Highlights
-
•
RT-LAMP test kits have good utility.
-
•
Detection sensitivity of 1.0 × 101 copies/μL can be obtained within 35 min.
-
•
RT-LAMP can be used as a point-of-care test because it is judged by turbidity under natural light.
Keywords: SARS-CoV-2, COVID-19, LAMP, RT-qPCR
Abstract
With the rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), there is an urgent need for more rapid and simple detection technologies at the forefront of medical care worldwide.
In this study, we evaluated the effectiveness of the Loopamp® 2019-SARSCoV-2 Detection Reagent Kit, which uses loop-mediated isothermal amplification (LAMP) technology. In this protocol, cDNA is synthesized from SARS-CoV-2 RNA using reverse transcriptase, followed by DNA amplification under isothermal conditions in one step. The RT-LAMP test kit amplified the targeted RNA of a SARS-CoV-2 isolate with a detection limit of 1.0 × 101 copies/μL, which was comparable to the detection sensitivity of quantitative reverse transcription PCR (RT-qPCR).
Comparison with the results of RT-qPCR for 76 nasopharyngeal swab samples from patients with suspected COVID-19 showed a sensitivity of 100 % and a specificity of 97.6 %. In the 24 RNA specimens derived from febrile Japanese patients with or without influenza A, no amplification was observed using RT-LAMP. RT-LAMP could be a simple and easy-to-use diagnostic tool for the detection of SARS-CoV-2.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), has emerged as a serious threat to human health worldwide [1,2]. With the rapid increase in the number of patients, a reliable, rapid, and simple detection system for SARS-CoV-2 is needed that can be used in all medical institutions as quickly as possible [3].
Loop-mediated isothermal amplification (LAMP)-based analysis, which can be performed without a thermal cycler, is suitable for the diagnosis of infectious diseases as a point-of-care test in resource-limited settings [4,5]. In particular, the use of bst DNA polymerase with high strand displacement activity, to which reverse transcriptase (RT) activity has also been added, makes amplification of specific viral RNA possible in one step at a constant temperature.
This study examined the usefulness of a commercially available RT-LAMP-based diagnostics kit for COVID-19 (Loopamp®︎ 2019-SARS-CoV-2 Detection Reagent Kit; http://loopamp.eiken.co.jp/), with the view that if the approach proves feasible, it could support the rapid detection of SARS-CoV-2.
2. Materials and methods
2.1. Standard RNA of SARS-CoV-2
To evaluate the analytical sensitivity of the RT-LAMP method, we used purified and quantified total RNA of SARS-CoV-2, which was provided by the National Institute of Infectious Diseases, Japan, as a standard specimen for the molecular diagnosis of COVID-19. Analytical sensitivity was determined using 10-fold serially diluted standard RNA ranging from 1.0 × 103 copies/μL to 1.0 copy/μL and stored at −30 °C until required.
2.2. Clinical specimens
Seventy-six nasopharyngeal swab samples were examined. The swabs were from patients with suspected COVID-19 who were admitted to Saitama Medical University Hospital, Japan, from February to March 2020. Viral RNA was extracted from the samples using a QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions, and RNA was eluted in 60 μL of the provided AVE buffer. Conventional quantitative reverse transcription PCR (RT-qPCR) for the specific amplification of the N gene of SARS-CoV-2 was performed using the previously reported TaqMan system with the following sets of primers and probe: 2.4 μM forward primer, 5′-AAA TTT TGG GGA CCA GGA AC-3′; 3.2 μM reverse primer, 5′-TGG CAG CTG TGT AGG TCA AC-3′; and 0.4 μM probe, 5′-FAM-ATG TCG CGC ATT GGC ATG GA-TAMRA-3′ [6]. Thermal cycling was carried out under the following conditions: reverse transcription at 50 °C for 30 min; initial denaturation at 95 °C for 15 min; and 40 cycles of denaturation at 94 °C for 15 s and annealing/extension at 60 °C for 60 s. This study was approved by Saitama Medical University Hospital Research Ethics Committee (Approval No. 19136).
2.3. Amplification of SARS-CoV-2 RNA with RT-LAMP
RT-LAMP for the specific detection of SARS-CoV-2 RNA was performed with a Loopamp®︎ 2019-SARS-CoV-2 Detection Reagent Kit (Eiken Chemical, Tokyo, Japan) at 62.5 °C for 35 min according to the manufacturer’s instructions. Positive amplification results were monitored by real-time measurement of turbidity with an LA-200 turbidimeter (Eiken Chemical). In this reaction, 10 μL RNA template was added to 15 μL of the provided master mix including a set of primers to make a final volume of 25 μL. The performance of the RT-LAMP kit was evaluated by comparing the results with those of conventional RT-qPCR.
2.4. Specificity of RT-LAMP
The specificity of the RT-LAMP reaction was evaluated using 12 RNA samples extracted from nasopharyngeal swabs taken from patients with influenza A at Saitama Medical University Hospital, Japan, from November to December 2019. In addition, 12 RNA samples negative for influenza A from febrile Japanese patients during the same period were also used in this study. During this sampling period, there were no cases of COVID-19 in Japan. The RT-LAMP reaction was performed using the same kit and conditions described above.
3. Results
3.1. Analytical sensitivity
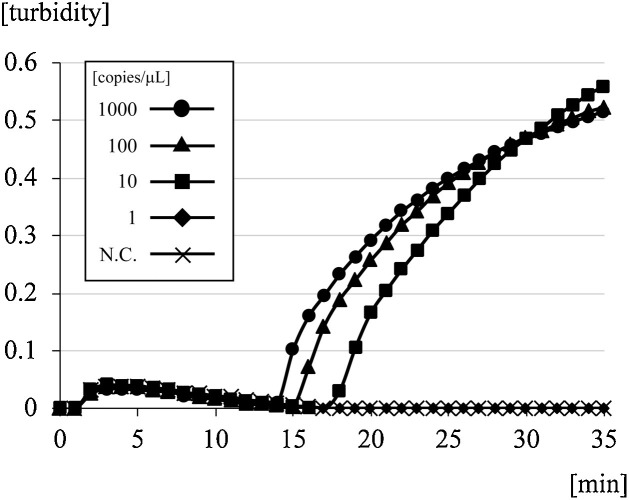
A 10-fold serial dilution of SARS-CoV-2 RNA was amplified to determine the lower limit of detection with the RT-LAMP kit. Fig. 1 shows the results for the detection of real-time turbidity with the LA-200 turbidimeter. The minimum amount of RNA detected was 1.0 × 101 copies/μL, which was achieved within 35 min in this procedure. After measurement, turbidity of the reaction solution could be observed visually under natural light (Fig. 2 ).
Fig. 1.
Detection limit of the RT-LAMP method for SARS-CoV-2 with 10-fold serial dilutions of standard RNA.
Fig. 2.
Visual confirmation of turbidity under natural light after completion of the LAMP reaction.
Photographs of microtubes after the end of the RT-LAMP reaction under natural light. N, negative control; P, positive control.
3.2. Utility of the RT-LAMP kit for clinical specimens
Based on the analysis of conventional RT-qPCR, 30 of the 76 patients were identified as positive for SARS-CoV-2; the median Ct value obtained was 30.85 (interquartile range, 25.31–36.08). Of the 76 patients who underwent conventional RT-qPCR, 32 were positive and 44 were negative by RT-LAMP. As shown in Table 1 , the agreement between RT-qPCR and RT-LAMP was 97.4 % (74/76). Among them, 2 patients were found to be negative with RT-qPCR but positive with RT-LAMP. In the 24 RNA specimens derived from febrile Japanese patients with or without influenza A, no amplification was observed using RT-LAMP.
Table 1.
Correlation between the results of RT-qPCR and Loopamp 2019-nCoV.
|
|
Loopamp 2019-nCoV |
|||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| RT-qPCR | Positive | 30 | 0 | 30 |
| Negative | 2 | 44 | 46 | |
| Total | 32 | 44 | 76 | |
4. Discussion
RT-qPCR is a sensitive and specific nucleic acid amplification method that can be used to diagnose emerging viral infections, including COVID-19. However, it requires trained personnel, expensive equipment, and an extended period of time to generate test results. Conversely, RT-LAMP is extremely convenient to use (the isothermal reaction requires a simple heating device) and produces rapid results (within 30–60 min). In addition to these advantages, the amplification products generated by the RT-LAMP test kit can be detected by turbidity under natural light.
In this study, we found that the detection limit of the RT-LAMP test kit was 1.0 × 101 copies/μL within 35 min with real-time detection of its amplification products. Previous studies using the same molecular diagnostic strategy reported sensitivity ranging from 2.0 × 101 to 1.0 × 102 copies/reaction, indicating that this RT-LAMP test kit has extremely high sensitivity and is also valuable for diagnosis, in terms of not only its convenience but also its detection sensitivity [7,8]. It is highly specific because it uses a set of four primers that recognize at least six different sequences in SARS-CoV-2 RNA. In the present study, it was considered that there was no nonspecific amplification with the RT-LAMP test kit. The results for two samples, re-collected at more than 1 week after the initial SARS-CoV-2-positive result, were inconsistent with those for RT-qPCR (RT-LAMP was positive, whereas RT-qPCR was negative). This phenomenon may depend on the concentration gradient or the aspiration rate in the RNA extracts because the amount of sample used for RT-qPCR was 5 μL, while that used for RT-LAMP was 10 μL.
Our findings support the use of the Loopamp®︎ 2019-SARS-CoV-2 Detection Reagent Kit for the early diagnosis of COVID-19 as a point-of-care test. The main limitations of our study are the small number of samples and the lack of validation of cross-reactivity with other respiratory pathogens.
Author contributions
TM designed the research; YK, YO, RK, KI, JS, and NT performed the research; MM, ST, and SM provided scientific guidance; YK and TM prepared the manuscript.
Funding
This study did not receive funding support.
Declaration of Competing Interest
All authors have no conflicts of interest to declare.
CRediT authorship contribution statement
Yutaro Kitagawa: Data curation, Formal analysis, Writing - original draft. Yuta Orihara: Data curation, Formal analysis. Rieko Kawamura: Data curation, Formal analysis. Kazuo Imai: Methodology, Project administration, Supervision, Writing - review & editing. Jun Sakai: Data curation. Norihito Tarumoto: Supervision, Validation, Writing - review & editing. Masaru Matsuoka: Supervision, Validation. Shinichi Takeuchi: Supervision, Validation. Shigefumi Maesaki: Supervision, Validation. Takuya Maeda: Conceptualization, Data curation, Formal analysis, Writing - review & editing.
Acknowledgment
None.
References
- 1.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Zhou M., Liu F. Exploring the reasons for healthcare workers infected with novel coronavirus disease 2019 (COVID-19) in China. J. Hosp. Infect. 2020 doi: 10.1016/j.jhin.2020.03.002. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehme C.C., Nabeta P., Henastroza G. Operational feasibility of using loop mediated isothermal amplification (LAMP) for the diagnosis of pulmonary TB in microscopy centers of developing countries. J. Clin. Microbiol. 2007;45:1936–1940. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitarai S., Okumura M., Toyota E. Evaluation of a simple loop mediated isothermal amplification test kit for the diagnosis of tuberculosis. Int. J. Tuberc. Lung Dis. 2011;l5:1211–1217. doi: 10.5588/ijtld.10.0629. [DOI] [PubMed] [Google Scholar]
- 6.National Institute of Infectious Diseases, Japan Manual for the Detection of Pathogen 2019-nCoV Ver.2.6. https://www.niid.go.jp/niid/ja/diseases/ka/corona-virus/2019-ncov/2484-idsc/9403-labo-manual.html
- 7.Park G.S., Ku K., Baek S.H. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. J. Mol. Diagn. 2020 doi: 10.1016/j.jmoldx.2020.03.006. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan C., Cui J., Huang L. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.04.001. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]