ABSTRACT
In the present study, a total of 7793 samples from 5 different types of hosts were collected and tested, with a seroprevalence of 2.4% (184/7793). Although the seroprevalence of human and animal brucellosis is relatively low, numbers of human brucellosis cases reported have increased continuously from 2004 to 2018. A total of 118 Brucella strains containing 4 biotypes were obtained, including Brucella melitensis bv.1 (n = 8) and bv.3 (n = 106), Brucella abortus bv.3 (n = 3) and bv.7 (n = 1). Twenty-one shared MLVA-16 genotypes, each composed of 2 to 19 strains obtained from different hosts, suggest the occurrence of a brucellosis outbreak epidemic with multiple source points and laboratory infection events. Moreover, 30 shared MLVA-16 genotypes were observed among 59.6% (68/114) B. melitensis isolates from Zhejiang and strains from other 21 different provinces, especially northern provinces, China. The analysis highlighted the imported nature of the strains from all over the northern provinces with a dominant part from the developed areas of animal husbandry. These data revealed a potential transmission pattern of brucellosis in this region, due to introduced infected sheep leading to a brucellosis outbreak epidemic, and eventually causing multiple laboratory infection events. It is urgent to strengthen the inspection and quarantine of the introduced animals.
KEYWORDS: B. melitensis, B. abortus, MLVA, trace-back, laboratory infection, Zhejiang province
Abbreviations
RBPT: Rose Bengal plate test
SAT: standard agglutination test
PCR: polymerase chain reaction
MLVA: multiple locus variable-number tandem repeat analysis
HGDI: Hunter–Gaston discrimination index
MST: minimum spanning tree
Introduction
Brucellosis is the most common zoonotic disease worldwide, with more than 500,000 new cases being reported annually, and a prevalence rate exceeding 10 cases per 100,000 in some countries [1]. The genus Brucella, a Gram-negative, facultative intracellular bacteria, infects a wide range of mammals, including domestic and wild animals as well as humans [2,3]. The Brucella bacteria infect humans typically through direct contact with infected animals or consumption of unpasteurized and unboiled milk or fresh cheese. Given this dependence on animal reservoirs [4], the veterinarians, farmers, employees of slaughter houses, and meat processing enterprises are the main threatened population [5]. The course of the disease may be acute, sub-acute, or chronic, and lead to serious damage to the physical and mental health of patients, mainly clinical syndrome, including high fever, night sweat, fatigue, joint pain, and headache [6]. Due to lack of proper treatments and reliable diagnosis methods, brucellosis seriously threatens the human health and is a global public health concern [7].
During 2004–2016, a total of 448,479 cases of brucellosis were confirmed in China, depicting a growing trend for an epidemic across all provinces [8]. Zhejiang Province, known as the “land of fish and rice”, is located on the southeast coast of China. Brucella abortus Brucellosis was prevalent in limited regions in the 1950s, due to imported cow from other regions. In 1963, the frequency of positive cows of a dairy farm in Hangzhou city was 22.28%. Subsequently, a comprehensive control strategy was launched, including surveillance quarantine, culling the infected herd, and environmental disinfection. The disease has been effectively controlled, and no human or animal brucellosis was reported until mid-1995s. However, sporadic human brucellosis was observed after the 2000s. With the increase of the livestock and the introduction of live sheep, the prevalence of brucellosis in livestock is increasing gradually, and human brucellosis exhibited a gradually increasing trend after 2005 [9]. Serology surveillance of human and animal brucellosis, identification and genotyping of the circulating Brucella species are crucial for the prevention and control of the disease [10,11]. Therefore, detailed knowledge of the epidemiological situation has become very important for the assessment of effective prevention and risk factors of public health. The purpose of this research is to better understand the zoonosis situation of Brucella in livestock and the human population. A systematic investigation in Brucellosis seroprevalence in humans and animals was performed from 2005 to 2015 in Zhejiang Province, China. To further illuminate the molecular characteristics of circulating Brucella, serology testing, bacterial isolation, and multiple locus variable-number tandem repeat analysis (MLVA) were also performed.
Methods
Data source
The human reported data in this study were extracted from medical cases occurring during 2004–2018. The numbers of human brucellosis cases obtained from CDC of the people in China must be reported to the Chinese CDC through the National Notifiable Disease Surveillance System. The reported human cases must be accompanied by clinical signs and confirmed by serology test (RBPT and SAT) or the isolation of the organism, in accordance with the case definition of the world health organization [12].
Sample collection and testing
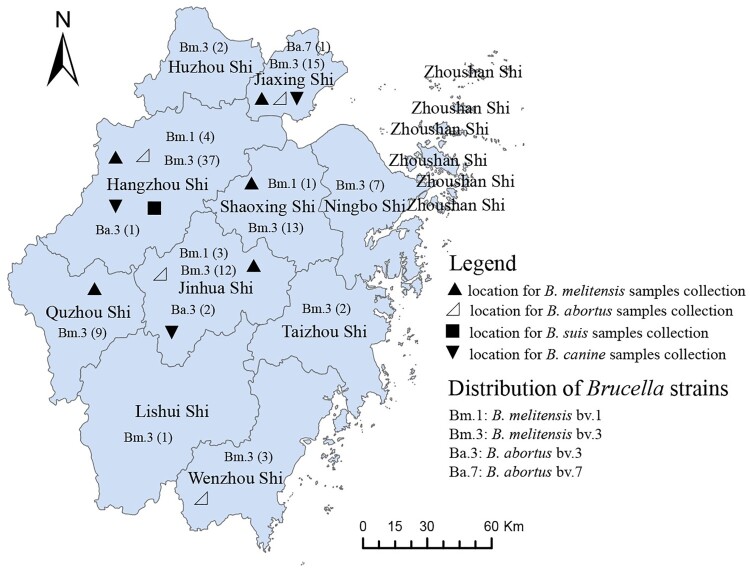
Serum samples and animal products were collected from humans and livestock in the Zhejiang Province, China, from 2006 to 2015. A total of 7793 samples, including humans (2656), sheep (4352), pigs (66), and canines (617), and 102 milk samples were collected from Zhejiang Province, China, of which 1204 were serum samples (292 in breeders, 605 in butchers, and 307 in salesmen) from humans with contact with sheep in seven regions (see Supplementary table 1), 4352 were serum samples of sheep from three regions (see Supplementary table 2), 740 were serum samples (547 in breeder, 42 in milkman, 64 in veterinary of cattle farm, and 87 in salesman) from humans in contact with cattle from four areas (see Supplementary table 3), 624 samples were from canine breeders, and 617 from canines. Eighty-eight serum samples were from pig breeders, and 66 from pigs were also collected (see Supplementary table 4). Brucella spp. strain isolation was performed in serum positive samples using a bacteriology method. Moreover, 274 animal products were collected and detected by the bacteriology procedure, including sheep liver, spleen, porcine blood, canine blood, canine liver, duck meat, chicken, and rats from food markets of Zhejiang (see Supplementary table 5). All samples were randomly collected, and the locations of the samples collected are shown in Figure 1. All serum samples were transported to the laboratory, where they were stored at −20°C until processing and detection by the Rose Bengal Plate Test (RBPT). The positive serum samples were reconfirmed by a Standard Agglutination Test (SAT) [13]. Subsequently, bacteriology experiments were performed [14]. All the reagents were purchased from the China Institute of Veterinary Drug Control, where the National Reference Laboratory for Animal Brucellosis is located.
Figure 1.
Location for the samples collection and distribution of Brucella strains in Zhejiang Province (note: the map does not represent the true borders of administrative regions of Zhejiang, China).
Isolation of Brucella strains
To study the molecular epidemiological characters, all samples with SAT positive were collected and subjected to a brucellosis purification process. The samples were cultured on Brucella serum dextrose agar composed of Brucella medium base (supplemented with Brucella selective antibiotic, OXOID, England) and 5–10% heat-inactivated horse serum. The plates were incubated with and without 5–10% carbon dioxide at 37°C after inoculation with sample materials. The plates were examined after 3–30 days for bacterial growth. A single clone was chosen for identification.
Bacterial strains and convention identification
A total of 118 isolates were isolated from Zhejiang Province from 2006 to 2015. These strains were isolated from human blood, dog, and animal products at the first line laboratory by the Hangzhou Center for Infectious Disease Control and Prevention. One hundred and ten samples were recovered from human blood, 6 from Hu sheep, 3 from sheep, 2 from goat, and 1 from dog. Forty-four strains were isolated from positive surveillance, and others were obtained from clinic brucellosis cases. The examined isolates were identified as Brucella species on the basis of morphology and conventional identification methods according to standard biotyping procedures, including requirement of CO2 for growth, H2S production, sensitivity to thionin (10 and 20 μg/ml), basic fuchsin (20 μg/ml), and agglutination with monospecific antiserum for A and M antigens and phage lysis test (Tbilisi, Tb; Berkeley, Bk2; Weybridge; Wb) [14,15]. B. melitensis 16M (BM), B. abortus 544 (BA), and B. suis 1330 (BS) were used as control strains. DNA was isolated using full-automatic nucleic acid extraction system (LLXBIO China Ltd, China) extraction from 48-h cultures according to the manufacturer's instructions. DNA extracted from all isolates was stored at −20°C. Subsequently, all strains were further identified by BCSP31-PCR [16] and AMOS-PCR [17].
MLVA-16 genotyping
All of isolates were further examined by MLVA, genotyping schedule, and a PCR amplification process as described previously [18,19]. The PCR products were preliminarily evaluated by 2% or 3% agarose gel electrophoresis. Then, positive products were denatured and resolved by capillary electrophoresis on an ABI Prism 3130 automated fluorescent capillary DNA sequencer (Applied Biosystems). Fragments were sized following comparison with a ROX (carboxy-X-rhodamine)-labelled molecular ladder (MapMaker 1000; Bioventures Inc., Murfreesboro, TN, USA) and Gene Mapper software version 4.0 (Applied Biosystems). The fragment sizes were converted to repeat unit numbers using a published allele numbering system [20]. B. melitensis bv. 1 16 M was used as control strain to calibrate the VNTR units.
Analysis of genotyping data
Statistical analysis
Throughout the process, Microsoft Excel (Microsoft, Redmond, CA, US) was used for data cleaning. Statistical analysis was performed with SPSS 19.0 (Chicago, IL, USA), P-values <0.05 were considered to be statistically significant. Hunter and Gaston diversity index (HGDI) for loci and MLVA panels were calculated to describe discriminatory capacity of each locus [21]. MLVA data were analysed using BioNumerics version 5.1 software (Applied Maths, Belgium). Both categorical coefficient and un-weighted pair group methods with arithmetic mean algorithm (UPGMA) were applied to MLVA clustering analysis (see Table S1). The resultant genotypes were compared using the online Brucella 2016 MLVAbank. Minimum spanning tree (MST) based on complete MLVA-16 was used to investigate molecular relationships between strains in this study and 1 344 B. melitensis isolates (see Table S2) from other provinces of China (MLVAbank Brucella_4_5) (http://microbesgenotyping.i2bc.paris-saclay.fr/databases/view/1156/).
Results
Seroprevalence characteristics of human and animal brucellosis
The incidence of Human brucellosis in Zhejiang Province increased continuously from 2004 to 2018. The incidence of Human brucellosis in 2002 was 0.0043/100,000, increasing to 0.263/100,000 in 2017, i.e. a value is 60 times higher than that in 2002 (see Supplementary figure 1). The seroprevalence of human brucellosis after contact with sheep differed significantly across the seven districts (χ2 = 28.756, P < 0.05), the range of the prevalence of human contact with sheep is 0–100% in the seven regions examined, the average seroprevalence is 5.2% (63/1,204), and 44 Brucella strains were obtained from these patients, 22 (50%) of which were from butchers; 20 strains were from Hangzhou, 11 from Tongxiang, 5 from Shangyu city, 3 from the Nanhu district, and 5 from Longyou county (see Supplementary table 1). A total of 4352 blood samples of sheep were tested in the 3 regions examined, and the average positive rate was 1.7% (72/4352). Ten Brucella strains were isolated from these samples, of which 7 were obtained from Tongxiang city, and the other 3 from Longyou county (see Supplementary table 2). Seven hundred and forty blood samples from humans contacting the infection from cows in 4 districts were collected and tested. The range of positive rate was 0–11.2%, the average serum positive rate was 5.8% (43/740). Seroprevalence of human brucellosis contracted from cows differed significantly across the four districts (χ2 = 35.921, P < 0.05) and the highest positive rate was observed in the Jindong district (see Supplementary table 3). A total of 624 blood samples of dog owners were tested, and only 1 positive sample was found, and one Brucella was isolated from this sample (Published) [22]. Meanwhile, 617 canine blood samples were detected, with a serum positive rate of 0.5% (3/617), and no Brucella strain was found (see Supplementary table 4). Moreover, 88 blood samples of pig farmers and 66 blood samples from pigs were collected and tested, which were all negative. A total of 102 raw milk samples were detected, and 2 samples were positive, but no strains were found (see Supplementary table 5). Finally, bacteriology tests were performed in 274 animal product samples, and only one Brucella strain was isolated from a liver sample of canine (see Supplementary table 5).
Characteristics and distributions of isolates
A total of 118 Brucella spp. strains were isolated in this study during 2005–2015, among which 118 strains exhibited a convex, circular, and translucent morphology profile. The growth characteristics, phage lysis experiments, dye bacteriostatic tests, and slide agglutination with monospecific anti-Brucella sera were used to characterize all isolates (Table 1). Species and biovars were discriminated based on standard bacteriological procedures. Finally, biotyping identified 114 strains as B. melitensis (biovar 3 (n = 106), biovar 1 (n = 8)) and 4 were B. abortus (biovar 3 (n = 3) and biovar 7 (n = 1)). A total of 106 strains were isolated from human blood, 6 in Hu Sheep, 3 in sheep, 2 in goats, and 1 in dogs. Except for Zhoushan city, these strains were widely distributed in all 10 other regions of Zhejiang Province (Figure 1), 42 of which were in Hangzhou, 17 in Jinhua city, 16 in Jiaxing city, and 14 in Shaoxing city.
Table 1. Biotyping characteristics of Brucella species isolates in Zhejiang, China.
| Strain No. | Growth characteristics | Monospecific Sera | Phages lysis testing | Interpreted | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CO2 requested | H2S | BF | TH | A | M | R | Tb | BK2 | Wb | ||
| BA | + | + | + | − | + | − | − | CL | CL | CL | B. abortus 544 |
| BM | − | − | + | + | − | + | − | NL | CL | NL | B. melitensis16M |
| BS | − | ++ | − | + | + | − | − | NL | CL | CL | B. suis 1330 |
| 8 | − | − | + | + | − | + | − | NL | CL | NL | B. melitensis bv. 1 |
| 106 | − | − | + | + | + | + | − | NL | CL | NL | B. melitensis bv. 3 |
| 3 | − | − | + | + | + | − | − | CL | CL | CL | B. abortus bv. 3 |
| 1 | − | − | + | + | + | + | − | CL | CL | CL | B. abortus bv. 7 |
Description of data: Strain No., the number conferred to isolates; BF, Basic fuchsin at 20 μg/ml (1/50,000 w/v); TH, Thionin at 20 μg/ml (1/50,000 w/v); Phages, Tb = Tbilisi, BK2 = Berkeley type 2, Wb = Weybridge; CL, Confluent Lysis; NL, No lysis; RTD, Routine test dilution; +, positive (serum agglutination positive); –, negative (serum agglutination negative).
MLVA genotyping characteristic of Brucella isolates
In B. melitensis strains, the HGDI value of three loci was >0.7, and the HGDI value of eight loci was <0.1191; the other five loci showed no diversity (HDGI = 0.0000). Moreover, the HGDI value of Panel1, MLVA-11, and MLVA-16 was 0.1667, 0.2141, and 0.9640, respectively (Table 2). In B. abortus strains, HGDI of four Panel 2B loci was >0.50, 0.6667 in bruce30, 0.8333 in bruce09, and 0.5000 in bruce55 and bruce18. The other 10 loci showed no diversity (HDGI = 0.0000), the HGDI values of Panel1, MLVA-11, and MLVA-16 were 0.5000, 0.5000, and 0.8333, respectively (Table 2).
Table 2. HGDI values of 114 B. melitensis isolates and 4 B. abortus isolates.
| Panels | B. melitensis strains (n = 114) | B. abortus strains (n = 4) | ||
|---|---|---|---|---|
| Panel 1 | Locus | HGDIa | Locus | HGDIa |
| bruce06 | 0.0000 | bruce06 | 0.0000 | |
| bruce08 | 0.0175 | bruce08 | 0.0000 | |
| bruce11 | 0.0000 | bruce11 | 0.0000 | |
| bruce12 | 0.0000 | bruce12 | 0.0000 | |
| bruce42 | 0.0846 | bruce42 | 0.0000 | |
| bruce43 | 0.0348 | bruce43 | 0.0000 | |
| bruce45 | 0.0175 | bruce45 | 0.0000 | |
| bruce55 | 0.0175 | bruce55 | 0.5000 | |
| Panel 2A | bruce18 | 0.0000 | bruce18 | 0.5000 |
| bruce19 | 0.0520 | bruce19 | 0.0000 | |
| bruce21 | 0.0000 | bruce21 | 0.0000 | |
| Panel 2B | bruce04 | 0.7316 | bruce04 | 0.5000 |
| bruce07 | 0.0846 | bruce07 | 0.5000 | |
| bruce09 | 0.1191 | bruce09 | 0.8333 | |
| bruce16 | 0.7576 | bruce16 | 0.0000 | |
| bruce30 | 0.7087 | bruce30 | 0.6667 | |
| MLVA | Panel1 | 0.1667 | Panel1 | 0.5000 |
| MLVA-11 | 0.2141 | MLVA-11 | 0.5000 | |
| MLVA-16 | 0.9640 | MLVA-16 | 0.8333 | |
aHGDI: Hunter Gaston Diversity Index, which measures the variation of the number of repeats at each locus and ranges from 0.0 (no diversity) to 1.0 (complete diversity).
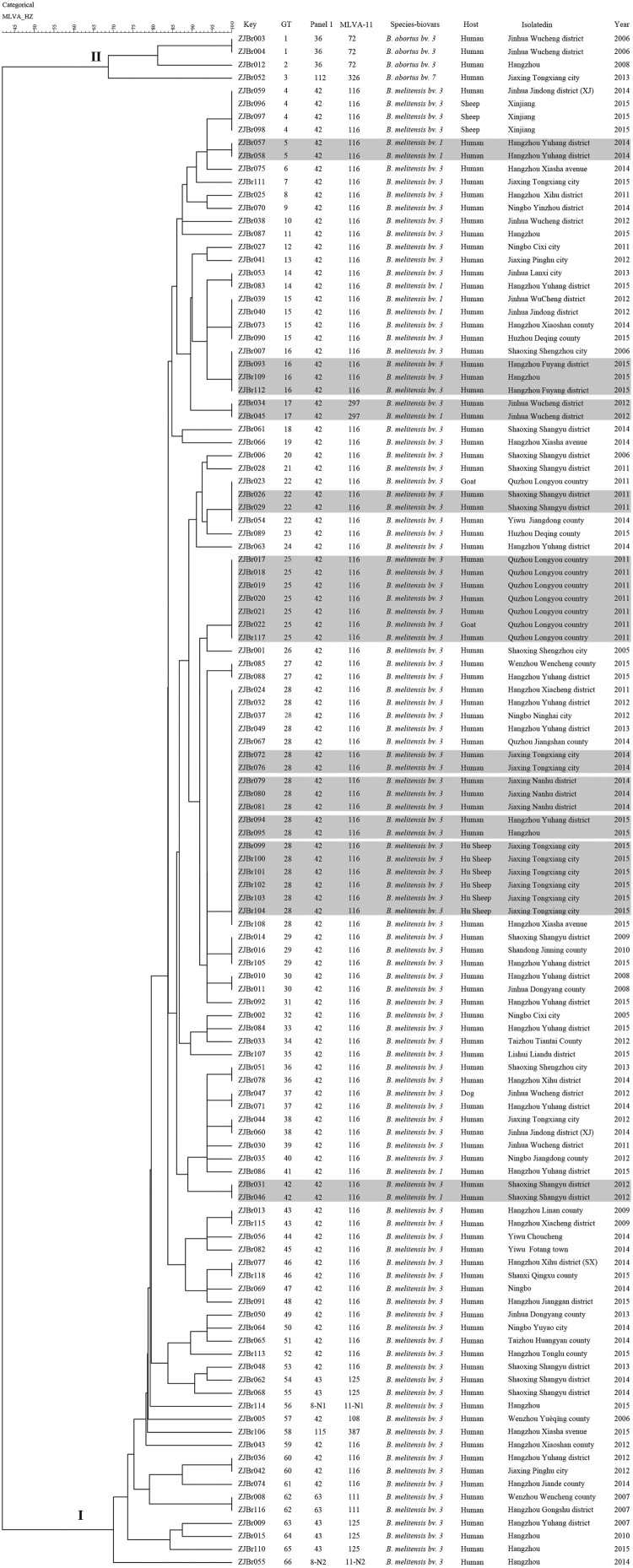
Based on the MLVA-16 assay, the 118 strains analysed were sorted into two groups (I and II), four B. abortus strains were clustered into group II and formed three MLVA-16 genotypes (GT1–3); the other 114 B. melitensis strains were clustered into group I and formed 63 MLVA-16 genotypes (GT4–66) with 70% similarity, of which 20 were shared genotypes that comprised 2–19 strains, and the remaining 43 genotypes were single genotypes, each genotype representing an independent strain (Figure 2). Using panel 1 markers, the present population was clustered into eight panel 1 genotypes, two of them in B. abortus strains (36 (1–4–5–3–12–2–2–3–1; N = 3) and 112 (3–4–5–3–12–2–2–3–3; N = 1)) and six in B. melitensis strains,42 (1–5–3–13–2–2–3–2; N = 104), 43 (1–5–3–13–3–2–3–2; N = 5), 63 (1–5–3–13–2–3–3–2; N = 2), 115 (1–4–3–13–2–2–3–2; N = 1), N1 (1–5–3–13–2–2–2–2; N = 1), N2 (1–5–3–13–2–2–3–1; N = 1), and 88.1% (104/118) of isolates belonging to genotype 42 and this panel 1 genotype plays a dominant role in B. melitensis (Figure 2). However, 10 MLVA-11 genotypes were identified in these populations, which of 2 MLVA-11 genotypes (72 (N = 3) and 326 (N = 1)) in B. abortus strains, the remaining 8 MLVA-11 genotypes in the B. melitensis population, including in 116 (N = 101), 125 (N = 5), 297 (N = 2), 111 (N = 2), 108 (N = 1), 387 (N = 1), N1 (N = 1), and N2 (N = 1), and MLVA-11 genotype 116 accounts for 88.6% (101/114) and is an overwhelmingly predominant population (Figure 2). Subsequently, comparison of these genotypes with MLVAbank suggested that all B. melitensis strains of this study belong to the East Mediterranean lineage.
Figure 2.
Dendrogram based on the MLVA-16 genotyping assay (UPGMA method), showing the relationships between the 118 Brucella isolates. The columns show the identification numbers, MLVA-16 genotypes (GT), panel 1 genotypes, MLVA-11 (panels 1 and 2A) genotypes, species-biovar, host, and the year of isolation of the strains.
Genetic relationship analysis of Brucella abortus strains
GT1 contained two strains (ZJBr003 and ZJBr004) obtained from two patients from the same farm. The strain had similar MLVA-16 genotypes with strain (2013Jiang#084), which had been collected from Xinjiang, China. ZJBr052 (GT3) was obtained from a patient who was a milkman in a cow farm, and this strain had a genotype similar to that of a strain from Inner Mongolia (2013Jiang#127) (MLVAbank), with three different loci observed among the two strains (see Table S3). ZJBr012 (GT2), which was isolated from a patient who was engaged in beef wholesale for a long time, had a genotype identical to strains (2013Jiang#093) (MLVAbank) from Chongqing, China (see Table S3).
Investigation of outbreak and laboratory infection of B. melitensis strains
Based on MLVA-16, seven shared (GT5, 16, 17, 22, 25, 28, and 42) (Figure 2, genotypes with black shadow) genotypes were from strains found in the same location and period (Figure 2). GT4 contained four B. melitensis strains, of which ZJBr059 was isolated from the blood of a human in Hangzhou who worked at sheep farm in Xinjiang Province before onset (Figure 2). GT25 was shared by seven strains, of which ZJBr022 was obtained from a goat in a farm of Longyou county, where an animal brucellosis outbreak had occurred that was caused by goats introduced from Northern China. The other five strains (ZJBr017–021) were from five farm employees, and the remaining strain (ZJBr117) was isolated from the staff of a microbiology laboratory of a hospital in this region. Interestingly, ZJBr017, which was obtained from one of farm worker, was identified as Brucella by this staff (Figure 2). GT28 was shared by 19 strains, of which 4 strains (ZJBr076, ZJBr079–ZJBr081) were isolated from four workers in a sheep farm in Jiaxing Tongxiang country, 6 strains (ZJBr099–ZJBr104) were obtained from Hu sheep in this farm, and the remaining 9 strains were isolated from a different region (Figure 2). GT29 was shared by three strains, of which ZJBr016 was isolated from a truck driver who had been trafficking sheep from Shandong Jining county to Hangzhou city many times. The other two strains were from Shaoxing and Hangzhou city (Figure 2). Moreover, GT37 comprised two strains, ZJBr047 and ZJBr071; the former was isolated from dog often fed with sheep offal and the latter from a human (Figure 2). GT46 included ZJBr077 and ZJBr11. ZJBr077 was isolated from a child from Hangzhou city who had drunk raw ewe's milk in grandma's home in Shanxi Qingxu county. ZJBr118 was obtained from a patient in Shanxi Qingxu county (Figure 2). GT43 and GT62 contained two strains each, of which ZJBr013 and ZJBr008 were isolated from the blood sample of two different patients, but ZJBr115 and ZJBr116 were obtained from two clinical laboratory staff members from two different hospitals in Hangzhou city. The two staff members had identified strains ZJBr013 and ZJBr008 without using biosafety protection facilities (Figure 2).
Molecular epidemiological investigation of 1, 344 Chinese B. melitensis strains
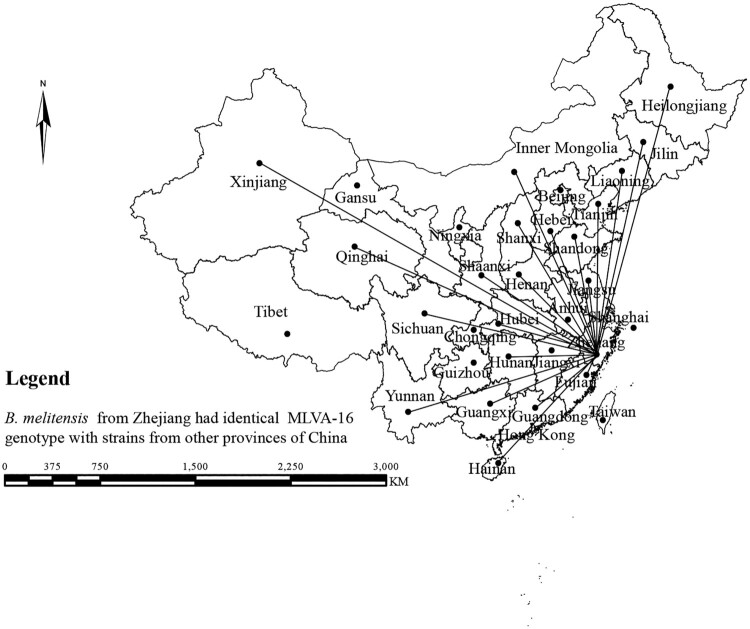
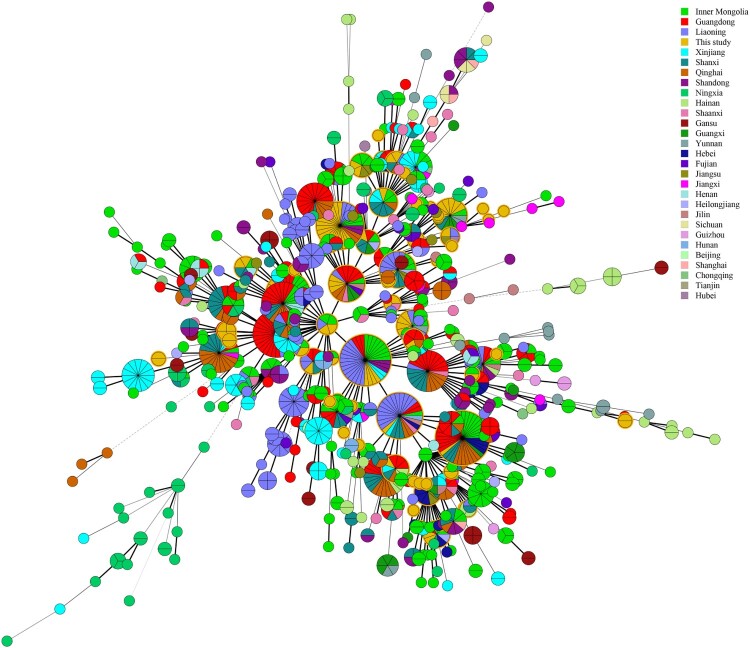
In this study, the MLVA-16 assay was used to investigate molecular relationships between this study's B. melitensis isolates and 1344 B. melitensis isolates from other provinces of China. Thirty shared genotypes were observed in this population (see Table S4). B. melitensis isolates from Zhejiang Province had genotypes identical to those of strains from 21 different provinces (Figure 3), including the Inner Mongolia, Xinjiang, and Qinghai, Shanxi, Heilongjiang, Jilin, Liaoning, Shaanxi, Henan, Hebei, Tianjin, Jiangsu, and Hunan Provinces, i.e. 59.6% (68/114) strains from this study shared MLVA-16 genotypes with strains from these provinces, especially strains from northern provinces (Figure 4). GT28 contained 11 strains from a brucellosis outbreak in a farm. These strains had identical genotype with strains from Inner Mongolia, Liaoning, Qinghai, and Shandong (see Table S4). GT37 comprised two strains from human and dog, which had a genotype identical to that of strains from Shaanxi (see Table S4). GT43 contained two strains from a patient and a laboratorian who had a genotype identical to strains from Inner Mongolia and Jiangsu Province (see Table S4). GT46 contained two strains obtained from a suspect infection event caused by raw ewe's milk. These strains have a genotype identical to that of strains from many northern provinces, including Liaoning, Qinghai, Shanxi, and Tianjin Provinces (see Table S4).
Figure 3.
The strain from this study has an MLVA-16 genotype identical to that of strains from 21 different provinces in China (note: the map does not represent the true borders of the administrative regions of China).
Figure 4.
Minimum spanning tree based on MLVA-16 data for Brucella melitensis in China (the strains in this study are indicated with yellow circles).
Discussion
Brucellosis is a common zoonotic disease of a public health menace that is still endemic in many countries and regions, including Zhejiang Province, China [23]. In this study, the seroprevalence of animal and human brucellosis was investigated in many regions, and we performed a phenotypic and molecular characterization of Brucella isolates from sheep, Hu sheep, goats, and human samples obtained from different geographical locations of Zhejiang Province. Human brucellosis case reports were rare in Zhejiang Province before the 2000s; however, the incidence rate of human brucellosis has increased continuously during 2004–2018; notably, the incidence rate in 2017 was 60 times higher than in 2002. The epidemic trend of human brucellosis in this region coincides with other southern provinces [9]. The main reason is that people are starting to promote livestock activities, especially sheep breeding in these regions. However, the livestock bred in these regions were introduced almost exclusively from Northern China, where animal husbandry has developed, and a prevalence of more than 90% brucellosis has been reported [24]. The rising incidence rate in this region could have a potential correlation to the trans-boundary transfer of infected animals from Northern China [25].
The average serum positive rate in humans contacting with sheep is 5.2% (63/1204), which is higher than the positive rate in sheep (1.7% (72/4352)). Interestingly, of the 44 Brucella strains obtained from these patients, 22 (around 50%) were from butchers, which suggests that butchers are a at high risk of infection. The Zhejiang Province is an economically developed region and numbers of livestock (small ruminate) is less than northern, China, and there is a great demand for mutton consumption. Thus thousands of live sheep have been introduced and slaughtered, and this is a reasonable explanation for the observation that seroprevalence in humans (butchers) contacting with sheep is higher than in sheep. The zoonotic risk of brucellosis posed by Brucella-infected slaughtered animals to abattoir workers cannot be ignored in emerging or non-epidemic regions of brucellosis [26]. Moreover, the highest positive rate (28.6%) of sheep brucellosis was found in a farm of Longyou county, and a scene survey showed that 200 Boer goats from Shandong Province had been introduced in this farm. The introduction of infected animals is a main risk factor for the outbreak of brucellosis in sheep herds [27].
The average serum positive rate of human infections contracted from cow was 5.8% (43/740), and the highest positive rate in a human population was observed in the Jindong district, home of the largest dairy farming base of Southern China and a district in which the dairy industry is the main economic source in the rural areas. This conclusion is in agreement with studies reporting that the seroprevalence of dairy cattle brucellosis in Southern China reached 5.5%; however, in Northern China, where the traditional agropastoral areas with the most developed animal breeding industry are located, the dairy cattle seroprevalence was >10% [28]. Our study showed that the serum positive rate of dog owners and dogs in this region was ≤ 0.5%. Although B. canis has limited epidemiological significance for the human populations, it remains a significant threat to the canine breeding industry and to humans who come into close contact with dogs [22, 29]. Moreover, no positive samples were observed in pig farmers and pigs, suggesting that the prevalence of brucellosis in pigs is low, but further investigation is needed. Moreover, not only there were few positive samples found in raw milk and animal products, but also one Brucella strain was isolated from a liver sample of canine (that often ate sheep viscus), which is a kind of food in local communities. These results revealed that consuming unpasteurized milk and unproperly cooked animal products is a significant public health concern in this region [30].
In the present study, a total of 118 Brucella spp. strains were isolated, of which 55 strains were obtained from active surveillance, and the remaining 63 strains were isolated from diagnosis of suspect patients. Brucella strains were widely distributed in 10 regions (a total of 11 cities), at least 4 species/biovars were found in human and animal hosts, and B. melitensis bv.3 was the predominant species. These data indicated that brucellosis in this province is becoming a serious health problem. Both B. abortus bv. 3 and bv.7 were first isolated from human contact with cow and beef in China. Although these strains cause infections less severe than those caused by B. melitensis or B. suis [31,32], they remain an important public health threat to human populations.
MLVA-16 assay displayed high resolution to 114 B. melitensis strains, with HGDI value of 0.9640, of which the three loci (bruce04, bruce16, and bruce30) from panel 2B were the most useful for genotyping analysis of isolates from Zhejiang. Based on panel1, two dominated genotypes in B. abortus and B. melitensis, 36 and 42, are common in Northern China [25]. Subsequently, four B. abortus strains were divided into two MLVA-11 genotypes (72 (N = 3) and 326 (N = 1)) revealed that the B. abortus strains in this study are abortus C group descent [33], which shares the geographic origin with strains from Chongqing, Xinjiang, and Inner Mongolia. All B. melitensis were clustered with eight MLVA-11 genotypes and belong to the East Mediterranean lineage, of which the MLVA-11 genotype 116 is the overwhelmingly predominant population. Strains belonging to this genotype have an important epidemiology for the human population [34]; these data are in agreement with serious situation of brucellosis in this province.
Based on the comparison of genetic similarly by MLVA-16, three B. abortus strains in this study had a MLVA-16 genotype similar to that of strains from developed areas of animal husbandry of Northern China, including Xinjiang and Inner Mongolia. Notably, ZJBr012 (GT2), found in this study, had MLVA-16 genotype completely matching that of a strain from Chongqing, China, historically an epidemic area of B. abortus brucellosis [32], suggesting that there are potentially molecular epidemiology links between B. abortus strains from this study and strains from Chongqing, Xinjiang, and Inner Mongolia, which may be the origin of the strains we detected. Certainly, genome analysis in these strains is essential to confirm their origin.
Based on MLVA-16 cluster analysis, seven shared genotypes (GT5, 16, 17, 22, 25, 28, and 42) (Figure 2, genotypes with black shadow) consisted of strains from the same location and period, suggesting the occurrence of a multipoint outbreak epidemic from multiple common sources [20]. GT4 contained four B. melitensis strains, of which ZJBr059 was isolated from the blood of a human in Hangzhou who worked at a sheep farm in Xinjiang Province before onset, and other three strains were from sheep in this farm, probably the source of infection of this patient from Xinjiang. GT25 was composed of seven strains, of which ZJBr022 was obtained from a goat from a goat farm in Longyou county, where an animal brucellosis outbreak had occurred that was introduced by goats from Northern China. The other five strains (ZJBr017–021) were from workers of this farm, and the remaining strain (ZJBr117) was isolated from staff of microbiology laboratory of hospital in this region. Interestingly, ZJBr017 was obtained from a farmer that was identified by this staff as being infected by Brucella. These data confirmed that the introduced goats from Northern provinces led to the brucellosis outbreak of Longyou county farm, and then caused laboratory infection events [35,36]. GT28 was shared by 19 strains, of which 4 strains (ZJBr076, ZJBr079–ZJBr081) were isolated from workers in a sheep farm in Jiaxing Tongxiang country, 6 strains (ZJBr099–ZJBr104) were obtained from Hu sheep in this farm, and the remaining 9 strains were isolated from a different region, suggesting an outbreak of animal and human brucellosis had started from a common source of infection [37]. Interestingly, strains from GT 28 had a genotype identical to that of strains from Inner Mongolia, Liaoning, Qinghai, and Shandong, suggesting a common source of infection outbreak in the farms from these provinces. This conclusion coincides with a field epidemiology survey, as it was found that more than 200 goats from northern provinces had been introduced into this farm. GT29 was composed of three strains, of which ZJBr016 was isolated from a truck driver who was trafficking sheep from Shandong Jining county to Hangzhou city, and the other two strains from Shaoxing and Hangzhou city, suggesting that Jining of Shandong Province is a potential source of infection for these cases. Moreover, GT37 consisted of two strains ZJBr047 and ZJBr071; the former was isolated from dog that was often fed with sheep offal and the latter from humans, suggesting that the two cases shared the source of infection [38]. However, strains from GT37 had a genotype identical to that of strains from Shaanxi, where they may have originated.
GT46 was composed of two strains, ZJBr077 and ZJBr118. ZJBr077 was isolated from a child in Hangzhou city that drank raw ewe's milk in grandma's home in Shanxi Qingxu county, whereas ZJBr118 was obtained from a patient in Shanxi Qingxu county. This result suggests that the source of infection of the child brucellosis was from Shanxi, a region where brucellosis is epidemic [39]. Indeed, strains from GT46 had a genotype identical to that of strains from many northern Provinces, including Liaoning, Qinghai, Shanxi, and Tianjin.
GT43 and GT62 contained two strains each and shared a genotype identical to that of MLVA-16. Two laboratory workers identified strains without using biosafety protection facilities, so these data indicated laboratory infection events [40,41]. Surprisingly, GT43 contained two strains, ZJBr013 and ZJBr115, which were obtained from a patient and laboratorian, respectively, and had the same genotype of strains from Inner Mongolia and Jiangsu Province, suggesting that the source of infection of these events maybe from Inner Mongolia [40, 42]. Because brucellosis is uncommon in Zhejiang Province and patients often present with nonspecific signs and symptoms, clinicians may not suspect brucellosis [43]. Brucellosis is one of the most commonly reported laboratory-acquired infections. The organism is easily aerosolized and has a low infectious dose [35, 44]. Also, laboratory workers may not be familiar with the strains, which can lead to exposures to Brucella spp. in clinical laboratories during culturing and isolation of clinical specimens. Working in microbiology laboratories and a lack of compliance with personal protective equipment and biosafety cabinets were the independent risk factors for the development of laboratory-acquired brucellosis. Increased adherence to personal protective equipment and use of biosafety cabinets should be priority targets to prevent laboratory-acquired brucellosis [45].
The 43 single MLVA-16 genotypes suggest that more than 36% (43/118) brucellosis cases are epidemiologically unrelated or sporadic. Moreover, 30 shared MLVA-16 genotypes were observed among 59.6% (68/114) B. melitensis isolates from Zhejiang and strains from 21 different provinces, especially northern provinces, China. The analysis highlighted the imported nature of the strains from all over the northern provinces, with a dominant part from the developed areas of animal husbandry. Most imported cases were associated with importation of infected animals. Travel or consumption of unpasteurized dairy products in endemic countries also occurred [46]. We consider that the Zhejiang Province imported animals from regions where brucellosis is epidemic, causing a communication chain leading to a serious brucellosis epidemic and laboratory infection events.
Our study has some limitations, such as the variability in the sample number of serum and strains collected among different regions, and the limited number of strains from animals that were analysed. Whole Genome Sequencing for tracing the geographical origin of the imported cases of human brucellosis is warranted.
Conclusion
In the present study, 7793 serum samples were tested to anti-Brucella, and we found a seropositive rate of 2.4%. A total of 118 Brucella spp. derived from humans and animals in Zhejiang from 2005 to 2015 were characterized by classical biotyping and MLVA. Although the numbers of human brucellosis cases reported have increased continuously from 2004 to 2018, the seroprevalence of human and animal brucellosis in this province is relatively low. Classical biotyping revealed the presence of at least four species and biovars. B. melitensis was obtained from dogs, sheep blood (food), and laboratory staff members, suggesting that B. melitensis is a crucial threat to the occupational population. The B. abortus bv.3 and bv.7 were first isolated from humans, and these strains had molecular epidemiology links with strains from Chongqing, Xinjiang, and Inner Mongolia. Circulating isolates were mainly B. melitensis, most of them belonging to biovar 3, which is the most abundant biovar in the Mediterranean region. MLVA genotyping confirmed the occurrence of multiple point outbreaks of human brucellosis and laboratory infection events caused by strains imported from Northern China. Our research provides a model for surveillance and control of animals and human brucellosis in other southern regions.
Declarations
Ethical approval and consent to participate
This study involves the investigation and isolation of Brucella spp. using serology, bacteriology, and molecular typing methods. All experiments in the study were approved by the Ethics Committee of the Hangzhou Center for Disease Control and Prevention, and were conducted strictly according to the guidelines. All patients gave written informed consent before participation and all animal samples used in this study were approved by the scientific ethical committee of our institute.
Consent to publish
Not applicable.
Availability of data and materials
All data generated or analysed during this study are included in this published article, and its supplementary information files will be freely available to any scientist wishing to use them for non-commercial purposes upon request via e-mail with the corresponding author.
Supplementary Material
Acknowledgments
We are grateful to all the staff in the Branch of Brucellosis Control and Prevention, both Zhejiang Province and Hangzhou Centers for Disease Control (CDC) and Prevention for their assistance in patient screening and patient epidemiology data collection. We are grateful to Bu-yun Cui, Hai Jiang, Dong-ri Piao, and Hong-yan Zhao from the China CDC for experimental guidance.
Funding Statement
This study was supported by Hangzhou Science and Technology Development Program (grant number 20160533B49 and 20140633B23), Hangzhou Health Science and Technology Program (grant number 2013A53), National Key R&D Program of China (grant number 2019YFC1200705), the China Special Grant for the Prevention and Control of Infectious Diseases (grant number 2018ZX10734401 and 2018ZX10734404), and the Nature Science Fund of Inner Mongolia Autonomous Region (No. 2018MS08004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authors’ contributions
WH and ZSJ performed most of the strain isolations and MLVA typing, WJ, YY, TY, JHY, ZKJ, SFY, TZL, and DF were in charge of the serology investigation in humans and animals. LZG performed the MLVA cluster analysis and drafted the manuscript; WH and XWM were in charge of the strain biotyping; ZHF and JNM prepared the DNA samples; LZG and LZJ participated in the design of the study and critically reviewed the manuscript; DH participated in the design of the study and also managed the project. All authors read and approved the final manuscript.
References
- 1.Pappas G, Papadimitriou P, Akritidis N, et al. The new global map of human brucellosis. Lancet Infect Dis. 2006;6:91–99. doi: 10.1016/S1473-3099(06)70382-6 [DOI] [PubMed] [Google Scholar]
- 2.Aghaali M, Mohebi S, Heydari H.. Prevalence of asymptomatic brucellosis in children 7 to 12 years old. Interdiscip Perspect Infect Dis. 2015;2015:187369. doi: 10.1155/2015/187369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira AC, et al. MLVA16 typing of Portuguese human and animal Brucella melitensis and Brucella abortus isolates. PLoS One. 2012;7:e42514. doi: 10.1371/journal.pone.0042514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Godfroid J, Garin-Bastuji B, Saegerman C, et al. Brucellosis in terrestrial wildlife. Rev Sci Tech. 2013;32:27–42. doi: 10.20506/rst.32.1.2180 [DOI] [PubMed] [Google Scholar]
- 5.Galinska EM, Zagorski J.. Brucellosis in humans–etiology, diagnostics, clinical forms. Ann Agric Environ Med. 2013;20:233–238. [PubMed] [Google Scholar]
- 6.Hasanjani Roushan MR, Ebrahimpour S.. Human brucellosis: An overview. Caspian J Intern Med. 2015;6:46–47. [PMC free article] [PubMed] [Google Scholar]
- 7.Khan MZ, Zahoor M.. An overview of brucellosis in cattle and humans, and its serological and molecular diagnosis in control strategies. Trop Med Infect Dis. 2018;2(3):65 doi: 10.3390/tropicalmed3020065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li K, Zhang L, Shahzad M, et al. Increasing incidence and changing epidemiology of brucellosis in China (2004-2016). Travel Med Infect Dis. 2019: 101464. doi: 10.1016/j.tmaid.2019.101464 [DOI] [PubMed] [Google Scholar]
- 9.Kong W. Brucellosis infection increasing in southern China. Eur J Intern Med. 2018;51:e16–e18. doi: 10.1016/j.ejim.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 10.Hanot Mambres D, et al. Imported human brucellosis in Belgium: Bio and molecular typing of bacterial isolates, 1996-2015. PLoS One. 2017;12:e0174756. doi: 10.1371/journal.pone.0174756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao X, et al. Enzootic situation and molecular epidemiology of Brucella in livestock from 2011 to 2015 in Qingyang, China. Emerg Microbes Infect. 2018;7(1):58. doi: 10.1038/s41426-018-0060-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang WY, et al. Human brucellosis, Inner Mongolia, China. Emerg Infect Dis. 2010;16:2001–2003. doi: 10.3201/eid1612.091081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Dahouk S, Tomaso H, Nockler K, et al. Laboratory-based diagnosis of brucellosis–a review of the literature. Part II: serological tests for brucellosis. Clin Lab. 2003;49:577–589. [PubMed] [Google Scholar]
- 14.Al Dahouk S, Tomaso H, Nockler K, et al. Laboratory-based diagnosis of brucellosis–a review of the literature. Part I: Techniques for direct detection and identification of Brucella spp. Clin Lab. 2003;49:487–505. [PubMed] [Google Scholar]
- 15.Cao X, et al. Molecular epidemiological characterization of Brucella isolates from sheep and yaks in northwest China. Transbound Emerg Dis. 2018;65:e425–e433. doi: 10.1111/tbed.12777 [DOI] [PubMed] [Google Scholar]
- 16.Baily GG, Krahn JB, Drasar BS, et al. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg. 1992;95:271–275. [PubMed] [Google Scholar]
- 17.Ewalt DR, Bricker BJ.. Validation of the abbreviated Brucella AMOS PCR as a rapid screening method for differentiation of Brucella abortus field strain isolates and the vaccine strains, 19 and RB51. J Clin Microbiol 2000;38:3085–3086. doi: 10.1128/JCM.38.8.3085-3086.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al Dahouk S, et al. Evaluation of Brucella MLVA typing for human brucellosis. J Microbiol Methods. 2007;69:137–145. doi: 10.1016/j.mimet.2006.12.015 [DOI] [PubMed] [Google Scholar]
- 19.Le Fleche P, et al. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 2006;6:9. doi: 10.1186/1471-2180-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu ZG, et al. MLVA genotyping characteristics of human Brucella melitensis isolated from Ulanqab of Inner Mongolia, China. Front Microbiol. 2017;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter PR. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990;28:1903–1905. doi: 10.1128/JCM.28.9.1903-1905.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu ZG, Wang H, Wang M, et al. Investigation of the molecular epizootiological characteristics and tracking of the geographical origins of Brucella canis strains in China. Transbound Emerg Dis. 2020;67:834–843. doi: 10.1111/tbed.13404 [DOI] [PubMed] [Google Scholar]
- 23.Piao D, et al. MLVA and LPS characteristics of Brucella canis isolated from humans and dogs in Zhejiang, China. Front Vet Sci. 2017;4:223. doi: 10.3389/fvets.2017.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai S, et al. Changing epidemiology of human brucellosis, China, 1955–2014. Emerg Infect Dis. 2017;23:184–194. doi: 10.3201/eid2302.151710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu ZG, et al. Investigation of the molecular characteristics of Brucella isolates from Guangxi Province, China. BMC Microbiol 2019;19:292. doi: 10.1186/s12866-019-1665-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolo FB, et al. Seroprevalence and characterization of Brucella species in cattle slaughtered at Gauteng abattoirs, South Africa. Vet Med Sci. 2019;5:545–555. doi: 10.1002/vms3.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musallam II, Abo-Shehada M, Omar M, et al. Cross-sectional study of brucellosis in Jordan: prevalence, risk factors and spatial distribution in small ruminants and cattle. Prev Vet Med. 2015;118:387–396. doi: 10.1016/j.prevetmed.2014.12.020 [DOI] [PubMed] [Google Scholar]
- 28.Ran X, et al. Brucellosis seroprevalence in dairy cattle in China during 2008–2018: a systematic review and meta-analysis. Acta Trop. 2019;189:117–123. doi: 10.1016/j.actatropica.2018.10.002 [DOI] [PubMed] [Google Scholar]
- 29.Hensel ME, Negron M, Arenas-Gamboa AM.. Brucellosis in dogs and public health risk. Emerging Infect Dis. 2018;24:1401–1406. doi: 10.3201/eid2408.171171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dadar M, Shahali Y, Whatmore AM.. Human brucellosis caused by raw dairy products: a review on the occurrence, major risk factors and prevention. Int J Food Microbiol. 2019;292:39–47. doi: 10.1016/j.ijfoodmicro.2018.12.009 [DOI] [PubMed] [Google Scholar]
- 31.Harrison ER, Posada R.. Brucellosis. Pediatr Rev. 2018;39:222–224. doi: 10.1542/pir.2017-0126 [DOI] [PubMed] [Google Scholar]
- 32.Deqiu S, Donglou X, Jiming Y.. Epidemiology and control of brucellosis in China. Vet Microbiol. 2002;90:165–182. doi: 10.1016/S0378-1135(02)00252-3 [DOI] [PubMed] [Google Scholar]
- 33.Shevtsov A, et al. Genetic diversity of Brucella abortus and Brucella melitensis in Kazakhstan using MLVA-16. Infect Genet Evol. 2015;34:173–180. doi: 10.1016/j.meegid.2015.07.008 [DOI] [PubMed] [Google Scholar]
- 34.Liu ZG, et al. Seroprevalence of human brucellosis and molecular characteristics of Brucella strains in Inner Mongolia Autonomous region of China, from 2012 to 2016. Emerg Microbes Infect. 2020;9:263–274. doi: 10.1080/22221751.2020.1720528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackelsberg J, et al. Brucella Exposure risk events in 10 clinical laboratories, New York city, USA, 2015 to 2017. J Clin Microbiol. 2020;58(2):e01096–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouza E, Sánchez-Carrillo C, Hernangómez S, et al. Laboratory-acquired brucellosis: a Spanish national survey. J Hosp Infect. 2005;61:80–83. doi: 10.1016/j.jhin.2005.02.018 [DOI] [PubMed] [Google Scholar]
- 37.Garofolo G, et al. Investigating genetic diversity of Brucella abortus and Brucella melitensis in Italy with MLVA-16. Infect Genet Evol. 2013;19:59–70. doi: 10.1016/j.meegid.2013.06.021 [DOI] [PubMed] [Google Scholar]
- 38.Hinić V, et al. Brucellosis in a dog caused by Brucella melitensis Rev 1. Vet. Microbiol. 2010;141:391–392. doi: 10.1016/j.vetmic.2009.09.019 [DOI] [PubMed] [Google Scholar]
- 39.Xiao P, et al. Genotyping of human Brucella melitensis biovar 3 isolated from Shanxi Province in China by MLVA16 and HOOF. PLoS ONE. 2015;10:e0115932. doi: 10.1371/journal.pone.0115932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marianelli C, et al. Use of MLVA-16 typing to trace the source of a laboratory-acquired Brucella infection. J Hosp Infect. 2008;68:274–276. doi: 10.1016/j.jhin.2008.01.003 [DOI] [PubMed] [Google Scholar]
- 41.Traxler RM, et al. Review of brucellosis cases from laboratory exposures in the United States in 2008 to 2011 and improved strategies for disease prevention. J Clin Microbiol. 2013;51:3132–3136. doi: 10.1128/JCM.00813-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiesch Sz, et al J.. Genomic tandem repeat analysis proves laboratory-acquired brucellosis in veterinary (camel) diagnostic laboratory in the United Arab Emirates. Zoonoses Public Health. 2010;57:315–317. [DOI] [PubMed] [Google Scholar]
- 43.Traxler RM, Lehman MW, Bosserman EA, et al. A literature review of laboratory-acquired brucellosis. J Clin Microbiol. 2013;51:3055–3062. doi: 10.1128/JCM.00135-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mense MG, Borschel RH, Wilhelmsen CL, et al. Pathologic changes associated with brucellosis experimentally induced by aerosol exposure in rhesus macaques (Macaca mulatta). Am J Vet Res. 2004;65:644–652. doi: 10.2460/ajvr.2004.65.644 [DOI] [PubMed] [Google Scholar]
- 45.Sayin-Kutlu S, et al. Laboratory-acquired brucellosis in Turkey. J Hosp Infect. 2012;80:326–330. doi: 10.1016/j.jhin.2011.12.020 [DOI] [PubMed] [Google Scholar]
- 46.Norman FF, Monge-Maillo B, Chamorro-Tojeiro S, et al. Imported brucellosis: a case series and literature review. Travel Med Infect Dis. 2016;14:182–199. doi: 10.1016/j.tmaid.2016.05.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article, and its supplementary information files will be freely available to any scientist wishing to use them for non-commercial purposes upon request via e-mail with the corresponding author.