703043 (black circles) and a clinical isolate of S. parauberis (SK451/04) (gray squares) (a) and the corresponding electrophoresis gel showing the band intensities correlating with the number of copies of the cps3K gene (b). Lanes: MM, Generuler DNA ladder 1-kb (Fermentas); 1, 2.67 × 108 copies; 2, 2.67 × 107 copies; 3, 2.67 × 106 copies; 4, 2.67 × 105 copies; 5, 2.67 × 104 copies; 6, 2.67 × 103 copies; 7, 2.67 × 102 copies
Table 3.
Determination of Cq values obtained from the amplification of tenfold dilutions of a purified amplicon of the cps3K gene of the strain NCIMB 703043 obtained by qPCR
| Intra-assay variance | Inter-assay variance | |||||
|---|---|---|---|---|---|---|
| DNA concentration (ng/μl) | Cq values | No. of target copies | %CV | Cq values | No. of target copies | %CV |
| 3.8 × 10−3 | 6.77 ± 0.51 | 2.67 × 108 | 7.53 | 6.48 ± 0.41 | 2.67 × 108 | 6.33 |
| 3.8 × 10−4 | 10.40 ± 0.10 | 2.67 × 107 | 0.96 | 10.71 ± 0.44 | 2.67 × 107 | 4.09 |
| 3.8 × 10−5 | 14.43 ± 0.08 | 2.67 × 106 | 0.55 | 13.97 ± 0.65 | 2.67 × 106 | 4.66 |
| 3.8 × 10−6 | 17.11 ± 0.21 | 2.67 × 105 | 1.23 | 16.98 ± 0.19 | 2.67 × 105 | 1.12 |
| 3.8 × 10−7 | 19.83 ± 0.06 | 2.67 × 104 | 0.30 | 19.90 ± 0.10 | 2.67 × 104 | 0.50 |
| 3.8 × 10−8 | 24.07 ± 0.04 | 2.67 × 103 | 0.17 | 23.80 ± 0.39 | 2.67 × 103 | 1.63 |
| 3.8 × 10−9 | 27.21 ± 0.11 | 2.67 × 102 | 0.40 | 27.13 ± 0.12 | 2.67 × 102 | 0.44 |
Applicability of the qPCR assay to detect S. parauberis serotype III in fish samples
Detection levels of 4.52 × 108 to 1.76 × 102 copies of the cps3K gene per μl were obtained using qPCR protocol and samples generated in the laboratory (extracted DNA, bacterial suspensions or artificially infected fish tissues) as template (Table 4). The bacterial load detected in tissues from diseased turbot with clinical signs of streptococcosis ranged from 1.25 × 104 to 3.02 × 102 copies of the cps3K gene (Cq values ranging from 21.58 to 27.02). Positive amplifications were also obtained using as template DNA extracted from tissues of fish experimentally infected without clinical signs of streptococcosis, detecting from 1.07 × 103 to 1.76 × 102 copies of the cps3K gene (Cq values ranging from 25.17 to 27.82). No amplifications were observed using as template DNA extracted from tissue samples from healthy fish, from fish infected with S. parauberis of other serotypes or with other bacterial pathogen. The β-actin gene was detected in all fish tissue samples, discarding false negative results.
Table 4.
Samples tested by qPCR for the detection of S. parauberis serotype III
| Sample | Concentration range | Results (Cq ranges) | No. of target copies |
|---|---|---|---|
| Genomic DNA | 3.8 × 10−3 – 3.8 × 10−9 (ng/μl) | + (6.27–27.21) | 4.52 × 108–2.67 × 102 |
| Bacterial | 1.0 × 108–1.0 × 101 (CFU/ml) | + (10.36–27.55) | 2.74 × 107–2.11 × 102 |
| Kidney and spleen | 5 × 107 - 5 × 103 (CFU/ml) | + (14.71–27.81) | 1.39 × 106–1.76 × 102 |
| Blood | 5 × 107–5 × 105 (CFU/ml) | + (23.94–25.71) | 2.49 × 103–7.37 × 102 |
Discussion
Accurate and rapid determination of the S. parauberis serotype is of crucial importance for surveillance studies and vaccine development. S. parauberis was originally described as phenotypically and serologically homogeneous (Toranzo et al. 1995), but several serological types have been reported over the years using conventional serotyping methods (Kanai et al. 2009, 2015; Torres Corral et al. 2019). Conventional serotyping based on the use of mono- or polyclonal sera and inactivated whole cell is the method more commonly used in clinical diagnostic laboratories. However, the use of this method is limited by the unavailability of specific commercial sera which difficult the comparison of the results within laboratories. PCR-based bacterial serotyping protocols have therefore emerged as a promising alternative to conventional serotyping. These mechanisms do not require the use of sera, but it is necessary to identify molecular markers that determine serotypes as a prerequisite for designing specific primers (Fratamico et al. 2016; Rochat et al. 2017).
In order to identify a molecular marker determining S. parauberis serotype III, whole genome of 15 strains of S. parauberis isolated from different hosts and geographical origins (Nho et al. 2011. 2013; Park et al. 2013; Oguro et al. 2014; Haines et al. 2016), were compared with the sequences of the loci cps described by Tu et al. (2015a) for serotypes I and II. A locus cps with a cassette-like structure, a variable region flanked by conserved regions, was found in 15 genomes of S. parauberis analyzed in the present study. This locus structure was previously described in the cps loci of S. parauberis strains belonging to serotypes I and II (Tu et al. 2015b), and other Streptococcus (Bentley et al. 2006; Jiang et al. 2006; Wang et al. 2011; Morais et al. 2018). In addition, in the present study, three variable regions (named III, IV, and V) within the loci cps, different from those described for serotypes I and II, were identified. These variable regions could represent serotype-specific regions of three different serotypes (serotypes III, IV, and V) of S. parauberis. In silico comparative analysis of the partial sequence of the cps3K gene of the variable region III obtained from three S. parauberis strains of the serotype III showed more than 99% homology with the variable region III detected in the genome of S. parauberis available in the NCBI database and less than 40% homology with genes encoding the same protein in other bacterial species. These results indicated that cps 3K gene is specific of the serotype III strains and suggest that strains isolated from turbot in Spain and striped bass in USA, which genomes are deposited in the NCBI database, could be included into the serotype III of S. parauberis. Based on these results cps 3K gene constitutes an excellent target for design of primers for the detection of strains of S. parauberis serotype III, thereby decreasing the possibility of false positive results. Thus, a primers pair SP3-130F and SP3-130R were designed based on the sequence of the cps3K gene for rapid and specific detection of strains of S. parauberis serotype III using PCR assays. Multiplex PCR assay was optimized for the detection and differentiation of the main serotypes of S. parauberis causing disease in fish (serotypes I, II and III) using the primers described by Tu et al. (2015b) and the primers SP3-130F and SP3-130R designed in the present study. The high correspondence observed between the results of conventional serotyping using antisera and molecular serotyping using multiplex PCR suggests that this rapid and specific typing method could be used in clinical diagnostic laboratories and/or epidemiological studies.
Typing of strains of S. parauberis of serotype III was also performed by qPCR using bacterial suspensions or DNA extracted from bacterial cultures or tissues from fish infected with strains of S. parauberis serotype III as template. PCR analysis demonstrated 100% specificity, since only DNA from strains of S. parauberis serotype III were amplified giving a specific peak at a Tm of 73 ± 0.33 °C. Similar Tm value was obtained using as template DNA from fish tissues experimentally or naturally infected with S. parauberis of serotype III. No amplifications were observed using bacterial suspensions or DNA extracted from other serotypes of S. parauberis or unrelated bacteria. The quantification of the bacterial load was assessed through a standard curve generated by plotting the values of Cq versus the logarithm of the amount (ng) of amplicon as previously described (Fernández-Álvarez et al. 2016; Torres-Corral et al. 2019). A detection limit of 2.67 × 102 copies of the amplicon per μl (equivalent to 3.8 × 10−9 ng/μl of genomic DNA) was obtained using the designed qPCR procedure.
The applicability of the qPCR protocol and SP3-130F and SP3-130R primers designed was tested using lethal (kidney and spleen) and non-lethal (blood) fish samples. High specificity and sensitivity were found using the described protocol and lethal fish samples, with detection levels of 1.25 × 104 to 1.76 × 102 gene copies of the cps3K gene in tissues of fish infected with strains of S. parauberis belonging to serotype III. High levels of detection (2.49 × 103–7.37 × 102 gene copies) were also observed using this qPCR protocol and DNA obtained from non-lethal fish samples (blood samples) as template, suggesting that this non-lethal diagnostic and typing method could be used without the need for bacterial isolation or the use of anti-S. parauberis sera. Detection levels using DNA extracted from infected fish tissues were lower than those obtained from pure bacterial cultures (bacterial suspensions or DNA extracted from bacterial cultures), possibly due to the presence of host DNA or tissue inhibitors such as hemoglobin or serum proteins (Wiklund et al. 2000; Cepeda et al. 2003; Fernández-Álvarez et al. 2019; Torres-Corral et al. 2019). Further studies using a high number of strains isolated from different fish species and other animals in different geographic area will help to determine the serological diversity of S. parauberis and to develop new diagnostic tools.
In conclusion, this study describes the predicted structure of the cps loci involved in CPS biosynthesis for S. parauberis of the serotypes III, IV, and V, suggesting the existence of new serotypes within this species. Serotype-specific regions were used to optimize a mPCR protocol capable of differentiating the main S. parauberis serotypes causing disease in fish (serotypes I, II, and III). In addition, a qPCR assay to identify and quantify strains of S. parauberis belonging to serotype III from bacterial cultures and fish tissues has been optimized. These PCR protocols could be useful tool in epidemiological surveillance studies or for diagnostic purposes using bacterial culture and lethal and non-lethal fish tissues as samples. The PCR methods described are inexpensive with low risk of contamination that can be implemented as a tool for diagnosis and typing in clinical laboratories, thus avoiding the disadvantages of conventional serological methods.
Electronic supplementary material
(PDF 110 kb)
Acknowledgments
This work was partially supported by the Proof of Concept Program “Acelerador de Transferencia” from the Universidade of Santiago de Compostela (Spain). Yolanda Torres Corral was supported by a grant from the Universidade of Santiago de Compostela under the program “Acelerador de Transferencia”. The authors are grateful to Dr. Kanai for supply the antisera against serotype I (subtypes Ia, Ib, I c) and II of S. parauberis and their representative strains and to Álvaro Robles, from the Aquaculture Division of Isidro 1952 SL, for supplying the fish used in this study.
Authors’ contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Yolanda Torres-Corral and Ysabel Santos. The first draft of the manuscript was written by Yolanda Torres-Corral and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding information
This work was partially supported by the Proof of Concept Program “Acelerador de Transferencia” from the Universidade of Santiago de Compostela (Spain).
Availability of data and material
The dataset used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The studies presented in this manuscript were approved by the USC Bioethics Committee.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yolanda Torres-Corral, Email: yolanda.torres.corral@gmail.com.
Ysabel Santos, Email: ysabel.santos@usc.es.
References
- Aoki T, Takami K, Kitao T. Drug resistance in a non-hemolytic Streptococcus sp. isolated from cultured yellowtail Seriola quinqueradia. Dis Aquat Org. 1990;8:171–177. doi: 10.3354/dao008171. [DOI] [Google Scholar]
- Austin B, Austin DA. Bacterial fish pathogens, Sixth Edit. Chichester: Springer; 2016. [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeck GW, Kim JH, Gomez DK, Park SC. Isolation and characterization of Streptococcus sp. from diseased flounder (Paralichthys olivaceus) in Jeju Island. J Vet Sci. 2006;7:53–58. doi: 10.4142/jvs.2006.7.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller N. Bacteria and fungi from fish and other aquatic animals, 2nd edit. Bentley: CABI Publishing; 2014. [Google Scholar]
- Cepeda, García-Márquez S, Santos Y. Detection of Flexibacter maritimus in fish tissue using nested PCR amplification. J Fish Dis. 2003;26:65–70. doi: 10.1046/j.1365-2761.2003.00431.x. [DOI] [PubMed] [Google Scholar]
- Cepeda C, García-Márquez S, Santos Y. Improved growth of Flavobacterium psychrophilum using a new culture medium. Aquaculture. 2004;238:75–82. doi: 10.1016/j.aquaculture.2004.05.013. [DOI] [Google Scholar]
- Cho MY, Il LJ, Kim MS, Choi HJ, Lee DC, Kim JW. Isolation of Streptococcus parauberis from starry flounder, Platichthys stellatus Pallas. J Fish Pathol. 2008;21:209–2017. [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704.tion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demczuk W, Martin I, Mulvey M, Mauffrey F, Bekal S, Domingo M, Martineau C, Le S, Doualla-bell F, Longtin J, Lefebvre B. Comparison of sequential multiplex PCR, sequetyping and whole genome sequencing for serotyping of Streptococcus pneumoniae. PLoS One. 2017;12:1–16. doi: 10.1371/journal.pone.0178040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech A, Fernandez Garayzabal JF, Pascual C, Garcia JA, Cutuli MT, Moreno MA, Collins MD, Dominguez L. Streptococcosis in cultured turbot, Scophthalmus maximus (L), associated with Streptococcus parauberis. J Fish Dis. 1996;19:33–38. doi: 10.1111/j.1365-2761.1996.tb00117.x. [DOI] [Google Scholar]
- Fernández-Álvarez C, González SF, Santos Y. Development of a SYBR green I real-time PCR assay for specific identification of the fish pathogen Aeromonas salmonicida subspecies salmonicida. Appl Microbiol Biotechnol. 2016;100:10585–10595. doi: 10.1007/s00253-016-7929-2. [DOI] [PubMed] [Google Scholar]
- Fernández-Álvarez C, González SF, Santos Y. Quantitative PCR coupled with melting curve analysis for rapid detection and quantification of Tenacibaculum maritimum in fish and environmental samples. Aquaculture. 2019;498:289–296. doi: 10.1016/j.aquaculture.2018.08.039. [DOI] [Google Scholar]
- Fratamico PM, Debroy C, Liu Y, Needleman DS, Baranzoni GM, Feng P. Advances in molecular serotyping and subtyping of Escherichia coli. Front Microbiol. 2016;7:1–8. doi: 10.3389/fmicb.2016.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines AN, Gauthier DT, Nebergall EE, Cole SD, Nguyen KM, Rhodes MW, Vogelbein WK. First report of Streptococcus parauberis in wild finfish from North America. Vet Microbiol. 2013;166:270–275. doi: 10.1016/j.vetmic.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Haines A, Nebergall E, Besong E, Council K, Lambert O (2016) Draft genome sequences for seven Streptococcus parauberis isolates from wild fish in the Chesapeake Bay 4:1–2 . 10.1128/genomeA.00741-16.Copyright [DOI] [PMC free article] [PubMed]
- Jiang S, Wang LEI, Reeves PR. Molecular characterization of Streptococcus pneumoniae type 4 ,6B, 8 and 18C capsular polysaccharide gene clusters. Infect Immun. 2006;69:1244–1255. doi: 10.1128/IAI.69.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai K, Yamada M, Meng F, Takakahashi I, Nagano T. Serological differentiation of Streptococcus parauberis strains isolated from cultured Japanese flounder in Japan. Fish Pathol. 2009;44:33–39. doi: 10.3147/jsfp.44.33. [DOI] [Google Scholar]
- Kanai K, Tu C, Katayama N, Suga K. Existence of subserotypes in Streptococcus parauberis serotype I. Fish Pathology. 2015;50:75–90. doi: 10.3147/jsfp.50.75. [DOI] [Google Scholar]
- Kannika K, Pisuttharachai D, Srisapoome P, Wongtavatchai J, Kondo H, Hirono I, Unajak S, Areechon N. Molecular serotyping, virulence gene profiling and pathogenicity of Streptococcus agalactiae isolated from tilapia farms in Thailand by multiplex PCR. J Appl Microbiol. 2017;122:1497–1507. doi: 10.1111/jam.13447. [DOI] [PubMed] [Google Scholar]
- Khan IU, Hassan AA, Lämmler AAC, Wolter W, Zschöck M. Identification and epidemiological characterization of Streptococcus uberis isolated from bovine mastitis. J Vet Sci. 2003;4:213–223. doi: 10.4142/jvs.2003.4.3.213. [DOI] [PubMed] [Google Scholar]
- Morais V, Dee V, Suárez N. Purification of capsular polysaccharides of Streptococcus pneumoniae : traditional and new methods. Front Microbiol. 2018;6:145. doi: 10.3389/fbioe.2018.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nho SW, Hikima JI, Cha IS, Bin PS, Bin JH, del Castillo CS, Kondo H, Hirono I, Aoki T, Jung TS. Complete genome sequence and immunoproteomic analyses of the bacterial fish pathogen Streptococcus parauberis. J Bacteriol. 2011;193:3356–3366. doi: 10.1128/JB.00182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nho SW, Hikima JI, Park SB, Jang HB, Cha IS, Yasuike M, Nakamura Y, Fujiwara A, Sano M, Kanai K, Kondo H, Hirono I, Takeyama H, Aoki T, Jung TS (2013) Comparative genomic characterization of three Streptococcus parauberis strains in fish pathogen, as assessed by wide-genome analyses. PLoS One 8:1–13. 10.1371/journal.pone.0080395 [DOI] [PMC free article] [PubMed]
- Oguro K, Yamane J, Yamamoto T, Ohnishi K, Oshima S-I, Imajoh M. Draft genome sequence of Streptococcus parauberis strain SK-417, isolated from diseased Sebastes ventricosus in Kagoshima, Japan. Genome Announc. 2014;2:e00453–e00414. doi: 10.1128/genomeA.00453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi K, Oinaka D, Hoai TD, Yoshida T, Nishiki I. PCR-mediated identification of the newly emerging pathogen Lactococcus garvieae serotype II from Seriola quinqueradiata and S. dumerili. Fish Pathol. 2017;52:46–49. doi: 10.3147/jsfp.52.46. [DOI] [Google Scholar]
- Okura M, Takamatsu D, Maruyama F, Nozawa T, Nakagawa I, Osaki M. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis : potential mechanisms for generation of capsular variation. Appl Environ Microbiol. 2013;79:2796–2806. doi: 10.1128/AEM.03742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MA, Kwon MG, Hwang JY, Jung SH, Kim D-W, Park J-Y, Kim J-S, Na Y-J, Kim M-Y, Kim D-S, Chae S-H, Seo JS. Genome sequence of Streptococcus parauberis strain KCTC11980, isolated from diseased Paralichthys olivaceus. Genome Announc. 2013;1:5–6. doi: 10.1128/genomeA.00780-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F, Santos Y, Macías AR, Núñez S, Toranzo AE. Evaluation of media for the successful culture of Flexibacter maritimus. J Fish Dis. 1996;19:193–197. doi: 10.1111/j.1365-2761.1996.tb00701.x. [DOI] [Google Scholar]
- Rochat T, Fujiwara-nagata E, Calvez S, Dalsgaard I, Madsen L, Calteau A, Lunazzi A, Nicolas P, Wiklund T, Bernardet J, Duchaud E, Lawrence ML. Genomic characterization of Flavobacterium psychrophilum serotypes and development of a multiplex PCR-based serotyping scheme. Front Microbiol. 2017;8:1752. doi: 10.3389/fmicb.2017.01752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CA, Xu D, García JC, Lafrentz BR. Capsular typing of Streptococcus agalactiae (Lancefield group B streptococci) from fish using multiplex PCR and serotyping. Bull Eur Assoc Fish Pathol. 2017;37:2017. [Google Scholar]
- Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toranzo AE, Cutrin JM, Nunez S, Romalde JL, Barja JL. Antigenic characterization of Enterococcus strains pathogenic for turbot and their relationship with other Gram-positive bacteria. Dis Aquat Org. 1995;21:187–191. doi: 10.3354/dao021187. [DOI] [Google Scholar]
- Torres Corral Y, Fernández Álvarez C, Santos Y. Proteomic and molecular fingerprinting for identification and tracking of fish pathogenic Streptococcus. Aquaculture. 2019;498:322–334. doi: 10.1016/j.aquaculture.2018.08.041. [DOI] [Google Scholar]
- Torres-Corral Y, Fernández-Álvarez C, Santos Y. High - throughput identification and quantification of Vagococcus salmoninarum by SYBR Green I - based real - time PCR combined with melting curve analysis. J Fish Dis. 2019;42:1359–1368. doi: 10.1111/jfd.13053. [DOI] [PubMed] [Google Scholar]
- Tu C, Suga K, Kanai K. Structure of genetic loci for capsular polysaccharide biosynthesis in Streptococcus parauberis isolated from japanese flounder. Fish Pathol. 2015;50:192–199. doi: 10.3147/jsfp.50.192. [DOI] [Google Scholar]
- Tu C, Suga K, Kanai K. A multiplex PCR assay for differentiation of Streptococcus parauberis serotypes. Fish Pathol. 2015;50:213–215. doi: 10.3147/jsfp.50.213. [DOI] [Google Scholar]
- Wang K, Fan W, Cai L, Huang B, Lu C. Genetic analysis of the capsular polysaccharide synthesis locus in 15 Streptococcus suis serotypes. FEMS Microbiol Lett. 2011;324:117–124. doi: 10.1111/j.1574-6968.2011.02394.x. [DOI] [PubMed] [Google Scholar]
- Wiklund T, Madsen L, Bruun MS, Dalsgaard I. Detection of Flavobacterium psychrophilum from fish tissue and water samples by PCR amplification. J Appl Microbiol. 2000;88:299–307. doi: 10.1046/j.1365-2672.2000.00959.x. [DOI] [PubMed] [Google Scholar]
- Williams AM, Collins MD. Molecular taxonomic studies on Streptococcus uberis types I and II. Description of Streptococcus parauberis sp nov. J Appl Bacteriol. 1990;68:485–490. doi: 10.1111/j.1365-2672.1990.tb02900.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 110 kb)
Data Availability Statement
The dataset used and/or analyzed during the current study are available from the corresponding author on reasonable request.