Summary
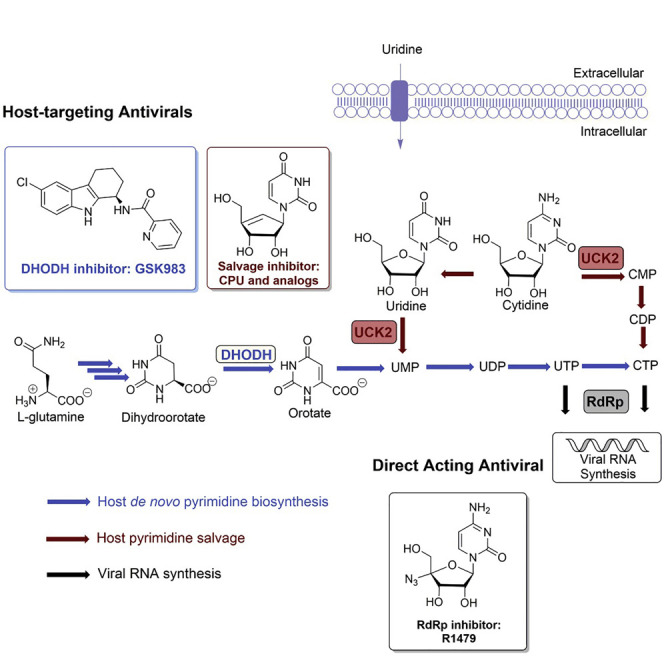
Genome-wide analysis of the mode of action of GSK983, a potent antiviral agent, led to the identification of dihydroorotate dehydrogenase as its target along with the discovery that genetic knockdown of pyrimidine salvage sensitized cells to GSK983. Because GSK983 is an ineffective antiviral in the presence of physiological uridine concentrations, we explored combining GSK983 with pyrimidine salvage inhibitors. We synthesized and evaluated analogs of cyclopentenyl uracil (CPU), an inhibitor of uridine salvage. We found that CPU was converted into its triphosphate in cells. When combined with GSK983, CPU resulted in large drops in cellular UTP and CTP pools. Consequently, CPU-GSK983 suppressed dengue virus replication in the presence of physiological concentrations of uridine. In addition, the CPU-GSK983 combination markedly enhanced the effect of RNA-dependent RNA polymerase (RdRp) inhibition on viral infection. Our findings highlight a new host-targeting strategy for potentiating the antiviral activity of RdRp inhibitors.
Keywords: antiviral therapy, combination, pyrimidine metabolism, RNA-dependent RNA polymerase, uridine-cytidine kinase, cytidine monophosphate kinase, dihydroorotate dehydrogenase, dengue
Graphical Abstract
Highlights
-
•
Cyclopentenyl uracil and analogs inhibit pyrimidine salvage in vitro and in cells
-
•
A host-targeting antiviral strategy combining DHODH inhibitors with CPU is explored
-
•
The strategy rescues the antiviral efficacy of a DHODH inhibitor in 20 μM uridine
-
•
Modulating pyrimidine metabolism boosts the antiviral activity of an RdRp inhibitor
Many RNA virus infections lack suitable treatments. Liu et al. identified a host-targeting antiviral strategy of modulating pyrimidine metabolism with cyclopentenyl uracil, an inhibitor of pyrimidine salvage, and GSK983, an inhibitor of de novo biosynthesis. This combination also increased the potency of an RNA-dependent RNA polymerase inhibitor, against dengue virus.
Introduction
Significant progress in the development of antiviral drugs has been achieved by targeting viral proteins with small molecules (Jordheim et al., 2013, Lou et al., 2014). For example, compounds like aciclovir and zidovudine block viral reverse transcriptase to treat herpes simplex virus and HIV infections, respectively, and RNA-dependent RNA polymerase (RdRp) inhibitors such as dasabuvir and sofosbuvir are used to treat hepatitis C virus infections (Nováková et al., 2018). More recently, the broad-spectrum RdRp inhibitor prodrug remdesivir (Gordon et al., 2020) has received U.S. Food and Drug Administration Emergency Use Authorization for the treatment of coronavirus disease 2019 (COVID-19). Meanwhile, targeting host proteins required for viral propagation is emerging as an attractive alternative that may circumvent the emergence of resistance (Bekerman and Einav, 2015). For example, maraviroc inhibits the human chemokine receptor CCR5, and is therefore used to treat multidrug-resistant HIV (Lieberman-Blum et al., 2008). More recently, pyrimidine biosynthesis has emerged as a potential host-targeting strategy for antivirals (Okesli et al., 2017). Here, we focus on devising a host-targeting antiviral approach for the treatment of RNA viruses, which cause many serious diseases such as viral hepatitis, influenza, Ebola, dengue, and COVID-19.
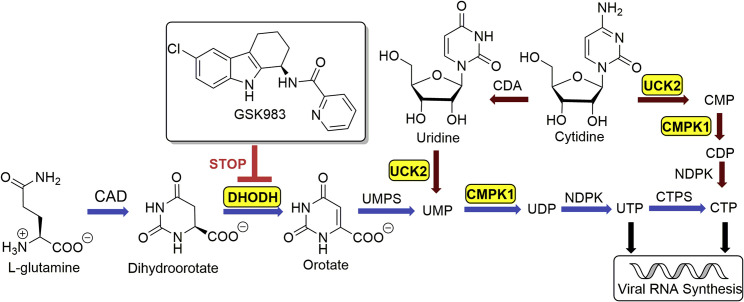
In mammalian cells, pyrimidine biosynthesis is a tightly regulated metabolic process. Two complementary pathways—de novo biosynthesis and pyrimidine salvage—are responsible for producing UTP and CTP for host as well as viral RNA synthesis (Figure 1 ). De novo pyrimidine biosynthesis is a resource-intensive process. In contrast, salvage occurs via phosphorylation of UMP and CMP derived from intracellular RNA degradation or via facilitated transport and phosphorylation of extracellular uridine, whose plasma concentration is tightly controlled in the low micromolar range (Traut, 1994). Recently, we discovered that GSK983, a broad-spectrum antiviral agent first reported in 2009 (Harvey et al., 2009), is a potent inhibitor of dihydroorotate dehydrogenase (DHODH), a rate-limiting step in de novo pyrimidine biosynthesis (Deans et al., 2016). In the course of those unbiased genome-wide studies, we also found that knockdown of uridine/cytidine kinase 2 (UCK2) and uridine monophosphate/cytidine monophosphate kinase 1 (CMPK1) in the pyrimidine salvage pathway strongly sensitized cells to growth inhibition by GSK983 (Deans et al., 2016). This finding was consistent with the observation that most DHODH inhibitors lack antiviral efficacy in vivo despite high potency in vitro presumably due to salvage metabolism of circulating uridine by virus-infected cells (Bonavia et al., 2011, Cheung et al., 2017, Grandin et al., 2016, Smee et al., 2012, Wang et al., 2011, Xiong et al., 2020).
Figure 1.
De Novo and Salvage Biosynthesis of Pyrimidine Nucleotides for Host and Viral RNA Synthesis
GSK983 is a DHODH inhibitor. Genes that sensitize cells to GSK983 are highlighted in yellow boxes. Reactions shown with blue arrows comprise the de novo biosynthetic pathway, whereas those with red arrows comprise the salvage pathway. CAD, carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase; CDA, cytidine deaminase; CMPK1, uridine monophosphate/cytidine monophosphate kinase 1; CTPS, CTP synthetase; DHODH, dihydroorotate dehydrogenase; NDPK, nucleoside diphosphate kinase; UCK2, uridine/cytidine kinase 2; UMPS, uridine monophosphate synthase.
To restore the antiviral efficacy of GSK983 in the presence of extracellular uridine, we therefore sought to inhibit pyrimidine salvage. Cyclopentenyl uridine (CPU) is a carbocyclic analog of uridine that has been shown to inhibit human UCK2 (Lim et al., 1984). To our surprise, we learned that the antiviral activity of CPU is due to its remarkable ability to deplete intracellular pyrimidine nucleotide pools via inhibition of salvage biosynthesis pathways. Our findings led us to redirect our search for a fundamentally new type of combination chemotherapy for RNA viruses, as described below.
Results
Diversity-Oriented Syntheses of CPU Analogs
Our search for lead inhibitors of pyrimidine salvage was inspired by earlier reports on the biological activity of CPU and cyclopentenyl cytosine (CPC), which were shown to block uridine salvage in vitro and in vivo (Cysyk et al., 1995, Lim et al., 1984, Moyer et al., 1985, Schimmel et al., 2007). In these and other reports, CPU was found to be remarkably well tolerated, whereas CPC was considerably more cytotoxic (Blaney et al., 1992, Ford et al., 1991, Song et al., 2001). This contrast is likely due to downstream inhibition of the CTP synthetase by CPC (Schimmel et al., 2007).
Due to the high toxicity of CPC, we undertook structure-activity relationship (SAR) analysis of CPU wherein uracil nucleobase was maintained intact or only modified at the C5 position. Meanwhile, the C-5′ substituent of CPU was modified because this is the site of UCK2-catalyzed phosphorylation. In order to rapidly access both nucleobase and carbocyclic moiety analogs, we implemented a diversity-oriented synthetic approach featuring a Mitsunobu reaction as the strategic transformation (Choi et al., 2012).
We first synthesized CPU analogs with nucleobase modifications (Figure 2 A). Mitsunobu reactions between the common cyclopentenyl moiety (1) (Choi et al., 2004) and the benzoyl protected uracil, C(5)-fluoro-uracil, C(5)-iodo-uracil or thymine (2a–2d) (Racine et al., 2014) furnished the carbon skeleton of C-5 analogs. Removal of acetal, benzoyl, and tert-butyldiphenyl silyl (TBDPS) groups then delivered CPU (4a) and analogs 4b–4d.
Figure 2.
Diversity-Oriented Syntheses of CPU Analogs
(A) Synthesis of CPU (4a) and uracil moiety analogs 4b–4d.
(B) Modification of the C5′ position to furnish carbocyclic moiety analogs 6, 7, 9, and 11. (a) DEAD, PPh3, THF; (b) NH3, MeOH; (c) HCl, THF; (d) TBAF, THF; (e) Ag2O, MeI, acetone; (f) DAST, CH2Cl2; (g) MsCl, Et3N, CH2Cl2; (h) NaN3, DMF; (i) AcCl, CH2Cl2; (j) Pd(OH)2, cyclohexene, ethanol. Ac, acetyl; Bz, benzoyl; DAST, diethylaminosulfur trifluoride; DEAD, diethyl azodicarboxylate; Ms, methanesulfonyl; TBAF, tetra-n-butylammonium fluoride; TBDPS, tert-butyldiphenyl silyl; THF, tetrahydrofuran.
Next, we turned to synthesis of CPU analogs modified at the C5′ position of the cyclopentenyl moiety (Figure 2B). Selective deprotection of TBDPS with tetra-n-butylammonium fluoride revealed the primary hydroxyl group (Song et al., 2001), which was then methylated in the presence of Ag2O (Francisco et al., 2001) to afford the protected 5′-methoxy-CPU. Alternatively, the alcohol could be converted to a terminal fluoride with diethylamino sulfurotrifluoride (Moon et al., 2004), resulting in protected 5′-fluoro CPU. Mesylation or acetylation of 5, followed by NaN3 substitution (Schaudt and Blechert, 2003) or Pd(OH)2/C-catalyzed hydrogenation (Bianco et al., 1989) afforded the protected 5′-azido- and 5′-deoxy-CPU, respectively. The resulting protected intermediates were treated with methanolic ammonia and/or HCl to provide analogs 6,7, 9, and 11.
Enzymatic Analysis of CPU Analogs Against UCK2 and CMPK1
In order to guide our SAR studies, we developed an enzymatic assay to evaluate the inhibitory effect of each CPU analog against recombinant human UCK2, which was expressed and purified in Escherichia coli. A continuous assay system was optimized by coupling UCK2 activity to the pyruvate kinase reaction, which in turn was coupled to lactate dehydrogenase (Tomoike et al., 2017). Overall reaction progress was continuously monitored by detecting the UV absorption change at 340 nm, which was correlated with the ATP consumption by UCK2.
The effect of 250 μM of each CPU analog on UCK2 activity was evaluated in the presence of 50 μM uridine. CPU and 5-F-CPU showed much higher ATP consumption compared with other analogs (Figure 3 A) even after the 50 μM uridine was presumably exhausted, suggesting these two compounds were substrates of UCK2. We then individually measured the steady-state kinetic parameters of UCK2 using uridine, CPU, and 5-F-CPU as substrates. Their K M values were 86, 25, and 41 μM, and their k cat /K M values were 2.6×105, 1.5×105, and 7.3×104 s−1⋅M−1 (Figure 3B). As predicted by the relative magnitude of these kinetic parameters, addition of CPU or 5-F-CPU to an assay mixture containing UCK2, uridine, and ATP resulted in a dose-dependent decrease in the rate of UMP synthesis (Figure 3C).
Figure 3.
Enzymatic Analysis of CPU Analogs
(A) CPU and 5-F-CPU are UCK2 substrates, as observed by addition of 250 μM of each CPU analog to a UCK2 reaction mixture that also contains 50 μM uridine. All other analogs tested are not UCK2 substrates.
(B) Steady-state kinetic analysis of uridine, CPU, and 5-F-CPU as UCK2 substrates. Error bars represent ±SD of three replicates.
(C) Comparative ATP consumption by UCK2 in the presence of 50 μM uridine and varying concentrations of CPU (blue bars) or 5-F-CPU (red bars). Error bars represent ±SD of three replicates.
(D) Initial velocities calculated from the data shown in (A).
(E and F) Lineweaver-Burk analysis of 5′-F-CPU (E) and 5′-deoxy-CPU (F) as UCK2 inhibitors.
(G) Comparative activity of CMPK1 on CMP, CPU-MP, and 5-F-CPU-MP. An authentic standard of CMP (std) was tested alongside a UCK2-synthesized sample of the same compound (syn).
Error bars represent ±SD of three replicates. See also Figures S1 and S2.
While CPU and 5-F-CPU were the only UCK2 substrates identified from our panel of carbocyclic nucleoside analogs, some of the other agents had measurable inhibitory activity against this enzyme (Figure 3D). The K i values of the two most potent competitive inhibitors, 5′-F-CPU and 5′-deoxy-CPU, were 170 μM and 230 μM, respectively (Figures 3E and 3F).
Given that CPU and 5-F-CPU could be phosphorylated by UCK2, we sought to establish whether the corresponding monophosphates were substrates of human CMPK1. For this purpose, human CMPK1 was expressed in E. coli and purified. We then used recombinant UCK2 to synthesize CMP, CPU-MP, and 5-F-CPU-MP and confirmed their identities by liquid chromatography tandem mass spectrometry (LC-MS/MS) (Figure S1). The results shown in Figure 3G demonstrate that both CPU-MP and 5-F-CPU-MP are substrates of CMPK1, but that the former is a better substrate. Identities of their corresponding products, CPU-DP and 5-F-CPU-DP, were also confirmed by LC-MS/MS (Figure S2).
LC-MS/MS Analysis of Intracellular Nucleotides
To understand the metabolic implications of the above biochemical findings, we developed an LC-MS/MS-based assay that measures the effects of CPU, 5-F-CPU, and 5′-F-CPU on intracellular pyrimidine nucleotide levels. To minimize the perturbation of sample quenching and analysis on the physiological concentrations of these metabolites, we adapted an established protocol for growing cells on glass coverslips to facilitate rapid washing (Figure S3) (Martano et al., 2014). LC-MS/MS of nucleotides (Table S1 for MS/MS transitions of selected precursors) was performed using a dynamic multiple reaction monitoring (dMRM) method (Sartain, 2016). Addition of micromolar concentrations of medronic acid into the mobile phase markedly increased the sensitivity for detecting di- and tri-nucleotides by this method (Hsiao et al., 2018).
With optimized sampling and the analysis method in hand, pyrimidine nucleotide levels were measured in cells cultured with either CPU, 5-F-CPU, or 5′-F-CPU in combination with 1 μM GSK983 and 5 μM uridine for 6 h (Figure 4 A). Inclusion of CPU or 5-F-CPU led to large decreases in UTP and CTP levels compared with cells treated with GSK983 alone; these effects grew more pronounced at 12 h (Figure S4). CPU depleted UTP and CTP concentrations more strongly than 5-F-CPU. Triphosphates of both CPU and 5-F-CPU could be detected by LC-MS/MS (Figure 4B), suggesting that nucleoside diphosphate kinase, a mammalian enzyme known to have broad substrate scope (Lascu and Gonin, 2000), could convert CPU-DP and 5-F-CPU-DP into their corresponding nucleoside triphosphate analogs. Inclusion of CPU or 5-F-CPU also led to moderate decreases in UMP, CMP, UDP, and CDP levels compared with cells treated with GSK983 alone at both 6 h and 12 h (Figure S4).
Figure 4.
LC-MS/MS Analysis of Intracellular Nucleotides
(A) LC-MS/MS analysis of intracellular uridine and cytidine nucleotide levels after 6 h treatments. In all assays, the culture medium was supplemented with 5 μM uridine and 1 μM GSK983. Error bars represent ±SD of three biological replicates with statistical comparisons with the untreated control denoted for UTP levels (blue) and CTP levels (red) as not significant (n.s.), ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 as calculated by one-way ANOVA, Dunnett's test.
(B) LC-MS/MS detection of mono-, di-, and triphosphates of CPU and 5-F-CPU in cells. The targeted multiple reaction monitoring (MRM) transitions were CPU-MP (319→79), 5-F-CPU-MP (337→79), CPU-DP (399→159), 5-F-CPU-DP (417→159), CPU-TP (479→159), and 5-F-CPU-TP (497→159) as detailed in Table S1.
See also Figures S3 and S4.
Despite its UCK2 inhibitory activity in vitro, 5′-F-CPU did not deplete intracellular nucleotide levels and even appeared to increase levels of some intracellular nucleotides (Figures 4A and S4). This contrast suggests that the activity of CPU and 5-F-CPU is derived primarily from their flux through pyrimidine metabolism pathways and not from their inhibition of UCK2.
Antiviral and Cytotoxic Activities of Selected CPU Analogs
From the above enzymological and metabolic data, we hypothesized that the combination of CPU and GSK983 would be more effective at inhibiting dengue virus replication than any other combination. Using an infectious clone of dengue serotype 2 (DENV-2) strain 16681 engineered to express a luciferase reporter (Marceau et al., 2016), the efficacy and cytotoxicity of each combination treatment was tested in infected or uninfected cultures of the A549 lung carcinoma cell line (Figure 5 A). In all assays, culture medium was supplemented with 20 μM uridine to mimic physiological plasma concentrations. In the presence of 20 μM exogenous uridine, 1 μM of the de novo pyrimidine synthesis inhibitor GSK983 alone did not significantly inhibit viral infection or cellular viability (Figure 5A). In line with our original hypothesis, combination of GSK983 with either 500 μM CPU and 5-F-CPU caused significant reductions in viral infection unlike the other cyclopentenyl compounds tested (Figure 5A). However, only CPU appeared to have a workable therapeutic index at this concentration and time point.
Figure 5.
Antiviral and Cytotoxic Activities of Modulators of Pyrimidine Metabolism in the Presence of Exogenous Uridine
(A) Effect of 1 μM GSK983 and 500 μM of individual CPU analogs on luminescence of luciferase-expressing DENV-2 virus (red) and on A549 cell viability (blue) at 72 h, relative to an untreated control. Error bars represent ±SD of three biological replicates with statistical comparisons with the untreated control denoted for viral infection (red) and cell viability (blue) as not significant (n.s), ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 as calculated by one-way ANOVA, Dunnett's test.
(B) Effects of CPU-GSK983 combination therapy on dengue virus replication and cell proliferation in the presence of exogenous uridine at 48 h. Each GSK983 titration curve is tested in the presence of 0 μM, 125 μM, 250 μM, 500 μM, and 1 mM CPU. Along the dotted line are CPU-only controls at 0 μM, 125 μM, 250 μM, 500 μM, and 1 mM CPU in the absence of GSK983. Error bars represent ±SD of two replicates. RLU, relative light units.
See also Figure S5.
To further examine the efficacy and cytotoxicity of CPU, we conducted a more comprehensive checkerboard analysis of the CPU-GSK983 combination. While neither GSK983 nor CPU was effective as a single agent in physiological uridine levels, the combination of both molecules successfully suppressed dengue virus infection (Figure 5B). For example, combining 0.2 μM GSK983 and 250 μM CPU inhibited ∼50% of virus replication (Figure 5B). At a CPU dose of 1 mM, virus replication was suppressed almost completely. Notably, the combination treatment had much less effect on A549 cell growth, suggesting that combinations of GSK983 and CPU could be selective for inhibition of virus but not host replication.
To further demonstrate that intracellular nucleotide depletion is correlated with antiviral activity, we also tested the effects of combining GSK983 with a broad inhibitor of nucleoside transport, dipyridamole (DPY). This drug significantly suppressed intracellular nucleotide levels (Figure S4) and consequently inhibited viral replication (Figure S5).
Improving the Therapeutic Index of RdRp Inhibitor R1479 by Combination Treatment Targeting Pyrimidine Biosynthesis
Given that the CPU-GSK983 combination was able to selectively inhibit viral infection through modulation of intracellular nucleotide pools, we hypothesized that the combination should potentiate the effects of an RdRp inhibitor on viral genome replication. Among RdRp inhibitors, the nucleoside inhibitor (NI) class is the largest (Klumpp et al., 2006) and has progressed the furthest in studies against dengue virus (Lim et al., 2015). Many NIs are nucleoside analogs that rely on intracellular phosphorylation to form triphosphate substrates that block RdRp (Sofia et al., 2012). For example, the cytidine analog 4′-azidocytidine (R1479) and its prodrug balapiravir (R1626) have been assessed for treating hepatitis C virus and later dengue virus infections (Chen et al., 2014, Klumpp et al., 2006). However, both compounds failed in clinical trials against dengue due to limited efficacy (Nguyen et al., 2013).
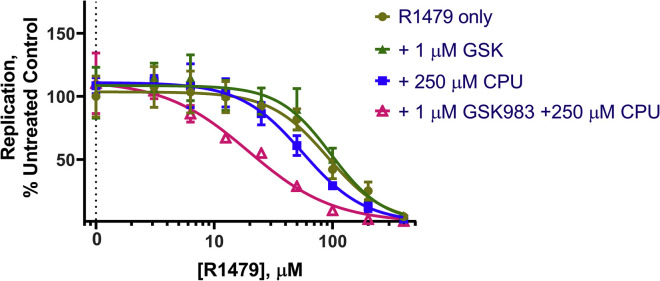
To test whether a combination therapy approach could improve the therapeutic index of R1479, we treated cells with R1479 in combination with GSK983 and CPU. As expected, R1479 showed dose-dependent inhibition of dengue virus replication with a half-maximal effective concentration (EC50) of ∼32 μM (Figure S6A). Inhibition of either de novo or salvage pyrimidine biosynthesis alone did not potentiate the antiviral activity of R1479 or cause additional cytotoxicity (Figure S6A). In contrast, inhibition of both the de novo and salvage pathways markedly enhanced the potency of R1479 (Figure 6 A). In the presence of 250 μM CPU, the EC50 of R1479 was lowered to ∼12 μM. Notably, such a three-component regimen had minimal impact on the cytotoxicity profile of R1479 (Figure 6B). Regarding other analogs, 5-F-CPU-GSK983 was able to slightly potentiate the antiviral activity of R1479, whereas 5′-F-CPU appeared to actually decrease antiviral efficacy of R1479 (Figures S6B and S6C).
Figure 6.
Potentiating the Effects of RdRp Inhibitor R1479 with CPU-GSK983 Combination Treatment
(A) Luminescence of luciferase-expressing DENV-2 virus in A549 cells at 48 h.
(B) Cell viability of A549 cells under the triple combination therapy at 48 h. In all assays, the culture medium was supplemented with 20 μM uridine to mimic plasma uridine concentration and 1 μM GSK983 to block de novo pyrimidine biosynthesis. Error bars represent ±SD of three replicates.
See also Figures S6 and S7.
As robust intracellular phosphorylation is critical for both R1479 and CPU activity, we confirmed that competition between these two compounds for intracellular kinases at the rate-limiting mono-phosphorylation step (Ford et al., 1991, Klumpp et al., 2008) should be minimal. Despite its 2′-α-OH group, R1479 is known to be solely phosphorylated by deoxycytidine kinase (dCK) (Klumpp et al., 2008) and was indeed not a UCK2 substrate in our hands (Figure S7). Similarly, cyclopentenyl compounds are substrates of UCK2 and not dCK (Ford et al., 1991).
To gain insight into the mechanism underlying the improvement of R1479 efficacy with CPU, we deployed a replicon assay that bypasses viral entry by electroporation of a reporter DENV-2 RNA construct (Alvarez et al., 2005, Marceau et al., 2016). Such replicon systems include the coding region needed for viral RNA translation and replication, while lacking the structural genes necessary for the production of viral particles. Consequently, viral genome replication is examined independently of viral entry and particle assembly (Kato and Hishiki, 2016). In this assay (Figure 7 ), the EC50 of R1479 alone was ∼90 μM. Inhibition of de novo pyrimidine biosynthesis alone with GSK983 did not alter this value (∼96 μM), whereas addition of 250 μM CPU shifted it to ∼56 μM. Targeting both the de novo and salvage pathways with a combination of 1 μM GSK and 250 μM CPU lowered the EC50 of R1479 by over 4-fold to ∼19 μM. These results suggest that intracellular nucleotide depletion is the major driver of CPU's antiviral activity. Together, these findings highlight the potential of combining modulators of pyrimidine metabolism with inhibitors of viral genome replication.
Figure 7.
Examining Viral Replication with Combinations of RdRp Inhibitor R1479 with CPU-GSK983 Treatment
Luminescence of luciferase-expressing DENV-2 replicon RNA in A549 cells at 48 h. In all assays, the culture medium was supplemented with 20 μM uridine and 1 μM GSK983. Error bars represent ±SD of three replicates.
Discussion
The flaviviruses, including dengue virus, are an important class of clinically relevant viral pathogens with limited treatment options (Boldescu et al., 2017). In particular, dengue causes hundreds of millions of symptomatic infections annually yet lacks any antiviral treatment, while the sole approved vaccine has limited use because of safety risks associated with vaccination of individuals not previously exposed to dengue (Lim et al., 2013, Sridhar et al., 2018). A series of high-throughput phenotypic cell-based screens have recently identified DHODH inhibition as a potent, broad-spectrum antiviral strategy in vitro (Hoffmann et al., 2011, Lucas-Hourani et al., 2013, Luthra et al., 2018, Wang et al., 2011). Furthermore, a DHODH inhibitor was recently shown to suppress SARS-CoV-2 in vitro (Xiong et al., 2020). However, DHODH inhibitors lose antiviral activity upon extracellular uridine addition due presumably to pyrimidine salvage (Deans et al., 2016). While a previous study has explored combining a DHODH inhibitor with guanosine analogs ribavirin or INX-08189 in a non-uridine supplemented cell culture dengue model (Yeo et al., 2015), the antiviral efficacy of a DHODH and a salvage inhibitor has not been previously reported despite interest (Luthra et al., 2018). Here, we found that a GSK983-CPU combination treatment effectively blocked dengue virus replication in the presence of physiological concentrations of extracellular uridine. At the same time, depletion of UTP and CTP led to a synergy between the CPU-GSK983 combination and RdRp inhibition.
Synthesis and evaluation of a series of CPU analogs revealed that CPU and 5-F-CPU were substrates of UCK2, and the resulting monophosphates were substrates of CMPK1. In combination with GSK983, the flux of CPU and 5-F-CPU to their triphosphate forms depleted intracellular pools of pyrimidine NTPs (Figure 4A). As the non-substrate analog 5′-F-CPU did not deplete intracellular pyrimidines despite in vitro UCK2 inhibitory activity, our results suggest that CPU's activity in host cells is driven by its ability to block multiple targets in pyrimidine salvage rather than solely UCK2 inhibition.
This distinctive antiviral activity of CPU could conceivably originate from host-targeting modulation of pyrimidine metabolism as well as potential direct-acting antiviral activity of its triphosphate on viral replication. Intracellular nucleotide depletion appears correlated with antiviral activity given that the structurally unrelated nucleoside transport inhibitor DPY also suppressed intracellular nucleotide levels and viral replication (Figure S5). In addition, while 250 μM CPU in the absence of GSK983 did slightly potentiate the effects of R1479 on viral replication (Figure 7), this did not translate to decreased viral infection at the same time point (Figure S6A). Nevertheless, further investigation of inhibitory effects of CPU-TP on dengue RdRp is warranted. Indeed, as noted previously, NIs are the largest class of RdRp inhibitors (Klumpp et al., 2008). A multi-factorial approach that incorporates information about interactions with multiple host and viral enzymes will likely be useful for future SAR around CPU as this compound remained the most efficacious in our panel. In addition, efforts to develop specific inhibitors of UCK2 are needed. We previously identified such inhibitors through a high-throughput screen (Okesli-Armlovich et al., 2019); however, these inhibitors appeared to lack sufficient potency to synergize with GSK983 in antiviral models (data not shown).
CPU is known to be safe in vitro (Blaney et al., 1992, Ford et al., 1991, Song et al., 2001) and does not show appreciable toxicity in mice (Cysyk et al., 1995) or non-human primates (Blaney et al., 1990). While the safety profile of GSK983 remains less clear, other DHODH inhibitors such as US Food and Drug Administration-approved arthritis drugs leflunomide and teriflunomide and the clinical-stage anti-cancer compound brequinar have been extensively studied in humans and are generally tolerated (Aly et al., 2017). We note that the potential immunosuppressive effects of DHODH inhibitors may be viewed as helpful (Xiong et al., 2020) or harmful (Bonavia et al., 2011) in the treatment of viral infection. Meanwhile, many RdRp inhibitors have been evaluated in clinical trials against dengue. While a preliminary study combining CPU with a de novo synthesis inhibitor in vivo resulted in toxicity (Cysyk et al., 1995), there is precedent for combining inhibitors of de novo pyrimidine synthesis with nucleoside transport inhibitors in humans (Casper et al., 1991). With careful dose optimization and an eye toward immune effects, our results suggest that a combination strategy targeting both host pyrimidine biosynthesis and viral RdRp holds promise as a potential therapy against RNA viruses.
Significance
RNA virus infections cause serious diseases such as viral hepatitis, influenza, Ebola, dengue, and coronavirus disease 2019, yet many of them lack suitable antiviral treatments. We identified a host-targeting antiviral strategy of modulating pyrimidine metabolism with analogs of cyclopentenyl uracil, an inhibitor of pyrimidine salvage, and GSK983, an inhibitor of de novo biosynthesis. This combination therapy also markedly increased the potency of R1479, an RNA-dependent RNA polymerase (RdRp) inhibitor, against dengue virus replication. At efficacious drug doses, the effect on the growth rates of uninfected cells was minimal. In light of the growing interest in RdRp inhibitors as antiviral agents, our findings shine light on a promising way to enhance their clinical utility by combining them with modulators of mammalian pyrimidine metabolism.
STAR★Methods
Key Resources Table
| REAGENTS OR RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli BL21(DE3) | Thermo Scientific | Cat#EC0014 |
| pDENV-Luc infectious clone | Marceau et al. (2016) | N/A |
| pDENV-Luc replicon | Marceau et al. (2016) | N/A |
| Chemicals, Peptides and Recombinant Proteins | ||
| DMEM | Gibco | Cat#11995065 |
| Trypsin/EDTA | Gibco | Cat#25300054 |
| Penicillin-Streptomycin | Gibco | Cat#15140122 |
| Glutamine | Gibco | Cat#25030081 |
| Fetal Bovine Serum | HyClone | Cat#SH30396.03 |
| Electroporation Buffer for replicon | Teknova | Cat#E0399 |
| XbaI Restriction Enzyme | New England BioLabs | Cat#R0145S |
| Lactate dehydrogenase, porcine heart (LDH) | EMD Millipore | Cat#427211 |
| Pyruvate kinase, rabbit muscle type III (PK) | Sigma-Aldrich | Cat#P9136 |
| Adenosine triphosphate disodium trihydrate (ATP) | VMR Life Science | Cat#0220 |
| β-Nicotinamide adenine dinucleotide, reduced disodium salt hydrate (NADH) | Santa Cruz | Cat# sc-205762A |
| Phosphoenolpyruvate monopotassium salt (PEP) | Sigma-Aldrich | Cat#P7127 |
| Cytidine 5’-monophospahte disodium salt (CMP) | Sigma-Aldrich | Cat#C1006 |
| Uridine | Sigma-Aldrich | Cat#U3750 |
| LB Broth | Fisher BioReagents | Cat#BP97235 |
| Isopropyl-1-thio-b-D-galacto-pyranoside (IPTG) | Thermo Fisher Scientific | Cat#R0392 |
| R1479 | MedChemExpress | Cat#HY-10444 |
| Dipyridamole (DPY) | MP Biomedicals | Cat#0215372205 |
| D-ribose | Chem Impex | Cat#50494709 |
| Tetrabutylammonium fluoride solution (TBAF), 1.0M | Sigma-Aldrich | Cat#216143 |
| Diethyl azodicarboxylate solution (DEAD), 40 wt.% | Sigma-Aldrich | Cat#563110 |
| Triphenylphosphine (PPh3) | Sigma-Aldrich | Cat#T84409 |
| Uracil | Sigma-Aldrich | Cat#U0750 |
| 5-Fluorouracil | Sigma-Aldrich | Cat#F6627 |
| Thymine | AK Scientific | Cat#J91030 |
| 5-Iodouracil | AK Scientific | Cat#K165 |
| Ammonia, ca. 7N solution in methanol | Sigma-Aldrich | Cat#499145 |
| Silver (I) oxide (Ag2O) | Sigma-Aldrich | Cat#226831 |
| Methyl iodide (MeI) | EMD Millipore | Cat#IX0185 |
| Diethylaminosulfur trifluoride (DAST) | Oakwood | Cat#002323 |
| Methanesulfonyl chloride (MsCl) | Sigma-Aldrich | Cat#471259 |
| Triethylamine (NEt3) | Thermo Fisher Scientific | Cat#04885 |
| Acetyl chloride (AcCl) | Sigma-Aldrich | Cat#114189 |
| Palladium hydroxide on carbon [Pd(OH)2/C], 5 wt.% | Acros Organic | Cat#199620100 |
| Cyclohexene | Sigma-Aldrich | Cat#29240 |
| Sodium azide (NaN3) | Sigma-Aldrich | Cat#71289 |
| 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) | Fisher BioReagents | Cat#BP310-100 |
| Tris(hydroxymethyl)aminomethane base (Tris) | Fisher BioReagents | Cat#BP152-1 |
| Dithiothreitol (DTT) | Fisher BioReagents | Cat#BP172-25 |
| Magnesium chloride (MgCl2) | Fisher Chemical | Cat#M33-500 |
| Potassium chloride (KCl) | Fisher Chemical | Cat#P217-500 |
| Medronic acid (InfinityLab Deactivator Additive) | Agilent | Cat#5191-3940 |
| Adenosine 15N5 5’-monophosphate disodium salt | Sigma-Aldrich | Cat#662658 |
| Uridine 15N2 5’-monophosphate sodium salt | Sigma-Aldrich | Cat#662666 |
| Critical Commercial Assays | ||
| MEGAscript T7 Transcription Kit | Invitrogen | Cat#AM1334 |
| m7G(5')ppp(5')G RNA Cap Structure Analog | New England BioLabs | Cat#S1404S |
| CellTiter-Glo | Promega | Cat#G7570 |
| Renilla-Glo | Promega | Cat#E2710 |
| Renilla | Promega | Cat#E2820 |
| Experimental Models: Cell Lines | ||
| A549 | ATCC | Cat#CCL-185 |
| Vero E6 | ATCC | Cat#CRL-1586 |
| Recombinant DNA | ||
| pET21a-UCK2 expression plasmid | Okesli-Armlovich et al. (2019) | N/A |
| pET28-CMPK1 expression plasmid | Deans et al. (2016) | N/A |
| Software | ||
| Prism 8.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ChemDraw Professional 17.1 | PerkinElmer | https://www.perkinelmer.com/category/chemdraw |
| MNova 12.0 | Mestrelab | https://mestrelab.com/download_file/mnova-12-0-0/ |
| Other | ||
| Centrifugal Filter Unit | Amicon | Cat#UFC900308 |
| Hi Trap Q anion exchange column | GE Healthcare Life Sciences | Cat#17115301 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chaitan Khosla (Khosla@stanford.edu)
Materials Availability
This study did not generate any unique biological reagents. Synthesis of CPU and related chemicals is detailed within the manuscript.
Data and Code Availability
This study did not generate any unique datasets or code not available in the manuscript files.
Experimental Models and Subject Details
Mammalian Cell Culture
All mammalian cell lines were maintained in a humidified incubator (37°C, 5% CO2). A549 cells (male) and Vero E6 cells (female) were obtained from the American Type Culture Collection and cultured in DMEM supplemented with 10% FBS, penicillin/streptomycin, and L-glutamine. Upon reaching 50-75% confluence, cells were detached from the growth surface using a trypsin/EDTA solution prior to analysis. Cells were maintained in logarithmic growth during all biological assays.
Bacterial Cell Culture
BL21(DE3) cells were obtained from ThermoFisher Scientific. Cells were cultured in LB Broth at 37°C with agitation or as required for recombinant protein production.
DENV-Luc Reporter Virus Generation
The design of the pDENV-Luc infectious clone derived from dengue serotype 2 (DENV-2) strain 16681 was detailed previously (Marceau et al., 2016). The plasmid was linearized with XbaI and in vitro transcribed into the genomic RNA of DENV-Luc virus using the T7 Megascript Kit (Ambion) in the presence of m7G(5')ppp(5')G RNA Cap Structure Analog (NEB). 5 μg DENV-Luc RNA was electroporated into 2X106 Vero E6 cells. The transfected cells were resuspended in DMEM with 10% FBS and 100 U/ml penicillin-streptomycin and transferred into a T-175 flask incubated at 37°C with 5% CO2. Supernatants were collected and replenished with fresh medium every 24 h from day 17 to 24 post-transfection, pooled together, clarified by centrifugation and stored in aliquots at −80°C. The amount of infectious DENV-Luc virus in the stock was titrated using the TCID50 assay and calculated by the Spearman & Kärber algorithm as described previously (Hierholzer and Killington, 1996).
pDENV-Luc Replicon Generation
The plasmid pDENV-Luc replicon containing a Renilla luciferase expressing DENV subgenomic replicon was described previously (Marceau et al., 2016). For in vitro transcription, 10 ug of plasmid DNA was linearized using the XbaI restriction enzyme. Replicon RNA was generated using the MEGAscript T7 Transcription Kit in the presence of 5mM m7G(5′)ppp(5′)G RNA Cap Structure Analogue. For assays, two million A549 cells were washed twice with PBS and re-suspended in electroporation buffer (Teknova, E0399), mixed with 4 ug of purified replicon RNA, and electroporated using Bio-Rad Gene Pulser Xcell electroporator using square wave protocol. Electroporated cells were resuspended in pre-warmed culture medium without antibiotics and seeded as required.
Method Details
Synthesis and Characterization of CPU Analogs
Unless otherwise noted, all reactions were performed under an argon atmosphere in flame- or oven- dried glassware. Reaction mixtures were stirred using Teflon-coated magnetic stirrer bars and monitored by thin layer silica gel chromatography (TLC) using 0.25 mm silica gel 60F plates with fluorescent indicator from Merck. Plates were visualized under UV or treated by KMnO4 stain with gentle heating. Products were purified on an AnaLogix IntelliFlash 280 Flash column chromatography system using the solvent gradients indicated. Anhydrous tetrahydrofuran (THF), dichloromethane (CH2Cl2), dimethylformamide (DMF), acetone, and Dimethylsulfoxide (DMSO) were obtained from Acros Organics. Diethyl ether (Et2O), ethyl acetate (EtOAc), hexanes, and methanol (MeOH) were from Fisher Scientific. DMSO used in bioassays and to prepare biological samples was from Fisher BioReagents. All other reagents were from commercial suppliers and were used as received without additional purification. Samples prepared for biological evaluation were purified via preparative HPLC in a water/acetonitrile (MeCN) gradient containing 0.1% (v/v) trifluoroacetic acid (TFA) using an Agilent 1260 Infinity system equipped with an Agilent Prep-C18 column (21.2 x 250 mm).
NMR spectra were measured on a Varian INOVA 500 (1H at 500 MHz, 13C at 125 MHz), a Varian 400 (1H at 400 MHz, 13C at 100 MHz), or a Varian INOVA 600 MHz (1H at 600 MHz, 13C at 150 MHz) magnetic resonance spectrometer, as noted. 1H chemical shifts were reported relative to the residual solvent peak (CDCl3 = 7.26 ppm; CD3OD = 3.31 ppm) as follows: chemical shift (δ) [multiplicity (s = singlet, brs = broad singlet, d = doublet, t = triplet, q = quartet, dd = doublet of doublet, m = multiplet), coupling constant(s) in Hz, integration]. 13C chemical shifts were reported relative to the residual deuterated solvent 13C signals (CDCl3 = 77.16 ppm, CD3OD = 49.00 ppm) and rounded to one decimal places. Infrared spectra were recorded on a Nicolet iS50 FT/IR Spectrometer at the Stanford Nano Share Facilities and were reported in wavenumbers (cm-1). Optical rotation data were obtained using a JASCO DIP were reported as [α] (c = grams/100 mL, solvent), where D indicates the sodium D line (589 nm). High resolution mass spectra were obtained on an Agilent 6545 QTOF mass spectrometer at the Metabolic Chemistry Analysis Center at Stanford University. The MNova and ChemDraw softwares aided analytical chemistry analysis.
3-benzoyl-1-((3aS,4R,6aR)-6-(((tert-butyldiphenylsilyl)oxy)methyl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-4-yl)pyrimidine-2,4(1H,3H)-dione (3a)
Compound 1 was prepared in 10 steps from D-ribose according to literature procedures (Choi et al., 2004). To a suspension of 1 (440 mg, 1.04 mmol, 1 equiv.), 2a (270 mg, 1.25 mmol, 1.2 equiv.), and PPh3 (410 mg, 1.56 mmol, 1.5 equiv.) in THF (12 mL) at 0°C was added DEAD solution in toluene (720 μL, 1.5 equiv., 40 w% in toluene). After the addition of DEAD, the reaction mixture turned from a white suspension to a yellow solution, which was slowly warmed up to room temperature and stirred overnight. The reaction mixture was then concentrated, loaded onto a 12g SiO2 flash cartridge, and purified with a linear gradient of 20-40% EtOAc in hexanes to afford 3a (446 mg, 0.72 mmol, 69%) as white powder. [α] –17.0 (c 1.3, MeOH); IR (film, cm-1): 3071, 2930, 2857, 1748, 1705, 1667, 1441, 1429, 1372, 1234, 1112, 905, 732, 704.1 H NMR (400 MHz, CDCl3) δ 7.95 (d, J = 7.4 Hz, 1H), 7.71 – 7.61 (m, 6H), 7.53 – 7.47 (m, 2H), 7.45 – 7.42 (m, 2H), 7.41 – 7.34 (m, 4H), 6.91 (d, J = 8.1 Hz, 1H), 5.77 (d, J = 8.1 Hz, 1H), 5.67 (s, 1H), 5.37 (s, 1H), 5.08 (d, J = 5.8 Hz, 1H), 4.60 (d, J = 5.8 Hz, 1H), 4.47 (d, J = 16.4 Hz, 1H), 4.41 (d, J = 16.7 Hz, 1H), 1.34 (s, 3H), 1.29 (s, 3H), 1.10 (s, 9H). 13 C NMR (100 MHz, CDCl3) δ 168.8, 162.3, 153.6, 149.8, 141.1, 135.6, 135.6, 135.3, 133.3, 133.1, 131.6, 130.7, 130.1, 129.3, 128.0, 128.0, 120.8, 112.9, 102.4, 84.6, 83.4, 68.4, 61.4, 27.3, 27.0, 25.9, 19.4. HRMS (ESI) m/z 623.2574 [(M+H)+; calcd for C36H39N2O6Si+: 623.2572].
1-((1R,4R,5S)-4,5-dihydroxy-3-(hydroxym ethyl)cyclopent-2-en-1-yl)pyrimidine-2,4(1H,3H)-dione (4a)
To compound 3a (30 mg, 0.048 mmol) was added 0.5 mL 7N NH3 solution in methanol. After 1 h, the reaction solvent was removed under positive N2 atmosphere and the resulting solid was further dried under high vacuum. The reaction crude was then treated with 30 μL HCl in 300 μL THF and stirred overnight. Excess NaHCO3 was added to neutralize the reaction, followed by the addition of 1 mL MeOH. The resulting suspension was filtered through a short Celite pad, rinsed with MeOH, and concentrated. The resulting crude was then resuspended with 1 mL H2O and subjected to HPLC purification with a linear gradient of 5-20% MeCN (0.1% TFA) in H2O (0.1% TFA) on a prep C18 column (Agilent 10 prep-C18 250 × 21.1 mm). Fractions containing desired product was then combined and lyophilized to afford the final product 4a (4.6 mg, 0.019 mmol, 40%) as white powder. [α] – 62.8 (c 3.2, MeOH); IR (film, cm-1): 3349 (br), 1667, 1465, 1390, 1258, 1202, 1114.1 H NMR (500 MHz, CD3OD) δ 7.42 (d, J = 7.9 Hz, 1H), 5.71 (s, 1H), 5.49 (s, 1H), 4.90 (s, 1H), 4.52 (d, J = 5.9, 1.2 Hz, 1H), 4.33 – 4.19 (m, 2H), 4.04 (t, J = 5.6 Hz, 1H). 13 C NMR (125 MHz, CD3OD) δ 166.4, 153.1, 152.1, 143.6, 125.5, 102.8, 78.4, 74.0, 67.4, 60.3. HRMS (ESI) m/z 263.0644 [(M+Na)+; calcd for C10H12N2NaO5 +: 263.0638 ].
In a similar manner, compound 1 (360 mg, 0.85 mmol, 1 equiv.) was reacted with 2b (238 mg, 1.02 mmol, 1.2 equiv.) to afford intermediate 3b as white powder (288 mg, 0.45 mmol, 53%). Intermediate 3b (32 mg, 0.05 mmol) was then deprotected to afford 4b (7.7 mg, 0.03 mmol, 60%) as white powder.
3-benzoyl-1-((3aS,4R,6aR)-6-(((tert-butyldiphenylsilyl)oxy)methyl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-4-yl)-5-fluoropyrimidine-2,4(1H,3H)-dione (3b)
[α] – 12.9 (c 1.4, CHCl3); IR (film, cm-1): 3072, 2932, 2857, 1753, 1712, 1665, 1662, 1448, 1373, 1234, 1106, 1088, 732, 702, 687. 1 H NMR (400 MHz, CDCl3) δ 7.97 – 7.92 (m, 2H), 7.73 – 7.63 (m, 5H), 7.55 – 7.49 (m, 2H), 7.48 – 7.42 (m, 2H), 7.42 – 7.35 (m, 4H), 7.06 (d, J = 5.8 Hz, 1H), 5.68 (brs, 1H), 5.41 (brs, 1H), 5.08 (d, J = 5.8 Hz, 1H), 4.59 (dd, J = 12.1, 6.4 Hz, 1H), 4.54 – 4.35 (m, 2H), 1.34 (s, 3H), 1.29 (s, 3H), 1.11 (s, 9H). 13 C NMR (100 MHz, CDCl3) δ 167.3, 154.3, 148.3, 141.4, 139.0, 135.6, 135.6, 133.2, 133.0, 131.2, 130.8, 130.2, 130.2, 129.4, 129.1, 128.0, 125.5, 125.1, 120.4, 113.0, 84.6, 83.2, 68.3, 61.4, 27.3, 27.0, 25.9, 19.4. 19 F NMR (376 MHz, CDCl3) δ -163.44 (d, J = 5.7 Hz). HRMS (ESI) m/z 663.2323 [(M+Na)+; calcd for C36H37FN2O6SiNa+:663.2297].
1-((1R,4R,5S)-4,5-dihydroxy-3-(hydroxymethyl)cyclopent-2-en-1-yl)-5-fluoropyrimidine-2,4(1H,3H)-dione (4b)
[α] – 87.8 (c 0.45, MeOH); IR (film, cm-1): 3375 (br), 1696, 1660, 1386, 1242, 1115, 1011.1 H NMR (400 MHz, CD3OD) δ 7.60 (d, J = 6.6 Hz, 1H), 5.68 (q, J = 1.8 Hz, 1H), 5.49 (brs, 1H), 4.52 (d, J = 5.6 Hz, 1H), 4.26 (d, J = 2.2 Hz, 2H), 4.03 (t, J = 5.5 Hz, 1H). 13 C NMR (100 MHz, CD3OD) δ 159.54 (d, J = 26.1 Hz), 152.64, 151.74, 142.07 (d, J = 233.0 Hz), 127.39 (d, J = 33.7 Hz), 125.24, 78.17, 73.94, 67.73, 60.26. 19 F NMR (376 MHz, CD3OD) δ -168.53 (dd, J = 6.8, 1.7 Hz). HRMS (ESI) m/z 259.0722 [(M+H)+; calcd for C10H12FN2O5 +: 259.0725].
In a similar manner, compound 1 (245 mg, 0.58 mmol, 1 equiv.) was reacted with 2c (238 mg,0.70 mmol, 1.2 equiv.) to afford intermediate 3c (160 mg, 0.21mmol, 36%) as white powder. Intermediate 3c (40 mg, 0.053 mmol) was then deprotected to afford 4c (9.6 mg, 0.026 mmol, 49%) as white powder.
3-benzoyl-1-((3aS,4R,6aR)-6-(((tert-butyldiphenylsilyl)oxy)methyl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-4-yl)-5-iodopyrimidine-2,4(1H,3H)-dione (3c)
[α] – 45.1 (c 1.3, CHCl3); IR (film, cm-1): 3071, 2932, 2857, 1749, 1703, 1666, 1608, 1420, 1233, 1112, 703.1 H NMR (400 MHz, CDCl3) δ 7.93 – 7.90 (m, 1H), 7.72 – 7.63 (m, 5H), 7.54 – 7.48 (m, 3H), 7.45 – 7.37 (m, 6H), 5.72 (brs, 1H), 5.37 (s, 1H), 5.13 (d, J = 5.8 Hz, 1H), 4.62 (d, J = 5.8 Hz, 1H), 4.51 – 4.34 (m, 2H), 1.34 (s, 3H), 1.29 (s, 3H), 1.09 (s, 9H). 13 C NMR (100 MHz, CDCl3) δ 167.8, 159.1, 154.1, 149.5, 145.8, 135.6, 135.4, 133.1, 133.1, 131.1, 130.7, 130.1, 129.4, 128.0, 128.0, 120.6, 113.0, 84.6, 83.4, 69.2, 68.1, 61.4, 27.3, 27.0, 25.9, 19.5. HRMS (ESI) m/z 749.1538 [(M+H)+; calcd for C36H38IN2O6Si+:749.1538].
1-((1R,4R,5S)-4,5-dihydroxy-3-(hydroxymethyl)cyclopent-2-en-1-yl)-5-iodopyrimidine-2,4(1H,3H)-dione (4c).
[α] –104.0 (c 0.6, MeOH); IR (film, cm-1): 3370 (br), 1682, 1608, 1424, 1260, 1111.1 H NMR (400 MHz, CD3OD) δ 7.78 (s, 1H), 5.70 (q, J = 1.8 Hz, 1H), 5.47 (brs, 1H), 4.52 (d, J = 5.5 Hz, 1H), 4.33-4.20 (m, J = 2.1 Hz, 2H), 4.04 (t, J = 5.7 Hz, 1H).13 C NMR (100 MHz, CD3OD) δ 163.0, 152.8, 152.4, 148.0, 125.5, 78.6, 73.9, 68.6, 67.9, 60.0.HRMS (ESI) m/z 366.9782 [(M+H)+; calcd for C10H12IN2O5 +: 366.9785].
In a similar manner, compound 1 (113 mg, 0.27 mmol, 1 equiv.) was reacted with 2d (74 mg, 0.32 mmol, 1.2 equiv.) to afford intermediate 3d as white powder (100 mg, 0.16 mmol, 59%). Intermediate 3d (30 mg, 0.047 mmol) was then deprotected to afford 4d (4.3 mg, 0.017 mmol, 36%) as white powder.
3-benzoyl-1-((3aS,4R,6aR)-6-(((tert-butyldiphenylsilyl)oxy)methyl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-4-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (3d)
[α] – 27.8 (c 1.2, CHCl3); IR (film, cm-1): 3071, 2932, 2857, 1748, 1699, 1656, 1429, 1371, 1235, 1111, 732, 702.1 H NMR (500 MHz, CDCl3) δ 7.97 – 7.89 (m, 2H), 7.71 – 7.61 (m, 5H), 7.53 – 7.46 (m, 3H), 7.45 – 7.42 (m, 1H), 7.40 – 7.35 (m, 4H), 6.90 (brs, 1H), 5.73 (s, 1H), 5.37 (s, 1H), 5.13 (d, J = 5.8 Hz, 1H), 4.65 (dd, J = 16.3, 4.7 Hz, 1H), 4.47 – 4.37 (m, 2H), 1.96 (s, 3H), 1.34 (s, 3H), 1.29 (s, 3H), 1.09 (s, 9H).13 C NMR (125 MHz, CDCl3) δ 169.1, 163.0, 152.9, 149.8, 137.2, 135.6, 135.1, 133.2, 133.1, 131.7, 130.6, 130.1, 129.3, 128.0, 121.3, 112.7, 111.0, 84.6, 83.5, 68.3, 61.4, 27.3, 27.0, 25.9, 19.5, 12.8. HRMS (ESI) m/z 637.2734 [(M+H)+; calcd for C37H41N2O6Si+: 637.2728].
1-((1R,4R,5S)-4,5-dihydroxy-3-(hydroxymethyl)cyclopent-2-en-1-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (4d)
[α] – 78.5 (c 0.38, MeOH); IR (film, cm-1): 3371, 1683, 1476, 1261, 1205, 1114, 1016.1 H NMR (400 MHz, CD3OD) δ 7.23 (d, J = 1.2 Hz, 1H), 5.68 (q, J = 1.8 Hz, 1H), 5.48 (brs, 1H), 4.52 (d, J = 5.9 Hz, 1H), 4.26 (q, J = 2.5 Hz, 2H), 4.04 (t, J = 5.6 Hz, 1H), 1.87 (d, J = 1.2 Hz, 3H). 13 C NMR (100 MHz, CD3OD) δ 166.5, 153.3, 151.8, 139.2, 126.0, 111.8, 78.3, 74.0, 67.2, 60.3, 12.3. HRMS (ESI) m/z 277.0799 [(M+Na)+; calcd for C11H14N2O5Na+: 277.0795].
3-benzoyl-1-((3aS,4R,6aR)-6-(hydroxymethyl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-4-yl)pyrimidine-2,4(1H,3H)-dione (5)
To a solution of 3a (300 mg, 0.48 mmol, 1 equiv.) in THF (5 mL) was added TBAF (580 μL, 0.58 mmol, 1.2 equiv., 1 M in THF) at 0°C. After 2 h, the reaction mixture was quenched with 5 mL of saturated NH4OH solution and extracted with 5 × 5 mL EtOAc. The combined organic layers were dried over anhydrous Na2SO4, concentrated, loaded onto a 4 g SiO2 flash cartridge, and purified with a linear gradient 80-95% EtOAc in hexanes to afford the free primary alcohol 5 (160 mg, 0.42 mmol, 87%) as white powder. [α] – 4.9 (c 4.5, CHCl3); IR (film, cm-1): 3473 (br), 3088, 2988, 2933, 1746, 1702, 1665, 1443, 1374, 1239, 1179, 1239, 1059.1 H NMR (500 MHz, CDCl3) δ 7.93 (d, J = 8.4 Hz, 2H), 7.70 – 7.61 (m, 1H), 7.50 (dd, J = 8.2, 7.4 Hz, 2H), 7.16 (d, J = 8.0 Hz, 1H), 5.81 (d, J = 8.0 Hz, 1H), 5.64 (s, 1H), 5.30 (s, 1H), 5.25 (d, J = 5.8 Hz, 1H), 4.70 (d, J = 5.8 Hz, 1H), 4.43 (d, J = 15.9 Hz, 1H), 4.38 (d, J = 15.9 Hz, 1H), 1.43 (s, 3H), 1.34 (s, 3H). 13 C NMR (125 MHz, CDCl3) δ 168.8, 162.2, 152.5, 149.7, 141.6, 135.3, 131.5, 130.7, 129.3, 121.7, 113.0, 102.5, 84.3, 84.0, 69.2, 60.2, 27.3, 25.8. HRMS (ESI) m/z 385.1391 [(M+H), calcd for C20H21N2O6 +: 385.1394].
1-((1R,4R,5S)-4,5-dihydroxy-3-(methoxymethyl)cyclopent-2-en-1-yl)pyrimidine-2,4(1H,3H)-dione (6)
To a solution of 5 (20 mg, 0.05 mmol, 1 equiv.) in dry acetone (0.5 mL) was added Ag2O (23 mg, 0.1 mmol, 2 equiv.) and MeI (31 μL, 0.5 mmol, 10 equiv.). The reaction mixture was stirred at room temperature for 24 h, filtered through Celite pad, and concentrated in vacuo to afford a residue oil 17 mg. In a similar manner as the synthesis of 3a from 4a, the crude was treated with 0.5 mL 7N NH3 in methanol, followed by 25 μL HCl in 250 μL of THF to furnish the analog 6 (2.5 mg, 0.01 mmol, 20% for three steps). [α] –64.7 (c 0.25, MeOH); IR (film, cm-1): 3369 (br), 2921, 2851, 1682, 1469, 1410, 1262, 1204, 1096.1 H NMR (600 MHz, CD3OD) δ 7.40 (d, J = 8.0 Hz, 1H), 5.72 (dd, J = 3.8, 1.7 Hz, 1H), 5.69 (d, J = 8.0 Hz, 1H), 5.51 – 5.43 (m, 1H), 4.50 (d, J = 5.8 Hz, 1H), 4.14 (ddd, J = 14.1, 2.5, 2.4 Hz, 1H), 4.08 (ddd, J = 14.2, 2.2, 1.8 Hz, 1H), 4.05 (dd, J = 5.7, 5.5 Hz, 1H), 3.40 (s, 3H). 13 C NMR (100 MHz, CD3OD) δ 166.4, 153.0, 148.8, 143.7, 127.5, 102.8, 78.1, 74.0, 70.3, 67.6, 59.0. HRMS (ESI) m/z 277.0799 [(M+Na)+; calcd for C11H14N2O5Na+: 277.0795].
3-benzoyl-1-((3aS,4R,6aR)-6-(fluoromethyl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-4-yl)pyrimidine-2,4(1H,3H)-dione (M1)

To alcohol 5 (50 mg, 0.13 mmol, 1 equiv.) in 1 mL CH2Cl2 at –78°C was added a stock solution of DAST (0.15 mmol, 1.2 equiv., 200 μL, prepared by diluting 100 μL of DAST with CH2Cl2 to 1 mL). The reaction mixture was warmed up to room temperature and stirred overnight before quenching with 1 mL saturated NaHCO3. The resulting mixture was then extracted with 3×3 mL EtOAc. The combined organic layers were dried over anhydrous Na2SO4, concentrated, loaded onto a 4 g SiO2 flash cartridge, and purified with a linear gradient 20-50% EtOAc in hexanes to afford the intermediate M1 (27 mg, 0.070 mmol, 54%) as white powder. [α] –12.8 (c 2.4, CHCl3); IR (film, cm-1): 3100, 2989, 2937, 1746, 1704, 1667, 1599, 1441, 1374, 1239, 1179, 1060, 989.1 H NMR (600 MHz, CDCl3) δ 7.93 (d, J = 8.1 Hz, 2H), 7.66 (t, J = 7.5 Hz, 1H), 7.51 (dd, J = 7.91, 7.13 Hz, 2H), 7.13 (d, J = 8.1 Hz, 1H), 5.83 (d, J = 8.1 Hz, 1H), 5.72 (s, 1H), 5.32 (s, 1H), 5.26 (d, J = 5.9 Hz, 1H), 5.14 (s, 1H), 5.07 (s, 1H), 4.73 (d, J = 5.7 Hz, 1H), 1.43 (s, 3H), 1.34 (s, 3H). 13 C NMR (125 MHz, CDCl3) δ 168.7, 162.2, 149.6, 141.5, 135.4, 131.5, 130.7, 129.4, 123.2, 113.2, 102.7, 95.7, 84.1, 83.2, 80.5 (d, J = 149 Hz), 78.8, 77.4, 77.2, 76.9, 69.2, 27.3, 25.8. 19 F NMR (376 MHz, CDCl3) δ – 224.4 (t, J = 46.5). HRMS (ESI) m/z 387.1366 [(M+H)+; calcd for C20H20FN2O5 +: 387.1351].
1-((1R,4R,5S)-3-(fluoromethyl)-4,5-dihydroxycyclopent-2-en-1-yl)pyrimidine-2,4(1H,3H)-dione (7)
In a similar manner as the synthesis of 4a from 3a, compound M1 (13 mg, 0.034 mmol, 1 equiv.) was treated with 0.5 mL 7N NH3 in methanol, followed by 25 μL HCl in 250 μL THF to furnish the analog 7 (4.4 mg, 0.018 mmol, 53% for two steps) as white powder. [α] – 86.0 (c 0.22, MeOH); IR (film, cm-1): 3365 (br), 2920, 2852, 1675, 1466, 1389, 1265, 1203, 1115.1 H NMR (600 MHz, CD3OD) δ 7.42 (d, J = 7.9 Hz, 1H), 5.85 (brs, 1H), 5.70 (d, J = 8.0 Hz, 1H), 5.48 (brs, 1H), 5.15 – 4.99 (m, 2H), 4.57 (d, J = 6.0 Hz, 1H), 4.10 (dd, J = 6.7, 6.0 Hz, 1H). 13 C NMR (150 MHz, CD3OD) δ 166.2, 152.8, 147.2, 143.6, 128.2, 102.8, 80.5 (d, J = 149 Hz), 77.6, 73.1, 67.4. 19 F NMR (376 MHz, CD3OD) δ – 224.9 (t, J = 46.8 Hz). HRMS (ESI) m/z 265.0592 [(M+Na)+; calcd for C10H11FN2O4Na+: 265.0595].
((3aS,4R,6aR)-4-(3-benzoyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-6-yl)methyl methanesulfonate (8)
To a solution of 5 (60 mg, 0.16 mmol, 1 equiv.) in anhydrous CH2Cl2 (2 mL) was added Et3N (44 μL, 0.32 mmol, 2 equiv.) at 0°C, followed by the dropwise addition of 0.2 mL MsCl stock solution (0.26 mmol, 1.6 equiv.; Stock solution was prepared by diluting 0.1 mL MsCl with CH2Cl2 to 1 mL). After 20 min at 0°C, the reaction mixture was quenched with 2 mL saturated aqueous solution of NH4Cl, extracted with 3 × 3 mL CH2Cl2, dried over anhydrous Na2SO4, concentrated, loaded on a 4g SiO2 flash cartridge, and purified with a linear gradient 50-95% EtOAc in hexanes to afford the compound 8 (53 mg, 0.11 mmol, 72%) as pale yellow oil. [α] – 35.3 (c 2.8, CHCl3); IR (film, cm-1): 2989, 2937, 1745, 1702, 1661, 1597, 1440, 1354, 1235, 1174, 1087, 959, 906, 729.1 H NMR (500 MHz, CDCl3) δ 7.92 (d, J = 7.2 Hz, 2H), 7.66 (dd, J = 8.0, 7.7 Hz, 1H), 7.50 (d, J = 7.8 Hz, 2H), 7.19 (d, J = 8.1 Hz, 1H), 5.83 – 5.67 (m, 2H), 5.30 (d, J = 4.8 Hz, 2H), 4.98 (d, J = 14.0 Hz, 1H), 4.83 (d, J = 14.0 Hz, 1H), 4.71 (d, J = 5.8 Hz, 1H), 3.04 (s, 3H), 1.41 (s, 3H), 1.33 (s, 3H). 13 C NMR (125 MHz, CDCl3) δ 168.7, 162.1, 149.6, 146.3, 141.8, 135.4, 131.4, 130.6, 129.4, 125.9, 113.1, 102.7, 83.8, 83.1, 77.4, 77.2, 76.9, 68.7, 65.0, 38.1, 27.3, 25.7. HRMS (ESI) m/z 463.1185 [(M+H)+; calcd for C21H23N2O8S+: 463.1170].
1-((3aS,4R,6aR)-6-(azidomethyl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-4-yl)pyrimidine-2,4(1H,3H)-dione (M2)
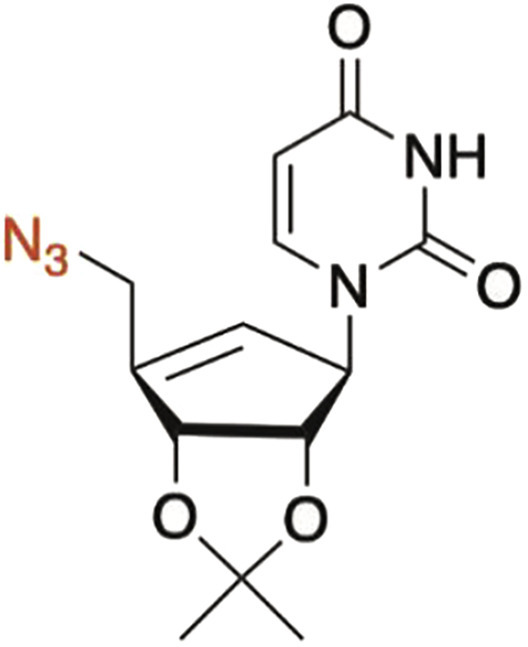
To the mesylate intermediate 8 (26 mg, 0.056 mmol, 1 equiv.) was added DMF (3 mL) and NaN3 (80 mg, 1.23 mmol, 22 equiv.). The resulting suspension was heated to 100°C and stirred for 18 hours, followed by the addition of 3 mL saturated solution of NH4Cl. The organic layer was extracted with 3 × 5 Et2O, washed with brine, and concentrated under reduced pressure. The crude material was purified through a 4g SiO2 flash cartridge with a linear gradient 40-95% EtOAc in hexanes to afford the azide intermediate S2 (14 mg, 0.046 mmol, 82%) as brown oil. [α] – 34.0 (c 0.7, CHCl3); IR (film, cm-1): 3197, 3059, 2988, 2929, 2102, 1687, 1456, 1380, 1243, 1082, 1058.1 H NMR (500 MHz, CDCl3) δ 8.78 (brs, 1H), 7.02 (d, J = 8.0 Hz, 1H), 5.71 (d, J = 8.0 Hz, 1H), 5.64 (brs, 1H), 5.31 (brs, 1H), 5.22 (d, J = 5.7 Hz, 1H), 4.65 (d, J = 5.8 Hz, 1H), 4.13 (d, J = 16.0 Hz, 1H), 4.04 (d, J = 16.0 Hz, 1H), 1.44 (s, 3H), 1.35 (s, 3H). 13 C NMR (125 MHz, CDCl3) δ 162.8, 150.5, 150.1, 141.2, 122.1, 112.6, 102.3, 86.2, 84.4, 68.0, 27.4, 26.0, 14.6. HRMS (ESI) m/z 306.1195 [(M+H)+; calcd for C13H16N5O4 +: 306.1197].
1-((1R,4R,5S)-3-(azidomethyl)-4,5-dihydroxycyclopent-2-en-1-yl)pyrimidine-2,4(1H,3H)-dione (9)
To intermediate S2 (6 mg, 0.023 mmol, 1 equiv.) was added THF (250 μL) and concentrated HCl (25 μL). The reaction mixture was stirred at room temperature for 5 h. An excess amount of NaHCO3 was added to neutralize the reaction, followed by the addition of 1 mL MeOH. The resulting suspension was then filtered through a short Celite pad, rinsed with 2 × 2 mL MeOH, and concentrated. The resulting crude was resuspended with 1 mL H2O and subjected to HPLC purification with a linear gradient of 5-20% MeCN (0.1% TFA) in H2O (0.01% TFA) on a prep C18 column (Agilent 10 prep-C18 250 × 21.1 mm). Fractions containing the desired product were then combined and lyophilized to afford the final product 9 (3.6 mg, 0.014 mmol, 59%) as white powder. [α] – 50.0 (c 0.36, MeOH); IR (film, cm-1): 3358 (br), 2921, 2104, 1683, 1467, 1393, 1261, 1205, 1116.1 H NMR (400 MHz, CD3OD) δ 7.39 (d, J = 8.0, 0.6 Hz, 1H), 5.81 (dd, J = 3.2, 1.9 Hz, 1H), 5.70 (d, J = 8.0 Hz, 1H), 5.52 – 5.40 (m, 1H), 4.52 (d, J = 5.8 Hz, 1H), 4.14 – 3.97 (m, 3H).13 C NMR (100 MHz, CD3OD) δ 166.4, 153.0, 146.7, 143.7, 128.5, 102.9, 77.9, 74.5, 67.9, 50.4. HRMS (ESI) m/z 288.0697 [(M+Na)+; calcd for C10H11N5O4Na+: 288.0703].
((3aS,4R,6aR)-4-(3-benzoyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-6-yl)methyl acetate (10)
Triethylamine (66 uL, 0.48mmol, 3 equiv.) and acetyl chloride (AcCl) (17 μL, 0.24 mmol, 1.5 equiv.) were sequentially added to a solution of 5 (60 mg, 0.16 mmol, 1 equiv.) in CH2Cl2 (2 mL) at 0°C. The reaction mixture was then warmed up to room temperature and stirred for another hour. A saturated solution of NH4Cl (2 mL) was added. The organic layer was extracted with 3 × 3 mL CH2Cl2, washed with H2O, and concentrated under reduced pressure. The crude material was purified through a 4g SiO2 flash cartridge with a linear gradient 65-95% EtOAc in hexanes to afford the intermediate 10 (56 mg, 0.13 mmol, 82%) as colorless oil. [α] – 62.8 (c 1.3, CHCl3); IR (film, cm-1): 3080, 2988, 2935, 1742, 1703, 1663, 1599, 1440, 1372, 1236, 1179, 1236, 1058, 904, 731.1 H NMR (400 MHz, CDCl3) δ 7.93 (d, J = 7.6 Hz, 2H), 7.78 – 7.61 (m, 1H), 7.50 (dd, J = 8.1, 7.7 Hz, 2H), 7.10 (d, J = 8.1 Hz, 1H), 5.81 (d, J = 8.1 Hz, 1H), 5.61 (brs, 1H), 5.34 (brs, 1H), 5.23 (d, J = 5.7 Hz, 1H), 4.79 (qt, J = 15.0, 1.8 Hz, 2H), 4.67 (d, J = 5.3 Hz, 1H), 2.12 (s, 3H), 1.42 (s, 3H), 1.34 (s, 3H). 13 C NMR (100 MHz, CDCl3) δ 170.7, 168.7, 162.2, 149.7, 148.3, 141.2, 135.3, 131.5, 130.7, 129.3, 123.2, 113.1, 102.6, 84.1, 83.7, 68.6, 60.9, 27.3, 25.9, 20.9. HRMS (ESI) m/z 427.1508 [(M+H)+; calcd for C22H23N2O7 +: 427.1500].
1-((1R,4R,5S)-4,5-dihydroxy-3-methylcyclopent-2-en-1-yl)pyrimidine-2,4(1H,3H)-dione (11)
To a solution of the allylic acetate 10 (56 mg, 0.13 mmol, 1 equiv.) in 95% ethanol (4 mL) and cyclohexene (2 mL) was added 20 w% Pd(OH)2 on carbon (20 mg, 1:3 catalyst substrate by weight). The resulting suspension was stirred under reflux overnight. The reaction mixture was then filtered, concentrated, and dried under high vacuum to afford 20 mg crude material as pale-yellow oil. In a similar manner as the preparation of 9, 6 mg of the crude product was treated with 25 μL HCl in 250 μL THF to afford the final product 11 (8 mg, 0.037 mmol, 28% for two steps) as white solid. [α] –78.0 (c 0.24, MeOH); IR (film, cm-1): 3364 (br), 2921, 2851, 1680, 1468, 1393, 1251, 1203, 1116.1 H NMR (400 MHz, CD3OD) δ 7.39 (d, J = 8.0 Hz, 1H), 5.67 (d, J = 8.0 Hz, 1H), 5.52 – 5.35 (m, 2H), 4.37 (d, J = 5.8 Hz, 1H), 3.99 (dd, J = 5.9, 5.4 Hz, 1H), 1.88 (s, 3H).13 C NMR (100 MHz, MeOH) δ 166.4, 153.1, 148.9, 143.5, 125.6, 102.7, 78.2, 77.2, 68.1, 15.1. HRMS (ESI) m/z 247.0685 [(M+Na)+; calcd for C10H12N2O4Na: 247.0689].
Purification of Recombinant Human UCK2
Previously, human UCK2 was cloned into the pET21a(+) vector resulting in an expression vector for C-terminally 6×His-tagged UCK2 (Okesli-Armlovich et al., 2019). As previously described, E. coli BL21(DE3) cells were electroporated with this pET21a-UCK2 expression plasmid, recovered and plated onto selective LB agar plates (carbenicillin) overnight at 37° C. A single colony was used to inoculate a 30 mL starter culture in LB Broth with carbenicillin and grown at 37°C overnight. The next day, this starter culture was used to inoculate 1 L LB Broth with carbenicillin and grown, with agitation, at 37°C. When cells reached an optical density (OD600) of 0.7, protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and cells were shaken for an additional 15 h at 18°C. Cell pellets were collected by centrifugation, flash-frozen and stored at -80°C as previously detailed (Okesli-Armlovich et al., 2019).
Pellets were thawed and resuspended in lysis buffer containing Tris (40 mM pH 7.5), NaCl (10 mM), imidazole (10 mM), dithiothreitol (DTT) (1 mM) and glycerol (10% v/v). Cells were lysed by sonication and centrifuged at 25,000g for 1 h. The supernatant was incubated with a slurry of Ni-NTA resin for 1 h at 4°C prior to column loading. The nickel column wash buffer was identical to lysis buffer with each wash step introducing increasing imidazole concentrations (10 mM, 50 mM, 250 mM). After SDS-PAGE confirmation of protein fractions, the protein was further purified using an Äkta Pure 25 FPLC with a 5 mL Hi Trap Q anion exchange column. Protein was eluted on a linear gradient from FPLC Buffer A (50 mM Tris-HCl pH 8, 1mM DTT, 10% glycerol) to FPLC buffer B (50 mM Tris-HCl pH 8, 1 mM DTT, 10% glycerol with 500 mM NaCl) from 2% to 100% Buffer B. Eluates containing recombinant UCK2 were concentrated and exchanged into storage buffer (50 mM Tris-HCl pH 7.5, 10% glycerol) using a centrifugal filter with a 3 KDa cut-off (Amicon). Enzyme was aliquoted, flash-frozen and kept at -80°C.
Purification of Recombinant Human CMPK1
Previously, human CMPK1 was cloned into the pET28 vector resulting in an expression vector for N-terminally 6×His-tagged CMPK1 (Deans et al., 2016). As previously described, E. coli BL21(DE3) cells were transformed with this pET28-CMPK1 expression plasmid, and single colony transformants were used to inoculate a 5 mL starter culture in LB Broth with kanamycin and grown at 37°C overnight. The next day, this starter culture was used to inoculate 1 L LB Broth with kanamycin and grown, with agitation, at 37°C until an OD600 of ∼0.65 was reached (Deans et al., 2016). Cell pellets were harvested by centrifugation and stored at -80°C (Deans et al., 2016).
Pellets were thawed and resuspended in lysis buffer containing Tris (40 mM pH 7.5), NaCl (10 mM), NaF (5 mM) and DTT (1 mM). Cells were lysed by sonication and supernatant was clarified at 25,000g for 1 h. This supernatant was incubated with a slurry of Ni-NTA resin for 1 h at 4°C prior to column loading. The nickel column wash buffer was identical to the lysis buffer with each wash step introducing increasing imidazole (0 mM, 10 mM, 40 mM, 200 mM) (Deans et al., 2016). After confirming Ni-NTA wash eluates by SDS-PAGE, relevant protein fractions were diluted as needed, in FPLC Buffer AA (50 mM Tris-HCl pH 8.0, 1 mM DTT) as previously detailed (Deans et al., 2016). The protein was further purified using an Äkta Pure 25 FPLC with a Hi Trap Q anion exchange column. Protein was eluted using a linear gradient from 0 to 95% FPLC Buffer BB (50 mM Tris-HCl pH 8.0, 1 mM DTT, 500 mM NaCl) under established conditions (Deans et al., 2016). Fractions containing CMPK1 were concentrated and exchanged into storage buffer (50 mM Tris-HCl pH 7.5, 10% glycerol) using a 10 kDa filter as previously detailed (Deans et al., 2016). Enzyme was then aliquoted, flash-frozen and kept at -80°C.
In Vitro Enzyme Activity Assays with UCK2
To continuously monitor reaction progress spectrophotometrically, ATP hydrolysis was coupled to NADH oxidation via pyruvate kinase (PK) and lactate dehydrogenase (LDH) (Tomoike et al., 2017). Reactions were conducted at room temperature in 100 μL in 96-well plates (Greiner Bio-One, UV-Star, Half Area). Mixtures contained 20 mM HEPES pH 7.2, 100 mM KCl, 2 mM MgCl2, 300 μM ATP, 0-500 μM uridine, 0-500 μM CPU analogs, 10 nM UCK2 (unless otherwise noted), 1 mM phosphoenolpyruvate, 500 μM NADH and 20 units/mL of PK and LDH. Progress was monitored in the linear region using a Biotek Synergy HT and kinetic and inhibition constants were determined using GraphPad Prism.
Enzymatic Synthesis of CMP, CPU-MP, and 5-F-CPU-MP
Reactions were conducted at room temperature in 100 μL in Eppendorf tubes. Mixtures contained 20 mM HEPES pH 7.2, 100 mM KCl, 2 mM MgCl2, 2.5 mM ATP, 5 mM substrates (cytidine, CPU and 5-F-CPU), and 2 μM UCK2. After gently mixing for 24 h on a rocking shaker, the reaction mixtures were heated for 3 minutes at 95°C to denature the UCK2 enzyme. Formation of CPU-MP and 5-F-CPU-MP were confirmed by LC-MS/MS (Figure S1).
In Vitro Enzyme Activity Assays with CMPK1
In a similar manner as the UCK2 assay, ATP hydrolysis was coupled to NADH oxidation via PK and LDH to continuously monitor reaction progress spectrophotometrically. Reactions were conducted at room temperature in 50 μL in 96-well plates (Greiner Bio-One, UV-Star, Half Area). The CMPK1 assay buffer contained 50 mM Tris-HCl pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT, 500 μM NADH, 1 mM PEP and20 units/mL of PK and LDH. To 39.5 μL of CMPK1 assay buffer, 5 μL were added of 0-1 mM stock solutions of substrates (CMP from the commercial vendor and denatured UCK2 reaction mixtures containing newly synthesized CMP, CPU-MP and 5-F-CPUMP) in UCK2 assay buffer. After UV-readouts at 340 nm stabilized (∼ 5 minutes), a mixture of ATP and CMPK1 in CMPK1 assay buffer was added to a final reaction volume of 50 μL, a final ATP concentration of 500 μM and a final CMPK1 concentration of 20-100 nM. Progress was monitored in the linear region using a Biotek Synergy HT, and kinetic constants were determined using GraphPad Prism. Generation of CPU-DP and 5-F-CPU-DP was confirmed by LC-MS/MS (Figure S2).
Sample Preparation for LC-MS/MS Analysis
A549 cells were plated overnight at 80,000 cells/well in 24-well plates in complete DMEM. The next day, cells were treated with dipyridamole or 250 μM or 500 μM CPU, 5-F-CPU or 5’-F-CPU in DMEM additionally supplemented with 1 μM GSK983 and 5 μM uridine. Six or twelve hours after drug addition, the cell culture medium was removed and the whole plate was rapidly rinsed twice by dipping vertically into a beaker containing 37°C Milli-Q water. The plate was then placed on dry ice, followed by the addition of 0.5 mL –20°C lysis buffer (MeCN:MeOH:H2O = 2:2:1) containing 0.5 M formic acid and 450 nM uridine 15N2 monophosphate sodium salt (Sigma Aldrich). The solution was then sonicated for 3 × 5 minutes on ice for the metabolite extraction. Subsequently, the sample was frozen with liquid nitrogen, freeze-dried, re-suspended with 150 μL Milli-Q water, and finally filtered through a MultiScreen 96-well filter plate prior to LC-MS analysis.
LC-MS/MS Analysis
LC-MS/MS was performed on an Agilent 1290 infinity II LC system tandem with 6470 triple quad mass spectrometer using ion pairing chromatography. LC separation was performed at 40°C with a solvent flow rate of 0.25 mL/min on a Zorbax RRHD Extended-C18 column (2.1 × 150 mm, 1.8 μm). Buffer A contained 97% H2O, 3% MeOH, 5 mM tributylamine, 5.5 mM Acetic acid and 1 μM medronic acid with pH=5.0. Buffer B contains ca. 100% MeOH, 5 mM tributylamine, 5.5 mM acetic acid and 10 μM medronic acid with pH = 7.0 The initial mobile phase composition was 100% solvent A, and the gradient (%B) after sample injection was as follows: 0% at 2.5 min, 20% at 7.5 min, 45% at 14 min, 99% at 20 to 23 min, and 0% from 23.1 to 27.1 min. The autosampler temperature was set to 4°C and the sample was injected at 5 μL. Samples were measured in the negative ESI mode with capillary voltage at −3.5kV. Further source settings were as follows: gas temperature, 250°C; gas flow, 13 L/min; nebulizer, 35 psi; sheath gas temperature, 325°C; sheath gas flow, 12 L/min; nozzle voltage, 500 V; and delta EMV, – 200. Acquisition was performed in dynamic multiple reaction monitoring (dMRM) mode with fragmentation pattern and retention time setting as Table S1.
DENV-2 Luciferase Reporter Assays
A549 cells were plated overnight at 5,000 cells/well in 96-well plates in complete DMEM and incubated for 24h at 37°C. Cells were then treated with pyrimidine de novo synthesis inhibitors, and/or salvage inhibitors, and/or R1479 in DMEM supplemented with 20 μM uridine. Four hours after drug addition, cells were infected with DENV-2 virus at a MOI = 0.04. After 48 h or 72 h, DENV-Luc replication was monitored by the production of Renilla luciferase, which was measured using the Renilla-Glo Luciferase Assay System (Promega) according to the specifications of the manufacturer.
For the accompanying cell viability assay, A549 cells were seeded at 5,000 cells/well in 96-well plates in complete DMEM and incubated for 24 h at 37°C. Cell were then treated with pyrimidine de novo synthesis inhibitors, and/or salvage inhibitors, and/or R1479 in DMEM supplemented with 20 μM uridine. Following 48 h or 72 h treatment, cell viability was monitored via ATP levels, which were measured using the CellTiter-Glo Luciferase Assay System (Promega) according to the specifications of the manufacturer.
For viral tests of the CPU-GSK983 combination (Figure 5B), A549 cells were plated overnight at 20,000 cells/well in 24-well plates in complete DMEM and incubated for 24 h at 37°C. Cell were then treated with pyrimidine de novo synthesis and/or salvage inhibitors in DMEM supplemented with 20 μM uridine. Four hours after drug addition, cells were infected with the DENV-2 virus infectious clone at a MOI = 0.01. After 48 h, DENV-Luc replication was monitored by the production of Renilla luciferase, which was measured using the Renilla-Glo Luciferase Assay System (Promega) according to the specifications of the manufacturer.
For cell viability tests of the CPU-GSK983 combination (Figure 5B), A549 cells were seeded into 24-well plates at a density of 20,000 cells/well incubated for 24 h at 37°C. Cell were then treated with pyrimidine de novo synthesis and/or salvage inhibitors in DMEM supplemented with 20 μM uridine. Following 48h treatment, cells were harvested, and the density of viable cells was determined by flow cytometry (FSC/SSC) using a BD Accuri C6 Flow Cytometer.
Replicon Assays
A549 cells transfected with the pDENV-Luc replicon were plated at 15,000 cells/well in 96-well plates, and immediately treated with R1479, and/or, pyrimidine de novo synthesis inhibitors, and/or salvage inhibitors in complete DMEM medium supplemented with 20 μM uridine without antibiotics (Marceau et al, 2016). Forty-eight hours after drug treatment, cells were lysed and subjected to Renilla luciferase detection, which was measured using the Renilla-Glo Luciferase Assay System (Promega) according to the specifications of the manufacturer.
Quantification and Statistical Analysis
Statistical analyses were performed using GraphPad Prism. In Figure 3, kinetic constants were calculated using this software. In Figures 4A, 5A, and S4, comparisons to an untreated control were calculated using one-way ANOVA, Dunnett’s test as detailed in Figure legends. In Figures 5B, 6, 7, S5, and S6, the IC50 curves visualized and IC50 values provided were generated using a non-linear fit (variable slope, four parameters, constrained to positive values). Additional experimental conditions are detailed in Figure captions.
Acknowledgments
The authors wish to thank Roberto Mateo, David Constant, and Khanh Nguyen for helpful discussions and technical advice. The authors also thank Maryline Dong and Nielson Weng for feedback on this manuscript. This research was supported by a grant from the National Institutes of Health (1U19 AI109662 from the National Institute of Allergy and Infectious Diseases) to C.K., M.C.B., and J.E.C. Also, J.E.C. acknowledges NIH (R01 AI141970) for financial support. A.G. is supported by a National Science Foundation Graduate Research Fellowship, a Stanford ChEM-H Chemistry/Biology Interface Predoctoral Training Program Fellowship, and an American Research College Scientists (ARCS) Fellowship.
Author Contributions
Q.L, A.G., and C.K. designed the research; Q.L., A.G., and W.Q. performed the research investigations; A.O.-A. and C.R.F. aided resource and method development. J.E.C., M.C.B., and C.K. provided supervision and acquired funding. Q.L., A.G., and C.K. wrote the paper (original draft) which was reviewed by all authors. M.S provided advice.
Declaration of Interests
A patent has been filed on this work by the Stanford University Office of Technology Licensing on behalf of Q.L, A.G, and C.K. with reference label S19-283. M.S. is a paid consultant for Riboscience LLD and Advisory Board Member for Tranquis Therapeutics and Angarus Therapeutics. The other authors declare no competing interests.
Published: May 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chembiol.2020.05.002.
Supplemental Information
References
- Alvarez D.E., De Lella Ezcurra A.L., Fucito S., Gamarnik A.V. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Aly L., Hemmer B., Korn T. From leflunomide to teriflunomide: drug development and immunosuppressive oral drugs in the treatment of multiple sclerosis. Curr. Neuropharmacol. 2017;15:874–891. doi: 10.2174/1570159X14666161208151525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekerman E., Einav S. Combating emerging viral threats. Science. 2015;348:282–283. doi: 10.1126/science.aaa3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A., Passacantilli P., Righi G. Mild hydrogenolysis process by catalytic transfer hydrogenation. Tetrahedron Lett. 1989;30:1405–1408. [Google Scholar]
- Blaney S.M., Balis F.M., Heideman R.L., McCully C., Murphy R.F., Poplack D.G., Hegedus L., Kelley J.A. Pharmacokinetics and metabolism of cyclopentenyl cytosine in nonhuman primates. Cancer Res. 1990;50:7915–7919. [PubMed] [Google Scholar]
- Blaney S.M., Balis F.M., Grem J., Cole D.E., Adamson P.C., Poplack D.G. Modulation of the cytotoxic effect of cyclopentenylcytosine by its primary metabolite, cyclopentenyluridine. Cancer Res. 1992;52:3503–3505. [PubMed] [Google Scholar]
- Boldescu V., Behnam M.A.M., Vasilakis N., Klein C.D. Broad-spectrum agents for flaviviral infections: dengue, Zika and beyond. Nat. Rev. Drug Discov. 2017;16:565–586. doi: 10.1038/nrd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia A., Franti M., Keaney E.P., Kuhen K., Seepersaud M., Radetich B., Shao J., Honda A., Dewhurst J., Balabanis K. Identification of broad-spectrum antiviral compounds and assessment of the druggability of their target for efficacy against respiratory syncytial virus (RSV) Proc. Natl. Acad. Sci. U S A. 2011;108:6739–6744. doi: 10.1073/pnas.1017142108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper E.S., Baselga J., Smart T.B., Magill G.B., Markman M., Ranhosky A. A phase II trial of PALA+dipyridamole in patients with advanced soft-tissue sarcoma. Cancer Chemother. Pharmacol. 1991;28:51–54. doi: 10.1007/BF00684956. [DOI] [PubMed] [Google Scholar]
- Chen Y.-L., Abdul Ghafar N., Karuna R., Fu Y., Lim S.P., Schul W., Gu F., Herve M., Yokohama F., Wang G. Activation of peripheral blood mononuclear cells by dengue virus infection depotentiates balapiravir. J. Virol. 2014;88:1740–1747. doi: 10.1128/JVI.02841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung N.N., Lai K.K., Dai J., Kok K.H., Chen H., Chan K.H., Yuen K.Y., Tsun Kao R.Y. Broad-spectrum inhibition of common respiratory RNA viruses by a pyrimidine synthesis inhibitor with involvement of the host antiviral response. J. Gen. Virol. 2017;98:946–954. doi: 10.1099/jgv.0.000758. [DOI] [PubMed] [Google Scholar]
- Choi W.J., Moon H.R., Kim H.O., Yoo B.N., Lee J.A., Shin D.H., Jeong L.S. Preparative and stereoselective synthesis of the versatile intermediate for carbocyclic nucleosides: effects of the bulky protecting groups to enforce facial selectivity. J. Org. Chem. 2004;69:2634–2636. doi: 10.1021/jo0356762. [DOI] [PubMed] [Google Scholar]
- Choi W.J., Chung H., Chandra G., Alexander V., Zhao L.X., Lee H.W., Nayak A., Majik M.S., Kim H.O., Kim J.-H. Fluorocyclopentenyl-cytosine with broad spectrum and potent antitumor activity. J. Med. Chem. 2012;55:4521–4525. doi: 10.1021/jm3004009. [DOI] [PubMed] [Google Scholar]
- Cysyk R.L., Malinowski N., Marquez V., Zaharevitz D., August E.M., Moyer J.D. Cyclopentenyl uracil: n effective inhibitor of uridine salvage in vivo. Biochem. Pharmacol. 1995;49:203–207. doi: 10.1016/0006-2952(94)00470-6. [DOI] [PubMed] [Google Scholar]
- Deans R.M., Morgens D.W., Ökesli A., Pillay S., Horlbeck M.A., Kampmann M., Gilbert L.A., Li A., Mateo R., Smith M. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat. Chem. Biol. 2016;12:361–366. doi: 10.1038/nchembio.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford H., Cooney D.A., Ahluwalia G.S., Hao Z., Rommel M.E., Hicks L.R., Dobyns K.A., Johns D.G. Cellular pharmacology of cyclopentenyl cytosine in Molt-4 lymphoblasts. Cancer Res. 1991;51:3733–3740. [PubMed] [Google Scholar]
- Francisco C.G., Freire R., González C.C., León E.I., Riesco-Fagundo C., Suárez E. Fragmentation of carbohydrate anomeric alkoxy radicals. synthesis of polyhydroxy piperidines and pyrrolidines related to carbohydrates. J. Org. Chem. 2001;66:1861–1866. doi: 10.1021/jo0057452. [DOI] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin C., Hourani M.L., Janin Y.L., Dauzonne D., Munier-Lehmann H., Paturet A., Taborik F., Vabret A., Contamin H., Tangy F. Respiratory syncytial virus infection in macaques is not suppressed by intranasal sprays of pyrimidine biosynthesis inhibitors. Antivir. Res. 2016;125:58–62. doi: 10.1016/j.antiviral.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Harvey R., Brown K., Zhang Q., Gartland M., Walton L., Talarico C., Lawrence W., Selleseth D., Coffield N., Leary J. GSK983: a novel compound with broad-spectrum antiviral activity. Antivir. Res. 2009;82:1–11. doi: 10.1016/j.antiviral.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer J.C., Killington R.A. Virus isolation and quantitation. In: Mahy B.W.J., Kangro H.O., editors. Virology Methods Manual. Academic Press; 1996. pp. 25–46. [Google Scholar]
- Hoffmann H.H., Kunz A., Simon V.A., Palese P., Shaw M.L. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc. Natl. Acad. Sci. U S A. 2011;108:5777–5782. doi: 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao J.J., Potter O.G., Chu T.-W., Yin H. Improved LC/MS methods for the analysis of metal-sensitive analytes using medronic acid as a mobile phase additive. Anal. Chem. 2018;90:9457–9464. doi: 10.1021/acs.analchem.8b02100. [DOI] [PubMed] [Google Scholar]
- Jordheim L.P., Durantel D., Zoulim F., Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013;12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- Kato F., Hishiki T. Dengue virus reporter replicon is a valuable tool for antiviral drug discovery and analysis of virus replication mechanisms. Viruses. 2016;8:1–11. doi: 10.3390/v8050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp K., Lévêque V., Le Pogam S., Ma H., Jiang W.R., Kang H., Granycome C., Singer M., Laxton C., Hang J.Q. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 2006;281:3793–3799. doi: 10.1074/jbc.M510195200. [DOI] [PubMed] [Google Scholar]
- Klumpp K., Kalayanov G., Ma H., Le Pogam S., Leveque V., Jiang W.R., Inocencio N., De Witte A., Rajyaguru S., Tai E. 2′-Deoxy-4′-azido nucleoside analogs are highly potent inhibitors of hepatitis C virus replication despite the lack of 2′-α-hydroxyl groups. J. Biol. Chem. 2008;283:2167–2175. doi: 10.1074/jbc.M708929200. [DOI] [PubMed] [Google Scholar]
- Lascu I., Gonin P. The catalytic mechanism of nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 2000;32:237–246. doi: 10.1023/a:1005532912212. [DOI] [PubMed] [Google Scholar]
- Lieberman-Blum S.S., Fung H.B., Bandres J.C. Maraviroc: a CCR5-receptor antagonist for the treatment of HIV-1 infection. Clin. Ther. 2008;30:1228–1250. doi: 10.1016/s0149-2918(08)80048-3. [DOI] [PubMed] [Google Scholar]
- Lim M.I., Moyer J.D., Cysyk R.L., Marquez V.E. Cyclopentenyluridine and cyclopentenylcytidine analogues as inhibitors of uridine-cytidine kinase. J. Med. Chem. 1984;27:1536–1538. doi: 10.1021/jm00378a002. [DOI] [PubMed] [Google Scholar]
- Lim S.P., Wang Q.Y., Noble C.G., Chen Y.L., Dong H., Zou B., Yokokawa F., Nilar S., Smith P., Beer D. Ten years of dengue drug discovery: progress and prospects. Antivir. Res. 2013;100:500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Lim S.P., Noble C.G., Shi P.Y. The dengue virus NS5 protein as a target for drug discovery. Antivir. Res. 2015;119:57–67. doi: 10.1016/j.antiviral.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Lou Z., Sun Y., Rao Z. Current progress in antiviral strategies. Trends Pharmacol. Sci. 2014;35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Hourani M., Dauzonne D., Jorda P., Cousin G., Lupan A., Helynck O., Caignard G., Janvier G., Andre-Leroux G., Khiar S. Inhibition of pyrimidine biosynthesis pathway suppresses viral growth through innate immunity. PLoS Pathog. 2013;9:1–18. doi: 10.1371/journal.ppat.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P., Naidoo J., Pietzsch C.A., De S., Khadka S., Anantpadma M., Williams C.G., Edwards M.R., Davey R.A., Bukreyev A. Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antivir. Res. 2018;158:288–302. doi: 10.1016/j.antiviral.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau C.D., Puschnik A.S., Majzoub K., Ooi Y.S., Brewer S.M., Fuchs G., Swaminathan K., Mata M.A., Elias J.E., Sarnow P. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martano G., Kiefer P., Christen P., Kentner D., Bumann D., Vorholt J.A. Fast sampling method for mammalian cell metabolic analyses using liquid chromatography–mass spectrometry. Nat. Protoc. 2014;10:1–11. doi: 10.1038/nprot.2014.198. [DOI] [PubMed] [Google Scholar]
- Moon H.R., Lee H.J., Kim K.R., Lee K.M., Lee S.K., Kim H.O., Chun M.W., Jeong L.S. Synthesis of 5′-substituted fluoro-neplanocin A analogues: importance of a hydrogen bonding donor at 5′-position for the inhibitory activity of S-adenosylhomocysteine hydrolase. Bioorg. Med. Chem. Lett. 2004;14:5641–5644. doi: 10.1016/j.bmcl.2004.08.047. [DOI] [PubMed] [Google Scholar]
- Moyer J., Karle J., Malinowski N.M., Marquez V.E., Salam M., Malspeis L., Cysyk R.L. Inhibition of uridine kinase and the salvage of uridine by modified pyrimidine nucleosides. Mol. Pharmacol. 1985;9:454–460. [PubMed] [Google Scholar]
- Nguyen N.M., Tran C.N.B., Phung L.K., Duong K.T.H., Huynh H., le A., Farrar J., Nguyen Q.T.H., Tran H.T., Nguyen C.V.V., Merson L. A randomized, double-blind placebo controlled trial of balapiravir, a polymerase inhibitor, in Adult dengue patients. J. Infect. Dis. 2013;207:1442–1450. doi: 10.1093/infdis/jis470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková L., Pavlík J., Chrenková L., Martinec O., Červený L. Current antiviral drugs and their analysis in biological materials – Part II: antivirals against hepatitis and HIV viruses. J. Pharm. Biomed. Anal. 2018;147:378–399. doi: 10.1016/j.jpba.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Okesli A., Khosla C., Bassik M.C. Human pyrimidine nucleotide biosynthesis as a target for antiviral chemotherapy. Curr. Opin. Biotechnol. 2017;48:127–134. doi: 10.1016/j.copbio.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okesli-Armlovich A., Gupta A., Jimenez M., Auld D., Liu Q., Bassik M.C., Khosla C. Discovery of small molecule inhibitors of human uridine-cytidine kinase 2 by high-throughput screening. Bioorg. Med. Chem. Lett. 2019;29:2559–2564. doi: 10.1016/j.bmcl.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine S., de Nanteuil F., Serrano E., Waser J. Synthesis of (carbo)nucleoside analogues by [3+2] annulation of aminocyclopropanes. Angew. Chem. Int. Ed. 2014;53:8484–8487. doi: 10.1002/anie.201404832. [DOI] [PubMed] [Google Scholar]
- Sartain M. The Agilent metabolomics dynamic MRM database and method. 2016. https://www.agilent.com/cs/library/technicaloverviews/public/5991-6482EN.pdf%20
- Schaudt M., Blechert S. Total synthesis of (+)-astrophylline. J. Org. Chem. 2003;68:2913–2920. doi: 10.1021/jo026803h. [DOI] [PubMed] [Google Scholar]
- Schimmel K.J.M., Gelderblom H., Guchelaar H.J. Cyclopentenyl cytosine (CPEC): an overview of its in vitro and in vivo activity. Curr. Cancer Drug Targets. 2007;7:504–509. doi: 10.2174/156800907781386579. [DOI] [PubMed] [Google Scholar]
- Smee D.F., Hurst B.L., Day C.W. D282, a non-nucleoside inhibitor of influenza virus infection that interferes with de novo pyrimidine biosynthesis. Antivir. Chem. Chemother. 2012;22:263–272. doi: 10.3851/IMP2105. [DOI] [PubMed] [Google Scholar]
- Sofia M.J., Chang W., Furman P.A., Mosley R.T., Ross B.S. Nucleoside, nucleotide, and non-nucleoside inhibitors of hepatitis C virus NS5B RNA-dependent RNA-polymerase. J. Med. Chem. 2012;55:2481–2531. doi: 10.1021/jm201384j. [DOI] [PubMed] [Google Scholar]
- Song G.Y., Paul V., Choo H., Morrey J., Sidwell R.W., Schinazi R.F., Chu C.K. Enantiomeric synthesis of D- and L-cyclopentenyl nucleosides and their antiviral activity against HIV and West Nile virus. J. Med. Chem. 2001;44:3985–3993. doi: 10.1021/jm010256v. [DOI] [PubMed] [Google Scholar]
- Sridhar S., Luedtke A., Langevin E., Zhu M., Bonaparte M., Machabert T., Savarino S., Zambrano B., Moureau A., Khromava A. Effect of dengue serostatus on dengue vaccine safety and efficacy. N. Engl. J. Med. 2018;379:327–340. doi: 10.1056/NEJMoa1800820. [DOI] [PubMed] [Google Scholar]
- Tomoike F., Nakagawa N., Fukui K., Yano T., Kuramitsu S., Masui R. Indispensable residue for uridine binding in the uridine-cytidine kinase family. Biochem. Biophys. Rep. 2017;11:93–98. doi: 10.1016/j.bbrep.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Wang Q.-Y., Bushell S., Qing M., Xu H.Y., Bonavia A., Nunes S., Zhou J., Poh M.K., Florez de Sessions P., Niyomrattanakit P. Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J. Virol. 2011;85:6548–6556. doi: 10.1128/JVI.02510-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong R., Zhang L., Li S., Sun Y., Ding M., Wang Y., Zhao Y., Wu Y., Shang W., Jiang X. Novel and potent inhibitors targeting DHODH, a rate-limiting enzyme in de novo pyrimidine biosynthesis, are broad-spectrum antiviral against RNA viruses including newly emerged coronavirus SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.03.11.983056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo K.L., Chen Y.L., Xu H.Y., Dong H., Wang Q.Y., Yokokawa F., Shi P.Y. Synergistic suppression of dengue virus replication using a combination of nucleoside analogs and nucleoside synthesis inhibitors. Antimicrob. Agents Chemother. 2015;59:2086–2093. doi: 10.1128/AAC.04779-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code not available in the manuscript files.