Abstract
COVID-19 has spread worldwide since 2019 and is now a severe threat to public health. We previously identified the causative agent as a novel SARS-related coronavirus (SARS-CoV-2) that uses human angiotensin-converting enzyme 2 (hACE2) as the entry receptor. Here, we successfully developed a SARS-CoV-2 hACE2 transgenic mouse (HFH4-hACE2 in C3B6 mice) infection model. The infected mice generated typical interstitial pneumonia and pathology that were similar to those of COVID-19 patients. Viral quantification revealed the lungs as the major site of infection, although viral RNA could also be found in the eye, heart, and brain in some mice. Virus identical to SARS-CoV-2 in full-genome sequences was isolated from the infected lung and brain tissues. Last, we showed that pre-exposure to SARS-CoV-2 could protect mice from severe pneumonia. Our results show that the hACE2 mouse would be a valuable tool for testing potential vaccines and therapeutics.
Keywords: SARS-CoV-2, human ACE2 transgenic mouse, COVID-19, pneumonia
Graphical Abstract
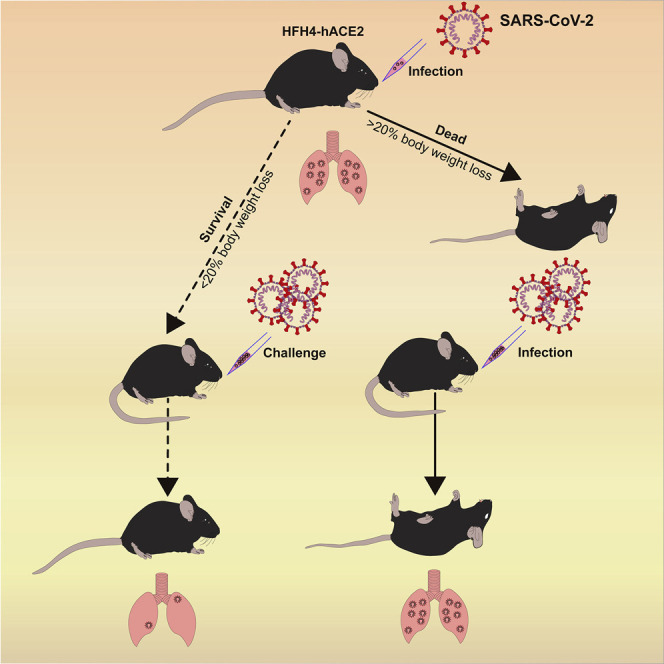
A SARS-CoV-2 hACE2 transgenic mouse infection model recapitulates a number of infection symptoms and pathology in COVID-19 patients. Pre-exposure to SARS-CoV-2 was able to protect mice from severe pneumonia.
Introduction
In December 2019, a coronavirus disease (named as COVID-19 later) was reported in Wuhan city, which rapidly spread throughout China and then to many other countries in the world (Li et al., 2020a, Zhu et al., 2020b). The World Health Organization (WHO) declared COVID-19 as a pandemic on March 11, 2020. As of April 26, 2020, 2,804,796 patients were confirmed from more than 200 countries or areas, leaving more than 193,710 casualties (WHO, 2020). We and other groups quickly identified the etiologic agent as a severe acute respiratory syndrome-related coronavirus (SARSr-CoV), named SARS-CoV-2 (Gorbalenya AE, 2020, Zhou et al., 2020, Zhu et al., 2020a). Hospital-admitted patients exhibited symptoms of pneumonia with abnormal findings in chest CT scans. Typical signs of diseases, including fever, cough, dyspnea, bilateral lung infiltration, and lymphopenia, were similar to that of SARS and Middle East respiratory syndrome (MERS) patients (de Wit et al., 2016, Guan et al., 2020).
Coronaviruses are enveloped non-segmented positive-sense RNA viruses. There are four coronavirus genera, Alphacoronavirus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus, in the subfamily Coronavirinae belonging to the family Coronaviridae. Human coronaviruses belong to the genera Alphacoronavirus and Betacoronavirus and usually cause mild respiratory illness except for the two highly pathogenic viruses, SARS-CoV and MERS-CoV, which cause a severe respiratory syndrome in humans (Cui et al., 2019). Thus, SARS-CoV-2 is the seventh human coronavirus and third highly pathogenic coronavirus identified. The SARS-CoV-2 shares 79.5% nt sequence identity with SARS-CoV-BJ01 and uses the same receptor, angiotensin-converting enzyme 2 (ACE2), for entry into cells (Hoffmann et al., 2020, Letko et al., 2020, Zhou et al., 2020). A previous study demonstrated that the SARS-CoV wild type replicated poorly in mice, requiring mouse adaptation by serial passage or the development of transduced or transgenic mouse models expressing the human ACE2 (Frieman et al., 2012, Menachery et al., 2016, Roberts et al., 2007, Tseng et al., 2007, Yang et al., 2007). Despite receptor-binding domain (RBD) structure homology between SARS-CoV-2 and SARS-CoV, several published SARS-CoV RBD monoclonal antibodies have been unable to neutralize SARS-CoV-2 (Walls et al., 2020, Wrapp et al., 2020), suggesting limited cross-neutralization protection between the two viruses. Currently, there is no specific antiviral therapy against SARS-CoV-2, the testing of which generally relies on animal infection models.
Animal models are vital for understanding viral pathogenesis, vaccine development, and drug screening. Non-human primates (NHPs) are instrumental for the preclinical evaluation. However, the application of NHPs is restricted by high costs, availability, and the complexity of husbandry facilities required. Thus, appropriate small animal models are essential for research and antiviral therapeutic development. Mouse models are popular because their affordability, availability, and clear genetic backgrounds and have been widely used for studying pathogenesis of human coronaviruses (Cockrell et al., 2018). Our previous study demonstrated that SARS-CoV-2 could use human, bat, or civet ACE2 as a cellular receptor but not the mouse ACE2 (Zhou et al., 2020). It appears that mice expressing hACE2 would be a reasonable choice. A transgenic mouse model (HFH4-hACE2 in C3B6 mice) expressing human ACE2 has been constructed under the control of a lung ciliated epithelial cell-specific HFH4/FOXJ1 promoter (Menachery et al., 2016, Ostrowski et al., 2003). HFH4-hACE2 mice expressed high levels of hACE2 in the lung, but at varying expression levels in other tissues, including the brain, liver, kidney, and gastrointestinal tract. These mice had been used to evaluate the pathogenesis of SARS-CoV and bat SARSr-CoVs. The infected mice that lost >20% body weight maintained robust replication viral RNA copies in the lung and brain, although some mice succumbed to lethal encephalitis (Menachery et al., 2016, Netland et al., 2008). Here, we evaluated the infectivity and pathological changes of HFH4-hACE2 mice following SARS-CoV-2 infection.
Results
SARS-CoV-2 Infection Induced the Pathological Features of Pneumonia in HFH4-hACE2 Transgenic Mice
SARS-CoV-2 uses human ACE2 as its cellular receptor but not the mouse ACE2 (Hoffmann et al., 2020, Letko et al., 2020, Zhou et al., 2020). To better study the pathogenesis of SARS-CoV-2, we initiated an experimental infection using HFH4-hACE2 (here after hACE2) transgenic mice. The HFH4-hACE2 mice were intranasally infected with 3 × 104 TCID50 virus each and sacrificed to collect tissue and blood samples at 1, 3, 5, and 7 days post-infection (DPI) (Figure 1 A). Four mice (two males and two females) were used as control animals. In the infected group, four mice were excluded for endpoint data analysis, in which two failed to be infected (negative for viral RNA in all tissues), and another two died resulting from surgical complications (Table S1). Mouse peripheral blood was used for routine blood and biochemistry analysis.
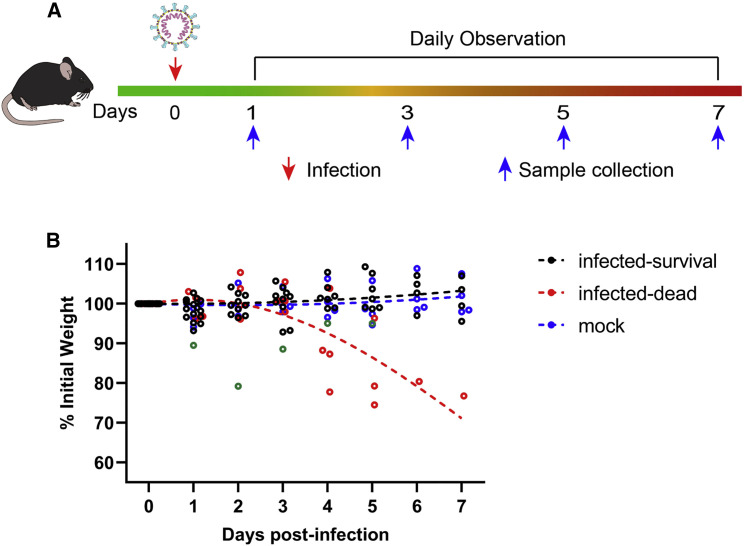
Figure 1.
Experiment Scheme and Body-Weight Changes in SARS-CoV-2-Infected HFH4-hACE2 Mice
(A) Twenty-four HFH4-hACE2 mice were intranasally infected with 3 × 104 TCID50 virus each and sacrificed to collect tissue and blood samples at 1, 3, 5, and 7 days post-infection (DPI). Mouse body weights were monitored for up to 7 days. Each dot represents one mouse at the indicated time point. One mouse showed a rapid body weight decrease with dyspnea beginning on day 2 and then recovered (green dot). Infected mice that died showed noticeable body weight decreases from day 4 to 6 (red dot).
(B) Mock-infected (blue dot) and survived mice (black dot) demonstrated normal statuses. The dotted lines represent the fitted curve of each color-indicated group.
There were no visible clinical signs during the initial 3 days of infection except in one male (G-D5-M2) that showed respiratory distress on day 2 but later recovered (Table S1). Mice showed two distinct clinical phenotypes from 4 DPI. One mouse (G-D5-M3) was found dead 4 DPI, and two lost weight 4 DPI and were humanely euthanized 5 DPI. The remaining mouse began to lose weight 6 DPI and was humanely euthanized 7 DPI. In total, these four mice showed significant body weight decreases from days 4 to 6 and generated noticeable respiratory distress and neurological symptoms before death or humane euthanization. All other mice survived to the endpoint without evident symptoms (Table S1; Figure 1B).
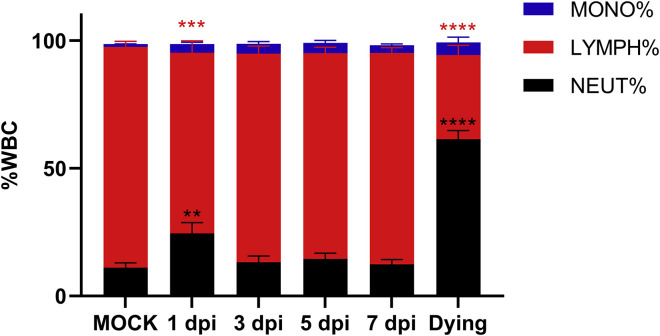
White blood cell (WBC) analysis indicated that the levels of most neutrophils and monocytes began to increase in the peripheral blood at day 1, peaked at day 3, and decreased 5 and 7 DPI in the infected group, compared with those of control mice (Figure S1 ). The infected mice demonstrated a reduced ratio of lymphocytes 1 DPI. However, lymphocytes ratio recovered, beginning on 3 DPI. All the dying mice (dead or lost >20% body weigh) showed significantly decreased lymphocyte numbers, which is one of the key clinical hallmarks of COVID-19 patients (Wang et al., 2020). Five mice showed significantly higher levels of creatine kinase compared with those of the control group, which may be linked to muscle fatigue, soreness, or myocarditis (Table S2). The deceased mice showed higher aspartate transaminase and creatine kinase levels than the control group. These data suggested that SARS-CoV-2-infected mice generate symptoms that partly resembled human infection symptoms (Chen et al., 2020, Guan et al., 2020, Huang et al., 2020, Wang et al., 2020).
Figure S1.
White Blood Cells in Peripheral Blood, Related to Figure 1 and Table S2
Peripheral blood was collected from mock-infected (n = 4) and, virus-infected mice 1 (n = 5), 3 (n = 3), 5 (n = 4) and 7 (n = 4) DPI and three dying (one from the infected group and the other two from the survival test group) mice 6 DPI. The WBC population was measured using ProCyte Dx Hematology Analyzer. Error bars indicate the standard error. Statistical significance was measured by two-way ANOVA compared with the mock infection group. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
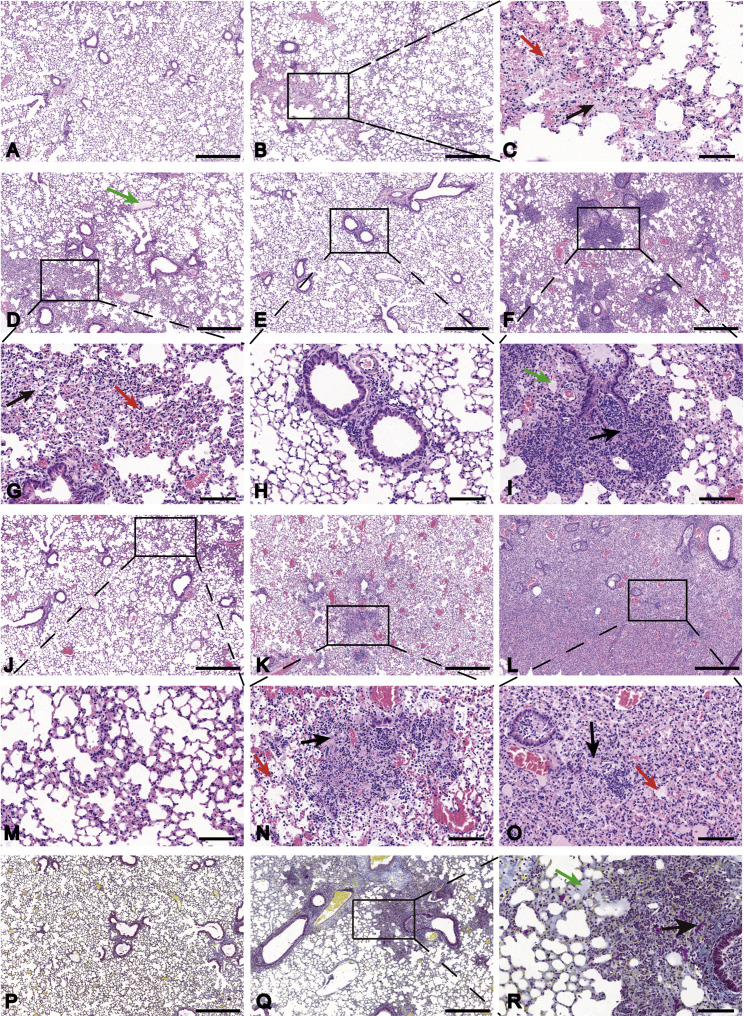
We then used histological assays to check whether these mice acquired pneumonia. The left lobe of infected mice was fixed, sectioned, and stained with hematoxylin and eosin. A similar sample from one healthy mouse was used as a negative control. Pathological sections showed different levels of pneumonia throughout the infection course. At 1 DPI, some minor changes were observed in the lung tissues of infected mice, including minor multifocal lesions, a few alveolar walls thickened with monocyte and lymphocyte infiltration, and increased numbers of macrophages and lymphocytes in some alveolar spaces with minor exudation of fibrin (Figures 2B and 2C) compared with those in the control (Figure 2A). Moderate interstitial pneumonia appeared at 3 DPI, including multifocal lesions, increased inflammatory cells (lymphocytes and monocytes) at peri-bronchial and peri-vascular infiltration and fibroblast hyperplasia with exudation of fibrin and protein edema in some alveoli (Figures 2D and 2G). Two distinct categories were observed after 5 DPI. Eight mice (72.7%) only show mild peri-bronchial and peri-vascular infiltration at 5 DPI (Figures 2E and 2H) and 7 DPI (Figures 2J and 2M), while the other three (27.3%) suffered from more severe pneumonia. In the latter category, the mouse lungs showed consistently massive peri-bronchial and peri-vascular mixed inflammatory cell infiltration, fibroplasia or organization, congestion, edema with hyaline membrane formation with some blocked terminal bronchioles, and dissolved and necrosis in some alveolar cells at 5 DPI (Figures 2F and 2I), 7 DPI (Figures 2K and 2N), and upon death (Figures 2L and 2O). Quantitative pathological scoring showed the same trends with the hematoxylin-eosin (H&E) pathology observation (Figure S2 ). Hyaline thrombus and fibrin were also observed in the lungs following Martius Scarlet Blue (MSB) staining of the tissue samples (Figures 2Q and 2R) compared with the control (Figure 2P). Since higher creatine kinase may result from myocardial or muscle damage, we examined the histology of heart tissue. The heart tissues of infected mice showed edema in some cardiomyocytes and several necrotic myocytes (Figure S3 B) compared with the control (Figure S3A). Taken together, SARS-CoV-2 infection successfully induced pneumonia in hACE2 mice, and the histological features of the lung are very similar to severe patients (Xu et al., 2020).
Figure 2.
Pathological Changes in HFH4-hACE2 Mouse Lung after SARS-CoV-2 Infection
Three to six euthanized mice were used to examine the pathological changes in the lungs 1, 3, 5, and 7 DPI, respectively.
(A) Mock-infected mice lacked lesions.
(B and C) Mouse lung 1 DPI showed multifocal lesion (B) with lymphocyte and macrophage infiltration (C, red arrow) and fibrin exudation (C, black arrow).
(D and G) Moderate interstitial pneumonia was observed 3 DPI with hyaline membrane formation (D, green arrow), inflammatory infiltration (G, red arrow), and fibrin exudation (G, black arrow).
(E, F, H, I, J–N) Mild peri-bronchial and peri-vascular infiltration were observed 5 DPI (E and H) and 7 DPI (J and M). Some infected mice 5 DPI (F), 7 DPI (K), and at death (L) suffered severe pneumonia with blocked terminal bronchioles, fibroplasia or organization, and hyaline membrane formation (I, green arrow; O, red arrow). Alveolar necrosis (I, N, and O, black arrow) and hyaline thrombus (N, red arrow) were observed in severely infected lungs. Martius Scarlet Blue (MSB) staining was performed to staining fibrin in the lung.
(P–R) Mock-infected mice served as control (P). Severely infected mice (Q) show hyaline membrane (R, green arrow) and hyaline thrombus (R, black arrow). Images were collected using a Pannoramic MIDI system.
Scale bar: 500 μm (A, B, D–F, J–L, P, and Q) and 100 μm (C, G–I, M–O, and R).
See also Figure S2.
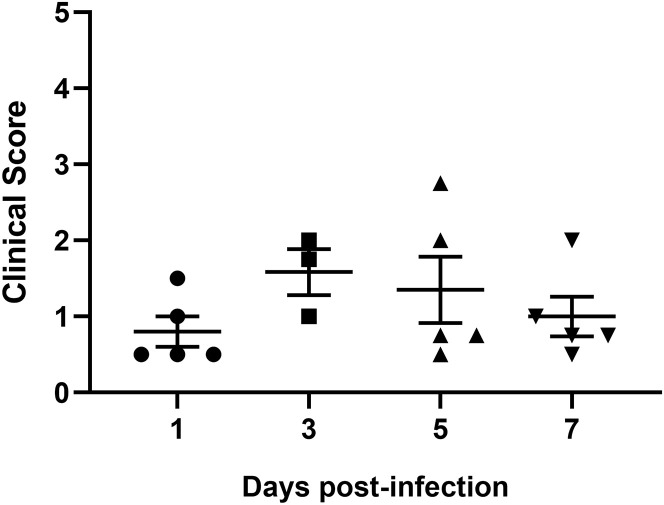
Figure S2.
Clinical Scores of Lung Pathology, Related to Figure 2
Quantitative scoring of pathology of lung tissues from infected mice 1 (n = 5), 3 (n = 3), 5 (n = 5) and 7 (n = 5) DPI. Error bars indicate the standard error.
Figure S3.
Pathological Changes of HFH4-hACE2 Mouse Heart after SARS-CoV-2 Infection, Related to Figure 3
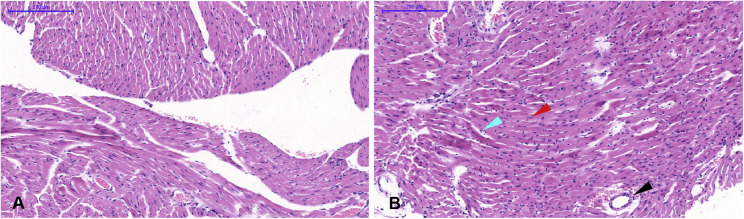
Mock-infected mice (A). Infected mice showed edema in some cardiomyocytes, lymphocyte proliferation (cyan arrow), several necrotic myocytes (red arrow), and vascular edema (black arrow) (B). Scale bar is 200 μm.
Tissue Tropism of SARS-CoV-2 in Infected Mice
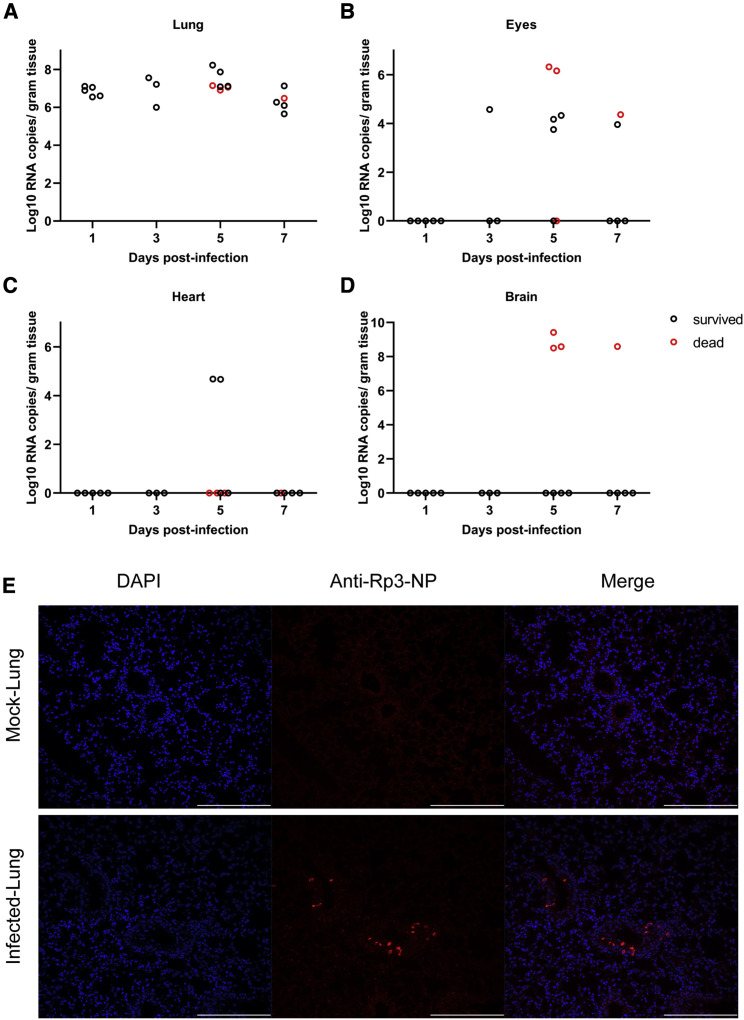
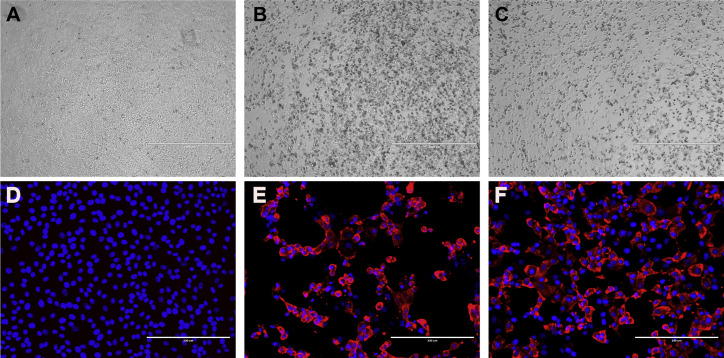
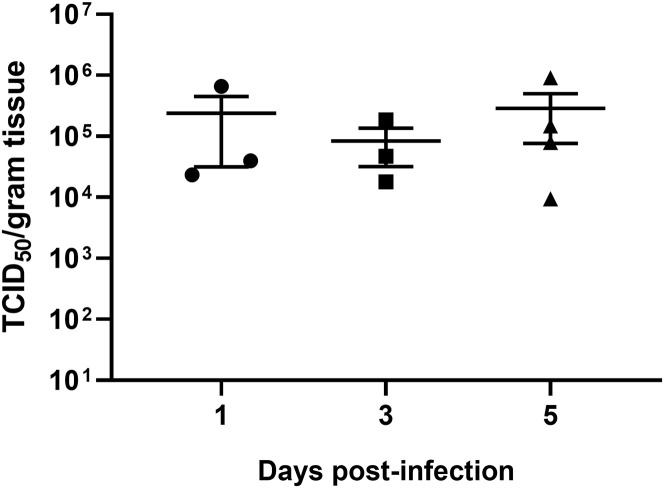
To better understand viral replication in hACE2 mice, viral RNA was quantified by RT-qPCR in different tissues from mice euthanized at 1, 3, 5, and 7 DPI (Figures 3 A–3D). The viral nucleic acid was detected in all lungs from 1 DPI and slowly increased until 5 DPI and then decreased beginning 7 DPI (Figure 3A). We also found low viral RNA copies in the eyes and hearts in eight and two infected mice, respectively (Figures 3B and 3C). No viral RNA was detected in other organs. Viral antigens were detected in bronchial epithelial cells and alveolar cells of the lungs using immunohistological assays with cross-reactive antibody that has been used for SARS-CoV-2 detection previously (Zhou et al., 2020) (Figure 3E). Notably, four of the animals demonstrated SARS-CoV-2 neuroinvasion of the brain but only in the deceased mice (Figure 3D). However, the exact details and mechanisms regulating SARS-CoV-2 neuroinvasion remain unclear, as this phenomenon has been previously described with this mouse model (Menachery et al., 2016). We then tried to re-isolate SARS-CoV-2 from infected animals. We inoculated Vero E6 cells with infected mouse (G-D7-M1) lung and brain homogenate supernatant and observed a noticeable cytopathic effect (CPE) in the infected cells (Figures S4 B and S4C) compared with the control (Figure S4A). The re-isolated virus from lung and brain of G-D7-M1 was confirmed by indirect imunofluorescence assays (IFAs) and genome sequencing (Figures S4D–S4F; Table S3). The lung isolates indicated two mutations, C14554T (nsp13, L372F) and C23525T (spike, H644Y), compared with the genome of SARS-CoV-2-WIV04 (GISAID: EPI_ISL_402124). The mutation at nsp13 was unique compared with published viral sequences identified by online BLAST. The mutation in the spike gene was located at the end of the S1 region of the spike gene. This mutation was identified using BLAST in SARS-CoV-2 genome sequence isolated from a patient from the United States (GenBank: MT163720). The brain isolates exhibited only one mutation, C23525T (H644Y), at the S1 region of the spike gene. The viral titers in the infected lung ranged from 9.4 × 103 to 9.1 × 105TCID50 per gram of tissue, as determined by serial dilution of VeroE6 cell lines (Figure S5 ). Conclusively, SARS-CoV-2 mainly replicated in mouse lung, although it may also target the brain, heart, and eyes of hACE2 mice. Brain infections were only observed in the deceased mice, suggesting that neuroinvasion is sporadic in this model and likely represents the cause of death. Similar findings have been reported in other transgenic mouse models infected with SARS-CoV (McCray et al., 2007, Menachery et al., 2016).
Figure 3.
Virus Quantification and Antigen Detection in Mice after SARS-CoV-2 Infection
(A–D) Viral RNA was quantified using RT-qPCR in mouse tissues collected 1, 3, 5, and 7 DPI. The virus was detected in the mouse lung (A), eyes (B), heart (C), and brain (D).
(E) Viral antigen was detected by anti-Rp3-CoV N protein polyclonal antibody (red) in lung bronchi and alveoli collected 1 DPI (G-D1-5) (E). Images were collected using FV1200 confocal microscopy. Scale bar is 200 μm.
See also Figures S3, S4, and S5 and Table S3.
Figure S4.
Virus Isolation from Infected Mouse Lung and Brain Tissues, Related to Figure 3 and Table S3
Infected mouse lung and brain tissues were homogenized with DMEM, and the supernatant was used to infect Vero E6 cells. CPE was observed in cells infected by the homogenized supernatant of lung (B) and brain (C) but not in mock-infected cells (A). Viral antigen was detected in the cells infected by homogenized supernatant of the lung (E) and brain (F) but not in mock-infected cells (D). Scale bar: 1000 μm (A, B, C) and 200 μm (D, E, F).
Figure S5.
Viral Titers in Lung Tissue of Mice after SARS-CoV-2 Infection, Related to Figure 3
Viral titers were determined by the TCID50 assay using VeroE6 cells for mouse lung tissues collected 1 (n = 3), 3 (n = 3) and 5 (n = 4) DPI. The lung samples collected 7 DPI were used for virus isolation. Error bars indicate the standard error.
Surviving Mice Were Resistant to Re-challenge with a High Dose of SARS-CoV-2
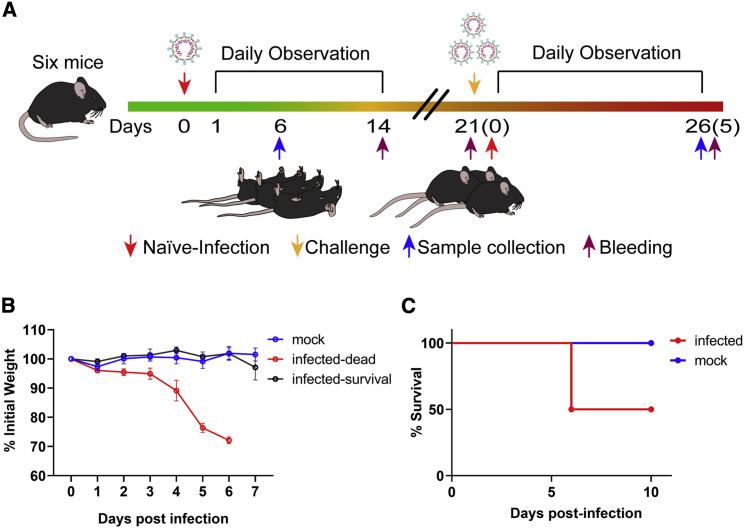
We next evaluated whether this hACE2 mouse model can be used as a platform for testing the performance of therapeutics against SARS-CoV-2 infection. We designed an experiment to test whether mice surviving SARS-CoV-2 infection developed resistance to reinfection (Figure 4 A). Six mice were infected using identical viral doses via the same route of infection at the same time with the infection group. Upon infection, three mice died 6 DPI. All the deceased mice were male, with total mortality of 50% (3/6). In general, there were two infection outcomes, those that lost more than 20% body weight from 4 or 5 DPI and died eventually, or those that lost less than 10% of body weight and survived (Figures 4B and 4C). Similarly, all deceased mice generated noticeable neurological symptoms before death.
Figure 4.
Experiment Scheme and Mortality of HFH4-hACE2 Mice after SARS-CoV-2 Infection
Six hACE2 mice (four males and two females) were intranasally infected with 3 × 104 TCID50 SARS-CoV-2. Body weights and survival were monitored. Three survived mice were observed at 21 days. Then, the three surviving mice and another three healthy mice were intranasally infected with 7 × 105 TCID50 SARS-CoV-2.
(A) The number in the parenthesis indicates the DPI of reinfection.
(B) Body weights and survival were monitored every day. Body weights decreased quickly, beginning at 4 DPI and led to death, or mice survived in the absence of significant body weight decreases.
(C) Three of four male mice died after infection, while two females survived.
Error bars indicate the standard error.
See also Table S4.
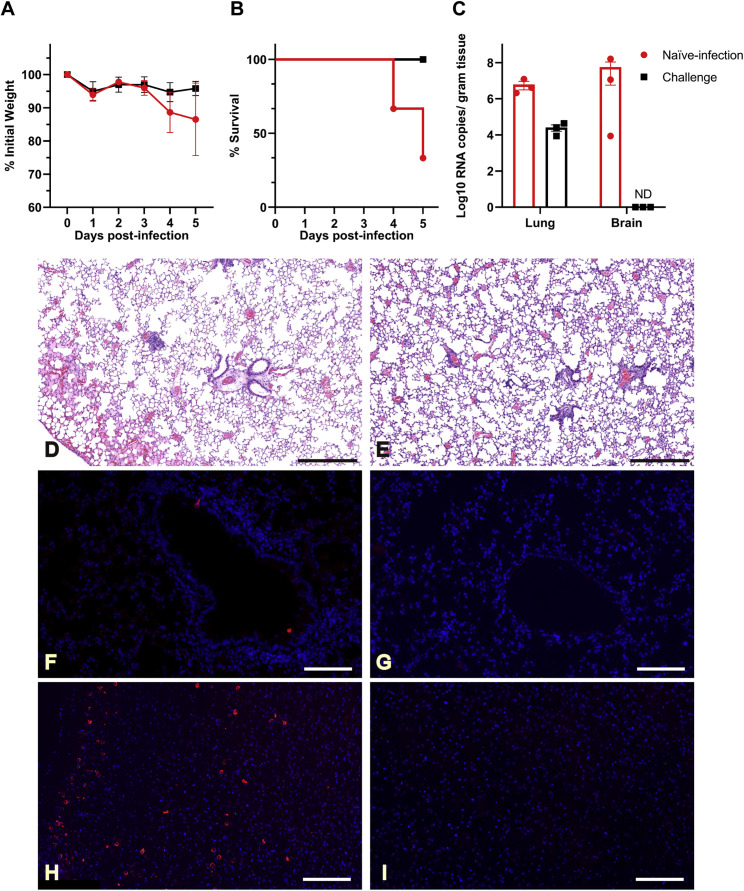
Three mice, one male and two females who survived the infection, were observed for 21 days. These mice generated antibodies against SARS-CoV-2 that could neutralize 100 TCID50 viruses at dilutions of 1:10 to 1:40 (Table S4). We then intranasally infected the three SARS-CoV-2 survivors and another three healthy mice with 7 × 105 TCID50 virus/each. All previous survivors also survived the second infection. At the same time, two of the three naive-infected mice demonstrated rapid bodyweight loss from 4 to 5 DPI with a male and female animal dead at 4 DPI and 5 DPI, respectively (Figures 5A and 5B). All mice were euthanized at 5 DPI to evaluate viral titers and pathologic changes in the lung and brain tissues. Survivors had markedly lower viral RNA copies in the lung and did not present any evidence of neuroinvasion compared with those of the naive-infected animals (Figure 5C). Histopathology of the lungs of the re-infected survivor cohort showed moderate inflammatory infiltration. In contrast, the peri-branchial and peri-vascular inflammation infiltration, hyaline membrane formation, and some blocked terminal bronchioles were observed in the naive-infected group (Figures 5D and 5E). Limited viral antigen was detected in the lung and brain tissues of the surviving cohort of mice compared with that of the naive infection group (Figures 5F–5I). The deceased mice showed systemic infection due to the high viral dose challenge (Table S5). These data demonstrate that pre-exposure to SARS-CoV-2 may protect mice from a lethal SARS-CoV-2 reinfection, which underscores the values of this model for future therapeutic studies.
Figure 5.
Previous Infection Protected Mice from a Higher Dose of Virus Reinfection
Three surviving mice 21 DPI and three previously mock-infected mice were infected with 7 × 105 TCID50 SARS-CoV-2 at 21 DPI, respectively.
(A and B) Body weights (A) and survival outcomes (B) were monitored for up to 5 days.
(C) Mice were euthanized 5 DPI, and the viral loads were measured using RT-qPCR.
(D) Naive-infected mouse lung showed peri-bronchial and peri-vascular infiltration and edema with hyaline membrane formation.
(E) In contrast, previously challenged mice presented mild peri-bronchial and peri-vascular infiltration.
(F–I) Viral antigen was widely present in the naive-infected mouse lung (F) and brain (H) but barely observed in previously challenged mouse lung (G) and brain (I).
Error bars indicate the standard error. ND, not detected. Scale bar: 500 μm (D and E), 100 μm (F and G), and 200 μm (H and I).
Discussion
As a cost-effective and easily manipulated animal model, mice are widely used in biomedical research. Various genetically modified mice have been generated to meet the demands of different projects. Wild-type mice have low levels of ACE2 expression, which limits their use as an infection model for SARSr-CoVs, which use ACE2 as a receptor. Previous studies indicated that HFH4-hACE2 mice are susceptible to SARS-CoV and bat SARSr-CoV WIV1 infection (Menachery et al., 2016), suggesting that this model can be used to evaluate the pathogenicity of all SARSr-CoVs using ACE2 as a receptor. In this study, we successfully established a SARS-CoV-2 infection model using these transgenic mice. In our results, the infected mice had two different outcomes, recovery and death. Significant weight loss was only observed in dead and severely diseased mice. Thus, weight loss could serve as a good indicator for monitoring disease progression. Our results provide strong evidence that SARS-CoV-2 infection could cause typical interstitial pneumonia in hACE2-expressing mice and that progeny virus could be re-isolated from diseased animal tissues.
COVID-19 patients commonly exhibit pneumonia with typical signs including fever, cough, dyspnea, and bilateral lung infiltration (Guan et al., 2020). Biopsy of the lungs showed evident desquamation of pneumocytes and hyaline membrane formation from one patient (Xu et al., 2020). After SARS-CoV-2 infection, the hACE2-experssing mice showed apparent interstitial pneumonia with inflammatory cell infiltration. Severe pneumonia in mice was accompanied by fibroplasia, organization, congestion, and edema with hyaline membrane formation. Some alveolar cells were dissolved and necrotic, and some terminal bronchioles were blocked, which simulated the pathological findings in COVID-19 patients (Xu et al., 2020). During routine blood examination, lymphopenia is a common feature in COVID-19 patients, and some patients had high-level aspartate amino transferase (AST) and creatine kinase (CK) (Chen et al., 2020, Guan et al., 2020). The blood routine examination results in SARS-CoV-2-infected mice were not entirely consistent with human patients, while the apparent lymphopenia was observed only in the dying mice. In our mouse model, the male mice showed a much higher death rate than the female mice, which partially reflects the observation infection outcome in humans (Channappanavar et al., 2017, WHO, 2020). Gender-based susceptibility differences have been reported in SARS-CoV-infected mice, which demonstrates that the male mice are more susceptible to SARS-CoV infection than the female mice (Channappanavar et al., 2017). Compared with SARS-CoV infection, HFH4-hACE2 mice infected with SARS-CoV-2 showed lower mortality rate but comparable viral titers in the lung tissues which may partially reflect the lower mortality in SARS-CoV-2 patients.
We also demonstrated that the lung is the primary target of this virus, although the eye, heart, and brain could also be infected. In human cases, it has been suspected that SARS-CoV or SARS-CoV-2 could also attack the eye, heart, and brain; however, only limited data are available (Gu et al., 2005, Li et al., 2020b, Lu et al., 2020, Xu et al., 2005). Our study cautioned that special care should be taken for these viral targets, especially as the brain and eyes can be persistently infected by other acute RNA viruses, such as Ebola virus (Shantha et al., 2018). There are few reports about organ tropism of virus infection in the patients of COVID-19, but multiple organ infections have been reported in SARS patients (Gu et al., 2005). In our mouse model, the viral RNA was mainly detected in lung tissues at lower virus titer infection. In comparison more positive tissues were detected with higher virus dose infection, in accordance with the tissue ACE2 expression in the HFH4-hACE2 mice. Mice experiencing rapid weight loss were found with high viral RNA in the brain, which is consistent with the results when HFH4-hACE2 mice were infected by the bat SARSr-CoV and SARS-CoV (Menachery et al., 2016). Considering that SARS-CoV has been reported in SARS patient brains, mouse brain infection after the SARS-CoV-2 challenge support the potential brain infection in patients (Gu et al., 2005, Menachery et al., 2016, Xu et al., 2005). Recently, virus was isolated from ocular fluids of SARS-CoV-2 patients (Colavita et al., 2020), which confirms our finding in mouse model and implies a potential eye-based infection and transmission route.
The surviving mice generated low titers of neutralization antibodies (1:10 to 1:40). The reinfection with much higher titers of virus (7 × 105 TCID50 SARS-CoV-2) only caused mild pneumonia in the surviving mice. In contrast, the naive-infected mice had severe disease with much higher viral RNA copies in the lung and brain tissues. It is unclear that the neutralizing antibody or humoral immunity caused the observed protection. Future studies are needed to determine whether cellular immune arms also contribute to the protection from reinfection.
In summary, a human ACE2 mouse model that partially simulated the pathology of COVID-19, will be a valuable platform for testing vaccines and other potential therapeutics. Although there are drawbacks associated with this model, such as lethal encephalitis, it is also a suitable choice for studying pathogenesis and therapeutic potential on this model before the central nervous syndromes. Other strains of mice, including mice of different ages, which had been previously tested with SARS-CoV and MERS-CoV infections, should also be verified with SARS-CoV-2 (Gretebeck and Subbarao, 2015, Song et al., 2019). During the revision of this manuscript, other researchers tested the susceptibility of SARS-CoV-2 in several animal species. Their results demonstrated that SARS-CoV-2 could infect hamsters, ferrets, and cats with resulting mild symptoms but not pigs, chickens, or ducks (Chan et al., 2020, Shi et al., 2020). The increasing global spread of SARS-CoV-2 promotes the urgent need for coronavirus vaccines and therapeutics. The successful establishment of an animal model for COVID-19 pathogenesis will be valuable for evaluating vaccines and therapeutics to combat SARS-CoV-2.
Limitations
The HFH4-hACE2 mice showed different susceptibility with the infection of SARS-CoV-2 based on gender. The LD50 of SARS-CoV-2 should be determined for different gender mice. The dead mice showed the brain infection that may not reflect the pathogenesis of SARS-CoV-2 in humans. Future studies should determine the persisting time of virus in infected mice. Aged mice should be tested the susceptibility of SARS-CoV-2 infection.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-RP3-CoV N protein polyclonal antibody | Zhou et al., 2020 | N/A |
| Cy3-conjugated goat-anti-rabbit IgG | Abcam | Cat#ab6939; RRID:AB_955021 |
| DAPI | Beyotime | Cat#C1002 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 | Isolated from patient Zhou et al., 2020 | IVCAS 6.7512 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Avertin | Sigma | Cat#T-48402-5g |
| Hematoxylin solution | Servicebio | Cat#G1005-1 |
| Eosin solution | Servicebio | Cat#G1005-2 |
| IDEXX Catalyst BUN | IDEXX | Cat#98-11072-01 |
| IDEXX Catalyst CK | IDEXX | Cat#98-11073-01 |
| IDEXX Catalyst AST | IDEXX | Cat#98-11069-01 |
| IDEXX Catalyst ALT | IDEXX | Cat#98-11067-01 |
| 4% paraformaldehyde | Boster | Cas#AR1068 |
| RNAlater Stabilization Solution | Invitrogen | Cat#AM7021 |
| Triton X-100 | Sigma | Cas#9002-93-1 |
| EDTA-Na2 | Sigma | Cas#6381-92-6 |
| Critical Commercial Assays | ||
| QIAamp 96 Virus QIAcube HT Kit | QIAGEN | Cat#57731 |
| HiSxript II One step qRT-PCR SYBR Green Kit | Vazyme | Cat#Q221-01 |
| Modified MSB staining Kit | Solarbio | Cat#G2040 |
| IDEXX ProCyte Dx Reagent Kit | IDEXX | Cat#99-26306-00 |
| Experimental Models: Cell Lines | ||
| Vero E6 | ATCC | ATCC CRL-1586; RRID:CVCL_0574 |
| HFH4-hACE2 mice | Ralph S. Baric’s lab | N/A |
| Oligonucleotides | ||
| RBD-qF1: 5′- CAATGGTTTAACAGGCACAGG-3′ | Zhou et al., 2020 | N/A |
| RBD-qR1: 5′- CTCAAGTGTCTGTGGATCACG-3′ | Zhou et al., 2020 | N/A |
| Software and Algorithms | ||
| Pannoramic Viewer | 3DHISTECH | https://www.3dhistech.com/software-downloads/ |
| StepOne Software | ABI | N/A |
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com |
| Geneious | Geneious 10.2.6 | https://www.geneious.com |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Zheng-Li Shi (zlshi@wh.iov.cn).
Materials Availability
All materials and reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The study did not generate unique datasets or code.
Experimental Model and Subject Details
Virus and Cell lines
The SARS-CoV-2 (IVCAS 6.7512) was isolated using bronchoalveolar lavage fluid sample of patient and was propagated and titrated in Vero E6 cell lines (Zhou et al., 2020). Vero E6 is a kidney epithelial cell line derived from female African green monkey (ATCC® CRL-1586). Vero E6 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Life Technologies) and 1% Anti-Anti (Invitrogen) at 37°C with 5% CO2. Viral titrations were performed with 10-fold serial dilutions in Vero E6 cells. Three days after inoculation, CPE was scored, and the Reed-Muench formula was used to calculate the TCID50.
HFH4-hACE2 mice
The transgenic mice (HFH4-hACE2 mice) expressing the human ACE2 protein (hACE2) under mixed genetic backgrounds (C3H, C57BL/6) were obtained from Ralph S. Baric’s lab (Menachery et al., 2016). The HFH4-hACE2 mice were bred and maintained in specific pathogen free (SPF) environment at the Laboratory Animal Center of Wuhan Institute of Virology, CAS. Twenty-two male and twelve female mice at eight to ten weeks-old were used in this study. Littermates of the same sex were randomly assigned to mock, 1, 3, 5, 7 DPI and survival groups. Viral infections were performed in a biosafety level 3 (BSL3) facility in accordance with recommendations for the care and use of laboratory animals and the Institutional Review Board of the Wuhan Institute of Virology, CAS (ethics number WIVA05202003).
Method Details
Mice infection
Mice were anaesthetised with tribromoethanol (Avertin) (250 mg/kg) and intranasally infected with 3∗104 TCID50 (for naive infection) or 7∗105 TCID50 (for the viral challenge) SARS-CoV-2 in 50 μL DMEM per mouse. The experiments were divided into three groups: the control group, two males and two females were inoculated with DMEM; the survival monitoring group, four males and two females were inoculated with the virus; the pathology progression group, sixteen males and eight females were inoculated with the virus (Table S1). Accidental death or uninfected mice were excluded from subsequent experiments. Infected mice negative for viral RNA in all tissues were regarded as a failed infection. Mice were weighted and observed for clinical signs daily throughout the study. Four to six mice were euthanised at 1, 3, 5 and 7 DPI, and mice presenting more than a twenty percent decrease in body weight were humane euthanised and considered as dead mice. The details regarding the number of male and female mice in each group are shown in Table S1 and Table S4. Tissues were harvested, including the blood, heart, liver, spleen, lung, brain, kidney, eyes, genital glands, and small intestines.
Blood sampling and biochemistry
Blood was collected retro-orbitally and transferred to a blood collection tube containing EDTA to prevent clotting. Cell classifications were analyzed using a ProCyte Dx Hematology Analyzer (IDEXX) followed with manufacturer’s instructions. Plasma was separated by centrifugation at 3,000 × g for 10 min at room temperature and stored at −80°C until use. Plasma was collected, diluted 1:6 with phosphate-buffered saline (PBS), and tested using Catalyst One (IDEXX) followed with manufacturer’s instructions.
Viral antibody testing and neutralisation assay
All serum samples were heat-inactivated by incubation at 56°C for 30 min before use. Mouse plasma (24 μL) was serially two-fold diluted from 1:10 to 1:1280, then an equal volume of virus solution with 100 TCID50 virus was added and incubated at 37°C for 30 min in a 5% CO2 incubator. Diluted negative sera were mixed with an equal volume of virus solution and were used as a negative control. After incubation, 50 μL mixtures were inoculated onto monolayer Vero E6 cells in a 96-well plate for 1 h. After removing the supernatant, the plate was washed twice with DMEM, cells were incubated with DMEM supplemented with 2% FBS for three days. Each serum concentration was duplicated. Cells were observed for CPE, and neutralising titers were recorded as the final serum dilution that could neutralise all inoculated wells.
Extraction of viral RNA and quantitative RT-PCR
Mouse organs were homogenized in RNALater, and viral RNA was isolated using the QIAamp ® 96 Virus QIAcube ® HT kit (QIAGEN). Two microliters of RNA were used as a template for the amplification of selected genes by real-time quantitative PCR using HiSxript ® II One step qRT-PCR SYBR® Green Kit (Vazyme). Average values from duplicates of each gene were used to calculate the viral genomic copies. The primers based on the SARS-CoV-2 S gene were designed as: RBD-qF1: 5′- CAATGGTTTAACAGGCACAGG-3′; RBD-qR1: 5′- CTCAAGTGTCTGTGGATCACG-3′. The PCR system was as follows: the 10 μL qPCR reaction mix contained 5 μL 2 × One Step SYBR Green mix, 1.9 μL nuclease free water, 0.5 μL One Step SYBR Green Enzyme mix, 0.2 μL 50 × ROX Reference Dye 1, 0.2 μL of each primer (10 μM) and 2 μL template RNA. Amplification was performed as follows: 50°C for 3 min, 95°C for 30 s followed by 40 cycles consisting of 95°C for 10 s and 60°C for 30 s, and a default melting curve step in an Step-One Plus Real-time PCR machine (ABI) (Zhou et al., 2020).
Histological Analysis
Lung and brain samples were fixed with 4% paraformaldehyde, paraffin embedded and cut into 3.5-μm sections. Fixed tissue samples were used for hematoxylin-eosin (H&E) staining and indirect immunofluorescence assays (IFA) for the detection of the SARS-CoV-2 antigen. For routine histology, tissue sections were stained with H&E. For IFA, slides were deparaffinised and rehydrated, followed by 15-min heat-induced antigen retrieval with EDTA pH 8.0 in a microwave oven. The slides were washed with PBS/0.02% Triton X-100 then blocked with 5% BSA at room temperature for 1 h. A primary antibody (rabbit anti-RP3-CoV N protein polyclonal antibody, 1:500, made in-house) was added dropwise to the sections and then washed in PBS. After the slides were slightly dried, tissues were covered with Cy3-conjugated goat-anti-rabbit IgG (Abcam, ab6939) at 1:200 dilution. After washing in PBS, slides were stained with DAPI (Beyotime) at 1:100 dilution. For MSB staining, fixed tissue sections were stained with hyposulfite solution, Celestine blue, and hematoxylin for 3-5 min, respectively. After acidic differentiation with acid alcohol, the sections were stained with Martius Yellow solution, carmoisine solution, phosphotungstic acid solution, and aniline blue staining solution, respectively. Tissue sections were rinsed with 1% acetic acid and then dehydrated, cleared, and mounted with neutral gum. The image information was collected using a Pannoramic MIDI system (3DHISTECH, Budapest) and FV1200 confocal microscopy (Olympus).
Virus isolation and sequencing
Infected lung and brain tissues from euthanised mice were homogenized with DMEM, and the supernatant was used for virus isolation. A total of 200 μL supernatant was inoculated onto monolayer Vero E6 cells in a 24-well plate. After 1 h inoculation, the supernatant was removed and replaced with fresh DMEM containing 2% FBS plus 1% Anti-Anti. CPE was monitored daily, and the supernatant was clarified from debris and subjected to viral RNA extraction after the onset of CPE. Viral RNA copies were quantified by qPCR, and viral antigens were detected using IFA. Images were collected by EVOS M5000 (Thermo). The Next-generation sequencing (NGS) was performed by using Illumina MiSeq 3000 sequencers and the sequences were analyzed by Geneious ®10.2.6 for Windows (Zhou et al., 2020).
Quantification and Statistical Analysis
Statistical Analysis
Statistical analyses were performed using PRISM 8.0.2 for Windows (GraphPad). Significant differences between groups were determined using a two-way analysis of variance (ANOVA). For contrasting two experimental groups, two-tailed Student’s t tests were performed to determine significant differences. Statistical significant: ∗p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001.
Acknowledgments
We thank He Zhao and Xue-Fang An from the animal center of the Wuhan Institute of Virology for the transgenic mouse breeding. We thank Tao Du and Ge Gao from the Center for Biosafety Mega-Science for their essential support. We also thank Ding Gao from the core facility of the Wuhan Institute of Virology for their technical support. We also thank the support of Shenzhen MGI Tech Co., Ltd. for their sequencing service using MGISEQ-2000RS FAST platform. This work was jointly supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29010101 to Z.-L.S.), China Natural Science Foundation (31770175 to Z.-L.S.), and a Youth innovation promotion association of CAS (2019328 to X.-L.Y.).
Author Contributions
Conceptualization, C.S., X.-L.Y., and Z.-L.S.; Methodology, R.-D.J., M.-Q.L., and C.S.; Investigation, R.-D.J., M.-Q.L., Y.C., X.-L.Y., C.S., X.-R.S., Q.L., L.Z., Y.Z., H.-R.S., Q.W., J.M., W.Z., B.L., and H.-J.Z.; Resources, J.M., X.W., and R.S.B.; Writing – Original Draft, R.-D.J., M.-Q.L., Y.C., and X.-L.Y.; Writing – Review & Editing, X.-L.Y., Y.-W.Z., R.S.B., P.Z., and Z.-L.S.; Funding Acquisition, X.-L.Y. and Z.-L.S.
Declaration of Interests
The authors declare no competing interests.
Published: May 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.05.027.
Supplemental Information
References
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa325. ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Mack M., Ten Eyck P.P., Meyerholz D.K., Perlman S. Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection. J. Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell A.S., Leist S.R., Douglas M.G., Baric R.S. Modeling pathogenesis of emergent and pre-emergent human coronaviruses in mice. Mamm. Genome. 2018;29:367–383. doi: 10.1007/s00335-018-9760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavita F., Lapa D., Carletti F., Lalle E., Bordi L., Marsella P., Nicastri E., Bevilacqua N., Giancola M.L., Corpolongo A. SARS-CoV-2 isolation from ocular secretions of a patient with COVID-19 in Italy with prolonged viral RNA detection. Ann. Intern. Med. 2020 doi: 10.7326/M20-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86:884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretebeck L.M., Subbarao K. Animal Models for SARS and MERS Coronaviruses. Curr. Opin. Virol. 2015;13:123–129. doi: 10.1016/j.coviro.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Bai W.Z., Hashikawa T. The neuroinvasive potential of SARS-CoV2 may be at least partially responsible for the respiratory failure of COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.W., Liu X.F., Jia Z.F. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395:e39. doi: 10.1016/S0140-6736(20)30313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., Meyerholz D.K., Moore S., Cassell M., Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski L.E., Hutchins J.R., Zakel K., O’Neal W.K. Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol. Ther. 2003;8:637–645. doi: 10.1016/s1525-0016(03)00221-1. [DOI] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shantha J.G., Mattia J.G., Goba A., Barnes K.G., Ebrahim F.K., Kraft C.S., Hayek B.R., Hartnett J.N., Shaffer J.G., Schieffelin J.S. Ebola virus persistence in ocular tissues and fluids (EVICT) study: reverse chain reaction and cataract surgery outcomes of Ebola survivors in Sierra Leone. EBioMedicine. 2018;30:217–224. doi: 10.1016/j.ebiom.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020 doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, Thrusting Coronaviruses Into the Spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.T., Huang C., Newman P., Wang N., Narayanan K., Watts D.M., Makino S., Packard M.M., Zaki S.R., Chan T.S., Peters C.J. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J. Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus disease (COVID-2019) situation reports.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhong S., Liu J., Li L., Li Y., Wu X., Li Z., Deng P., Zhang J., Zhong N. Detection of severe acute respiratory syndrome coronavirus in the brain: potential role of the chemokine mig in pathogenesis. Clin. Infect. Dis. 2005;41:1089–1096. doi: 10.1086/444461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.H., Deng W., Tong Z., Liu Y.X., Zhang L.F., Zhu H., Gao H., Huang L., Liu Y.L., Ma C.M. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007;57:450–459. [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.B., Zhong C.K., Zhang K.X., Dong C., Peng H., Xu T., Wang A.L., Guo Z.R., Zhang Y.H. [Epidemic trend of corona virus disease 2019 (COVID-19) in mainland China] Zhonghua Yu Fang Yi Xue Za Zhi. 2020;54:E022. doi: 10.3760/cma.j.cn112150-20200222-00163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The study did not generate unique datasets or code.