Abstract
Envelope protein of coronaviruses is a structural protein existing in both monomeric and homo-pentameric form. It has been related to a multitude of roles including virus infection, replication, dissemination and immune response stimulation. In the present study, we employed an immunoinformatic approach to investigate the major immunogenic domains of the SARS-CoV-2 envelope protein and map them among the homologue proteins of coronaviruses with tropism for animal species that are closely inter-related with the human beings population all over the world. Also, when not available, we predicted the envelope protein structural folding and mapped SARS-CoV-2 epitopes. Envelope sequences alignment provides evidence of high sequence homology for some of the investigated virus specimens; while the structural mapping of epitopes resulted in the interesting maintenance of the structural folding and epitope sequence localization also in the envelope proteins scoring a lower alignment score. In line with the One-Health approach, our evidences provide a molecular structural rationale for a potential role of taxonomically related coronaviruses in conferring protection from SARS-CoV-2 infection and identifying potential candidates for the development of diagnostic tools and prophylactic-oriented strategies.
Keywords: SARS-CoV-2, COVID-19, Coronavirus, Immunoinformatics, Diagnosis, One-health
The begin of 2020 has seen the rapid diffusion of a novel betacoronavirus strain, SARS-CoV-2, as the causal agent of the COVID-19. Over boundaries diffusion of SARS-CoV-2 resulted in the pandemic declaration by WHO and the consequent adoption of a series of containment measures to promptly control the virus spread and COVID-19 diffusion [1,2]. To date, COVID-19 diffusion seems to be approaching to an end in the outbreak epicentre and the early affected countries (e.g. Italy), whereas lately affected countries are likely to diminish the COVID-19 incidence in the very near future [3,4]. Nevertheless, prospective studies are predicting the cyclic recurrence of epidemic waves of SARS-CoV-2 over a very short period of time [3], raising the need for the timely development of effective diagnostic tools and prophylactic strategies, other than the design of adequate guidelines for patients’ treatments in the diverse stages of the infection.
The circulating SARS-CoV-2 is a novel strain of the coronavirus genus. Coronaviruses are spherical viruses of large dimension with a positive-sense unsegmented RNA genome of approximately 30 kilobases. Coronavirus specimens are already known in veterinary medicine since capable of infecting an extended spectrum of both wild (e.g. bat, palm civet) and domestic animals (e.g. bovine) including the companion ones (e.g. dogs). The infection is commonly manifested as respiratory illness along with other mild signs such as enteritis [5,6]. Acknowledged their zoonotic potential, coronaviruses have also represented a public health issue with mild manifestation commonly referable to flu-like symptoms. Exceptions are represented by the SARS-CoV epidemic in China in 2002 [7] and the Middle-East Respiratory Syndrome coronavirus (MERS-CoV) registered in Saudi Arabia in 2012 [8] when the more severe manifestation of the infection occurred as a consequence of increased pathogenicity of the circulating viral specimens. Unfortunately, no vaccines or therapy were made available from the previous outbreaks and the recent warningly predictions justify the overwhelming effort being performed worldwide under diverse perspectives.
According to the One-Health concept, extending the target of studies to the major animals the humans are most frequently interacting with would provide valuable information for a better comprehension of the virus biology and pathogenetic mechanisms. This is of pivotal importance for the adequate design of effective diagnostic tools and control measures. Elucidating immunological properties of the circulating SARS-CoV-2 along with the tracking of the epitopes distribution among coronaviruses with tropism for the most common synanthropic animals might provide a clear depiction of the co-shared epitopes. Such approach might, from one side, suggest the occurrence of partial protection arising from the human-animal interrelation while, on the other hand, it might provide a valuable list of epitope candidates to be used for the differential diagnosis of SARS-CoV-2 infection. Investigation of the major viral structural proteins (i.e. Spike, Membrane, Nucleocapsid and Envelope) provides unevaluable knowledge for a comprehensive depiction of the antigenic determinants of the circulating virus [[9], [10], [11]]. Our previous studies investigated epitope diffusion of the Spike and Nucleocapsid SARS-CoV-2 proteins, highlighting some epitopes sequences as conserved among a restricted group of taxonomically-related specimens while other epitopes sequences were shared between coronavirus with tropism for the animal’s human are continuously interacting with [9,10].
Along with Spike (S) and Nucleocapsid (NC) protein, Envelope (E) is a structural protein involved in diverse phases of the virus infection. E protein of SARS-CoV-2 is a 75 amino acids long protein existing in both monomeric and homo-pentameric form. Approximately 20 copies of the protein have been found in the viral particle and previous mutagenesis-based studies demonstrated its pivotal role in the onset and development of the viral infection [12]. Specifically, E protein lacking virions are associated with incapability of infecting host cell or very low viral titers [13]. Other functions have been predicted for E protein within the infected cells. E protein localizes within the secretory pathways at the interspace between the endoplasmic reticulum and Golgi apparatus. Here, the arrangement of the hydrophobic transmembrane domain into a homo-pentamer has been predicted to form an ion channel (i.e. virioporin) in several coronavirus strains. Whereas, PDZ binding domain (PBD) at the C-terminal of the amino acid polymer is supposed to hinder the host cell functionality to favor viral replication and dissemination within the host body [12,14,15]. Besides its multifunctional properties, E protein is also highly immunogenic attracting the attention of diverse research groups that target this protein for the development of antigen-based studies including those aimed at developing immunodiagnostic tools and prophylactic oriented studies [11,16]. To date, immunoinformatic is among the most promising approaches to survey the antigenic determinants of novel and unknown molecules. It is based on the use of computational resources (e.g. software, algorithms and statistic models) to investigate protein sequences and predict the antigenicity of molecule portions (epitope prediction). It is based on different parameters such as the physicochemical properties of the amino acids, their spatial localization, and the complementarity and affinity with models of the immune response effectors such as the B-cells and T-cells [[16], [17], [18]].
In the present study, we employed the immunoinformatic approach to evaluate the antigenicity potential of the SARS-CoV-2 E protein. Major epitopes of the SARS-CoV-2 E protein are predicted and resulting sequences compared with the homologue immunological domains of taxonomically related coronaviruses with tropism for the synanthropic animal species that are most commonly interacting with the human beings population all over the world. The epitope mapping and comparison among the cross-species presented in the present study might provide a keystone for assessing the potential occurrence of partial protection arising from the human-animal interaction besides their importance for the development of immunodiagnostic tools and/or the design further efficient prophylactic-oriented studies.
1. Material and methods
1.1. Data collection and protein sequence processing
The virus species and accession number of the proteins employed in this survey are provided in Table 1 . Envelope protein sequences were retrieved from NCBI Protein repository (https://www.ncbi.nlm.nih.gov/protein/) and UniProt KB (https://www.uniprot.org).
Table 1.
Selected virus overview. The table lists the viral specimens employed in this study along with the relative details.
| Virus | NCBI TaxID | NCBI Genome | Protein GI |
|---|---|---|---|
| SARS-CoV-2 | 2697049 | NC_045512.2 | YP_009724392.1 |
| Bat CoV RaTG13 | 693998 | MN996532 | QHR63302.1 |
| SARS-CoV | 694009 | NC_004718.3 | NP_828854.1 |
| Pangolin CoV | 2708335 | MT084071 | QIQ54050.1 |
| Camel CoV | 1335626 | MK967708 | QGV13485.1 |
| MERS-CoV | 1335626 | NC_019843.3 | YP_009047209.1 |
| Dromedarius CoV | 1335626 | MH259486 | QCI31485.1 |
| H-Enteric CoV | 166124 | FJ415324 | ACJ35487.1 |
| Canine CoV | 215681 | KX432213 | AQT26502.1 |
| Bovine CoV | 11128 | NC_003045 | NP_150081.1 |
| Avian CoV | 11120 | NC_001451 | NP_040834.1 |
Protein sequences as of Table 1 were analyzed via the Basic Local Alignment Search Tool for protein sequences (pBLAST) [19]. This tool implements BLOSUM62 algorithm to compare protein sequences and calculates the statistical significance of matches as means of e-values. Multiple sequence alignment was performed by keeping default settings. Only statistically significant matches were considered for drawing the similarity tree. Fast minimum evolution algorithm was employed to produce a tree from given distances between sequences by setting a maximum fraction of mismatch between pairs of sequences of 0.75.
1.2. Envelope protein epitopes prediction
SARS-CoV-2 E protein epitopes prediction was performed by means of the IEDB prediction tools. Both B-cell and T-cell predictions were performed. Prediction of linear B-cell epitopes was performed through BepiPred algorithm. It predicts the location of linear B-cell epitopes using a combination of a hidden Markov model and a propensity scale method [20]. Both default threshold settings (0.350) and a lower threshold value (0.250) were used. T-cell epitopes were predicted by using the artificial neural network (ANN) algorithm. The list of epitopes binding to MHC class I and class II molecules was obtained via MHC–I Binding and MHC–II Binding tools respectively. In line with a very recent study [16], epitopes sequences with a minimal length of 10 amino acids were selected and immunogenic domains binding to MHC I and II molecules scoring respectively a median inhibitory concentration (IC50) equal or less than 100 and 1000, were employed for the sequence alignment against the E protein sequence of the taxonomically related coronaviruses with tropism for other synanthropic and/or domestic animals. The BLAST-based alignment was performed using BLOSUM62 algorithm to compare protein sequences and calculate the statistical significance of matches as means of e-values. Only the best hits were considered for each virus and hits with alignment length minor or equal to 4 amino acids were manually removed.
1.3. Tridimensional (3D) folding prediction of the E proteins
Tridimensional structure of the SARS-CoV E protein has been recently elucidated and made available in the RCSB PDB database under the identifier 5 × 29 [21]. Acknowledged the high similarity occurring among the E protein sequences of SARS-CoV, SARS CoV-2, bat CoV and pangolin CoV we considered 5 × 29 as unique reference structure for the E proteins of these viral specimens. The E proteins of the other coronaviruses whose 3D structure has not yet been elucidated were computationally predicted by using homology modeling tool [22]. Predicted models were exported as PDB files to be employed in the next step of structural mapping of the epitopes.
1.4. Structural mapping of epitopes to PDB structures
The protein epitopes of SARS-CoV E protein were mapped on the available PDB structure (5 × 29) whereas predicted PDB files were used for the proteins whose high definition crystallographic structure was not available. PyMOL Molecular Graphics System, Version 2.3.4 (Schrödinger, LLC.) was employed for the structural mapping of the selected epitopes over the coronavirus E protein structures.
2. Results
The full protein sequence of the E proteins from the selected coronavirus specimens was employed to assess the virus similarity on the basis of the conserved protein sequences. The multiple sequence alignment of the entire E protein sequences reveals that SARS-CoV-2 E protein is 100% identical to the bat coronavirus and the pangolin coronavirus counterparts as also supported by the nucleotide-based investigations performed at the level of the whole genome sequence. Slightly less than 95% of sequence similarity is observed between the query sequence (i.e. SARS-CoV-2 E protein) and the E protein sequence of SARS-CoV, the causal agent of the previous outbreak. Minor sequence similarity was observed in regard to the E protein of the causal agent of MERS, MERS-CoV, and the (Camelus bactrianus) and dromedaries (Camelus dromedarius) coronavirus, scoring an amino acid sequence similarity below 40%. Finally, the whole sequence similarity between the SARS-CoV-2 E protein and the homologues from the human enteric coronavirus as well as those with tropism for the bovine and dogs is slightly below 31%. Even minor is the sequence similarity observed with the chicken coronavirus, where 20% of the overall protein sequence is shared with SARS-CoV-2, Table 2 .
Table 2.
Whole protein sequence alignment between the selected coronavirus representatives.
| Virus | Protein GI | Identity (%) | Alignment length | Mismatches | e-value |
|---|---|---|---|---|---|
| Pangolin CoV | QIQ54050.1 | 100 | 75 | 0 | 2.15E-51 |
| Bat CoV | QHR63302.1 | 100 | 75 | 0 | 2.15E-51 |
| SARS-CoV | NP_828854.1 | 94.737 | 76 | 3 | 2.22E-47 |
| Dromedarius CoV | QCI31485.1 | 36 | 75 | 44 | 4.01E-12 |
| MERS-CoV | YP_009047209.1 | 36 | 75 | 44 | 4.01E-12 |
| Camel CoV | QGV13485.1 | 36 | 75 | 44 | 4.01E-12 |
| Canine CoV | AQT26502.1 | 30.769 | 39 | 27 | 3.30E-06 |
| Bovine CoV | NP_150081.1 | 30.769 | 39 | 27 | 3.40E-06 |
| H-Enteric CoV | ACJ35487.1 | 30.769 | 39 | 27 | 3.40E-06 |
| Avian CoV | NP_040834.1 | 20 | 75 | 53 | 0.066 |
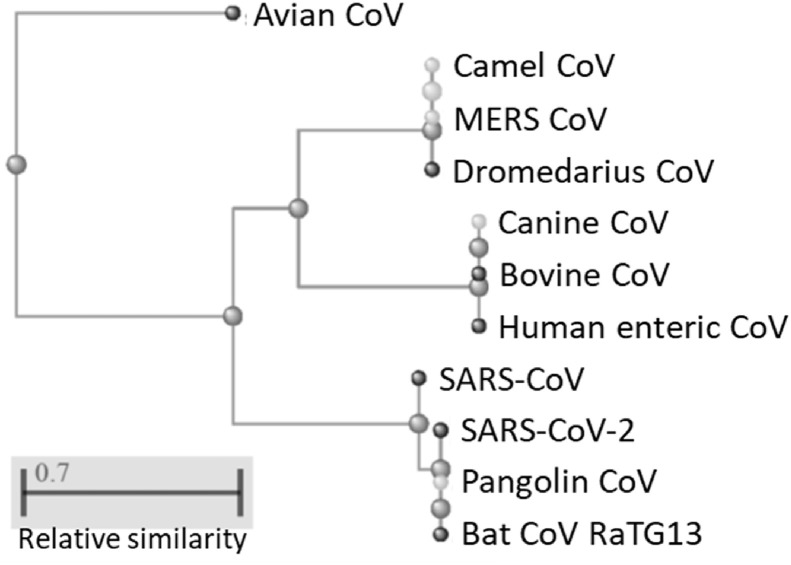
Depiction of the virus specimens by means of the distance matrix calculated from the E protein sequences comparison is in line with the observation summarized in Table 2. Indeed, the bat coronavirus RaTG13, pangolin coronavirus and the circulating SARS-CoV-2 are grouped in the same clade, supporting the occurrence of a very strong similarity. These are followed by the SARS-CoV, whose E protein sequence enables a close classification between the past and the current epidemic causal agents. A separate clade is reserved to the E protein of the MERS-CoV and the coronaviruses with tropism for camels and dromedaries. These are displayed as the sister group of another clade including the human enteric coronavirus along with the coronaviruses with tropism for dog and bovine; whilst avian coronavirus is placed apart since featured by a lower degree of similarity with respect to SARS-CoV-2 E protein sequence, Fig. 1 .
Fig. 1.
Phylogenetic classification of the selected coronaviruses on the basis of the envelope protein sequence.
Once assessed the overall sequence similarity among the diverse coronavirus representatives, the major immunogenic domains of the SARS-CoV-2 E protein were predicted by following an immunoinformatic approach. Prediction of the B-cell epitopes failed to predict epitopes sequence that meets the quality filter prefixed (score threshold of 0.350 and minimum 10 amino acid length). A further prediction employing a lower score threshold (0.250) predicted one epitope sequence, but it was subsequently discarded since redundant with the prediction of T-cell epitopes.
Prediction of T-cell epitopes sequences capable of binding both class I and class II MHC are listed in Table 3 and the supplementary file S1.
Table 3.
Selected envelope protein epitopes.
| Epitope | Virus | % Identity | Length | MHC Class | Alignment length | Position | e-value |
|---|---|---|---|---|---|---|---|
| SLVKPSFYV | Pangolin CoV | 100 | 9 | I | 9 | 50–58 | 7.71E-08 |
| Bat CoV | 100 | I | 9 | 50–58 | 7.71E-08 | ||
| SARS-CoV-2 | 100 | I | 9 | 50–58 | 7.71E-08 | ||
| SARS-CoV | 77.8 | I | 9 | 50–58 | 2.32E-04 | ||
| Bovine CoV | 80 | I | 5 | 56–60 | 0.048 | ||
| H-Enteric CoV | 80 | I | 5 | 56–60 | 0.048 | ||
| Canine CoV | 80 | I | 5 | 56–60 | 0.048 | ||
| LTALRLCAY | SARS-CoV | 100 | 9 | I | 9 | 34–42 | 4.94E-08 |
| Pangolin CoV | 100 | I | 9 | 34–42 | 4.95E-08 | ||
| Bat CoV | 100 | I | 9 | 34–42 | 4.95E-08 | ||
| SARS-CoV-2 | 100 | I | 9 | 34–42 | 4.95E-08 | ||
| Dromedarius CoV | 85.7 | I | 7 | 34–40 | 0.002 | ||
| Camel CoV | 85.7 | I | 7 | 34–40 | 0.002 | ||
| MERS-CoV | 85.7 | I | 7 | 34–40 | 0.002 | ||
| Avian CoV | 100 | I | 4 | 15–18 | 0.09 | ||
| LVKPSFYVY | Pangolin CoV | 100 | 9 | I | 9 | 51–59 | 1.93E-08 |
| Bat CoV | 100 | I | 9 | 51–59 | 1.93E-08 | ||
| SARS-CoV-2 | 100 | I | 9 | 51–59 | 1.93E-08 | ||
| SARS-CoV | 77.8 | I | 9 | 51–59 | 5.70E-05 | ||
| Bovine CoV | 66.7 | I | 6 | 56–61 | 0.023 | ||
| H-Enteric CoV | 66.7 | I | 6 | 56–61 | 0.023 | ||
| Canine CoV | 66.7 | I | 6 | 56–61 | 0.023 | ||
| KPSFYVY SRVKNLNS |
Pangolin CoV | 100 | 15 | II | 15 | 53–67 | 2.11E-14 |
| Bat CoV | 100 | II | 15 | 53–67 | 2.11E-14 | ||
| SARS-CoV-2 | 100 | II | 15 | 53–67 | 2.11E-14 | ||
| SARS-CoV | 86.7 | II | 15 | 53–67 | 5.88E-11 | ||
| Bovine CoV | 62.5 | II | 8 | 56–63 | 0.004 | ||
| H-Enteric CoV | 62.5 | II | 8 | 56–63 | 0.004 | ||
| Canine CoV | 62.5 | II | 8 | 56–63 | 0.004 | ||
| LLVTLAIL TALRLCA |
Pangolin CoV | 100 | 15 | II | 15 | 27–41 | 2.37E-13 |
| Bat CoV | 100 | II | 15 | 27–41 | 2.37E-13 | ||
| SARS-CoV-2 | 100 | II | 15 | 27–41 | 2.37E-13 | ||
| SARS-CoV | 100 | II | 15 | 27–41 | 2.37E-13 | ||
| Dromedarius CoV | 71.5 | II | 14 | 27–40 | 1.06E-05 | ||
| Camel CoV | 71.5 | II | 14 | 27–40 | 1.06E-05 | ||
| MERS-CoV | 71.5 | II | 14 | 27–40 | 1.06E-05 | ||
| Avian CoV | 100 | II | 4 | 15–18 | 0.27 | ||
| CNIVNVS LVKPSFYV |
Pangolin CoV | 100 | 15 | II | 15 | 44–58 | 1.49E-14 |
| Bat CoV | 100 | II | 15 | 44–58 | 1.49E-14 | ||
| SARS-CoV-2 | 100 | II | 15 | 44–58 | 1.49E-14 | ||
| SARS-CoV | 86.7 | II | 15 | 44–58 | 4.17E-11 | ||
| H-Enteric CoV | 50 | II | 16 | 49–60 | 0.023 | ||
| Canine CoV | 50 | II | 16 | 49–60 | 0.023 | ||
| LIVNSVLLF LAFVVFLLV TLAILTAL RLCAY |
SARS-CoV | 100 | 31 | II | 31 | 12–42 | 7.76E-16 |
| Pangolin CoV | 100 | II | 31 | 12–42 | 8.65E-16 | ||
| Bat CoV | 100 | II | 31 | 12–42 | 8.65E-16 | ||
| SARS-CoV-2 | 100 | II | 31 | 12–42 | 8.65E-16 | ||
| Dromedarius CoV | 71.5 | II | 14 | 27–40 | 0.01 | ||
| Camel CoV | 71.5 | II | 14 | 27–40 | 0.01 | ||
| Canine CoV | 35.7 | II | 14 | 29–42 | 1.1 | ||
| Bovine CoV | 35.7 | II | 14 | 29–42 | 1.2 | ||
| H-Enteric CoV | 35.7 | II | 14 | 29–42 | 1.2 | ||
| Avian CoV | 40 | II | 10 | 20–29 | 78 |
Mapping of the epitopes between the selected coronavirus with tropism for synanthropic and/or domestic animals reveal that all the predicted epitopes are “shared” between the closest relatives in taxonomic terms, complete identity. This consideration includes the SARS-CoV that, although showing matches for all the predicted epitope sequences, scored a minor percentage of similarity in a handful of epitopes predicted to be capable of class II MHC binding.
Nevertheless, some epitopes are also shared between the circulating SARS-CoV-2 and the causal agent of a previous outbreak such as the MERS-CoV and the related camel-CoV and dromedaries-CoV. Despite the relatively weak similarity observed at the level of the whole E protein sequence (Fig. 1, Table 2), bovine CoV, canine CoV, human enteric CoV and avian-CoV are also sharing interesting epitope sequences, as supported by the epitope length and the relative e-value (Table 3). In addition, structure prediction of the E protein of the human enteric coronavirus, bovine coronavirus and dog coronavirus resulted in a very similar 3D structure among each other and, in turn, with the high-definition 3D structure of SARS-CoV-2 E protein suggesting that the low sequence similarity might not hinder the antigenic properties of the E protein from diverse coronaviruses (Fig. 2 ).
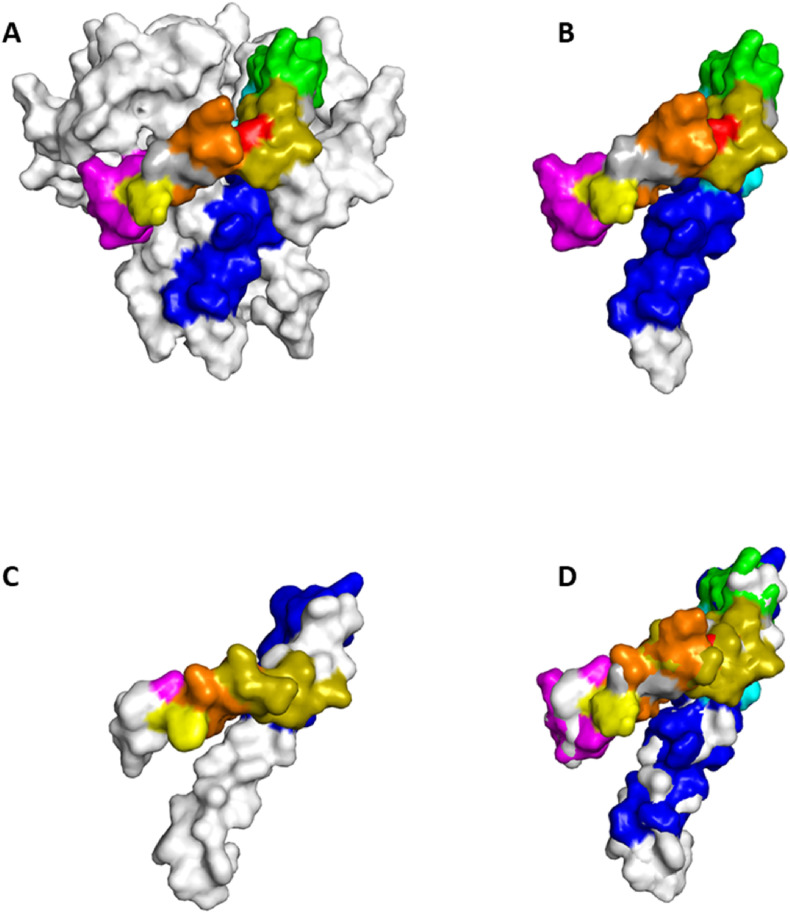
Fig. 2.
Tridimensional structure of envelope proteins from coronavirus representatives and epitope mapping. Epitope sequences are coloured as follow: blue: LIVNSVLLFLAFVVFLLVTLAILTALRLCAY; cyan: LLVTLAILTALRLCA; green: LTALRLCAY; olive green: CNIVNVSLVKPSFYV; red: SLVKPSFYV; orange: LVKPSFYVYSRVKNL; yellow: LVKPSFYVY; magenta: KPSFYVYSRVKNLNS. A) Homopentameric form of the envelope protein from SARS-CoV (PDB database identifier 5 × 29). Selected epitopes sequences are mapped in each monomer. B) Monomer for of the SARS-CoV-2 E protein coloured according to the selected epitopes. C) Predicted 3D structure (monomeric form) of the bovine CoV, dog CoV and human enteric CoV. Selected epitopes sequences are mapped in the monomer. D) Consensus structural mapping of the selected epitopes by overlaying the monomeric form of the E proteins coloured as in the panels B and C.
Structural mapping of selected epitopes highlights a plurality of epitopes adjacent each other, forming a conformational immunogenic domain of the protein that might exist either in its monomeric form or associate as a homo-pentamer (Fig. 2). Interestingly, structural mapping of the SARS-CoV-2 E epitopes in the 3D structure of the bovine CoV, dog CoV and the human enteric CoV highlights conserved structure and mapping position of some immunogenic domains, suggesting a potential interest for conferring partial cross-protection against the circulating SARS-CoV-2.
3. Discussion
The ongoing COVID-19 is a pandemic issue that solicited interest and expertise from the global scientific community, on the attempt to elucidating multiple aspects of this novel virus and drawing the most appropriate measures for rapid and effective control of the SARS-CoV-2 spread and patients treatments. According to the One Health approach, extending the focus of studies to the environment surrounding humans and the animals they are frequently interacting with might represent a valid strategy for obtaining an all-embracing view on this phenomenon and provide suggestions for novel research directions e.g. aimed at developing innovative diagnostic and/or prophylactic interventions.
In the present study, we employed an immunoinformatic approach to investigate the major SARS-CoV-2 E protein distribution among the pool of animals most frequently interacting with the human population all over the world. Coronavirus members were already known as the responsible agents of past epidemics in both humans and animals [7,8]. This aspect has also been taken into account while choosing the viral specimens to be included in this survey. Being all selected coronavirus taxonomically related, we believe that some immunological features can be shared between these viruses with tropism for different animals and, at the same, time this can be exploited for obtaining a partial cross-protection against the circulating SARS-CoV-2. Such thought might be partially supported by previous studies reporting early detection of IgG in the very first phase of the infection along with the heterogeneous clinical manifestation of the SARS-CoV-2 infections, other than preventing epitopes-exposed subjects from the severe form of COVID-19 [23]. On the other hand, the “shared” immunostimulant domains can be used for the design of a more accurate and efficient immunodiagnostic tool, enabling a detailed tracking of the infected patients and the fair discrimination of the SARS-CoV-2 infected ones from the others that came in contact with the antigens carried by other coronaviruses. This might be of great help, especially among the population of individuals diagnosed as infected but without clinical signs.
Structural proteins are among the most antigenic components of the coronaviruses. Previous studies performed by this group already highlighted the diffusion of the Spike and Nucleocapsid protein of the circulating SARS-CoV-2 among the taxonomically related coronaviruses [9,10]. Envelope protein is featured by multiple functions in the diverse phases of the infection. Although present in a relatively low number of copies per viral particle, it has shown important immunostimulant activity. Multiple sequence alignment of the E protein sequences reveals bat coronavirus and pangolin coronaviruses as the most similar viral specimens to SARS-CoV-2. Although very high, SARS-CoV has scored a minor sequence homology than the E protein of the virus with tropism for bat and pangolin. This is also true when analyzing the diffusion of the predicted SARS-CoV-2 sequence epitopes and the relative matching scores. Although all predicted sequence epitopes have been matched in the SARS-CoV E protein sequence, some of the epitopes capable of binding the class II MHC molecules scored a lower identity percentage when compared with bat and pangolin coronavirus, supporting our hypothesis that investigating synanthropic and/or domestic animals might be a promising strategy that provides interesting research suggestions.
Envelope proteins of the human enteric coronavirus along with the bovine coronavirus and canine respiratory coronavirus have scored a low percentage of identity at the level of the whole E protein sequences. Nevertheless, also these viral specimens are sharing a number of SARS-CoV-2 epitopes in their E protein amino acid chain. Also, the structural comparison performed between the newly released high-definition 3D structure of the SARS-CoV E-protein and the predicted 3D model of the E protein of human enteric-, bovine- and canine-coronaviruses confirms the maintaining of the conformational structure of the overall monomer regardless the amino acid sequence of each protein.
Moreover, the structural mapping of the predicted epitopes sequences highlighted the adjacent mapping of some of the SARS-CoV-2 E domains suggesting the most immunogenic portion of the E protein in both its monomeric and homopentameric forms. In addition, structural mapping of some of the investigated epitopes in the human enteric CoV, bovine CoV and canine CoV demonstrated a consensus in both the location of the sequence along the aminoacidic chain and their 3D folding. Such an evidence suggests, once again, their potential immunogenicity and possible implication for the design of immunodiagnostic tools and/or the plan of prophylactic-oriented investigations, even though showing reduced sequence homology with the SARS-CoV-2 E counterpart.
To conclude, we strongly believe that synergistic cooperation elucidating multiple aspects of the virus biology and pathology enables the success in a fast and effective control of the SARS-CoV-2 infections. The thorough survey of the SARS-CoV-2 epitopes and their mapping in the diverse geographical areas and among the major animal species involved in animal–human interactions represents a valuable approach. Providing such knowledge is indeed of pivotal importance for fighting the current COVID-19 pandemic through innovative solutions such as the personalized medicine and the design of specific community-based prophylactic measures.
Declaration of Competing Interest
The Authors declare no conflict of interest.
Acknowledgement
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micinf.2020.05.013.
Contributor Information
Andrea Urbani, Email: andrea.urbani@unicatt.it.
Paola Roncada, Email: roncada@unicz.it.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen S.F., Ho Y.C. SARS-CoV-2: a storm is raging. J Clin Invest. 2020;130:2202–2205. doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020 doi: 10.1126/science.abb5793. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlickeiser R., Schlickeiser F. A Gaussian model for the time development of the Sars-Cov-2 corona pandemic disease. Predictions for Germany made on March 30, 2020. MedRxiv. 2020 doi: 10.1101/2020.03.31.20048942. 03.31.20048942. [Online ahead of print] [DOI] [Google Scholar]
- 5.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microb Infect. 2020;22:72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;3:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tilocca B., Soggiu A., Musella V., Britti D., Sanguinetti M., Urbani A. Molecular basis of COVID-19 relationships in different species: a one health perspective. Microb Infect. 2020 doi: 10.1016/j.micinf.2020.03.002. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tilocca B., Soggiu A., Sanguinetti M., Musella V., Britti D., Bonizzi L. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microb Infect. 2020 doi: 10.1016/j.micinf.2020.04.002. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Pascarella S. Sars-CoV-2 Envelope and Membrane proteins : differences from closely related proteins linked to cross-species transmission ? Preprints. 2020 doi: 10.20944/preprints202004.0089.v1. 2020040089, [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stodola J.K., Dubois G., Le Coupanec A., Desforges M., Talbot P.J. The OC43 human coronavirus envelope protein is critical for infectious virus production and propagation in neuronal cells and is a determinant of neurovirulence and CNS pathology. Virology. 2018;515:134–149. doi: 10.1016/j.virol.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nieto-Torres J.L., DeDiego M.L., Álvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres J., Wang J., Parthasarathy K., Liu D.X. The transmembrane oligomers of coronavirus protein E. Biophys J. 2005;88:1283–1290. doi: 10.1529/biophysj.104.051730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelmageed M.I., Abdelmoneim A.H., Mustafa M.I., Elfadol N.M., Murshed N.S., Shantier S.W. Design of multi epitope-based peptide vaccine against E protein of human 2019-nCoV: an immunoinformatics approach. BioRxiv. 2020 doi: 10.1101/2020.02.04.934232. 02.04.934232. 2020.02.04.934232, [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backert L., Kohlbacher O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med. 2015;7:119. doi: 10.1186/s13073-015-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomar N., De R.K. Immunoinformatics: a brief review. Methods Mol Biol. 2014;1184:23–55. doi: 10.1007/978-1-4939-1115-8_3. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S.F., Wootton J.C., Gertz E.M., Agarwala R., Morgulis A., Schäffer A.A. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen J.E.P., Lund O., Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surya W., Li Y., Torres J. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim Biophys Acta Biomembr. 2018;1860:1309–1317. doi: 10.1016/j.bbamem.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30:S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Clinical Infectious Diseases; 2019. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease. ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.