Abstract
Aberrant expression of PDGFR-β is associated with a number of diseases. The G-quadruplexes (G4s) formed in PDGFR-β gene promoter are transcriptional modulators and amenable to small molecule targeting. The major G4 formed in the PDGFR-β gene promoter was previously shown to have a broken G-strand. Herein, we report that the PDGFR-β gene promoter sequence forms a vacancy G-quadruplex (vG4) which can be filled-in and stabilized by physiologically relevant guanine metabolites, such as dGMP, GMP, and cGMP, as well as guanine-derivative drugs. We determined the NMR structure of the dGMP-fill-in PDGFR-β vG4 in K+ solution. This is the first structure of a guanine-metabolite-fill-in vG4 based on a human gene promoter sequence. Our structure and systematic analysis elucidate the contributions of Hoogsten hydrogen bonds, sugar, and phosphate moieties to the specific G-vacancy fill-in. Intriguingly, an equilibrium of 3′- and 5′-end vG4s is present in the PDGFR-β promoter sequence and dGMP favors the 5′-end fill-in. Guanine metabolites and drugs were tested and showed a conserved selectivity for the 5′-vacancy, except for cGMP. cGMP binds both the 3′- and 5′-end vG4s and forms two fill-in G4s with similar population. Significantly, guanine metabolites are involved in many physiological and pathological processes in human cells, thus our results provide a structural basis to understand their potential regulatory functions by interaction with promoter vG4s. Moreover, the NMR structure can guide rational design of ligands that target the PDGFR-β vG4.
Keywords: Vacancy G-quadruplex, PDGFR-β promoter, Guanine metabolite, Guanine drug, NMR structure
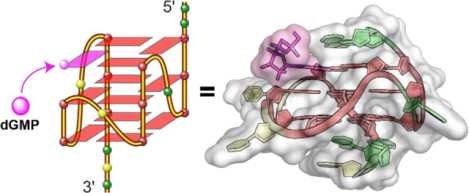
Graphical Abstarct
INTRODUCTION
G-quadruplexes (G4s) are DNA or RNA secondary structures formed by stacked Hoogsteen hydrogen-bonded G-tetrads and stabilized by K+ or Na+.1–2 G4s have been visualized in chromosomes of human cells and are found in human regulatory chromatin.3–5 G4-forming sequences are accumulated in specific regions of the human genome, such as oncogene promoters, telomeres, and 5′-UTR, and affect gene expression and genome stability.2, 6–8 Thus the corresponding G4 structures have emerged as novel therapeutic targets.9–12
The platelet-derived growth factor receptor beta (PDGFR-β) is a cell-surface-receptor tyrosine kinase, which is essential for cellular growth, proliferation, survival, motility, and differentiation.13–14 Aberrant expression of PDGFR-β is associated with a number of pathologies including cancer, atherosclerosis, rheumatoid diseases, and fibrotic disorders.15–17 In the PDGFR-β gene promoter, a nuclease hypersensitivity element (NHE) is located in the highly GC-rich proximal promoter region −165 to −132 nt upstream of the transcriptional start site (TSS), which is crucial for basal promoter activity and can form different G4s (Figure 1a).14 Small molecules can target the PDGFR-β G4s to inhibit gene transcription.14–15 Intriguingly, all the G4s formed in the PDGFR-β promoter sequence contain a broken-strand core.13, 17 We previously determined the major G4 in the PDGFR-β promoter region using the 22-mer Pu22m1 DNA sequence (Figure 1a). It is an intramolecular broken-strand G4 in which the 3′-tetrad is filled-in by the 3′-terminal G22 connected with a 5-nt fold-back loop (Figure 1b).13 The fill-in by the discontinued G22 was shown to be more dynamic in 3′-extended sequences in which G22 is not a terminal residue.13 Previous studies reported a unique type of intramolecular vacancy G-quadruplex (vG4) that is formed by three G3 tracts and one G2 tract and has an incomplete G-tetrad with a G-vacancy, which can be filled-in by a guanine derivative, such as GMP or GTP (Figure 1c and Table S1).18–21 The Pu22 (Table S2) PDGFR-β promoter sequence also has one G2 tract and can potentially form a vG4 that may be filled-in by guanine metabolites or derivatives.
Figure 1.
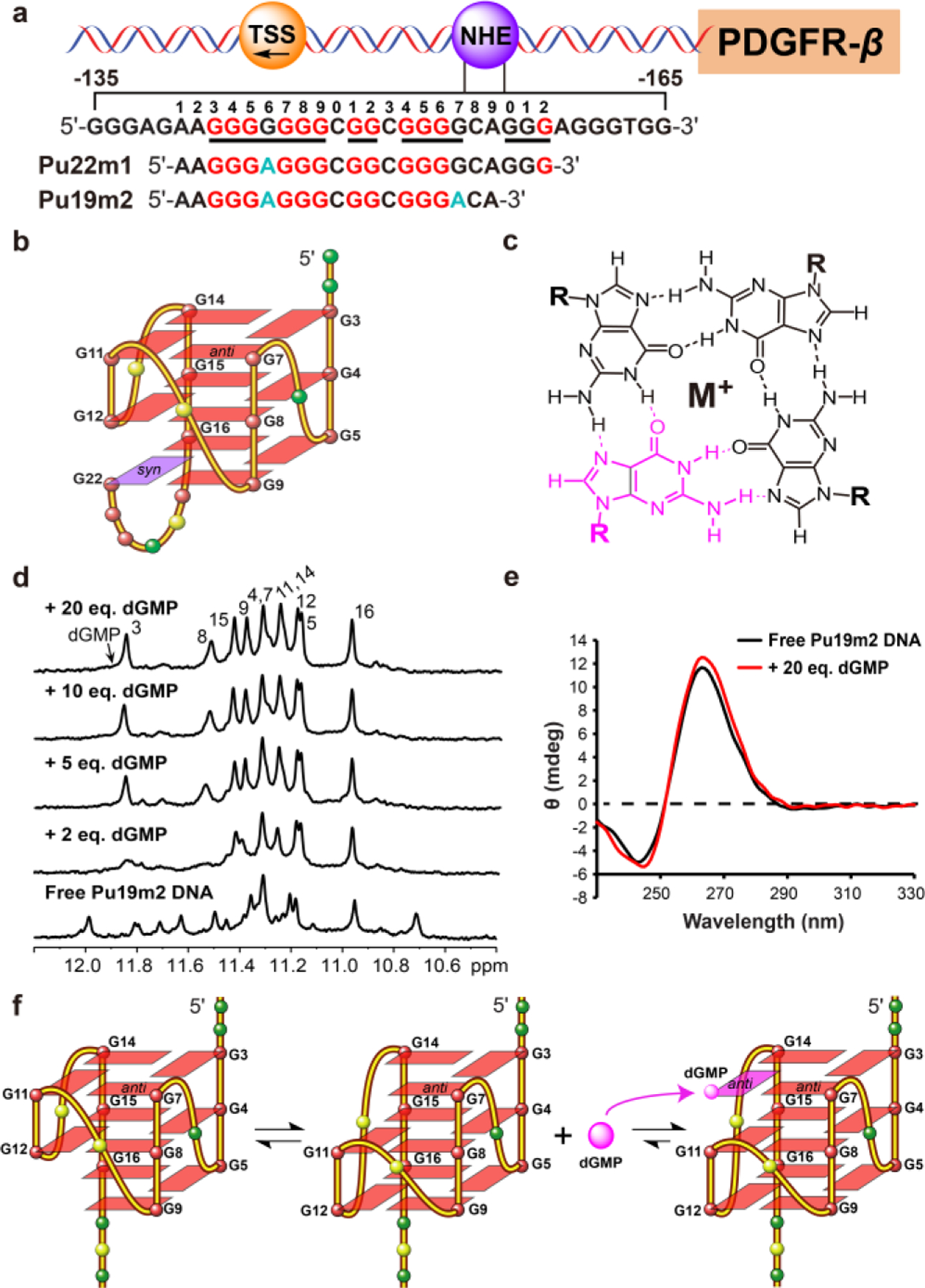
a) The schematic of the human PDGFR-β gene promoter. The G4-forming region of NHE sequence and its modifications are shown. The guanine runs involved in the formation of the major G4 and mutations are highlighted in red and cyan, respectively. b) Schematic structure of the major PDGFR-β G4 with the sequence of Pu22m1. c) An intact G-tetrad layer including the bound guanine derivative (pink). Hoogsteen hydrogen bonds are indicated by dashed lines. d) 1D 1H NMR titration of Pu19m2 DNA by dGMP with assignment of complex at 25 °C. e) CD spectra of Pu19m2 DNA in the presence and absence of 20 dGMP equivalents. f) A schematic model of Pu19m2 vG4s and dGMP fill-in.
In this study, we report that the human PDGFR-β gene promoter sequence forms a vG4 which can be filled-in by guanine metabolites, such as dGMP, GMP, cGMP, and guanine-derivative drugs. We determined the NMR structure of the dGMP-fill-in PDGFR-β vG4 in K+ solution. In addition, we systemically tested the fill-in activity of a series of guanine metabolites and drugs to the PDGFR-β vG4. Our results provide a structural basis to understand the potential regulatory functions of guanine metabolites by interaction with promoter vG4s and insights into design of ligands targeting the PDGFR-β vG4.
RESULTS AND DISCUSSION
PDGFR-β Gene Promoter Sequence Forms a vG4 That Can be Filled-in and Stabilized by dGMP.
We examined the binding of deoxyguanosine monophosphate (dGMP) (Table S1) to the potential vG4s of different Pu22m1 variants using 1H NMR titration experiments in K+-containing solution. Pu22m1 forms a broken-strand G4 with twelve characteristic tetrad-guanine imino protons between 10−12 ppm (Figures 1b and S1). No changes were observed upon dGMP titration to Pu22m1 (Figure S1), indicating that the dGMP cannot compete with the intramolecular 3′-terminal G22 in Pu22m1 for 3′-tetrad fill-in. We then truncated the Pu22m1 sequence by 2 and 3 nucleotides at the 3′-terminus (Table S2) to de-stabilize the formation of the fold-back loop for intramolecular fill-in. Both truncated sequences Pu20m1 and Pu19m1 do not form a single well-defined intramolecular G4 and can be filled-in by dGMP in a similar way as shown by NMR titration spectra (Figures S2 and S3). The fill-in by dGMP was supported by the DMS foot-printing of the wild-type Pu19 (Table S2), which showed that the addition of dGMP stabilize a major G4 formation as shown by clearly protected 11 guanines in the sequence (Figure S4). In addition, G6 and G17 were shown not to be involved in the major G4 formation (Figure S4). Therefore, we prepared Pu19m2 (Figure 1a) with G6-to-A and G17-to-A mutations.
The free Pu19m2 DNA adopts two stable G4s as indicated by ~ 20 imino peaks between 10.5−12.0 ppm characteristic of tetrad-guanines (Figure 1d). This is likely due to the coexistence of the 3′- and 5′-end vG4s, as the G2 tract can move up and down (Figure 1f). Upon titration with dGMP, the number of peaks decreased and new peaks appeared. At ratios over 1:5, a total of 11 well-resolved tetrad-guanine imino protons was observed at 25 °C (Figures 1d and S5). At 5 °C, one additional imino peak was seen for the fill-in dGMP (Figure S10). These results showed a well-defined dGMP-Pu19m2 complex with high-quality NMR spectra that is suitable for structure determination (Figures 1d and S5). This dGMP-Pu19m2 complex is unimolecular, whereas the free Pu19m2 adopts monomeric and dimeric topologies, as shown by a native EMSA gel (Figure S6). An increase of ~15 °C in Tm of Pu19m2 was observed in the dGMP complex (~60 °C) as compared to the unimolecular free vG4 (~45 °C) in 50 mM K+-containing solution (Figure S7). The CD spectra of both free Pu19m2 and its dGMP complex showed the characteristic 264 nm positive band and 240 nm negative band for a parallel G4, with enhanced CD bands in the presence of 20 eq. dGMP (Figure 1e).
NMR Structure Determination of the 5′-end dGMP-fill-in PDGFR-β vG4.
The H1 (Figure S8) and H8 protons (Figure S9) of each Pu19m2 guanine were unambiguously assigned using site-specific 15N-labeled DNA and 1D 15N-edited NMR experiments in presence of 20 eq. dGMP at 25 °C. In addition, the H1 and H8 proton for the bound dGMP were determined using 13C,15N-labeled dGMP and 15N-edited or 13C-edited experiments at 5 °C and 25 °C, respectively, as well as a 1H-13C-HSQC experiment (Figures S8–S11). Subsequently, the complete proton assignment of the dGMP-Pu19m2 G4 was achieved using 2D TOCSY, HSQC, and NOESY spectra at different mixing times and temperatures in both H2O and D2O. Overall, the dGMP-Pu19m2 complex structure from the PDGFR-β promoter is a 3-tetrad, parallel-stranded G4 with three 1-nucleotide double-chain reversal loops (Figure 1f). The nuclear Overhauser effect (NOE) cross-peaks of guanine H1−H1 and H1−H8 protons determined the arrangement and topology of three G-tetrad planes: G3-G7-dGMP-G14, G4-G8-G11-G15, and G5-G9-G12-G16 (Figures 1f, 2b, and S12). Surprisingly, dGMP fills only into the 5′-tetrad, in contrast to the 3′-tetrad fill-in by the 3′-terminal G22 in the intramolecular broken-strand G4 (Figures 1b and 1f). Apparently, dGMP modulates the equilibrium of 3′- and 5′-end vG4s and favors the formation of the 5′-fill-in G4. All the tetrad-guanines adopt anti-glycosidic torsion angles including the bound-dGMP, as shown by medium intensities of intra-guanine H8-H1 NOE cross-peaks and relatively upfield-shifted C8 chemical shifts (Figures 2a and S11).22–23 Most loop and flanking residues also adopt anti-glycosidic conformations except for the 5′- terminal A1. A strong intra-residue H8−H1′ NOE cross-peak in combination with a downfield-shifted C8 reveals a syn orientation of the A1 (Figures 2a and S11).24
Figure 2.
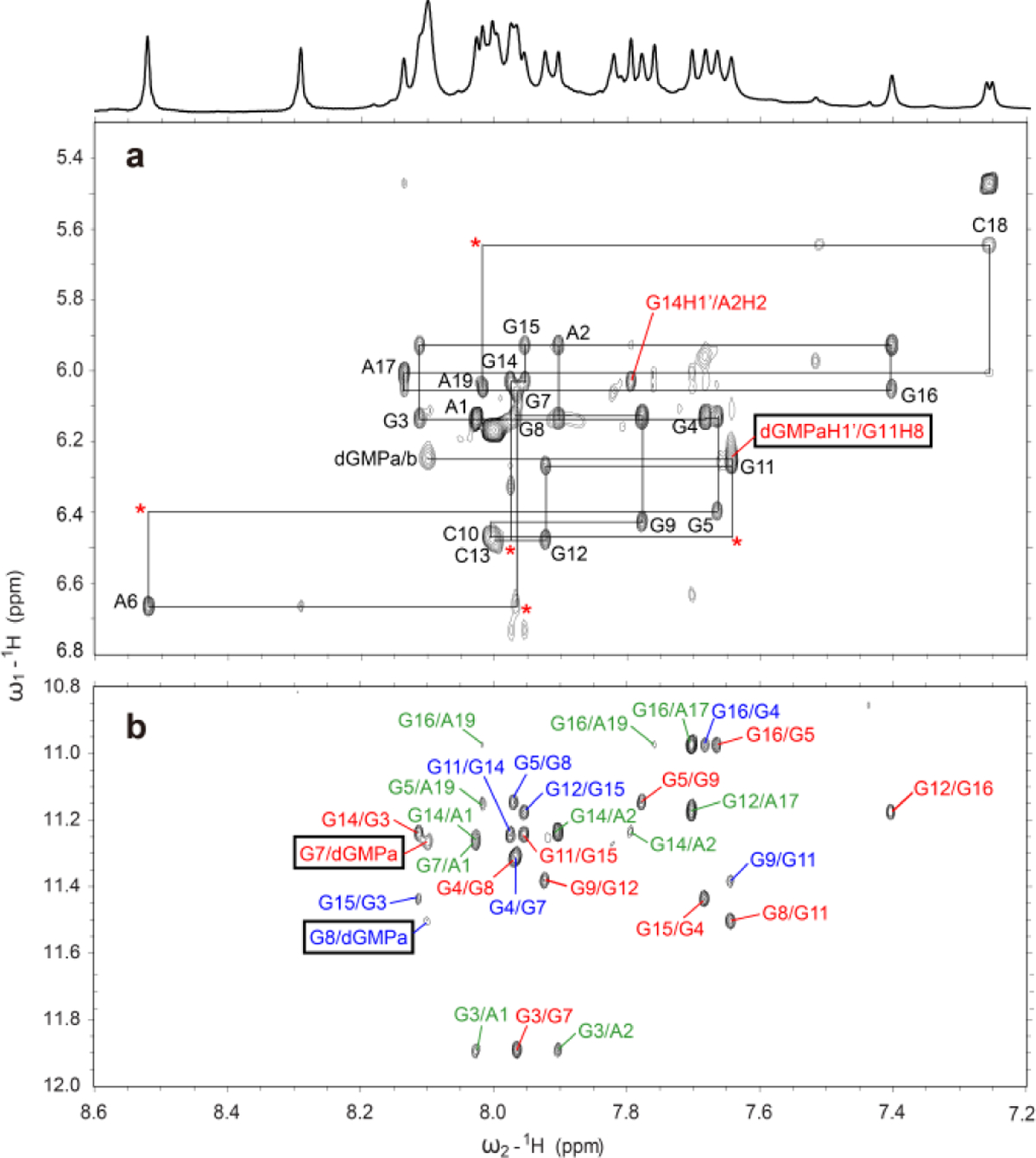
a) The H1′-H8 region from the 2D-NOESY spectrum of dGMP-Pu19m2 complex in H2O at 15 °C with the sequential assignment pathway, mixing time of 250 ms. Missing connectivities are labeled with asterisks. The G14H1′/A2H2 NOE cross-peak is labeled in red. b) The H8-H1 region. Intra-tetrad, inter-tetrad, and NOEs with flanking bases are labeled in red, blue, and green, respectively. The intermolecular NOE cross-peaks of bound-dGMP (dGMPa) and Pu19m2 DNA residues are marked by black boxes.
Numerous intermolecular and inter-residue NOEs were observed (Figure 2, Tables S5–S7) that clearly define the dGMP binding site and the 5′ and 3′ capping structures. Using NOE-restrained molecular dynamics (MD) calculations, we determined the molecular structure of the 1:1 dGMP-Pu19m2 complex in K+-containing solution (Figures 3 and 4). A total of 520 NOE distance restraints were used, including 11 intermolecular dGMP-Pu19m2 restraints (Tables 1 and S5). The 10 lowest energy structures are well-con-verged with an overall root-mean-square-deviation (RMSD) of 0.99 ± 0.22 Å for all heavy atoms and 0.74 ± 0.16 Å for the G-core (Figure 3 and Table 1). The base and sugar of dGMP is well con-verged in contrast to the more flexible phosphate moiety (Figure 3). This is likely due to the fact that intermolecular NOEs were observed for the dGMP sugar moiety (Table S5) but not the phosphate group.
Figure 3.
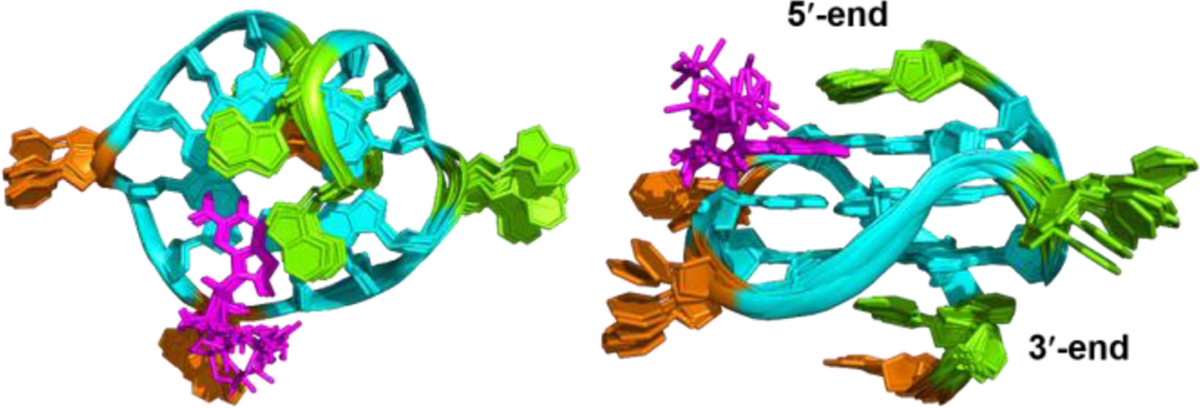
Superposition of 10 lowest energy structures of the dGMP-Pu19m2 complex by NOE-restrained structure calculation: top view (left) and side view (right). Cyan indicates anti-guanine; magenta, dGMP; green, adenine; and orange, cytosine.
Figure 4.
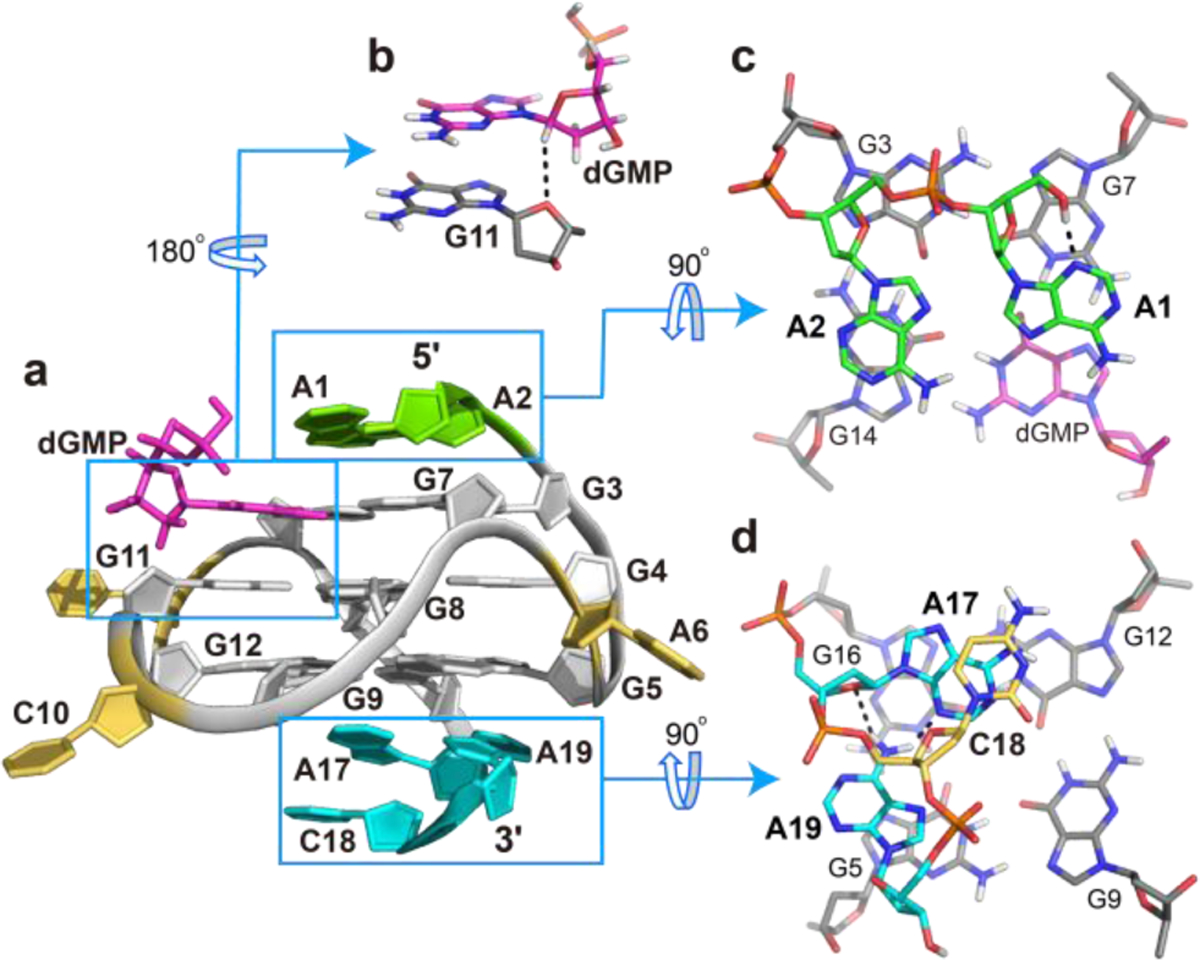
a) Cartoon representation of the 1:1 dGMP-Pu19m2 complex (PDB ID: 6V0L). b) Side view of base stacking of dGMP to G11. A potential C1′−H1′⋯O4′ hydrogen bond is shown in dashed line. c) 5′-end and d) 3′-end views of the capping structures. Potential hydrogen-bonds are shown as dashed lines.
Table 1.
NMR restraints and structural statistics for the 1:1 dGMP-Pu19m2 G-quadruplex.
| NOE-Based Distance Restraints | |
| Total | 520 |
| Intra-residue | 353 |
| Inter-residue | 167 |
| Sequential | 93 |
| dGMP-Pu19m2 | 11 |
| Long-range | 63 |
| Other Restraints | |
| Hydrogen bond restraints | 48 |
| Torsion angle restraints | 20 |
| G-quartet planarity restraints | 48 |
| Structural Statistics | |
| Pairwise heavy atom RMSD (Å) | |
| Overall | 0.99 ± 0.22 |
| G-tetrad core | 0.74 ± 0.16 |
| Violations | |
| Max. NOE restraint violation (Å) | 0.07 |
Like intramolecular guanines, dGMP connects via four Hoogsteen hydrogen bonds to its neighbors in the tetrad (G7 and G14) and can be stabilized by the coordination of potassium cations as well as base stacking interaction with the central-tetrad (Figure 4). Notably, the glycosidic torsion angle of dGMP deviates by about 20° from the angle of other tetrad guanines, which enables the formation of a potential C1′−H1⋯′O4′ hydrogen bond between dGMP and G11 (Figure 4b). This is supported by a relative strong dGMP H1′/G11H8 NOE cross-peak and an extremely weak dGMP H3′/G11H8 NOE cross-peak (Figure 2, Table S5). Such non-con-ventional CH⋯O hydrogen bonds have been widely accepted as important factor in biomolecular structures,25–26 including the stabilization of parallel G4s.27–28 Therefore, the C1′−H1⋯′O4′ hydrogen bond in this dGMP-Pu19m2 complex might contribute to the dGMP
In the 5′-crecognition and binding. apping structure, the two flanking adenines stack upon the 5′-tetrad and their bases point into opposite grooves. While the terminal A1 covers the dGMP-G7 interface, A2 stacks upon G14 (Figure 4c), as supported by numerous NOE cross-peaks such as A1H8/G3H1, A1H8/G7H1, A1H8/G14H1, A2H8/G3H1, A2H8/G14H1, and A2H2/G14H1′ (Figure 2 and Table S6). This arrangement may also contribute to the different solvent exchange rates of the G7 and dGMP imino protons, because the latter is not covered by the capping structure and only observable at low temperatures (Figures 3 and S11). Notably, truncations or
On the other side, the 3′-ACA sequence adopts a fold-back con conformation with A17 and the terminal A19 stacking upon the 3′-tetrad (Figure 4d). Thereby, an A17-A19 base pair is formed that is stabilized by two hydrogen bonds between the amino group of A19 and the sugar edge of A17 (O4′ and N3). Both adenines are connected by C18, which stacks upon A17, as supported by numerous sequential NOE contacts, such as G16H8/A17H8, A17H8/C18H6, and A17H8/C18H5 (Figure 4d and Table S7).
The dGMP is stably incorporated in the G-tetrad as tested by unrestrained MD simulation of 650 ns in explicit water. No notable structural changes occurred, in particular for the dGMP, with an overall RMSD of 1.75 ± 0.43 Å for all heavy atoms during the simulation (Figure S15).
Extended PDGFR-β Promoter Sequence Forms a vG4 That Can be Filled-in by dGMP.
Whereas dGMP cannot compete with the 3′-terminal G22 in Pu22m1 (Figures 1b and S1), we wanted to test whether dGMP can better compete in 3′-extended sequences in which G22 is not a terminal residue. Therefore, we extended the 3′-terminus of the Pu22m1 sequence with one, two, and three wild-type residues (AGG), to Pu23m1, Pu24m1, and Pu25m1, respectively (Table S2), and titrated each sequence with dGMP (Figures S16−S18). The NMR spectral quality for the free DNA decreased along with the increased 3′-extension. Interestingly, dGMP was able to fill in the Pu25m1 sequence with arising of a new set of imino protons (Figure S18). This result supports our hypothesis that the wild-type PDGFR-β promoter sequence forms a vG4, which may be responsive to guanine metabolites.
Binding Affinity of Guanine Derivatives to PDGFR-β vG4.
Microscale Thermophoresis (MST) experiments were performed to determine the binding affinity of guanine derivatives to the Pu19m2 vG4. The 3’-FAM labeled Pu19m2 DNA was used to avoid interference of the fluorophore with the 5’-vacancy binding site. Guanine derivatives had dissociation constants (Kd) from 3.1 to 510.6 μM (Table 2 and Figure S19). The strongest binding was observed for dG and acyclovir with a Kd value of 3.1 and 4.2 μM, respectively. The guanine base binds with a Kd value of 19.5 μM. The monophosphate nucleotides, dGMP, GMP, and cGMP, had similar binding affinity with a Kd value of 59.3, 90.5, and 59.3 μ M, respectively. The binding of triphosphate nucleotides, dGTP and GTP, is weaker with a higher Kd value of 510.6 and 222.8 μ M, respectively. As shown by the results, the presence of a sugar moiety gives rise to a stronger binding, whereas the phosphate group has a negative effect on the binding.
Table 2.
Dissociation Constants (Kd) of Pu19m2-ligand complexes determined by MST experiments.
| Substrate | Ligand | Kd (μM) |
|---|---|---|
| Guanine | 19.5 ± 3.1 | |
| dG | 3.1 ± 0.4 | |
| dGMP | 59.3 ± 4.1 | |
| Pu19m2 DNA | dGTP | 510.6 ± 44.2 |
| GMP | 90.5 ± 9.2 | |
| GTP | 222.8 ± 29.1 | |
| cGMP | 59.3 ± 7.7 | |
| Acyclovir | 4.2 ± 0.5 |
The concentration of guanine metabolites, such as GTP, in eukaryotic cells was found to be ~300 μM.29–30 At such concentration, the binding of guanine metabolites to physiologically relevant vG4s is possible (Table 2), suggesting potential regulatory functions (e.g. transcriptional regulation) of guanine metabolites. In addition, the Pu19m2 DNA could serve as a template to design bio-sensors for the detection of guanine metabolites.20
NMR Study of G-vacancy Fill-in Activity of Various Gua-20 nine Metabolites, Drugs, and Derivatives.
A series of guanine metabolites, drugs, and derivatives (Table S1) was investigated for their ability to fill in the Pu19m2 vG4 by 1H NMR titration experiments. All analogs mimicking the guanine hydrogen bond donor and acceptor pattern can fill into the vG4 and form the essential Hoogsteen hydrogen bonds, but 2-aminopurine, mercaptopurine, and dAMP fail due to their deviating chemical structures (Figures 5 and S20–S32). The NMR titration data are in good agreement with the MST binding data (Figure 5 and Table 2). The free Pu19m2 DNA showed the coexistence of the 3′- and 5′-end vG4s. The strong preference for the 5′-vacancy fill-in is observed for most binders, except for cGMP, although an alternative minor species is often observed likely corresponding to a 3′-fill-in (Figure 5). The 5’-end fill-in is preferred likely due to several factors, including stacking interaction of the guanine base, electrostatic repulsion and steric collision resulting from the sugar-phosphate group. Guanine without any sugar moiety shows a preference to the 5’-end fill-in suggesting a preferred 5’-stacking interaction (Figures 5 and S23). The presence of a sugar moiety of dG clearly enhances the fill-in binding (Figures 5, S25, and Table 2), likely due to intermolecular hydrogen bonds as observed between dGMP C1′-H1′ and G11 O4′ (Figure 4b). An increasing number of phosphate groups (dGMP, dGDP, and dGTP) reduces the binding affinity, suggesting a negative effect from an increased electrostatic repulsion between negatively charged phosphate groups and the polyanionic G4 (Table 2, Figures 5, S5, S26, and S27).20 The strong binding to the 5′-vacancy G4 of the two guanine-derivative drugs, acyclovir and ganciclovir, is likely facilitated by their flexible N9 side chains ca-pable of forming intermolecular hydrogen bonds (Figures 5, S28, and S29). Interestingly, the presence of the 2′-OH group in the case of RNA, such as GMP and GTP, decreases the 5′-selectivity as shown by a weaker 5′-vacancy fill-in and a larger population of the 3′-fill-in species (Figures 5, S30, and S31). The 5’-end selectivity is further attenuated in the case of cGMP, suggesting the additional steric or electrostatic interference of the 3’-shifted cyclic phosphate group with the vG4 backbone (Figures 5 and S32). cGMP binds equally to both the 5′- and 3′-vacancy sites and forms two co-exist-ing fill-in G4s, as shown by two sets of imino protons with compa-rable intensity upon cGMP titration. This observation may explain the recent report that a dinucleotide and a cyclic dinucleotide cGAMP can fill into the 3′-vacancy site of an artificial vG4.31 Whereas dGTP cannot bind to that artificial vG4, all tested guanine metabolites and guanine-derivative drugs can bind to our biologi-cally relevant PDGFR-β vG4.
Figure 5.
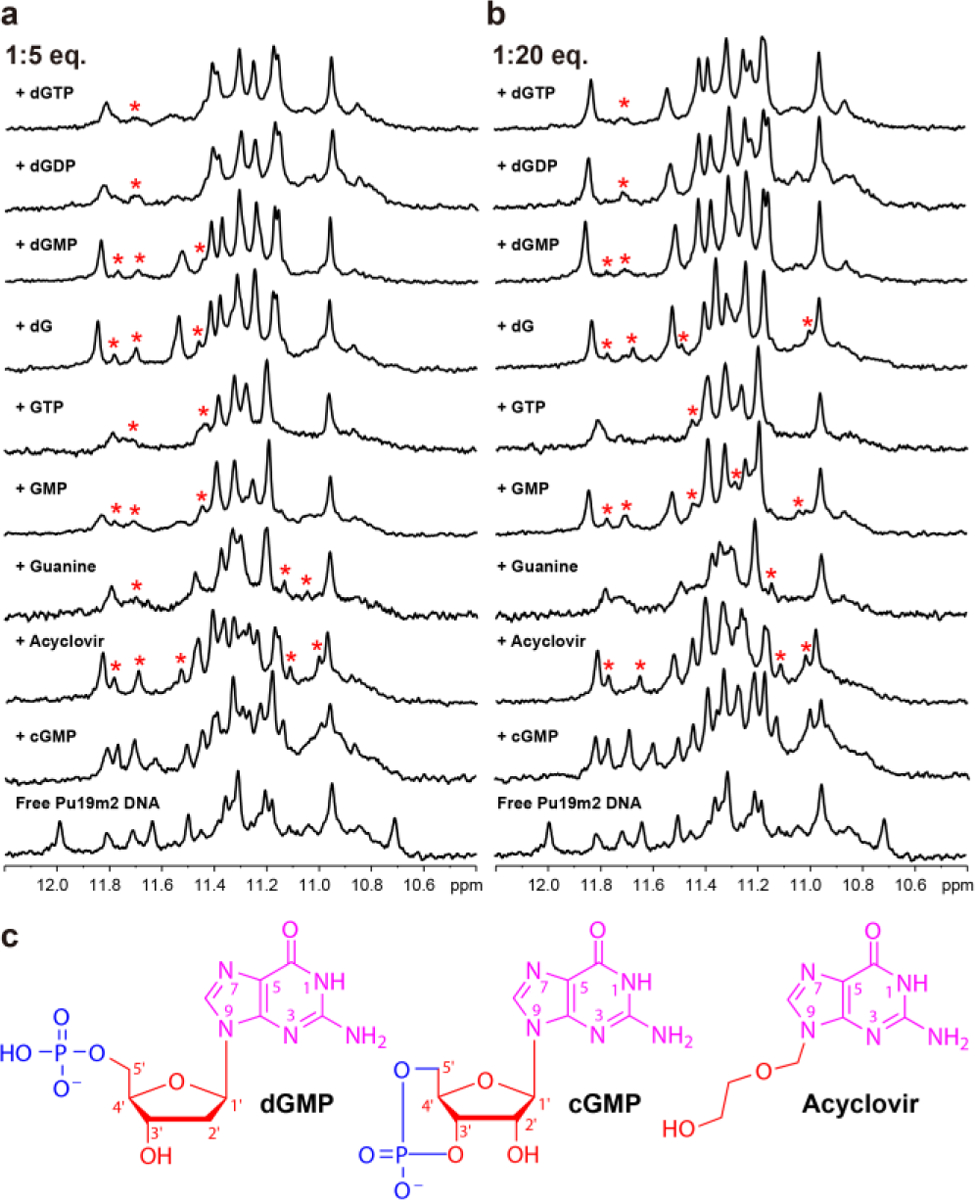
a) and b) 1D 1H NMR titrations of Pu19m2 DNA with various guanine derivatives. Imino protons corresponding to minor species are labeled with red asterisks. c) Chemical structures of dGMP, cGMP, and Acyclovir.
CONCLUSION
In conclusion, we report the first high-resolution NMR structure of a physiological relevant dGMP-fill-in G4 formed in the PDGFR-β promoter sequence. An equilibrium of 3′- and 5′-end vG4s is present in the PDGFR-β promoter sequence and dGMP favors the 5′-end fill-in. The study of various guanine metabolites and drugs elucidates a conserved selectivity for the 5′-vacancy, except for cGMP. cGMP binds both the 3′- and 5′-end vG4s and forms two fill-in G4s with similar population. The dGMP-fill-in vG4 structure and systematic analysis of guanine metabolites and drugs demonstrate the contributions of Hoogsteen hydrogen bonds, sugar, and phosphate moieties to the specific G-vacancy fill-in. Significantly, guanine metabolites are involved in many physiological and pathological processes in human cells.29–30, 32 Their binding and consequent stabilization of physiologically relevant vG4s may play a regulatory role in biological processes. The characteristic broken-strand G4s formed in the PDGFR-β promoter sequence may make it sensitive to guanine metabolites for transcriptional regulation. Our results thus provide a structural basis to understand the potential regulatory functions of guanine metabolites and drugs by interaction with vG4s. In addition, the NMR structure can guide rational design of ligands that target the PDGFR-β vG4.
MATERIAL AND METHODS
Sample Preparation.
Oligonucleotides were synthesized and purified using commercially available reagents as previously described.10, 33–34 The sequences are listed in Table S2. 13C-15N-labeled dGMP was purchased from Cambridge Isotope Laboratories, Inc. DNA concentrations were quantified by UV/Vis absorption at 260 nm using their calculated extinction coefficients. DNA oligonucleotides were dissolved in 10/90% D2O/H2O solution with 12.5 mM potassium phosphate and 37.5 mM potassium chloride at pH 7 to final concentrations of 0.15−2.8 mM. The relative molar ratio of the dGMP and Pu19m2 was about 3:1 in the 2.8 mM Pu19m2 DNA sample (Figure S14). Guanine derivatives were purchased from Sigma-Aldrich and their stock solutions were dissolved in d6DMSO or 50 mM K+-containing buffer to 75 or 150 mM.
Nuclear Magnetic Resonance (NMR) Spectroscopy Experiments.
NMR experiments were conducted using a Bruker AV-500 spectrometer (with Prodigy cryoprobe), Bruker DRX-600 spectrometer, or Bruker AV-800 spectrometer (with QCI Prodigy cryoprobe). Watergate water suppression scheme with a w5 element was used for all experiments unless otherwise stated. The spectra were processed using Topspin 3.5 (Bruker) and analyzed with Sparky (UCSF) software. The guanine H1 imino proton, one-bond coupled to N1, and H8 proton, two-bond coupled to N7, can be unambiguously assigned by 1D 15N-edited HMQC experiments.35–36 For this purpose, site-specifically labeled DNA with 6% 15N-labeled-guanine phosphoramidite was synthesized and used.37 The 1D GE-JRSE HMQC experiments were used for measuring 15N-edited spectra to identify guanine H1 and H8 protons. The similar HMQC experiment was used for measuring 13C-edited spectra to identify the dGMP H8 proton. The 1D variable temperature (VT) proton NMR experiments were done in the range from 5 °C to 70 °C. NOESY spectra were acquired with mixing times of 80 and 250 ms with temperature of 5, 15, and 25 °C, respectively, in both H2O and D2O. TOCSY experiments with 30 and 80 ms mixing times. HSQC experiments were executed with a 3–9-19 water suppression scheme and was optimized for a 1J(C,H) of 180 Hz. Chemical shift referencing was done directly for 1H based on the water signal relative to TSP and indirectly for 13C relative to DSS.
Structure Calculation.
NOE-based distance restraints were obtained by defining NOESY cross-peaks as strong (2.9 ± 1.1 Å), medium (4.0 ± 1.5 Å), weak (5.5 ± 1.5 Å), and very weak (6.0 ± 1.5 Å) based on the spectra acquired with mixing times of 80 and 250 ms. Exchangeable protons were classified as medium (4.0 ± 1.2 Å), weak (5.0 ± 1.2 Å), and very weak (6.0 ± 1.2 Å). A distance of 5.0 ± 2.0 Å was applied in case of overlapped and thus ambiguous resonances. Dihedral restraints were assigned for glycosidic torsion angles with 170°−310° and 200°−280° for anti-conformations in loop regions and within the G-tetrad, respectively. The syn-conformation of A1 was restricted to 0°−120°. A distance geometry simulated annealing protocol was used in Xplor-NIH 2.48 to generate 100 1:1 dGMP-Pu19m2 starting structures.38 dGMP topology and parameter files were generated using Amber software package. The sander module of the Amber 16 package was employed for simulated annealing of the 100 starting structures in implicit water.39 The OL15 version of the Amber force field for DNA was used that further modifies the parmbsc0 version.40–42 Structures were initially equilibrated for 5 ps at 100 K and then heated to 1000 K during 10 ps. The system was cooled down to 100 K during 45 ps after 30 ps of 1000 K equilibration. Finally, a cool down to 0 K was performed in the last 10 ps. The force constants for NOE-based distance−1 and hydrogen bond restraints were set to 20 and 50 kcal·mol−1·Å−2, respectively. In addition, G-tetrad planarity restraints of 30 kcal·mol1·Å−2 and glycosidic angle restraints of 200 kcalm· ol−1·rad−1 were applied. Subsequently, the 20 lowest-energy structures were selected and solvated with TIP3P water molecules in a truncated octahedral box with a minimal distance of 10 Å from the DNA to the box border. The system was neutralized with K+ cations with two of them placed between the tetrads. For initial equilibration, the DNA position was fixed with 25 kcal·mol−1·Å−2 and 500 steps each of steepest descent and conjugated gradient minimization were performed. Then, the system was heated from 100 to 300 K in 20 ps under constant volume, and the force constant on the DNA was slowly decreased going from 5, 4, 3, 2, 1, and 0.5 kcal·mol−1·Å−2 during steps of each 10 ps. The final production run of 4 ns used the pmemd module of Amber 16 under constant pressure, and a snapshot was taken every picosecond. NMR-derived distance and hydrogen bond restraints were employed with a force constant of 10 and 25 kcal·mol−1·Å−2, respectively. A G-tetrad planarity restraint of 5 kcal·mol−1·Å−2 was also included. Finally, the last 100 ps of the trajectories were averaged and energy-minimized for 100 steps in vacuum, and the 10 lowest energy structures were selected for the final ensemble. Structures were analyzed and visualized with PyMOL and the VMD software.43–44
DMS Footprinting.
Gel-purified oligonucleotides (with 7T flanking ends) 5’-end labeled using [γ-32P]-ATP in the presence of T4 polynucleotide kinase. The samples were then methylated by treatment with 0.5% (final concentration) DMS, 1 μg calf thymus DNA at desired conditions for 7 mins at room temperature. The reaction was stopped by addition of β-mercaptoethanol. The methylated DNA was subsequently purified by native PAGE. The cleavage at methylated guanines was induced by treatment with 10% piperidine for 18 min at 90 °C. A Speedvac was used to remove the piperidine and two successive water washes. The cleaved products were analyzed on a 16% sequencing (denaturing) PAGE gel.
Native Gel Electrophoretic Mobility Shift Assay (EMSA).
Native PAGE experiments were performed with a 1.5 mm thick 10×7 cm native gel containing 18% acrylamide (acrylamide:bisacrylamide 29:1) in 1X TBE buffer, pH 8.0, supplemented with 12.5 mM KCl. Free Pu19m2 DNA and 20:1 dGMP-Pu19m2 complex were prepared in 37.5 mM KCl, 12.5 mM phosphate buffer. Each well contained 5 μL of 50 μM DNA. DNA bands were visualized using ultraviolet (UV) light absorption at 260 nm.
Circular Dichroism (CD) Spectroscopy Experiments.
Circular dichroism spectra were recorded using a Jasco-1100 spectropolarimeter (Jasco Inc.) equipped with a temperature controller. Samples were prepared in 37.5 mM KCl, 12.5 mM phosphate buffer at a DNA concentration of 15 μM in the absence and presence of the different guanine derivatives. CD measurements were taken through a quartz cell with a 1 mm path length, 1 nm bandwidth, and 1 s response time for spectra at 25 °C. Spectra were obtained using three averaged scans between 230 and 330 nm. The baseline was corrected by subtracting the buffer spectrum.
MicroScale Thermophoresis (MST) Experiments.
MST experiments were performed on a Monolith NT.115 system (NanoTemper Technologies) as previously described.45–47 The 3’-FAM labeled Pu19m2 DNA solutions were prepared in 37.5 mM KCl, 12.5 mM phosphate buffer at pH 7 and annealed by heating to 70 °Cf or 1 min and then slowly cooled down to room temperature. To determine the Kd values of each ligand to Pu19m2 DNA, 200 nM 3’-FAM labeled Pu19m2 DNA was incubated with increasing concentrations of ligands (16 points of a 2-fold serial dilution starting from 1, 1.3 or 4 mM) for 30 min at room temperature in 37.5 mM KCl, 12.5 mM phosphate buffer. The MST measurement was then car-ried out at 40% (medium) IR power and 20% blue LED excitation power. Normalized fluorescence values at a given time were used as the measure of fraction of bound population analyzed using GraphPad Prism (GraphPad Software, San Diego, CA) and the Kd values were determined by fitting the data to the following equation assuming a 1:1 binding model: FB= FBmax[(M + LT + Kd) – ((M + LT + Kd)2 – (4MLT))1/2] / (2MT), where FB represents the fraction bound population. FBmax is fitted in addition to Kd value. The labelled Pu19m2 DNA concentration, M, was held constant, and LT, the total ligand concentration is an independent variable, varying with each titration step.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the National Institutes of Health R01CA177585 (DY), P30CA023168 (Purdue Center for Cancer Research), and the Deutsche Forschungsgemeinschaft (German Research Foundation) Projektnummer 427347592 (JD). We thank Dr. Clement Lin for his comments and help on NMR experiments. We thank Dr. Lan Chen at Chemical Genomics Facility, Purdue Institute for Drug Discovery at Purdue University for helping on MST experiments.
Footnotes
SUPPORTING INFORMATION
The Supporting Information is available free of charge on the ACS Publications website at DOI: Chemical structures of tested guanine metabolites, drugs, and derivatives, DNA sequences, tables of proton chemical shifts and NOEs, 1H NMR titration spectra of various guanine derivatives, DMS footprinting, EMSA, variable temperature 1H NMR spectra, 15N-edited and 13C-edited NMR spectra, 1H-13C-HSQC, 2D-NOESY, superposition of 10 lowest energy structures, 650 ns MD simulation, MST data, and CD spectra (PDF).
The coordinates of the dGMP-fill-in PDGFR-β vG4 structure were deposited in the Protein Data Bank (6V0L).
The authors declare no competing interests.
REFERENCES
- 1.Yang D; Okamoto K, Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med. Chem 2010, 2 (4), 619–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubramanian S; Hurley LH; Neidle S, Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug. Discov 2011, 10 (4), 261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biffi G; Di Antonio M; Tannahill D; Balasubramanian S, Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem 2014, 6 (1), 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biffi G; Tannahill D; McCafferty J; Balasubramanian S, Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem 2013, 5 (3), 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hänsel-Hertsch R; Beraldi D; Lensing SV; Marsico G; Zyner K; Parry A; Di Antonio M; Pike J; Kimura H; Narita M, G-quadruplex structures mark human regulatory chromatin. Nat. Genet 2016, 48 (10), 1267–1275. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes D; Lipps HJ, G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res 2015, 43 (18), 8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen MC; Tippana R; Demeshkina NA; Murat P; Balasubramanian S; Myong S; Ferré-D’ Amaré AR, Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 2018, 558 (7710), 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenarčič Živković M; Rozman J; Plavec J, Adenine-driven structural switch from a two- to three-quartet DNA G-quadruplex. Angew. Chem., Int. Ed 2018, 130 (47), 15621–15625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neidle S, Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem 2016, 59 (13), 5987–6011. [DOI] [PubMed] [Google Scholar]
- 10.Wang KB; Elsayed MS; Wu G; Deng N; Cushman M; Yang D, Indenoisoquinoline topoisomerase inhibitors strongly bind and stabilize the MYC promoter G-quadruplex and downregulate MYC. J. Am. Chem. Soc 2019, 141 (28), 11059–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin C; Wu G; Wang K; Onel B; Sakai S; Shao Y; Yang D, Molecular recognition of the hybrid-2 human telomeric G-quadruplex by Epiberberine: insights into conversion of telomeric G-quadruplex structures. Angew. Chem., Int. Ed 2018, 130 (34), 11054–11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang KB; Li DH; Hu P; Wang WJ; Lin C; Wang J; Lin B; Bai J; Pei YH; Jing YK; Hua HM, A series of β-carboline alkaloids from the seeds of Peganum harmala show G-quadruplex interactions. Org. Lett 2016, 18 (14), 3398–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y; Agrawal P; Brown RV; Hatzakis E; Hurley L; Yang D, The major G-quadruplex formed in the human platelet-derived growth factor receptor beta promoter adopts a novel broken-strand structure in K+ solution. J. Am. Chem. Soc 2012, 134 (32), 13220–13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin Y; Fortin JS; Tye D; Gleason-Guzman M; Brooks TA; Hurley LH, Molecular cloning of the human platelet-derived growth factor receptor beta (PDGFR-beta) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-beta expression. Biochemistry 2010, 49 (19), 4208–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown RV; Wang T; Chappeta VR; Wu G; Onel B; Chawla R; Quijada H; Camp SM; Chiang ET; Lassiter QR, The consequences of overlapping G-quadruplexes and i-motifs in the platelet-derived growth factor receptor β core promoter nuclease hypersensitive element can explain the unexpected effects of mutations and provide opportunities for selective targeting of both structures by small molecules to downregulate gene expression. J. Am. Chem. Soc 2017, 139 (22), 7456–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betsholtz C; Karlsson L; Lindahl P, Developmental roles of platelet-derived growth factors. Bioessays 2001, 23 (6), 494–507. [DOI] [PubMed] [Google Scholar]
- 17.Onel B; Carver M; Agrawal P; Hurley LH; Yang D, The 3’end region of the human PDGFR-β core promoter nuclease hypersensitive element forms a mixture of two unique end-insertion G-quadruplexes. Biochim. Biophys. Acta, Gen. Subj 2018, 1862 (4), 846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li XM; Zheng KW; Zhang JY; Liu HH; He YD; Yuan BF; Hao YH; Tan Z, Guanine-vacancy-bearing G-quadruplexes responsive to guanine derivatives. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (47), 14581–14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heddi B; Martín-Pintado N; Serimbetov Z; Kari TMA; Phan AT, G-quadruplexes with (4n-1) guanines in the G-tetrad core: formation of a G-triad · water complex and implication for small-molecule binding. Nucleic Acids Res 2015, 44 (2), 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XM; Zheng KW; Hao YH; Tan Z, Exceptionally selective and tunable sensing of guanine derivatives and analogues by structural complementation in a G-quadruplex. Angew. Chem., Int. Ed 2016, 128 (44), 13963–13968. [DOI] [PubMed] [Google Scholar]
- 21.Kolesnikova S; Srb P; Vrzal L; Lawrence MS; Veverka V; Curtis EA, GTP-dependent formation of multimeric G-quadruplexes. ACS Chem. Biol 2019, 14 (9), 1951–1963. [DOI] [PubMed] [Google Scholar]
- 22.Greene KL; Wang Y; Live D, Influence of the glycosidic torsion angle on 13C and 15N shifts in guanosine nucleotides: investigations of G-tetrad models with alternating syn and anti bases. J. Biomol. NMR 1995, 5 (4), 333–338. [DOI] [PubMed] [Google Scholar]
- 23.Dickerhoff J; Weisz K, Flipping a G-tetrad in a unimolecular quadruplex without affecting its global fold. Angew. Chem., Int. Ed 2015, 127 (19), 5680–5683. [DOI] [PubMed] [Google Scholar]
- 24.Dickerhoff J; Onel B; Chen L; Chen Y; Yang D, Solution structure of a MYC promoter G-quadruplex with 1:6:1 loop length. ACS Omega 2019, 4 (2), 2533–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger I; Egli M; Rich A, Inter-strand CH⋯O hydrogen bonds stabilizing four-stranded intercalated molecules: stereoelectronic effects of O4’ in cytosine-rich DNA. Proc. Natl. Acad. Sci. U. S. A 1996, 93 (22), 12116–12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horowitz S; Trievel RC, Carbon-oxygen hydrogen bonding in biological structure and function. J. Biol. Chem 2012, 287 (50), 41576–41582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martín-Pintado N; Yahyaee-Anzahaee M; Deleavey GF; Portella G; Orozco M; Damha MJ; González C, Dramatic effect of furanose C2’ substitution on structure and stability: directing the folding of the human telomeric quadruplex with a single fluorine atom. J. Am. Chem. Soc 2013, 135 (14), 5344–5347. [DOI] [PubMed] [Google Scholar]
- 28.Dickerhoff J; Appel B; Müller S; Weisz K, Sugar−edge interactions in a DNA-RNA G-quadruplex: evidence of sequential C−H⋯O hydrogen bonds contributing to RNA quadruplex folding. Angew. Chem., Int. Ed 2016, 55 (48), 15162–15165. [DOI] [PubMed] [Google Scholar]
- 29.Curtis EA; Liu DR, Discovery of widespread GTP-binding motifs in genomic DNA and RNA. Chem. Biol 2013, 20 (4), 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traut TW, Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem 1994, 140 (1), 1–22. [DOI] [PubMed] [Google Scholar]
- 31.Winnerdy FR; Das P; Heddi B; Phan AT, Solution structures of a G-quadruplex bound to linear-and cyclic-dinucleotides. J. Am. Chem. Soc 2019, 141 (45), 18038–18047. [DOI] [PubMed] [Google Scholar]
- 32.Mathews CK, Deoxyribonucleotide metabolism, mutagenesis and cancer. Nat. Rev. Cancer 2015, 15 (9), 528–539. [DOI] [PubMed] [Google Scholar]
- 33.Dai J; Carver M; Hurley LH; Yang D, Solution structure of a 2: 1 quindoline-c-MYC G-quadruplex: insights into G-quadruplexinteractive small molecule drug design. J. Am. Chem. Soc 2011, 133 (44), 17673–17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onel B; Carver M; Wu G; Timonina D; Kalarn S; Larriva M; Yang D, A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J. Am. Chem. Soc 2016, 138 (8), 2563–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szewczak AA; Kellogg GW; Moore PB, Assignment of NH resonances in nucleic acids using natural abundance 15N-1H correlation spectroscopy with spin-echo and gradient pulses. FEBS Lett. 1993, 327 (3), 261–264. [DOI] [PubMed] [Google Scholar]
- 36.Agrawal P; Hatzakis E; Guo K; Carver M; Yang D, Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res 2013, 41 (22), 10584–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao H; Pagano AR; Wang W; Shallop A; Gaffney BL; Jones RA, Use of a 13C atom to differentiate two 15N-labeled nucleosides. Syntheses of [15NH2]-Adenosine, [1, NH2-15N2]-and [2-13C-1, NH2-15N2]-Guanosine, and [1, 7, NH2-15N3]-and [2-13C-1, 7, NH215N3]-2’-Deoxyguanosine. J. Org. Chem 1997, 62 (22), 7832–7835. [Google Scholar]
- 38.Schwieters CD; Kuszewski JJ; Tjandra N; Clore GM, The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson 2003, 160 (1), 65–73. [DOI] [PubMed] [Google Scholar]
- 39.Case, D. A. B. RM;Cerutti DS; Cheatham ITE; Darden TA; Duke RE; Giese TJ; Gohlke H; Goetz AW; Homeyer N; Izadi S; Janowski P; Kaus J; Kovalenko A; Lee TS; LeGrand S; Li P; Lin C; Luchko T; Luo R; Madej B; Mermelstein D; Merz KM; Monard G; Nguyen H; Nguyen HT; Omelyan I; Onufriev A; Roe DR; Roitberg A; Sagui C; Simmerling CL; Botello-Smith WM; Swails J; Walker RC; Wang J; Wolf RM; Wu X; Xiao L; Kollman PA, Amber 2016; University of California: San Francisco, 2016 2016. [Google Scholar]
- 40.Krepl M; Zgarbová M; Stadlbauer P; Otyepka M; Banáš P; Koca J; Cheatham TE III; Jurecka P; Šponer J. i., Reference simulations of noncanonical nucleic acids with different χ variants of the AMBER force field: quadruplex DNA, quadruplex RNA, and Z−DNA. J. Chem. Theory Comput 2012, 8 (7), 2506–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zgarbová M; Luque FJ; Šponer J. í.; Cheatham TE III; Otyepka M; Jurecka P, Toward improved description of DNA backbone: revisiting epsilon and zeta torsion force field parameters. J. Chem. Theory Comput 2013, 9 (5), 2339–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zgarbová M; Sponer J; Otyepka M; Cheatham TE III; Galindo−Murillo R; Jurecka P, Refinement of the sugar-phosphate backbone torsion beta for AMBER force fields improves the description of Z-and B-DNA. J. Chem. Theory Comput 2015, 11 (12), 5723–5736. [DOI] [PubMed] [Google Scholar]
- 43.Humphrey W; Dalke A; Schulten K, VMD: Visual molecular dynamics. J. Mol. Graph 1996, 14 (1), 33–38. [DOI] [PubMed] [Google Scholar]
- 44.Schrödinger L, The PyMOL Molecular Graphics System, version 2.1 2018.
- 45.Wienken CJ; Baaske P; Rothbauer U; Braun D; Duhr S, Protein−binding assays in biological liquids using microscale thermophoresis. Nat. Commun 2010, 1, 100. [DOI] [PubMed] [Google Scholar]
- 46.Moon MH; Hilimire TA; Sanders AM; Schneekloth JS Jr, Measuring RNA−ligand interactions with microscale thermophoresis. Biochemistry 2018, 57 (31), 4638–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baaske P; Wienken CJ; Reineck P; Duhr S; Braun D, Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew. Chem. Int. Ed 2010, 49 (12), 2238–2241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


