Highlights
-
•
Interaction of PEDV M protein with host cellular proteins eIF3L, CDC42 and Rab11A was confirmed.
-
•
PEDV replication may be regulated by eIF3L expression.
-
•
218 host cell proteins were designated putative PEDV M protein interacting proteins.
Keywords: Porcine epidemic diarrhea virus (PEDV), Membrane (M) protein, Interaction, Immunoprecipitation
Abstract
Porcine epidemic diarrhea virus (PEDV) is a coronavirus that causes severe diarrhea in pigs of all ages and a high fatality rate in neonates. The PEDV membrane protein (M) plays crucial roles in viral assembly, viral budding and host immune regulation, most likely by interacting with host cell proteins that have yet to be identified. In this study, co-immunoprecipitation (Co-IP) using an M-specific monoclonal antibody, coupled with LC-MS/MS, was employed to identify M protein-interacting proteins in PEDV-infected cells. Three viral proteins (S, E and ORF3) and 218 host cell proteins were identified as putative M-interacting partners. Bioinformatic analysis showed that the identified host cell proteins were related to 131 signal pathways and 10 biological processes. In addition, interaction between translation initiation factor 3(eIF3L) and M protein was validated by Co-IP. Down-regulation of eIF3L expression significantly increased viral production, which suggests that eIF3L could be a negative regulator in PEDV replication. This interactome study of the PEDV M protein will serve to clarify its function during viral replication.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is the etiologic agent of porcine epidemic diarrhea (PED), a highly contagious disease with a high mortality rate in neonatal pigs. PED was first reported among feeder and fattening pigs in England and Belgium during the 1970s (Wood, 1977). The disease was first recorded in China during the 1980s, and a more virulent form of the virus appeared in 2010 (Wang et al., 2013). PEDV is an alphacoronavirus assigned to the family Coronaviridae with a single-stranded positive-sense RNA genome of approximately 28 kb. The genome consists of seven open reading frames (ORFs) encoding four structural proteins (spike [S], envelope [E], membrane [M], nucleocapsid [N]), two non-structural proteins (pp1a and pp1ab), and an accessory gene (ORF3) located between the S and E genes. The M protein, a component of the outer envelope of the viral particle, is categorized as a type III glycoprotein consisting of a short amino-terminal ectodomain, three successive transmembrane domains, and a long carboxy-terminal inner envelope domain (De Haan et al., 2000). The protein participates in viral assembly through interaction with the S and N proteins, viral budding (De Haan et al., 2000; Klumperman et al., 1994; Arndt et al., 2010), and may also mitigate the host innate immune responses. For instance, M protein is reported to induced cell cycle arrest at the S-phase via the cyclin A pathway (Xu et al., 2015) and may modulate the host innate immune responses by inhibiting the IFN-β and IRF3 promoter activities (Zhang et al., 2016; Song and Park, 2012). As obligate parasites, viruses rely on host: pathogen protein-protein interactions to modulate the ongoing biochemical activities of the host cells so that they benefit virus proliferation (Crua et al., 2017). However, the interactome profile of PEDV M protein in host cells is still unclear although, in order to fulfill the aforementioned functions of M proteins, their interaction with a large number of host cell proteins would be expected to occur. In this study, co-immunoprecipitation (Co-IP) coupled with liquid chromatography-mass spectrometry (LC-MS/MS) was used to generate an interactome profile of PEDV M protein and to identify the viral and host cell protein interactions.
2. Materials and methods
2.1. Cells, virus, and plasmids
Vero and Hela cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco BRL, Gaithersburg, MD, USA), supplemented with 10% fetal bovine serum (FBS) (Gibco BRL, Gaithersburg, MD, USA), 100 U/mL of penicillin and 100 μg/mL of streptomycin, at 37 ℃ and in a 5% CO2-enriched atmosphere. The cell culture-adapted PEDV DR13att strain (JQ023162; isolated from a commercial vaccine of Green Cross, South Korea) was propagated and titrated in Vero cells with DMEM supplemented with 2% FBS.
M protein, the L subunit of eukaryotic translation initiation factor 3(eIF3L), and Rab11A, CDC42 and H2AFY expression plasmids containing tags were generated using the homologous recombinant method. All the primer sequences in this study will be made available upon request. M-HA gene was amplified by RT-PCR using PEDV DR13att RNA as template, and cloned into vector pCAGGS-HA with homologous recombinant reagent (one-step cloning kit, Vazyme, China) to form the recombinant plasmid pCAGGS-M-HA. RT-PCR using full-genome RNA of Vero cells as template was employed to amplify the eIF3L/Rab11A/CDC42/H2AFY-Flag genes, which were cloned into vector pCAGGS with homologous recombination reagent to obtain the recombinant plasmids pCAGGS-eIF3L/Rab11A/CDC42/H2AFY-Flag, respectively. All the plasmids were verified by sequencing.
2.2. Antibodies and reagents
PEDV M monoclonal antibody (mAb) was a gift from Shaobo Xiao, Huazhong Agricultural University. Anti-GAPDH mouse mAb was purchased from Abclonal Co. (Abclonal, USA), anti-Flag mAb was from Abcam (Abcam, England), and anti-HA mAb was obtained from Cell Signaling Technology (CST, USA). Alexa Fluor 488 and 647 antibody were purchased from Beyotime Co. (Beyotime, China). Anti-Rab11A and anti-CDC42 rabbit polyclonal antibodies were obtained from Sang Biotech Co. (BBI, China). Anti-eIF3L rabbit polyclonal antibody was purchased from Ango Biotechnology Co. (Proteintech, China), Protein A/G Plus-Agarose SC-2003 was from Santa Cruz Biotechnology (Santa Cruz, USA), and lipofectamine 2000 was purchased from Life Technologies (Invitrogen, USA).
2.3. Detection of M protein expression
Vero cells in a six-well plate were infected with PEDV DR13att at a multiplicity of infection (MOI) of 0.1. Cells were collected at 24, 36, 48, 60 h post-infection (hpi) and subjected to Western blot analysis with anti-M protein mAb.
2.4. Immunoprecipitation
Vero cells seeded in 10-cm-diameter culture dishes were infected with PEDV DR13att at an MOI of 0.1. After 36 hpi, the infected cells were lysed in RIPA lysis buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1% protease inhibitor cocktail by incubation at 4 ℃ on a shaker for 30 min, followed by centrifugation at 14,000×g for 10 min. Supernatants were precipitated with anti-M protein mAb and incubated with gentle rocking overnight at 4 ℃. Protein A/G beads washed with cell lysate were added to supernatant fractions and incubated with gentle rocking for 12 h at 4 ℃. The beads were washed four times with cold cell lysate and boiled with 1 × SDS loading buffer for 10 min, followed by SDS-PAGE and silver staining.
2.5. LC-MS/MS analysis
Immunoprecipitated proteins were separated by electrophoresis on 10% gels and sliver stained. For mass spectrometry, pieces of SDS-PAGE gel were destained with 30% ACN/100 mM NH4HCO3 and dried in a vacuum centrifuge. The in-gel proteins were reduced with dithiothreitol (10 mM DTT, 100 mM NH4HCO3) for 30 min at 56 ℃, then alkylated with iodoacetamide (200 mM IAA, 100 mM NH4HCO3) in the dark at room temperature for 30 min. Gel pieces were then washed sequentially with 100 mM NH4HCO3 and ACN, and digested overnight in solution of 25 mM NH4HCO3 (containing 12.5 ng/μl trypsin). Peptides were extracted three times with 60% ACN/0.1% TFA and dried in a vacuum centrifuge.
LC-MS/MS analysis was performed using a Q Exactive mass spectrometer coupled to Easy nLC (Thermo Fisher Scientific). The peptide mixture was loaded onto a C18-reversed phase column (15 cm long, 75 μm internal diameter) packed in-house with RP-C18 5 μm resin in buffer A (0.1% formic acid in HPLC-grade water) and separated with a linear gradient of buffer B (0.1% formic acid in 84% acetonitrile) over 60 min at a flow rate of 250 nl/min controlled by IntelliFlow Technology. MS data was acquired using a data-dependent top 10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. Determination of the target value was based on predictive Automatic Gain Control (pAGC). Dynamic exclusion duration was 20 s. Survey scans were acquired at a resolution of 70,000 at m/z 200 and resolution for HCD spectra was set to 17,500 at m/z 200. Normalized collision energy was 27 eV and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
2.6. Bioinformatic analysis
MS data were analyzed using MaxQuant software version 1.3.0.5. The peptides and proteins were blasted against the uniprot chlorocebus database (20,655 total entries, downloaded 06/07/2018) and the uniprot PEDV database (uniprot- taxonomy-28295-PEDV-20180713). Carbamidomethylation of cysteines was defined as fixed modification, while protein N-terminal acetylation and methionine oxidation were defined as variable modifications for database searching. The cutoff of global false discovery rate (FDR) for peptide and protein identification was set to 0.01. Cellular proteins identified in mock-infected cells were removed and cellular proteins identified in PEDV-infected cell supernatants were analyzed using the Gene Ontology (GO) program Blast2GO (https://www.blast2go.com/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (http://www.genome.jp/kaas-bin/kaas_main).
2.7. Co-immunoprecipitation (Co-IP) assay
Hela cells, seeded in 10 cm diameter culture dishes, were transfected with pCAGGS-M-HA and cellular target gene expression plasmids. Transfected cells were harvested 36 h post-transfection (hpt), and lysis buffer containing 1 mM protease inhibitor was added. After incubation for 30 min on ice, the lysed cell solutions were centrifuged at 14,000×g for 10 min and the lysate supernatants, containing 1−2 mg total protein, were incubated overnight at 4 ℃ with anti-Flag mouse mAb. Protein A/G beads washed with cell lysate were added to the supernatants and incubated with gentle rocking for 12 h at 4 ℃. The beads were washed four times with cold cell lysate and boiled with 1 × SDS loading buffer for 10 min, followed by SDS-PAGE.
2.8. Western blot analysis
Whole cell extracts and immunoprecipitated (IP) samples were separated by 10% SDS-PAGE and then transferred to PVDF membranes. After blocking with 10% non-fat milk in Tris-buffered saline-Tween (TBST), the membranes were incubated with the specified primary antibodies at room temperature for 1 h, washed with TBST, and then treated with HRP-conjugated goat anti-rabbit IgG or goat anti-mouse IgG. Specific protein bands were visualized by enhanced chemiluminescence using ECL plus western blot detection reagents (GE Healthcare, Chalfont St Giles, UK) according to the manufacturer’s instructions.
2.9. Immunofluorescence assay (IFA)
Vero cells were transfected with plasmids as specified using lipofectamine 2000 according to the manufacturer’s instructions. After fixing with 5% paraformaldehyde 24 hpt, cells were permeabilized with Triton X-100, followed by incubation with anti-HA or anti-Flag antibodies and then with HRP-conjugated goat anti-rabbit IgG or goat anti-mouse IgG antibodies. Cells were stained with DAPI and observed with a Zeiss Scope A1 microscope (Zeiss Microsytems, Germany).
2.10. RNA interference and virus infection test
Double-stranded siRNA sequences targeting eIF3L, Rab11A and CDC42 genes and a negative control were designed and synthesized by Genepharma Co. (Shanghai China) (Table 1 ). The siRNAs (175 nM) were then used to transfect Vero cells which were inoculated 16 hpt with PEDV DR13att at an MOI of 1.0. After 1 h of infection, the inoculum was removed and the cells were washed 3x times and cultured in fresh DMEM. Virus titers in cell supernatants were determined after 18 hpi.
Table 1.
Sequences of host cell gene siRNAs.
| Gene | siRNA sequence (5′-3′) | |
|---|---|---|
| eIF3L | Sense | GCAGAGGUUUGAAUCCUAUTT |
| antisense | AUAGGAUUCAAACCUCUGCTT | |
| Rab11A | Sense | UGUCAGACAGACGUGAAAATT |
| antisense | UUUUCACGUCUGUCUGACAUU | |
| CDC42 | Sense | GAUCCAAAUUGGCCUCAGATT |
| antisense | UCUGAGGCCAAUUUGGAUCTT | |
| Negative control |
Sense | UUCUCCGAACGUGUCACGUTT |
| antisense | ACGUGACACGUUCGGAGAATT | |
2.11. RNA extraction and real time RT-PCR
Total RNA was extracted from cells using the total RNA miniprep kit (Axygen, China) and subjected to reverse transcription with reverse transcription reagent (Promega, US). Real-time RT-PCR was performed on a ABI 7500-fast Real-time PCR systems (ABI, US). Each 20 μl qPCR reaction contained 2 μl reverse transcription sample, 10 μl TliRNaseH Plus (2×), 0.4 μl forward and reverse primers (10 μM), 0.4 μl ROX Reference Dye Ⅱ (50×), and 6.8 μl sterile purified water. Amplification conditions were: 95 ℃ for 30 s, followed by 40 cycles of 95 ℃ for 5 s and 60 ℃ for 34 s. Each reaction was performed in triplicate using GAPDH as the internal control, and the qRT-PCR data was analyzed using the 2−△△CT method.
2.12. Statistical analysis
Statistical analyses (one-way ANOVA) were performed using SPSS software (SPSS 21.0 for Windows). Data are expressed as mean values ± standard error (SEM). The t-test was employed to determine the statistical significance (P < 0.05) of the differences between the means. Data relating to viral RNA copies and virus titer levels were converted to log10 to maintain a normal distribution.
3. Results
3.1. Surveillance of PEDV M protein synthesis
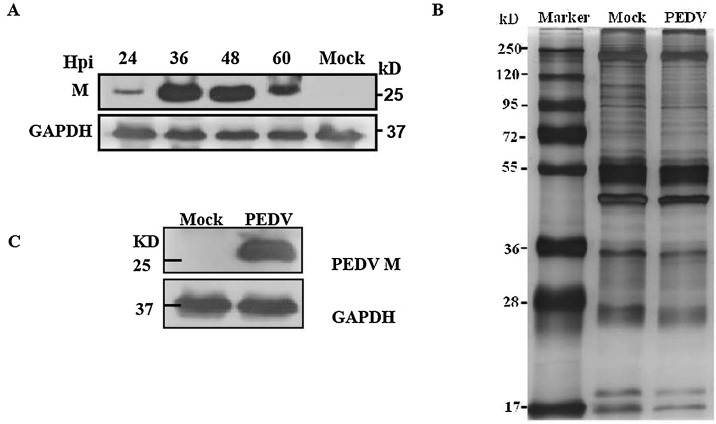
Vero cells were infected with PEDV DR13att and M protein synthesis determined by Western blot analysis with anti-M protein mAb between 24 and 60 hpi using GAPDH as the internal reference. M protein synthesis increased during PEDV infection and reached a peak between 36 and 48 hpi (Fig.1 A). Samples were collected at 36 hpi for interaction analyses.
Fig. 1.
Identification of host cell proteins that interact with PEDV M protein using Co-IP. A. Western blot analysis of M protein synthesis in PEDV DR13att-infected Vero cells at different post-infection times (hpi) using anti-M mAb. B. Cell lysates from PEDV-infected or mock-infected Vero cells were immune -precipitated with anti-M mAb. Immuno-precipitated proteins were separated by 10% SDS-PAGE and visualized by silver staining. C. Western blot analysis of immune-precipitated M proteins in PEDV- and mock-infected Vero cells using anti-M mAb.
3.2. Preliminary identification of 218 host cell proteins interacting with PEDV M protein
Vero cells were infected with PEDV DR13att at an MOI of 0.1, and cell samples were harvested after 36 hpi and immune-precipitated with anti-M protein mAb. The immune-precipitated proteins were separated by 10% SDS-PAGE and visualized by silver staining (Fig. 1B) and Western blot (Fig. 1C). Immunoprecipitated samples of PEDV-infected cells and mock-infected cells were checked for protein composition using SDS-PAGE electrophoresis and Coomassie brilliant blue staining, and these two lanes of the gel were subjected to MS analysis. Comparisons of the LC-MS/MS data to the chlorocebusuniprot database identified a total of 670 host cell proteins, among which 452 could also be found in the mock-infected cell samples. Therefore, the remaining 218 host cell proteins were designated putative M protein interacting host cell proteins, of which 62 shared a high confidence level (Unique Peptide≥2) (Table 2 ). A search of the PEDV proteins database also identified PEDV S, ORF3 and E proteins among the MS data pool (Table 3 ).
Table 2.
Putative M protein-interacting host cell proteins identified from PEDV-infected cells.
| Protein ID | Name | Unique peptides | Sequence coverage (%) | iBAQ intensity (%) | Protein ID | Name | Unique peptides | Sequence coverage (%) | iBAQ intensity (%) |
|---|---|---|---|---|---|---|---|---|---|
| A0A0D9QUJ7 | KARS | 2 | 3.8 | 1,912,600 | A0A0D9RNF7 | CSRP1 | 2 | 16.6 | 11,263,000 |
| A0A0D9QXJ1 | PRDX3 | 2 | 8.2 | 0 | A0A0D9RNG4 | IGF2BP2 | 2 | 4.7 | 3,265,400 |
| A0A0D9QZQ5 | HSD17B12 | 2 | 9.6 | 5,759,600 | A0A0D9RPF0 | RAB2A | 2 | 12.7 | 0 |
| A0A0D9R011 | CYB5R3 | 2 | 10.3 | 5,541,000 | A0A0D9SC62 | N/A | 2 | 15.4 | 39,325,000 |
| A0A0D9R0H6 | ACO2 | 2 | 4.1 | 1,729,200 | A0A0D9RR51 | PSMC6 | 2 | 7.2 | 4,413,000 |
| A0A0D9R354 | SLC3A2 | 2 | 4.3 | 1,264,300 | A0A0D9RRD3 | RTRAF | 2 | 13.8 | 3,382,200 |
| A0A0D9R3K0 | EIF3L | 3 | 136 | 2,801,700 | A0A0D9RS97 | PSMA2 | 3 | 17.5 | 7,083,600 |
| A0A0D9R3Q2 | DNM2 | 3 | 4.5 | 1,763,100 | A0A0D9RSE1 | BLVRA | 2 | 10.5 | 5,070,000 |
| A0A0D9RYP7 | RAC1 | 2 | 10.4 | 10,820,000 | A0A0D9RSM7 | PSMA6 | 2 | 12.3 | 8,606,500 |
| A0A0D9R6M6 | AIFM1 | 2 | 3.9 | 3,306,300 | A0A0D9RSX3 | ERLIN2 | 2 | 7.9 | 3,056,000 |
| A0A0D9R6P7 | Rab11A | 4 | 17 | 5,409,500 | A0A0D9RTZ7 | IPO4 | 3 | 4 | 2,005,100 |
| A0A0D9R7C5 | SAR1A | 2 | 15.7 | 18,015,000 | A0A0D9S247 | RAB5C | 2 | 9.8 | 22,669,000 |
| A0A0D9R7D9 | PPA1 | 2 | 10.8 | 6,033,500 | A0A0D9S338 | RUVBL2 | 3 | 7.6 | 6,205,700 |
| A0A0D9R8E8 | RAB7A | 2 | 13.5 | 7,068,500 | A0A0D9S3R1 | HYOU1 | 3 | 5.9 | 2,352,500 |
| A0A0D9RAB6 | RPS24 | 2 | 20.8 | 43,934,000 | A0A0D9S3W2 | TAGLN2 | 3 | 21.1 | 8,782,600 |
| K7 × 429 | TNPO3 | 2 | 3.3 | 1,591,600 | A0A0D9S4H7 | RPLP0 | 3 | 20.1 | 48,573,000 |
| A0A0D9RDI7 | RPL26 | 3 | 12.4 | 59,140,000 | A0A0D9S4H8 | PXN | 2 | 5.9 | 1,837,000 |
| A0A0D9RDT3 | NCAPD2 | 2 | 1.9 | 577,950 | A0A0D9S4I8 | NPLOC4 | 3 | 7.7 | 4,529,700 |
| A0A0D9RDV7 | ARPC4 | 3 | 18.5 | 19,474,000 | A0A0D9S4L8 | N/A | 2 | 13.9 | 3,280,800 |
| A0A0D9RF48 | MIF | 2 | 17.4 | 24,952,000 | A0A0D9S5Z6 | SEC22B | 2 | 10.2 | 7,615,300 |
| A0A0D9RG57 | MPDU1 | 2 | 8.1 | 19,791,000 | A0A0D9S6E6 | TRIM28 | 2 | 5.6 | 2,154,900 |
| A0A0D9RGJ5 | RPS14 | 2 | 16.6 | 129,360,000 | A0A0D9S885 | HMGCL | 2 | 7.4 | 3,384,000 |
| A0A0D9RGR4 | KIF5B | 4 | 7.9 | 2,494,400 | A0A0D9SDS3 | N/A | 2 | 13.2 | 61,616,000 |
| A0A0D9RHD6 | PSMB1 | 2 | 7.9 | 1,749,900 | A0A0D9S8E4 | UBR4 | 2 | 0.4 | 99,260 |
| A0A0D9RIA0 | ECPAS | 3 | 2.1 | 1,475,500 | A0A0D9S8A9 | CDC42 | 2 | 8.9 | 1,849,200 |
| A0A0D9RIN4 | LARS | 3 | 2.9 | 1,388,700 | A0A0D9SD70 | Trx-1 | 2 | 21.6 | 73,275,000 |
| Q5D0W6 | ABCB1 | 2 | 2.7 | 975,110 | A0A0D9SB94 | N/A | 2 | 14.4 | 28,687,000 |
| A0A0D9RKC1 | DECR1 | 2 | 7.8 | 4,006,100 | A0A0D9SCB9 | RPL27 | 3 | 18.4 | 65,946,000 |
| A0A0D9RLR8 | PCNA | 2 | 7.3 | 7,638,200 | A0A0D9SCH7 | N/A | 2 | 24.1 | 44,490,000 |
| A0A0D9RLW4 | ATP5F1C | 3 | 11.1 | 25,949,000 | A0A0D9SCR9 | N/A | 2 | 20 | 142,290,000 |
| A0A0D9SB72 | N/A | 2 | 19.4 | 18,120,000 | Q6SSD8 | Rli1 | 2 | 5.3 | 0 |
N/A: protein not fully verified.
Table 3.
PEDV proteins that interact with M protein identified by LC-MS/MS.
| Protein | Unique peptides | Sequence coverage (%) | LFQ intensity |
|---|---|---|---|
| S | 1 | 13.7 | 37,578,000 |
| E | 2 | 26.1 | 86,356,000 |
| ORF3 | 1 | 13.4 | 0 |
3.3. Assignment of 218 putative M protein-interacting host cell proteins to various signal pathways and biological processes
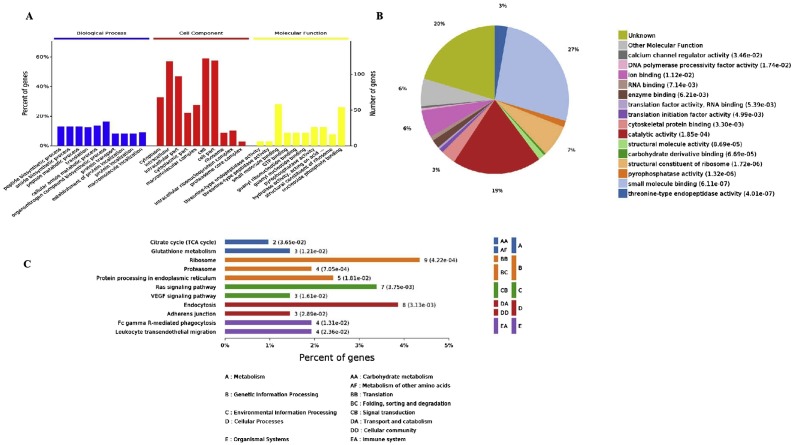
All 218 putative M protein-interacting host cell proteins were assigned to one of the three categories (Biological Processes, Cellular Components or Molecular Functions) delineated at the Gene Ontology (GO) website (Fig. 2 A). Among the proteins clustered in Biological Processes, 34 (15.6%) were involved in organonitrogen compound biosynthetic process, 27 (12.4%) in peptide metabolic processes, 28 (12.8%) in cellular amide metabolic processes, 27 (12.4%) in amide biosynthetic processes, 26 (11.9%) in translation, and 17 (7.8%) in protein transport. The most enriched subclasses in cellular components were macromolecular complex (26.1%), cell (56.0%) and cell part (93.0%). Enrichments based on molecular function were mainly in small molecule binding, structural constituent of ribosome and nucleoside phosphate binding (Fig. 2B). A KEGG database search assigned the putative M protein-interacting host cell proteins to 11 pathways including translation (9 proteins), signal transduction (7 proteins) and transportation and catabolism (8 proteins) (Fig. 2C).
Fig. 2.
Bioinformatic analysis of host cell proteins putatively identified as interacting with PEDV M protein. A. Gene Ontology annotation of the proteins; B. Detailed molecular functions involved; C. KEGG analysis of the proteins.
3.4. Validation of interactions between M protein and three host cell proteins
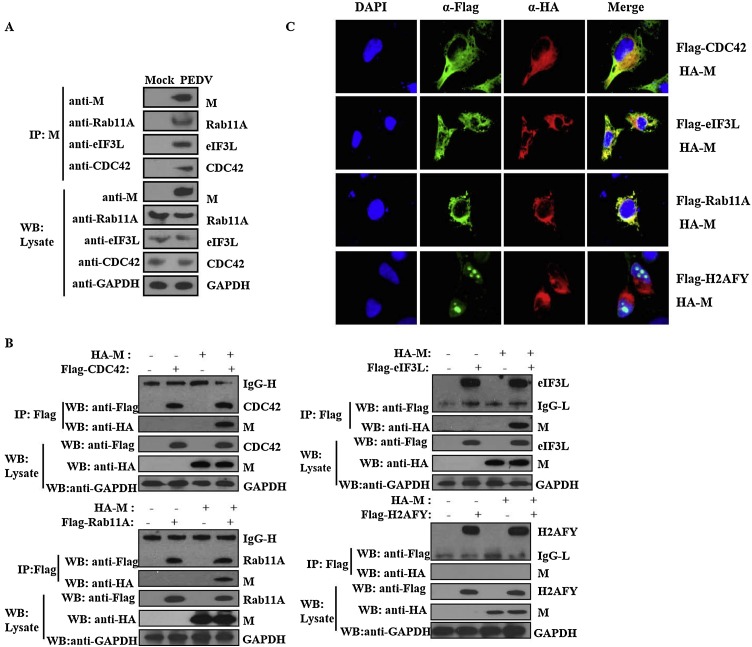
To confirm the interaction of M protein and the host cell proteins identified within the interactome networks, we selected three host cell proteins (CDC42, eIF3L and Rab11A) for validation. Separation by SDS-PAGE of immunoprecipitated (IP) protein samples from lysates of PEDV-infected Vero and mock-infected cells and visualization by western blot analysis with anti-CDC42, -eIF3L or -Rab11A rabbit polyclonal antibodies confirmed that M protein interacted with CDC42, eIF3L and Rab11A in the PEDV-infected cells (Fig. 3 A). These interactions were further validated using the Co-IP assay and overexpression of M, CDC42, eIF3L and Rab11A. Core histone macro-H2A (H2AFY) was used as the negative control as this protein could not be co-immunoprecipitated with the M protein. Following co-transfection of Hela cells with pCAGGS-M-HA and pCAGGS-CDC42/eIF3L/Rab11A/H2AFY–Flag, Co-IP was performed with anti-Flag mAb to capture protein complexes. The immune complexes were applied to 10% SDS-PAGE and probed for the presence of M protein using anti-HA mAb. M protein was detected in the Co-IP samples of CDC42, eIF3L and Rab11A but not in the Co-IP sample of H2AFY (Fig. 3B) further confirming that M protein was able to interact with CDC42, eIF3L and Rab11A proteins. In addition, IFA analysis indicated that CDC42, eIF3L and Rab11A all co-localized with M protein (Fig. 3C).
Fig. 3.
Validation of interactions between PEDV M protein and host cell proteins. A. Immunoblot of M protein and host cell proteins precipitated with mock- or PEDV-infected Vero cell lysates using anti-M-mAb and probed with anti-Rab11A, -eIF3L or -CDC42 polyclonal antibodies. B. Immunoblot of M protein and host cell proteins precipitated using anti-Flag mAb from Hela cells co-transfected with pCAGGS-M-HA and pCAGGS-CDC42/eIF3L/Rab11A/H2AFY-Flag. C. Co-localization analysis of M protein with CDC42, eIF3L, Rab11A and H2AFY in Hela cells co-transfected with pCAGGS-M-HA and pCAGGS-CDC42/eIF3L/ Rab11A-Flag, which were immune-stained with corresponding anti-HA and Flag mAb.
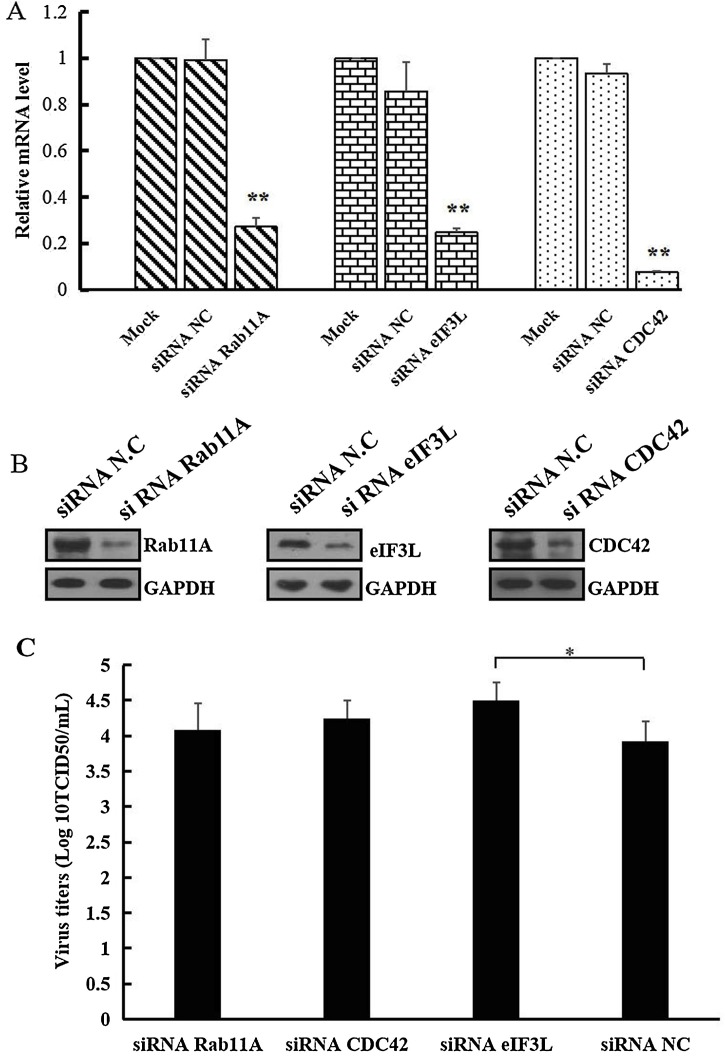
3.5. Knockdown of eIF3L promoted PEDV production
The knockdown efficiencies of CDC42, eIF3L and Rab11A siRNAs proteins on PEDV replication in Vero cells were validated by real time RT-PCR (Fig. 4 A) and western blot (Fig. 4B). Viral titers in cell suspensions with eIF3L knockdown were significantly higher compared with negative controls (Fig. 4C), demonstrating that knockdown of eIF3L increased PEDV multiplication. However, there were no significant differences between the viral titers recorded in the CDC42 and Rab11A knockdown groups and negative controls.
Fig. 4.
Effects of eIF3L knockdown on PEDV production in Vero cells. A. Relative mRNA levels of Rab11A, eIF3L or CDC42 in cells transfected with siRNAs against Rab11A,CDC42, eIF3L and control siRNA analyzed by qRT-PCR. The data represent the means ± SD for three independent experiments and were subjected to one-way ANOVA for statistical significance (P < 0.01). B. Relative protein expression levels of Rab11A, eIF3L or CDC42 in cells transfected with siRNAs against Rab11A,CDC42, eIF3L and control siRNA analyzed by western blot analysis with anti-Rab11A, -eIF3L or -CDC42 polyclonal antibodies. C. Comparison of PEDV titers in cells transfected with siRNAs against Rab11A, eIF3L, CDC42 and control siRNA. The data represent the means ± SD for three independent experiments and were subjected to one-way ANOVA for statistical significance (*P < 0.05).
4. Discussion
In this study, Co-IP coupled with LC-MS/MS was used to screen host cell proteins interacting with PEDV M after PEDV infection. PEDV mainly infects pig small intestinal epithelial cells. However, attempts at PEDV proliferation in a porcine intestinal epithelial cell line were unsuccessful. Consequently, we selected Vero cells, previously shown to be highly susceptible and permissive to PEDV infection (Hofmann and Wyler, 1988), to investigate the M protein interactome. Co-IP and LC-MS/MS identified three viral proteins (Spike-S, Envelope-E and ORF3) and 218 host cell proteins that appeared to interact with M protein in PEDV DR13att-infected Vero cells. Coronavirus (CoV) studies have shown that the M protein interacts with E protein and S proteins during the assembly of the CoV envelope (Nguyen and Hogue., 1997; Opstelten et al., 1995). In the present study, both the PEDV S and E proteins were immunoprecipitated together with the M protein, indicating an interaction with the latter. Our data also showed that anti-M mAb immunoprecipitated truncated ORF3 protein in PEDV DR13att-infected cells, which suggested an interaction between the ORF3 and M proteins. Accessory proteins from other coronaviruses such as SARS-CoV and hCoV-NL63 have also been shown to interact with M, S and E proteins (Tan et al., 2004; Muller et al., 2010). hCoV-NL63 ORF3 is a structural viral protein incorporated into viral particles (Muller et al., 2010) while PEDV ORF3 is involved in many host cell processes and in viral replication (Si et al., 2020; Ye et al., 2015; Kaewborisuth et al., 2018; Wongthida et al., 2017). The likelihood of interaction between PEDV M and ORF3 proteins suggests that the latter cooperates with M to take part in the process of PEDV assembly and budding.
It was notable that 6 Rab family members (Rab2A, Rab5C, Rab6B, Rab7A, Rab9 and Rab11A) were among the 218 host cell proteins identified. Rab proteins are a subfamily of the Ras superfamily of small GTPases that are known to regulate specific intracellular membrane trafficking pathways (Spearman, 2018). Among the six subfamily members, Rab11A has been studied most extensively in terms of its role in viral budding and release. Rab11 is reported to be essential for viral budding and release in RSV and HIV-1 (Brock et al., 2003; Varthakavi et al., 2006). Rab11A also played a role in influenza virus morphogenesis and budding, and depletion of Rab11 decreased viral titers by up to 100-fold (Bruce et al., 2010). Furthermore, Rab11A promoted porcine reproductive and respiratory syndrome virus (PRRSV) replication in a manner that correlated with autophagy (Wang et al., 2017). Rab11A may serve a similar function in the proliferation of PEDV, PRRSV and influenza viruses. However, our siRNA studies failed to reveal any correlation between Rab11A expression levels and PEDV production although the knockdown of the Rab11A gene may not have been sufficiently strong enough to interfere with the Rab11A protein-M protein interaction. Alternatively, other routes may exist that replace the function of Rab11A, and future studies will further investigate the role of Rab11A in PEDV infection.
In the present study, we also confirmed interaction between the M protein and translation factor eIF3L. A reduction in the expression level of eIF3L significantly increased PEDV production compared to negative controls, indicating that the factor acted as a negative regulator in viral replication. eIF3L is one of the subunits of the eukaryotic translation initiation factor eIF3 (Morris-Desbois et al., 2001) and has been shown to inhibited yellow fever virus (YFV) replication by binding to the viral NS5 protein (Morais et al., 2013). Other eIFs, such as eIF3F, play an inhibiting role in HIV-1 replication by interfering with the 3′ end processing of viral mRNAs (Valente et al., 2009).
CDC42, another host cell protein identified as interacting with M protein, belongs to the Rho family, an important subgroup of small RAS-like GTPases (Ha and Boggon, 2018). Studies have shown that many RNA viruses such as respiratory syncytial virus (RSV) (San-Juan-Vergara et al., 2012), rotavirus (RV) (Diaz-Salinas et al., 2014) and Ebola virus (EBOV) (Quinn et al., 2009) depend on CDC42 signaling pathway to infect target cells. However, in this study, interference with CDC42 expression did not influence the proliferation of PEDV, and the significance of the interaction between M protein and CDC42 needs to be further explored.
In conclusion, a total of 218 host cell proteins that potentially interact with PEDV M protein in PEDV-infected cells were identified by combined Co-IP and LC/MS-MS. These proteins were enriched in a variety of signaling pathways, including immune response, cell cycle and apoptosis, and are involved in a wide diversity of biological processes. Among the identified proteins, eIF3L influenced the proliferation of PEDV. Our findings should serve to elucidate novel functions of M and host cell proteins in PEDV-infected cells.
Acknowledgements
We thank Dr. Shaobo Xiao for the generous gift of anti-M monoclonal antibody, Dr. John Buswell for linguistic revision of the manuscript and Dr. Peter Rottier for advice on the manuscript. This research was supported by a grant from the Natural Science Foundation of China (31602060) and the Shanghai Key Project on Agricultural Development (No. 2015-6-1-9).
Contributor Information
Shijuan Dong, Email: dsjnm@163.com.
Zhen Li, Email: zhenli60@163.com.
References
- Arndt A.L., Larson B.J., Hoque B.G. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 2010;84:11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock S.C., Goldenring J.R., Crowe J.E., Jr. Apical recycling systems regulate directional budding of respiratory syncytial virus from polarized epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15143–15148. doi: 10.1073/pnas.2434327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce E.A., Digard P., Stuart A.D. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 2010;84:5848–5859. doi: 10.1128/JVI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crua A.N., Munoz G.E., de Groot N.S., Torrent B.M. Centrality in the host-pathogen interactome is associated with pathogen fitness during infection. Nat. Commun. 2017;8:14092–14098. doi: 10.1038/ncomms14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haan C.A.M., Vennema H., Rottier P.J.M. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Salinas M.A., Silva-Ayala D., López S., Arias C.F. Rotaviruses reach late endosomes and require the cation-dependent mannose-6-phosphate receptor and the activity of cathepsin proteases to enter the cell. J. Virol. 2014;2014(88):4389–4402. doi: 10.1128/JVI.03457-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha B.H., Boggon T.J. CDC42 binds PAK4 via an extended GTPase-effector interface. Proc. Natl. Acad. Sci. U.S.A. 2018;115:531–536. doi: 10.1073/pnas.1717437115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewborisuth C., He Q., Jongkaewwattana A. The accessory protein ORF3 contributes to porcine epidemic diarrhea virus replication by direct binding to the spike protein. Viruses. 2018;10:399–412. doi: 10.3390/v10080399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J.M. Coronavirus M proteins accumulate in the golgi complex beyond the site of virion budding. J. Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais A.T.S., Terzian A.C.B., Duarte D.V.B., Bronzoni R.V.M., Madrid M.C.F.S., Gavioli A.F., Gil L.H.V.G., Oliveira A.G., Zanelli C.F., Valentini S.R., Rahal P., Nogueira M.L. The eukaryotic translation initiation factor 3 subunit L protein interacts with Flavivirus NS5 and may modulate yellow fever virus replication. Virol. J. 2013;10:205–218. doi: 10.1186/1743-422X-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Desbois C., Réty S., Ferro M., Garin J., Jalinot P. The human protein HSPC021 interacts with Int-6 and is associated with eukaryotic translation initiation factor 3. J. Biol. Chem. 2001;276:45988–45995. doi: 10.1074/jbc.M104966200. [DOI] [PubMed] [Google Scholar]
- Muller M.A., van der Hoek L., Voss D., Bader O., Lehmann D., Schulz A.R., Kallies S., Suliman T., Fielding B.C., Drosten C., Niedrig M. Human coronavirus NL63 open reading frame 3 encodes a virion-incorporated N-glycosylated membrane protein. Virol. J. 2010;7:6–18. doi: 10.1186/1743-422X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.P., Hogue B.G. Protein interactions during coronavirusassembly. J. Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opstelten D.J., Raamsman M.J., Wolfs K., Horzinek M.C., Rottier P.J. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn K., Brindley M.A., Weller M.L., Kaludov N., Kondratowicz A., Hunt C.L., Sinn P.L., McCray P.B.Jr., Stein C.S., Davidson B.L., Flick R., Mandell R., Staplin W., Maury W., Chiorini J.A. Rho GTPases modulate entry of Ebola virus and vesicular stomatitis virus pseudotyped vectors. J. Virol. 2009;83:10176–10186. doi: 10.1128/JVI.00422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Juan-Vergara H., Sampayo-Escobar V., Reyes N., Cha B., Pacheco-Lugo L., Wong T., Peeples M.E., Collins P.L., Castano M.E., Mohapatra S.S. Cholesterol-rich microdomains as docking platforms for respiratory syncytial virus in normal human bronchial epithelial cells. J. Virol. 2012;86:1832–1843. doi: 10.1128/JVI.06274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si F., Hu X., Wang C., Chen B., Wang R., Dong S., Yu R., Li Z. Porcine epidemic diarrhea virus (PEDV) ORF3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses. 2020;12:214–232. doi: 10.3390/v12020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Park B. Porcine epidemic diarrhea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman P. Viral interactions with host cell Rab GTPases. Small GTPases. 2018;9:192–201. doi: 10.1080/21541248.2017.1346552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. Novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente S.T., Gilmartin G.M., Mott C., Falkard B., Goff S.P. Inhibition of HIV-1 replication of eIF3F. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4071–4078. doi: 10.1073/pnas.0900557106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varthakavi V., Smith R.M., Martin K.L., Derdowski A., Lapierre L.A., Goldenring J.R., Spearman P. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic. 2006;7:298–307. doi: 10.1111/j.1600-0854.2005.00380.x. [DOI] [PubMed] [Google Scholar]
- Wang X.M., Niu B.B., Yan H., Gao D.S., Yang X., Chen L., Chang H.T., Zhao J., Wang C.Q. Genetic properties of endemic Chinese porcine epidemic diarrhea virus strains isolated since 2010. Arch. Virol. 2013;158:2487–2494. doi: 10.1007/s00705-013-1767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li S., Worku T., Hao X., Yang L., Zhang S. Rab11a is required for porcine reproductive and respiratory syndrome virus induced autophagy to promote viral replication. Biochem. Biophys. Res. Commun. 2017;492:236–242. doi: 10.1016/j.bbrc.2017.08.057. [DOI] [PubMed] [Google Scholar]
- Wongthida P., Liwnaree B., Wanasen N., Narkpuk J., Jongkaewwattana A. The role of ORF3 assessory protein in replication of cell-adapted porcine epidemic diarrhea virus (PEDV) Arch. Virol. 2017;162:2553–2563. doi: 10.1007/s00705-017-3390-5. [DOI] [PubMed] [Google Scholar]
- Wood E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977;100:243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- Xu X.G., Zhang H.L., Zhang Q., Dong J., Huang Y., Tong D.W. Porcine epidemic diarrhea virus M protein blocks cell cycle progression at S-phase and its subcellular localization in the porcine intestinal epithelial cells. Acta Virol. 2015;59:265–275. doi: 10.4149/av_2015_03_265. [DOI] [PubMed] [Google Scholar]
- Ye S., Li Z., Chen F., Li W., Guo X., Hu H., He Q. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes. 2015;51:385–392. doi: 10.1007/s11262-015-1257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of typeⅠinterferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]