ABSTRACT
Staphylococcus aureus is a major human pathogen that causes a great diversity of community- and hospital-acquired infections. Rsp, a member of AraC/XylS family of transcriptional regulators (AFTRs), has been reported to play an important role in the regulation of virulence determinants in S. aureus via an agr-dependent pathway. Here we demonstrated that Rsp could bind to the rsp promoter to positively regulate its own expression. We then constructed an isogenic rsp deletion strain and compared the haemolysis in the wild-type and rsp mutant strains. Our results indicated that the rsp mutant strain displayed decreased haemolytic activity, which was correlated with a dramatic decrease in the expression of hla and psm. Furthermore, we analysed the regulatory effects of Rsp in the agr mutant strain and found that they are agr-independent. Electrophoretic mobility shift assay indicated that Rsp can directly bind to the promoter regions of hla and psm. The mouse model of subcutaneous abscess showed that the rsp mutant strain displayed a significant defect in virulence compared to the wild-type strain. These findings reveal that Rsp positively regulates the virulence of S. aureus by promoting the expression of hla and psm through direct binding to their promoter regions.
KEYWORDS: Staphylococcus aureus, Rsp, Agr, haemolysis, Hla, PSMs, virulence
Introduction
Staphylococcus aureus is a detrimental and versatile human pathogen responsible for a wide diversity of the community- and hospital-acquired infections, ranging from innocuous skin infections to life-threatening conditions like pneumonia, osteomyelitis and infective endocarditis [1,2]. The pathogenicity of the bacterium is a sophisticated process involving multiple virulence determinants, such as exotoxins, enzymes, and surface protein adhesins [3,4]. The expression of virulence factors is coordinately controlled by three global regulators, agr, sarA, and sae [5], which are believed to enable S. aureus to survive and to elicit subsequent infection in different environments.
The AraC/XylS family [6] is a group of transcriptional regulators with a highly conserved 99 amino acid at the C-terminal region. Members of this family are widely distributed in numerous species of gram-negative and gram-positive bacteria and mainly involved in metabolism of carbon sources, responses to environmental stress and pathogenesis. In S. aureus, 6 AraC/XylS-like proteins are predicted [7] and four of which, Rbf, Rsp, AryK and HptR, have been experimentally characterized. Rbf was initially reported to participate in biofilm formation in response to sodium chloride and glucose [8], and subsequent studies demonstrated that Rbf can promote biofilm formation via repression of icaR [9] and activation of sarX [10]. Rsp originally was described to modulate biofilm formation by repressing surface proteins [11], and further studies showed that Rsp was essential for the expression of virulence factors and the development of pneumonia and skin infections in mouse models [12,13]. AryK was shown to potentiate toxin expression and virulence of S. aureus JDK6159, a highly virulent strain, by a loss-of-function point mutation [14]. HptR, a response regulator protein of three-component regulatory system HptRSA, was found to facilitate the uptake of glucose-6-phosphate (G6P) in S. aureus and support the bacterial survival and proliferation in host cells [15,16]. Therefore, the regulatory effects of the AraC/XylS family proteins in S. aureus are varied, and much of them still remains to be explored.
The agr locus is the most investigated quorum-sensing system of staphylococci and consists of two divergent transcripts, RNAII and RNAIII, driven by P2 and P3 promoters, respectively [17]. The RNAII transcript encodes a typical two-component signal-transduction system, which comprises the sensor histidine kinase AgrC and the response regulator AgrA in response to the extracellular concentration of the autoinducing peptide (AIP) encoded and modified by the proteins AgrD and AgrB [18,19]. Induction of agr results in the amplification of quorum-sensing signal and the expression of 514-nucleotide transcript RNAIII, the major effector of the agr system, mediating the expression of agr regulon by an antisense mechanism [20]. The agr system is critical for the pathogenicity of S. aureus and can modulate the expression of virulence factors in both RNAIII-dependent and RNAIII-independent patterns. In the RNAIII-independent manner, the agr system regulates the transcription of virulence genes by direct binding of AgrA to the promoters of target genes [21].
Hla and PSMs are the two prominent and well-characterized cytotoxins in S. aureus, and both of them are pore-forming toxins. Hla is a hydrophilic polypeptide that presents in culture supernatant as a monomer [22]. Oligomerization into hexamers or heptamers on the host membrane mediated by its cellular receptor ADAM10 triggers the pore formation and the membrane lysis [23]. Thus, with the ability to lyse a wide array of cell types, this toxin has been recognized as an important contributor to the pathogenesis of pneumonia, sepsis, mastitis, and skin infections [24–28]. PSMs are small, amphipathic peptides consisting of five shorter α-type peptides (PSMα1-PSMα4 and δ-toxin) and two longer β-type peptides (PSMβ1-PSMβ2) [29]. PSMαs are transcribed from the psmα operon, PSMβs are transcribed from the psmβ operon, and the δ-toxin is transcribed from the agrP3 promoter [29,30]. Unlike Hla, the ability of PSMs to lyse eukaryotic cells is receptor-independent [31]. Among the 7 peptides produced by S. aureus, the PSMα peptides, especially PSMα3 showed by far the strongest cytolytic activity [32]. Deletion of the psmα operon in community-associated methicillin-resistant S. aureus (CA-MRSA) significantly decreases the ability to cause skin and soft-tissue infections in mice and the capacities to attract and lyse neutrophils [33,34]. Compared with hospital-associated (HA)-MRSA, the CA-MRSA shows much higher in vitro expression of PSMs, giving the fact that PSMs peptides contribute to a great extent to the enhanced virulence of CA-MRSA [29,35]. The expression of hla and psm is strictly regulated by global virulence regulators. hla is tightly controlled by the positive regulators sRNA RNAIII, SarA and Sae [36–38], and the negative regulators Rot and SarT [39,40]. The expression of psm is positively regulated by the agr system through the direct binding of AgrA to the promoter regions of psm operons [21]. Although the functions and regulations of hla and psm have been studied extensively, it is necessary to identify the potential transcriptional regulators of hla and psm to provide a greater insight into pathogenic mechanisms.
Previously, Rsp has been reported to regulate the expression of virulence genes via an agr-dependent pathway [12]. However, whether Rsp directly regulates the expression of these genes remains unknown. In this study, we demonstrated that Rsp could bind to the rsp promoter to upregulate its own expression. Additionally, we found that Rsp can positively regulate the expression of hla, psmα, and psmβ by directly binding to their promoter regions in an agr-independent manner. The contribution of Rsp to the pathogenicity of S. aureus was further confirmed by using a mouse subcutaneous abscess model.
Materials and methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultivated with shaking (220 rpm) in lysogeny broth (LB) medium (Oxoid) or on lysogeny broth agar (LA) at 37°C. S. aureus strains were grown with shaking (220 rpm) in tryptic soy broth (TSB) medium (Difco) or on tryptic soy agar (TSA) at 37°C. When required, appropriate antibiotics were used for plasmid selection and maintenance at the following concentrations: for E. coli, ampicillin at 150 μg/ml and kanamycin at 50 μg/ml; for S. aureus, chloramphenicol at 15 μg/ml.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Relevant genotypea | Reference or source |
|---|---|---|
| S. aureus strains | ||
| RN4220 | 8325-4 r-, initial recipient for modification of plasmids which are introduced into S. aureus from E. coli | NARSAb |
| NCTC8325 | HA-MSSA, wild-type | NARSA |
| Δrsp | 8325 strain deletion of rsp | This study |
| Crsp | rsp chromosomal complementation of Δrsp | This study |
| Δagr | 8325 strain deletion of agr | This study |
| ΔagrΔrsp | 8325 agr rsp double mutant | This study |
| N315 | HA-MRSA, agr-deficient | NARSA |
| N315Δrsp | N315 strain deletion of rsp | This study |
| Newman | HA-MSSA, α- and β-toxin-deficient | [41,42] |
| NewmanΔrsp | Newman strain deletion of rsp | This study |
| E. coli strains | ||
| Trans1-T1 | Clone host strain, F- ϕ80 (lacZ) ΔM15ΔlacX74 hsdR (rK- mK+) ΔrecA1398 endA1 tonA | TransGen |
| BL21(DE3) | Express strain, F- ompT hsdSB (rB- mB-) gal dcm (DE3) | TransGen |
| Plasmids | ||
| pBTs | Shuttle vector, temp sensitive, ampr cmr | [43] |
| pBTsΔrsp | pBTs derivative, for rsp deletion, ampr cmr | This study |
| pBTs-up-rsp-down | pBTs derivative, for rsp chromosomal complementation, ampr cmr | This study |
| pBTsΔagr | pBTs derivative, for agr deletion, ampr cmr | This study |
| pLI50 | Shuttle vector, ampr cmr | [44] |
| pLIrsp | pLI50 derivative, harbouring ORF of rsp and its promoter, ampr cmr | This study |
| pRMC2 | Shuttle vector, anhydrspetracycline inducible, ampr cmr | [45] |
| pRMCrsp | pRMC2 derivative, harbouring ORF of rsp, ampr cmr | This study |
| pOS1-lacZ | Shuttle vector, with lacZ ORF lacking first 6 amino acids, ampr cmr | [46] |
| pOSrsp | POS1-lacZ derivative, harbouring 301-bp region of rsp promoter and 18 bp of rsp coding sequence from strain NCTC8325, ampr cmr | This study |
| pOShla | POS1-lacZ derivative, harbouring 235-bp region of hla promoter and 18 bp of hla coding sequence from strain NCTC8325, ampr cmr | [47] |
| pOSpsmα | POS1-lacZ derivative, harbouring 308-bp region of psmα promoter and 18 bp of psmα coding sequence from strain NCTC8325, ampr cmr | This study |
| pOSpsmβ | POS1-lacZ derivative, harbouring 226-bp region of psmβ promoter and 18 bp of psmβ coding sequence from strain NCTC8325, ampr cmr | This study |
| pRSF-Duet | Expression vector with a hexa-histidine tag, kanr | Novagen |
| pRSF-Duet-Rsp | pRSF-Duet derivative, with ORF of rsp, kanr | This study |
ar−, restriction system negative; kanr, kanamycin resistant; ampr, ampicillin resistant; cmr, chloramphenicol resistant.
bNARSA, Network on Antimicrobial Resistance in Staphylococcus aureus.
Construction of the rsp mutant, agr mutant, and agr rsp double mutant strains
To construct the rsp mutant strain, the plasmid pBTs was used as described previously [43]. The upstream and downstream regions of rsp, which were amplified from genomic DNA of S. aureus strains NCTC8325, N315, and Newman, respectively, using primer pairs rsp-up-F/rsp-up-R and rsp-down-F/rsp-down-R. The products were ligated by overlap PCR to form an up–down fragment. The resultant fragment was digested with KpnI and SalI, and cloned into the KpnI/SalI-digested plasmid pBTs, which containing a temperature-sensitive S. aureus origin of replication, a chloramphenicol resistance cassette, and a suicide gene for plasmid maintenance or selection. The resulting plasmid, pBTsΔrsp, was first electroporated into S. aureus strain RN4220 for modification and subsequently transformed into S. aureus strains NCTC8325, N315, and Newman. An allelic replacement mutant was selected using a previously described method [48] and was further confirmed by PCR and sequencing. The agr mutant and agr rsp double mutant strains were constructed using a similar strategy by introducing the plasmid pBTsΔagr into the wild-type (WT) and rsp mutant strains, respectively. All primers used in this study are listed in Table 2.
Table 2. Primers used in this study.
| Primer | Sequence (5′–3′)a | Application |
|---|---|---|
| rsp-up-F-KpnI | GCGggtaccAAGGTGTTAATTCATTGATG | rsp deletion |
| rsp-up-R | ATTTCTCTCTCCTGCCTAAT | rsp deletion |
| rsp-down-F | ATTAGGCAGGAGAGAGAAATATACTAACAGTCCTCTTGTG | rsp deletion |
| rsp-down-R-SalI | ACGCgtcgacAACTAATACCAAGACCAAAA | rsp deletion |
| Crsp-F-KpnI | GCGggtaccTTACGGCAATAACTAGATGG | rsp chromosomal complementation |
| Crsp-R-SalI | ACGCgtcgacTGCAGTATTTATATTACATA | rsp chromosomal complementation |
| agr-up-F-KpnI | GCGggtaccAAGAGGTTGAACAAGCATTT | agr deletion |
| agr-up-R | GCAGCGATGGATTTTATTTT | agr deletion |
| agr-down-F | AAAATAAAATCCATCGCTGCGATGAATAATTAATTACTTTCAT | agr deletion |
| agr-down-R-SalI | ACGCgtcgacGTCATTGGAACTAATAGCAC | agr deletion |
| pLI50-rsp-F-SacI | TCCgagctcTATTGTTTTTTGAAATACATAGG | rsp overexpression |
| pLI50-rsp-R-KpnI | CGGggtaccTTGAATTGCTTGTGAGTTAC | rsp overexpression |
| pRMC2-rsp-F-KpnI | CGGggtaccATTAGGCAGGAGAGAGAAAT | rsp induced expression |
| pRMC2-rsp-R-EcoRI | CCGgaattcAGGACTGTTAGTATTTAGCT | rsp induced expression |
| RT-rsp-F | ACGATATTCACGATATTGAGATT | qRT-PCR |
| RT-rsp-R | TCTTGCTTGTCTTATAGTCTTG | qRT-PCR |
| RT-agrA-F | TATGGCGATTGACGACAA | qRT-PCR |
| RT-agrA-R | GCAGTAATTCAGTGTATGTTCA | qRT-PCR |
| RT-psmα-F | GTATCATCGCTGGCATCA | qRT-PCR |
| RT-psmα-R | AAGACCTCCTTTGTTTGTTATG | qRT-PCR |
| RT-psmβ-F | TGGACTAGCAGAAGCAATC | qRT-PCR |
| RT-psmβ-R | TAGTAAACCCACACCGTTAG | qRT-PCR |
| RT-hla-F | GGTATATGGCAATCAACTT | qRT-PCR |
| RT-hla-R | CTCGTTCGTATATTACATCTAT | qRT-PCR |
| RT-hu-F | AAAAAGAAGCTGGTTCAGCAGTAG | qRT-PCR |
| RT-hu-R | TTTACGTGCAGCACGTTCAC | qRT-PCR |
| Prsp-F-EcoRI | CCGgaattcGTATTGTTTTTTGAAATACATAG | pOSrsp |
| Prsp-R-BamHI | CGCggatccGGTTTAAGTTGGCATGTCATAT | pOSrsp |
| Phla-F-EcoRI | CCGgaattcTTCAACTTTGACTAACCCTC | pOShla |
| Phla-R-BamHI | CGCggatccGGGACTATACGTGTTTTCATTT | pOShla |
| Ppsmα-F-EcoRI | CCGgaattcTCTGTTCAATTCATCTTCATA | pOSpsmα |
| Ppsmα-R-BamHI | CGCggatccGGGCCAGCGATGATACCCATTAAG | pOSpsmα |
| Ppsmβ-F-EcoRI | CCGgaattcGGCAACTTAATTGTGTTAAA | pOSpsmβ |
| Ppsmβ-R-BamHI | CGCggatccGGGTTAAATAAACCTTCCATTG | pOSpsmβ |
| E-rsp-F-BamHI | CGCggatccGATGACATGCCAACTTAAAAT | Rsp expression |
| E-rsp-R-EcoRI | CCGgaattcTTAGCTTGGTTTAAAGCAAA | Rsp expression |
| probe-prsp-F319 | GTATTGTTTTTTGAAATACATAG | EMSA |
| probe-prsp-F139 | TTCACTATTACCTTTTCAAAA | EMSA |
| probe-prsp-F89 | TGATTTTAAAAATTTCCATG | EMSA |
| probe-prsp-Biotin-R | TTTAAGTTGGCATGTCATAT | EMSA |
| probe-phla-F310 | TTAGCTATGTCTTTTCCTTG | EMSA |
| probe-phla-F239 | CCTTTCAAATTTTAAATAAA | EMSA |
| probe-phla-F189 | CATCATCACTCAGTAATTTA | EMSA |
| probe-phla-F110 | TGCAATATTCTAAATTGACA | EMSA |
| probe-phla-Biotin-R | TAGTGTTGTTGTTACTGAG | EMSA |
| probe-ppsmα-F326 | TCTGTTCAATTCATCTTCATA | EMSA |
| probe-ppsmα-F166 | ACCTGTACAGTTAGGCAGTA | EMSA |
| probe-ppsmα-F85 | AAAATCAATTACGCACAAGA | EMSA |
| probe-ppsmα-Biotin-R | GCCAGCGATGATACCCATTAAG | EMSA |
| probe-ppsmβ-F253 | ACTTAAATACGAATTCAGGCAACT | EMSA |
| probe-ppsmβ-F162 | CATTATAAGATGTTGTGCGG | EMSA |
| probe-ppsmβ-F139 | AACAAACTAATTGCATCAAA | EMSA |
| probe-ppsmβ-F89 | AAGCGAATAACATTTCGGTG | EMSA |
| probe-ppsmβ-Biotin-R | AACCTTCCATTGAAAACACTCC | EMSA |
aLowercase letters indicate restriction sites. Letters in bold indicate complementary sequences used for overlap pcr ligation.
Construction of the complementation and overexpression strains
For rsp chromosomal complementation, the DNA fragment covering the open reading frame (ORF) of rsp and the flank upstream and downstream regions was amplified from S. aureus strain NCTC8325 genomic DNA using the primer pair Crsp-F/Crsp-R. The PCR product was digested with KpnI and SalI, and then cloned into the pBTs vector. The resulting plasmid, pBTs-up-rsp-down, was first electroporated into S. aureus strain RN4220 for modification and subsequently transformed into the rsp mutant strain. The allelic replacement complementation strain was selected using the same method described above and was further confirmed by PCR and sequencing.
For rsp overexpression, both the plasmid pLIrsp and pRMCrsp were employed. To construct pLIrsp, the fragment encompassing the ORF of rsp and its native promoter was amplified from S. aureus strain NCTC8325 genomic DNA using the primer pair pLI50-rsp-F/pLI50-rsp-R. The fragment was digested with SacI and KpnI, and then cloned into the shuttle plasmid pLI50 to derive the plasmid pLIrsp. To construct pRMCrsp, the fragment encompassing the ORF of rsp was amplified from S. aureus strain NCTC8325 genomic DNA using the primer pair pRMC-rsp-F/pRMC-rsp-R. The fragment was digested with KpnI and EcoRI, and then cloned into the shuttle plasmid pRMC2 with an anhydrotetracycline (ATC) inducible promoter. The combinant plasmids were first electroporated into S. aureus strain RN4220 for modification and subsequently transformed into the WT and agr mutant strains to derive the overexpressed strains.
Total RNA isolation, cDNA generation, and real-time quantitative reverse transcription-PCR
Overnight cultures of S. aureus were diluted 1:100 in TSB and then grown to the indicated cell density until being collected. S. aureus cells were collected by centrifugation and processed with 1 ml of RNAiso plus (TaKaRa) in combination with 0.1-mm-diameter silica-zirconia beads in a FastPrep-24 automated system (MP Biomedicals), and the residual DNA was removed with RNase-free DNase I (TransGen, 3 U/μl). The concentration of total RNA was adjusted to 200 ng/μl. For the reverse transcription, the cDNAs were synthesized with a PrimeScript 1st Strand cDNA synthesis kit (TaKaRa) using random primers and real-time quantitative reverse transcription-PCR (qRT-PCR) was performed with SYBR Premix Ex Taq (TaKaRa) using the StepOne Real-Time PCR System (Applied Biosystems) and LC96 Real-Time PCR System (Roche). The quantity of cDNA measured by real-time PCR was normalized to the abundance of hu cDNA [49], and the corresponding control sample as the run calibrator. All qRT-PCR assays were repeated at least three times.
Determination of haemolytic activity
Qualitative evaluation of haemolytic activity was evaluated on sheep blood agar (SBA). The OD600s of overnight culture were measured and adjusted to the same level. Two-microliter culture volumes were then spotted onto SBA plates. The plates were left to dry for about 2 min and incubated at 37°C for 24 h, and visually evaluated for zones of lysis.
Quantitative evaluation of haemolytic activity was determined by incubating samples with sheep red blood cells. Overnight cultures were collected by centrifugation, and supernatants (100 μl) were mixed with 900 μl phosphate-buffered saline (PBS) buffer containing 3% sheep red blood cells, and the mixtures were incubated at 37°C for proper time. The absorption of supernatant at 543 nm was measured after centrifugation. A mixture with 1000 μl ddH2O containing 3% sheep red blood cells was used as the positive control, and a mixture with 1000 μl PBS containing 3% sheep red blood cells was used as the negative control. The percentage of haemolytic activity was calculated relative to the positive control, which was regarded as 100% haemolytic activity. For hemolysin-deficient strains, the supernatants were mixed with PBS containing 15% sheep red blood cells. A mixtures with 1000 μl ddH2O and 1000 μl PBS containing 15% sheep red blood cells were used as the positive control and the negative control, respectively.
Expression and purification of Rsp
The 6-His-tagged Rsp was expressed and purified using standard procedures. The fragment of the full-length rsp ORF was amplified by PCR with the primer pair E-rsp-F/E-rsp-R from S. aureus strain NCTC8325 genomic DNA, cloned into the expression vector pRSF-Duet to generate the plasmid pRSF-Duet-Rsp, and transformed into E. coli BL21 (DE3). The transformant was grown in LB at 37°C to an OD600 of 0.6 and induced with 0.5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 16°C for additional 12 h. The cells were harvested and lysed by sonication in a lysis buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl). The hexahistidine-tagged Rsp protein was purified with a nickel-nitrilotriacetic acid-agarose solution (Qiagen) by following the manufacturer’s recommendation. The bound protein was eluted with an elution buffer (200 mM imidazole, 50 mM Tris-HCl, pH 8.0, 300 mM NaCl). The imidazole in the eluent was removed using a Centrifuge Biomax-5 column (Millipore). The purity of the protein was analysed using SDS-PAGE, and the protein concentration was determined using the bicinchoninic acid (BCA) assay with bovine serum albumin as the standard.
Electrophoretic mobility shift assay
The biotin-labelled DNA fragments containing the putative promoter regions of rsp (319 bp), which was predicted using BPROM (http://linux1.softberry.com/berry.phtml) and hla (310 bp), psmα (326 bp) or psmβ (253 bp), which were based on our previous studies [47,50], were amplified from S. aureus strain NCTC8325 genomic DNA. The biotin-labelled probe was incubated at 25°C for 30 min with various amounts of Rsp in 10 μl of incubation buffer (50 mM Tris–HCl, pH 8.0, 300 mM NaCl). After incubation, the mixtures were electrophoresed in a 5% native polyacrylamide gel in 1 × Tris-borate-EDTA (TBE) buffer and then transferred to a nylon membrane in 0.5 × TBE buffer. The band shifts were detected using the Chemiluminescent Nucleic Acid Detection Module Kit (ThermoFisher), and imaged with the ImageQuant LAS 4000 (GE Healthcare). The unlabelled fragments of each promoter were added to the labelled fragments at a ratio of approximately 100:1 or 200:1 as specific competitors. The unlabelled DNA fragment of the hu ORF (125 bp) (150-fold or 200-fold) was added as a nonspecific competitor.
Construction of the LacZ reporter vector
To construct the reporter plasmid pOSrsp for detection of rsp expression, a fragment containing the 301-bp promoter of rsp and the first 18-bp region of the rsp coding sequence was amplified from S. aureus strain NCTC8325 genomic DNA with primer pair Prsp-F/R. The fragment was digested with BamHI and EcoRI and cloned into the shuttle vector pOS1-lacZ to generate the reporter plasmid pOSrsp. The same protocol was followed to construct the reporter plasmids pOShla, pOSpsmα and pOSpsmβ using primer pairs Phla-F/R, Ppsmα-F/R and Ppsmβ-F/R, respectively. These reporter plasmids were first transformed into S. aureus strain RN4220, and then the WT and rsp mutant strains.
β-Galactosidase activity assay
For β-galactosidase assay with o-nitrophenyl-β-D-galactopyranoside (ONPG) as the substrate, overnight cultures of the WT and rsp mutant strains containing different reporter plasmids were diluted 1:100 into TSB with chloromycetin (15 μg/ml) and grown to the indicated cell density until being collected. The collected cells were resuspended in 100 μl of ABT-LSA buffer (60 mM K2HPO4, 40 mM KH2PO4, 100 mM NaCl, 0.1% Triton X-100, 50 μg/ml lysostaphin) and incubated at 37°C until thoroughly lysed. Then, 100 μl ABT buffer and 100 μl 4 mg/ml ONPG were added to initiate the reaction. After incubation at 37°C for the proper time (0–1 h), the reactions were terminated by the addition of l ml Na2CO3 (1 M). Sample absorbance was read at 420 nm and units were calculated as the following formula: units = (1000 × OD420)/(T × V × OD600). T (measured in minutes) was the incubation time and V (in millilitres) was the volume of culture used in the assay.
mRNA half-lives
For mRNA half-life determination, overnight culture of S. aureus was inoculated at 1:100 into fresh medium and grown to mid-exponential, transcription was arrested by the addition of rifampin (200 µg/ml), and aliquots were removed at 0-, 3-, 5-, 10-, 15-, 30- and 60- min post-transcriptional arrest. RNA was isolated from each aliquot and the mRNA levels of prsp-lacZ and rsp were measured by qRT-PCR.
Western blot analysis
To detect the production of Hla, stationary-phase culture supernatant was collected and heated for 10 min at 95°C. The samples were then separated by 12% SDS-PAGE and electrotransferred onto a polyvinylidene difluoride membrane (GE, Piscataway, NJ). After blocking with 5% (w/v) nonfat milk in TBST buffer at room temperature for 1 h, the membrane was incubated with a rabbit anti-Hla IgG antibody (Sigma) at a 1/2500 dilution. Bound antibody was detected with the goat anti-rabbit conjugated to horseradish peroxidase (ThermoFisher) at a 1/5000 dilution. The images were obtained with ImageQuant LAS 4000 (GE Healthcare).
Mouse skin infection model
Outbred, immunocompetent female BALB/c mice with age of 6 weeks were purchased from Beijing Vital River Laboratory Animal Technology Company, and housed in isolated cages in an animal facility under specific pathogen-free. The hair on the back was removed by an animal shaver. Overnight cultures were diluted 1:100 into TSB and grown to the indicated cell density until being collected. The collected cells were washed twice and diluted in sterile PBS, and viable cells were counted via their colony-forming units (CFU) on TSA plates in order to quantify the infectious dose. Mice were anesthetized with 1% pentobarbital sodium and inoculated by subcutaneous injection in both flanks of the back with 5 × 107 live S. aureus cells in 50 μl PBS. Abscess formation and skin lesion area were monitored at 24-h intervals for 7 days. The sizes of the skin lesions were calculated by the maximal length × width, as previously described [51]. The skin lesions were excised 7 days after infection and homogenized in PBS. The CFU recovered from each individual lesion was determined by serial dilution and plated onto TSA plates.
Biofilm formation and analysis
Biofilm formation under static conditions was determined by the microtiter plate assay based on the method described previously [52]. Briefly, overnight cultures were diluted 1:200 in fresh TSB medium and dispensed into wells of sterile flat-bottom 96-well polystyrene plates (Costar) at 200 μl per well. The plates were incubated at 37°C for 24 h, and the wells were washed gently three times with water to remove non-adherent cells. The plates were stained with 0.5% crystal violet for 20 min, and then again washed three times with water. After that, the plates were dried, and the optical density at 560 (OD560) was determined with an enzyme-linked immunosorbent assay reader ELX800 (Bio-Tek) in a 3 × 3 scan model. To investigate the biofilm formation of the S. aureus strains N315 and Newman, the TSB medium was supplemented with 0.25% glucose.
Ethics statement
The use and care of mice in the present study followed strictly the guidelines adopted by the Ministry of Health of the People’s Republic of China in June 2004. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Science and Technology of China (USTCACUC192001037).
Statistical analyses
F tests of two samples for variance were performed. Unpaired two-tailed t tests for equal or unequal variance were then performed to calculate the significant differences (P values). A value of P < .05 was considered statistically significant.
Results
Rsp promotes the transcription of rsp through a self-activation manner
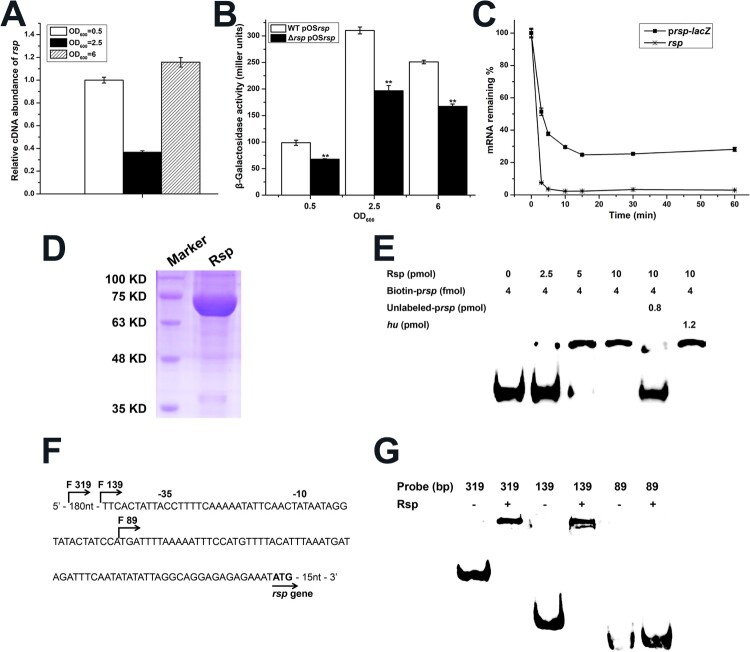
Transcriptional autoregulation has been reported for some members of AraC/XylS family and seems to be essential for the transcription of their target genes under stress conditions [53–55]. Our previous work also found that Rbf can specifically bind to the rbf promoter to upregulate its own expression [56]. To determine if Rsp has a similar effect on its own transcription, we first detected rsp transcription at different growth phases by qRT-PCR, and observed that the transcript level of rsp was significantly decreased at the mid-logarithmic phase (Figure 1(A)). Then, we constructed the lacZ fusion reporter plasmid pOSrsp with the promoter region of rsp and detected the β-galactosidase activities in the WT and rsp mutant strains. As shown in (Figure 1(B)), the β-galactosidase activity of rsp was significantly decreased in the rsp mutant strain throughout the growth phases, and the β-galactosidase activities in the WT and rsp mutant strains were the highest at mid-logarithmic phase. We examined the mRNA half-lives of prsp-lacZ and rsp (Figure 1(C)), the results showed although the mRNA half-lives of prsp-lacZ and rsp are both short, about 20% of prsp-lacZ mRNA could survive for more than 1 h. Due to the continuous translation of these RNAs and the stability of LacZ protein, we assumed that the accumulation of LacZ from early exponential phase leads to the high β-galactosidase activity at mid-logarithmic phase.
Figure 1.
Rsp positively regulates its own expression. (A) The transcript levels of rsp at different growth phases were measured by qRT-PCR. (B) The β-galactosidase activities of the WT and rsp mutant strains with a plasmid pOSrsp were detected in the indicated time points. (C) Analysis of mRNA half-lives of prsp-lacZ and rsp. Cells were collected after rifampin (200 μg/ml) treatment for RNA isolation, and then the mRNA half-lives were measured by qRT-PCR. (D) SDS-PAGE analysis of Rsp purified from the pRSF-Duet expression vector. The gel was stained with Coomassie blue. (E) EMSA of purified Rsp with the biotin-labelled DNA fragment containing the putative promoter region of rsp. Increasing concentrations of purified Rsp and 4 fmol of the biotin-labelled probe were used in the reactions. The unlabelled probe was added as a specific competitor, and the unlabelled fragment of the hu ORF region was added as a nonspecific competitor. (F) Promoter sequence of the rsp gene. The start points of the truncated probes are marked by arrows. (G) EMSA assay of Rsp with rsp truncated probes. The nonspecific competitor was added to the reactions. The error bars indicate the standard errors of the means of three biological replicates. **P < .01.
To test whether the transcription of rsp is under the direct control of Rsp, we purified the recombinant Rsp-His6 (Figure 1(D)) and employed the 319-bp biotin-labelled putative promoter fragment to perform electrophoretic mobility shift assay (EMSA). The result showed that Rsp could specifically bind to its own promoter region, and the shifted band disappeared when an approximately 200-fold excess of unlabelled rsp promoter fragment was added, but not influenced by the addition of 300-fold excess of unlabelled coding sequence fragment of hu (Figure 1(E)). Then, to identify the Rsp binding region on its own promoter, we designed the truncated probes of rsp (Figure 1(F)). Binding of Rsp was completely inhibited when the probe length was truncated from 139 to 89 bp (Figure 1(G)), suggesting that the Rsp binding sites may located within the 50-bp region. Collectively, these data indicated that Rsp could bind to its own promoter to positively regulate its expression.
Rsp contributes to haemolytic activity of S. aureus NCTC8325
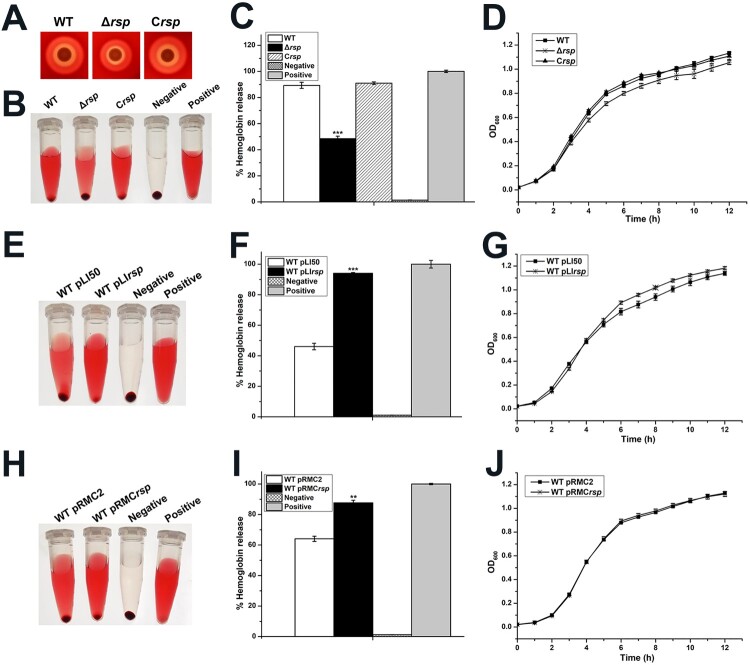
Rsp has been shown to be involved in the positive regulation of haemolysis in staphylococci [12,13]. To further investigate whether Rsp modulates the haemolysis of S. aureus NCTC8325, the haemolytic activities of the WT strain, rsp mutant, and rsp chromosomal-complemented strains were compared using qualitative and quantitative methods. Qualitative evaluation of haemolytic activity was performed on sheep blood agar plates. As shown in Figure 2(A), the haemolytic activity of the rsp mutant strain was much weaker than the WT strain after 24 h of incubation. Quantitative test was performed with the sheep red blood cells, and the percentage of haemolytic activity was calculated relative to the positive control (100% haemolytic activity) by measuring the optical density at 543 nm. The rsp mutant strain displayed significantly decreased haemolytic activity compared with the WT strain after incubation for 35 min at 37°C (Figure 2(B,C)). These changes could be restored by the rsp chromosomal-complemented strain. In addition, we compared the growth rates of the WT, rsp mutant, and rsp chromosomal-complemented strains, and found no remarkable difference (Figure 2(D)).
Figure 2.
Rsp positively regulates the haemolytic activity of S. aureus NCTC8325. (A) Haemolytic activities of the WT, rsp mutant, and rsp chromosomal-complemented strains were evaluated on SBA plates. (B) Haemolytic activities of the WT, rsp mutant, and rsp chromosomal-complemented strains were determined by incubating samples with 3% sheep red blood cells, PBS and ddH2O were used as negative control and positive control, respectively. (C) Haemolytic activities of the WT, rsp mutant, and rsp chromosomal-complemented strains were determined by measuring the absorption of supernatants at 543 nm. (D) Comparison of the growth rates of the WT, rsp mutant, and rsp chromosomal-complemented strains in TSB medium. (E) Haemolytic activities of the WT strains containing plasmids pLI50 and pLIrsp were determined by incubating samples with 3% sheep red blood cells. (F) Haemolytic activities of the WT strains containing plasmids pLI50 and pLIrsp were determined by measuring the absorption of supernatants at 543 nm. (G) Comparison of the growth rates of the WT strains containing plasmids pLI50 and pLIrsp in TSB medium containing 15 μg/ml Cm. (H) Haemolytic activities of the WT strains containing plasmids pRMC2 and pRMCrsp were determined by incubating samples with 3% sheep red blood cells. (I) Haemolytic activities of the WT strains containing plasmids pRMC2 and pRMCrsp were determined by measuring the absorption of supernatants at 543 nm. (J) Comparison of the growth rates of the WT strains containing plasmids pRMC2 and pRMCrsp in TSB medium containing 15 μg/ml Cm and 100 ng/ml ATC. The error bars indicate the standard errors of the means of three biological replicates. **P < .01, ***P < .001.
To confirm the regulatory role of Rsp in haemolysis, we overexpressed rsp in the WT strain by using a constitutive expression plasmid pLI50 and an ATC inducible expression plasmid pRMC2, and measured the haemolytic activity with the sheep red blood cells. As shown in Figure 2(E,F), the WT strain containing pLIrsp exhibited significantly increased haemolytic activity compared with the WT strain containing pLI50 after incubation for 15 min at 37°C. Similarly, the haemolytic activity of the WT strain containing pRMCrsp was significantly increased compared with the WT strain containing pRMC2 induced by 100 ng/ml ATC after incubation for 25 min at 37°C (Figure 2(H,I)). Meanwhile, we compared the growth curves of the overexpression strains with their corresponding WT strains, the results showed no significant difference (Figure 2(G,J)). Taken together, our data suggested that the haemolytic activity of S. aureus NCTC8325 was modulated by Rsp.
Rsp positively regulates the transcription of virulence genes
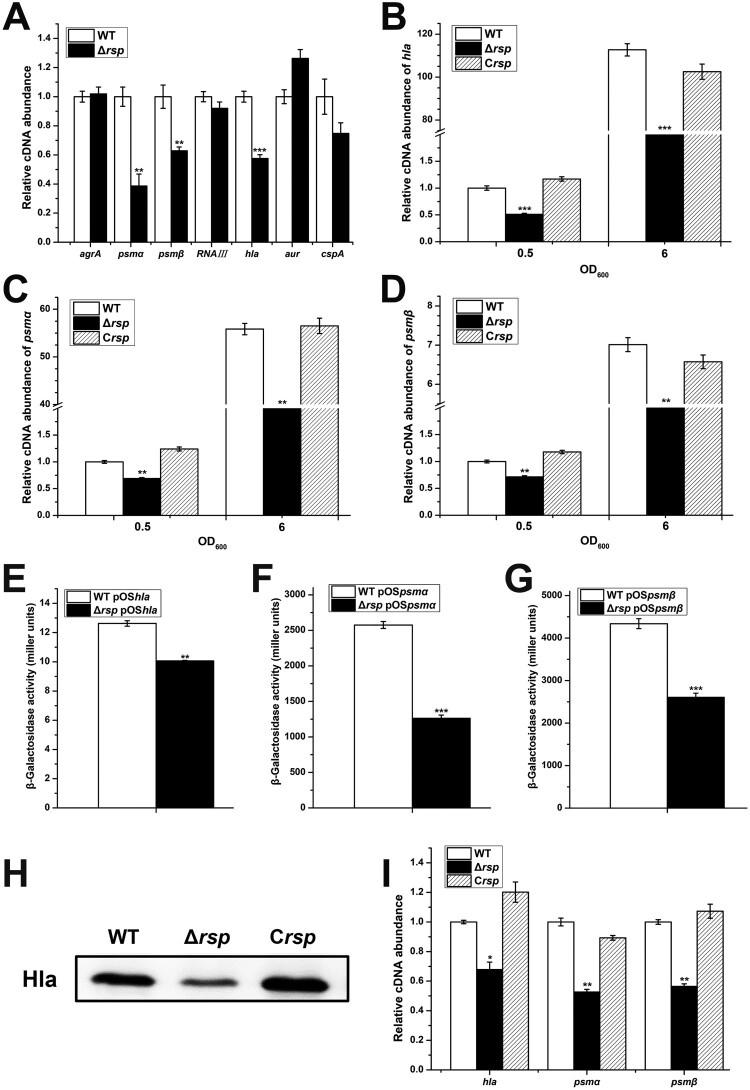
The data presented above clearly demonstrated that Rsp is involved in the control of haemolysis in S. aureus NCTC8325. Many virulence determinants participated in pathogenesis of S. aureus infections can influence the haemolysis. To determine whether the expression of virulence factors was altered in the rsp mutant, we performed qRT-PCR to examine the mRNA levels of 7 potential target genes with RNA isolated from the WT and rsp mutant strains (Figure 3(A)). The transcript levels of hla, psmα, and psmβ were significantly decreased in the rsp mutant strain. Notably, the transcript levels of agrA and RNAIII were not altered in the rsp mutant strain, implying that the transcriptional changes of hla and psm is agr-independent. To verify the transcriptional control by Rsp, we examined the mRNA levels of hla, psmα, and psmβ at different growth phases. The qRT-PCR results showed that the transcript levels of hla, psmα, and psmβ were significantly decreased in the rsp mutant strain at early exponential and stationary phases compared with those of the WT strain, and these effects could be fully restored by chromosomal complementation (Figure 3(B–D)).
Figure 3.
Rsp positively regulates the transcription of virulence genes. (A) The transcript levels of 7 virulence-related genes were detected by qRT-PCR in the WT and rsp mutant strains. The transcript levels of hla (B), psmα (C), and psmβ (D) in the WT, rsp mutant, and rsp chromosomal-complemented strains at different growth phases. The β-galactosidase activities of hla (E), psmα (F), and psmβ (G) promoter in the WT and rsp mutant strains. Cells were collected at early stationary phase, and the β-galactosidase activity was detected with ONPG. (H) Hla protein levels of the WT, rsp mutant, and rsp chromosomal-complemented strains were measured by Western blot. (I) The transcript levels of hla, psmα, and psmβ in the WT, rsp mutant, and rsp chromosomal-complemented strains in the process of haemolysis. Cells were collected after incubated with 3% sheep red blood cells for 30 min, and the mRNA levels were analysed by qRT-PCR. The error bars indicate the standard errors of the means of three biological replicates. *P < .05, **P < .01, ***P < .001.
To further confirm the positive effect of Rsp on the expression of hla, psmα, and psmβ, the β-galactosidase assay was performed. The lacZ fusion reporter plasmids pOShla, pOSpsmα, and pOSpsmβ were constructed and transformed into the WT and rsp mutant strains, respectively. The results showed that the β-galactosidase activities in the rsp mutant strain were significantly lower than those in the WT strain (Figure 3(E–G)), which are consistent with the results of qRT-PCR. Western blot analysis indicated that the production level of Hla was also significantly decreased in the rsp mutant strain (Figure 3(H)). To determine the pivotal role of Hla and PSMs in haemolysis, the bacterial cells were collected after incubated with sheep red blood cells, and the mRNA levels were analysed by qRT-PCR. The transcript levels of hla, psmα, and psmβ were decreased significantly in the rsp mutant strain (Figure 3(I)). These data suggested that the decrease of haemolytic activity of the rsp mutant strain is mainly due to the decreased expression levels of hla and psm.
Rsp modulates the haemolytic activity of S. aureus and the transcription of virulence genes in an agr-independent manner
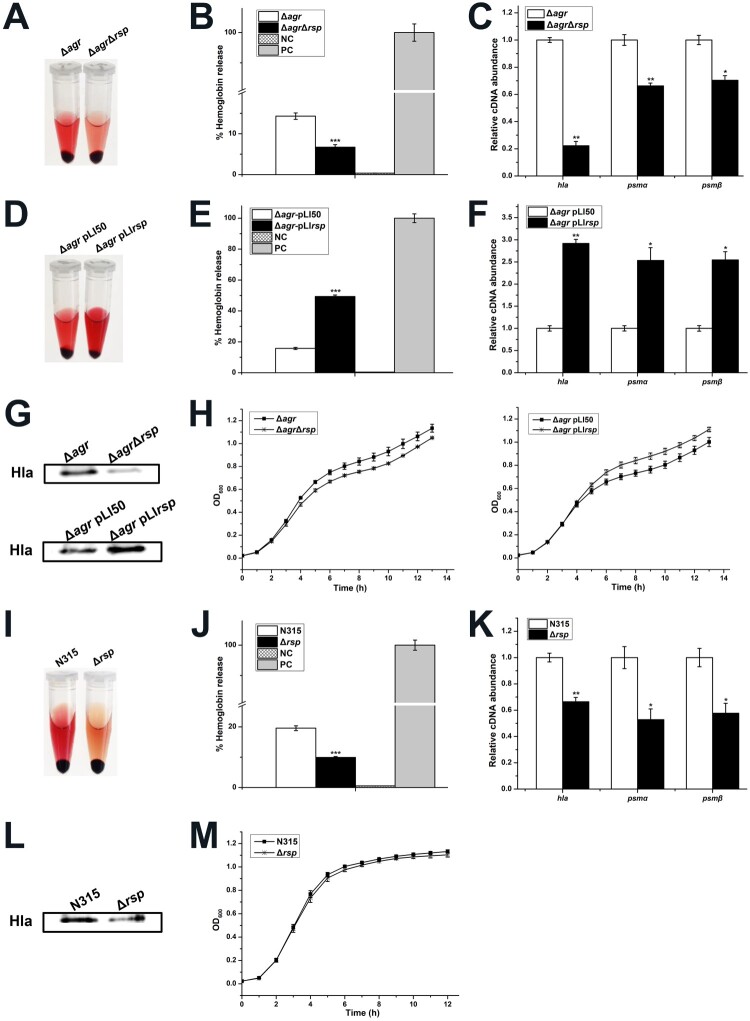
The agr has been recognized as a global regulator involved in the regulation of virulence factors [21] and has been proposed to be positively regulated by Rsp [12]. However, as mentioned above, we did not observe the transcriptional change of agrA in the rsp mutant strain (Figure 3(A)). We further detected the transcript levels of agrA at different growth phases, and no difference was found between the WT and rsp mutant strains (data not shown). Thus, it is reasonable to assume that Rsp modulates the haemolysis and the expression of virulence genes independent of agr. To investigate whether the regulatory effects described here exist in S. aureus NCTC8325, we constructed the agr mutant and agr rsp double mutant strains, and measured the haemolytic activity with the sheep red blood cells. As shown in Figure 4(A,B), the haemolytic activity of the agr rsp double mutant strain was significantly decreased compared with the agr mutant strain after incubation for 1.5 h at 37°C. We also performed overexpression of the agr mutant with the overexpressed plasmid pLIrsp. The results showed that transformation of the agr mutant with pLIrsp increased the haemolytic activity (Figure 4(D,E)). In addition, the growth rates of these strains showed no significant difference (Figure 4(H)).
Figure 4.
Rsp modulates the haemolytic activity of S. aureus and the transcription of virulence genes in an agr-independent manner. (A) Haemolytic activities of the agr mutant and agr rsp double mutant strains were determined by incubating samples with 15% sheep red blood cells, PBS and ddH2O were used as negative control and positive control, respectively. (B) Haemolytic activities of the agr mutant and agr rsp double mutant strains were determined by measuring the absorption of supernatants at 543 nm. (C) The transcript levels of hla, psmα, and psmβ in the agr mutant and agr rsp double mutant strains. (D) Haemolytic activities of the agr mutant strains with the empty plasmid pLI50 and rsp overexpressed plasmid pLIrsp were determined by incubating samples with 15% sheep red blood cells. (E) Haemolytic activities of the agr mutant strains with the empty plasmid pLI50 and rsp overexpressed plasmid pLIrsp were determined by measuring the absorption of supernatants at 543 nm. (F) The transcript levels of hla, psmα, and psmβ in the agr mutant strains with the empty plasmid pLI50 and rsp overexpressed plasmid pLIrsp. (G) Western blot analysis of Hla. The protein levels of Hla in the agr mutant and agr rsp double mutant strains as shown in the upper panel. The lower panel shows the protein levels of Hla in the agr mutant strains with the empty plasmid pLI50 and rsp overexpressed plasmid pLIrsp. (H) Comparison of the growth rates of the agr-deficient strains. (I) Haemolytic activities of the N315 and rsp mutant strains were determined by incubating samples with 15% sheep red blood cells. (J) Haemolytic activities of the N315 and rsp mutant strains were determined by measuring the absorption of supernatants at 543 nm. (K) The transcript levels of hla, psmα, and psmβ in the N315 and rsp mutant strains. (L) Hla protein levels of the N315 and rsp mutant strains were measured by Western blot. (M) Comparison of the growth rates of the N315 and rsp mutant strains. The error bars indicate the standard errors of the means of three biological replicates. *P < .05, **P < .01, ***P < .001.
To confirm if the changes of haemolytic activity were associated with the expression of virulence genes, we examined the transcript levels of hla, psmα, and psmβ. In the agr rsp double mutant strain, the expression of hla, psmα, and psmβ were significantly decreased (Figure 4(C)). Introduction of pLIrsp into the agr mutant increased the transcript levels of hla, psmα, and psmβ (Figure 4(F)). Moreover, we measured the Hla production in these strains by Western blot assay. The results showed that the production level of Hla in the agr rsp double mutant strain was much lower than the agr mutant strain and transformation of the agr mutant with pLIrsp displayed a distinctly higher production level of Hla than that in the agr mutant with plasmid pLI50 (Figure 4(G)). These results indicated that Rsp modulates the haemolytic activity of S. aureus and the transcription of virulence genes in an agr-independent manner.
Since Rsp has been reported to regulate the expression of virulence genes in CA-MRSA strains MW2 and BD02-25 via an agr-dependent pattern, we were interested in whether the agr-independent regulatory pathway mediated by Rsp is specific to HA S. aureus. We constructed the rsp mutant in HA S. aureus strains N315 and Newman and performed haemolytic activity assay. The results showed that the haemolytic activities of the rsp mutant strains were significantly decreased compared with those of the WT strains (Figure 4(I,J) and Figure S1(A,B)). Similar with S. aureus NCTC8325, no significant difference was observed in the growth rates of the WT and rsp mutant strains (Figure 4(M) and Figure S1(E)). We then examined the transcript levels of virulence genes in these strains and the results indicated that the expression of hla, psmα, and psmβ were significantly decreased in the rsp mutant strains (Figure 4(K) and Figure S1(C)). Western blot analysis also showed that the rsp mutant strains exhibited decreased production of Hla (Figure 4(L) and Figure S1(D)). Additionally, the expression of agrA was significantly decreased in the rsp mutant strain of S. aureus Newman (Figure S1(C)), suggesting that Rsp regulates the transcription of virulence genes in this strain might through an agr-dependent manner. Taken together, these results corroborated the regulatory effects of Rsp on the transcription of virulence genes and also proved that they are variable in different S. aureus strains.
Rsp directly binds to the promoter regions of hla, psmα, and psmβ
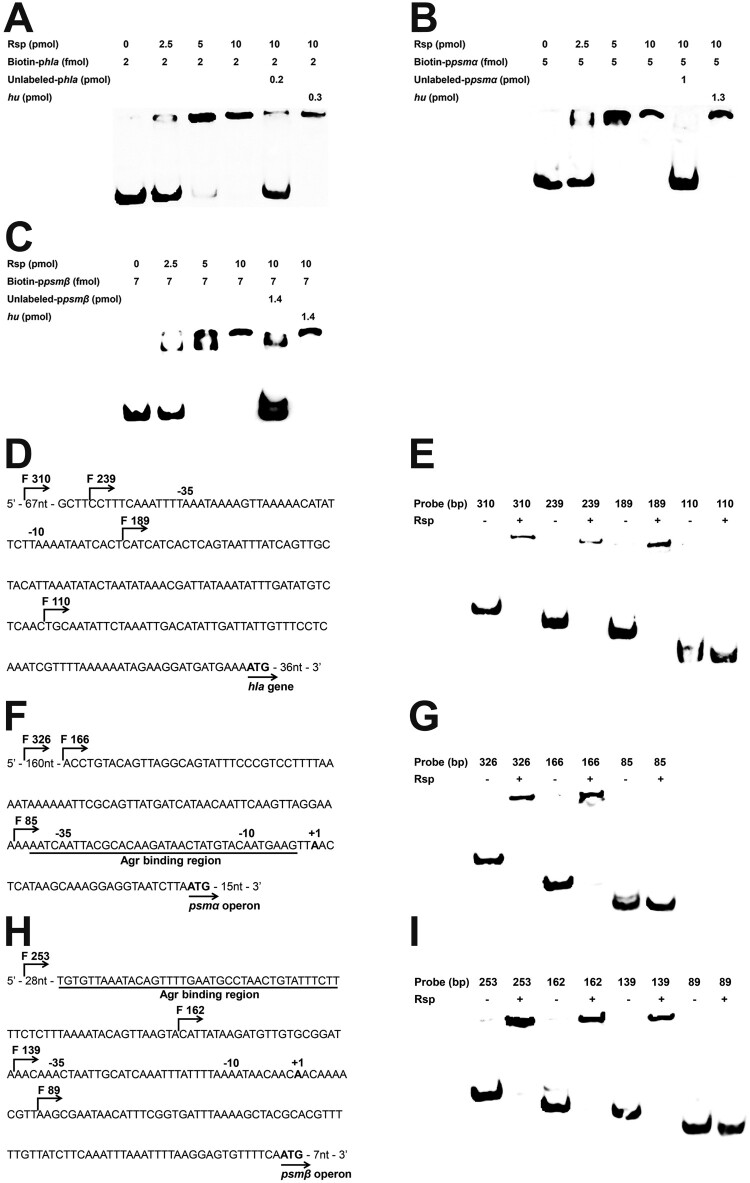
To determine whether hla, psmα, and psmβ are under the direct control of Rsp, we performed EMSA assay. The result showed that Rsp can retard the mobility of the hla promoter in a dose-dependent manner (Figure 5(A)). This binding can be disrupted with an approximately 100-fold of unlabelled hla promoter fragment, while a 150-fold excess of unlabelled coding sequence fragment of hu did not have the same effect. We also employed the psmα and psmβ promoter regions upstream of the initiation codon to perform an EMSA, and similar binding patterns were observed with the psmα and psmβ promoters (Figure 5(B,C)). To further identify the Rsp binding regions, EMSA was performed using the truncated probes of hla, psmα, and psmβ (Figure 5(D,F,H)). Concerning hla, the binding of Rsp was abolished when the probe length was truncated from 189 to 110 bp (Figure 5(E)). For psmα, the shifted bind disappeared when the probe length was truncated from 166 to 85 bp (Figure 5(G)). For psmβ, when the probe length was truncated from 139 to 89 bp, the binding was completely inhibited (Figure 5(I)). Additionally, EMSAs with truncated probes of psmα and psmβ showed that the Rsp binding regions were located upstream and downstream of the AgrA binding sites, respectively [21]. These results strongly suggested that Rsp can directly bind to the promoter regions of hla, psmα, and psmβ to upregulate their expression. Interestingly, no conserved sequence has been found in the Rsp binding regions of rsp, hla, psmα, and psmβ promoters through sequence alignment.
Figure 5.
Rsp specifically binds to the hla, psmα, and psmβ promoter regions. EMSA assay of the purified Rsp with the biotin-labelled DNA fragments of hla (A), psmα (B), or psmβ (C). Increasing concentrations of purified Rsp and 2 fmol (hla), 4 fmol (psmα), or 7 fmol (psmβ) of the biotin-labelled probe were used in the reactions. The unlabelled probes were added as a specific competitors, and the unlabelled fragment of hu ORF region was added as a nonspecific competitor. (D) Promoter sequence of the hla gene. The start points of the truncated probes are marked by arrows. (E) EMSA assay of Rsp with hla truncated probes. The nonspecific competitor was added to the reactions. (F) Promoter sequence of the psmα operon. The start points of the truncated probes are marked by arrows. The AgrA binding region is underlined in black. (G) EMSA assay of Rsp with psmα truncated probes. The nonspecific competitor was added to the reactions. (H) Promoter sequence of the psmβ operon. The start points of the truncated probes are marked by arrows. The AgrA binding region is underlined in black. (I) EMSA assay of Rsp with psmβ truncated probes. The nonspecific competitor was added to the reactions.
Rsp contributes to virulence of S. aureus in a subcutaneous abscess model of mice
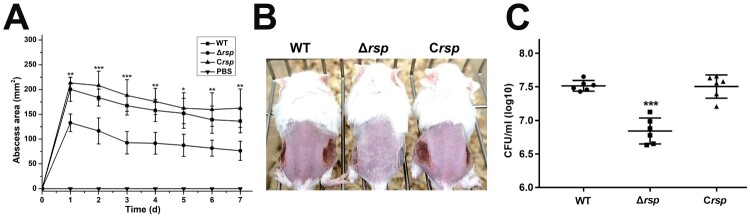
Hla and PSMs are well-characterized toxins, and previous studies have indicated that they play a significant role in S. aureus skin and soft-tissue infection [28,29]. To investigate the contribution of Rsp to the virulence of S. aureus, the mouse subcutaneous abscess model was used. Mice were administered 50 μl of PBS containing 5 × 107 live S. aureus, and the area of abscesses was measured daily after infection. As shown in Figure 6(A,B), the ability of rsp mutant strain to cause skin abscesses in mice was significantly decreased compared with the WT and rsp chromosomal-complemented strains. The bacterial recovery from the skin abscesses was also significantly decreased in mice infected with the rsp mutant strain, and the effect could be fully restored by chromosomal complementation (Figure 6(C)). These results indicated that Rsp can positively regulate the expression of virulence genes to affect the pathogenicity of S. aureus.
Figure 6.
Rsp contributes to virulence of S. aureus in a subcutaneous abscess model of mice. The mice were inoculated with 50 μl PBS containing 5 × 107 CFU of the WT, rsp mutant, and rsp chromosomal-complemented strains, or PBS alone as control, in both flanks of the back by subcutaneous injection. (A) (n = 6–8) Abscess area was measured daily with a caliper. (B) The photographic images of representative abscesses in mice 7 days after infection. (C) (n = 6) CFU recovered from each abscess harvested 7 days after infection was determined by serial dilution and plated onto TSA plates. The error bars indicate the standard errors of the means of three biological replicates. *P < .05, **P < .01, ***P < .001.
Rsp has no influence on the virulence of S. aureus Δagr strain in a subcutaneous abscess model of mice
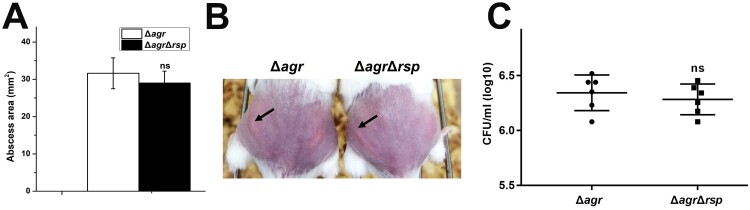
Since Rsp can directly regulate the transcription of virulence genes, the mouse subcutaneous abscess model was further used to assess the virulence of the agr mutant and agr rsp double mutant strains. As shown in Figure 7(B), abscesses without dermonecrosis were present in mice infected with the agr mutant strain, which is consistent with previous observation [57]. The area of abscesses (Figure 7(A)) and the bacterial loads (Figure 7(C)) were similar in mice administered by 5 × 107 CFU of the agr mutant and agr rsp double mutant strains. In addition, transformation of the agr mutant with pLIrsp also did not affect its pathogenicity, as measured by abscess formation abilities and bacterial loads (data not shown). These data suggested that Rsp has no influence on the virulence of the agr mutant strain in mouse skin infection model. We figured that the dramatic impact of Agr on the expression of virulence genes and pathogenicity of S. aureus might weaken the difference in virulence between these strains.
Figure 7.
Virulence of the agr mutant and agr rsp double mutant strains in a subcutaneous abscess model of mice. The mice were inoculated with 50 μl PBS containing 5 × 107 CFU of the agr mutant and agr rsp double mutant strains, or PBS alone as control, in both flanks of the back by subcutaneous injection. (A) (n = 6) Abscess area was measured 4 days after infection. (B) The photographic images of representative abscesses in mice 4 days after infection. Arrowheads indicate the abscesses of mice. (C) (n = 6) CFU recovered from each abscess harvested 4 days after infection was determined by serial dilution and plated onto TSA plates. The error bars indicate the standard errors of the means of three biological replicates. NS, not significant (P > .05).
Discussion
The success of S. aureus as an opportunistic human pathogen is typically attributed to its ability to cause a diverse array of infections and syndromes, which requires the coordination of various virulence factors and global regulators. The studies on the global virulence regulators, including Agr system, Sar family, and Sae two-component system have shown that the virulence regulation network of this bacterium is highly complex [58,59]. Thus, much remains to be elucidated to fully understand the molecular mechanisms of the virulence and pathogenesis of S. aureus. In this study we attempted to further investigate the physiological roles of Rsp in S. aureus NCTC8325.
Rsp belongs to the AraC/XylS family transcriptional regulators, which are involved in the regulation of carbon source utilization, stress responses, pathogenesis and general metabolism, either as a transcriptional activators or a transcriptional repressors [6]. The results of β-galactosidase activity assay and EMSA revealed that Rsp can specifically bind to the rsp putative promoter region to upregulate its own expression. Moreover, except HptR, all of the AraC/XylS family members of S. aureus have an atypical features with the conserved 99 amino acid region containing two helix-turn-helix (HTH) DNA binding motifs at the N-terminal and the divergent C-terminal region, which seems to be involved in environmental signals recognition and protein multimerization [6]. Recently, Dasa et al. [13] found that the transcript levels of rsp and rsp regulon were rapidly up-regulated in response to hydrogen peroxide, which is produced by neutrophils. Thus, it is reasonable to assume that Rsp can be modulated by interacting with effectors that accumulate in the infection sites and then facilitate S. aureus to adapt to the specific, often hostile environment by altering the expression of large pools of genes.
Previous studies have demonstrated that Rsp is a positive regulator of S. aureus haemolysis [12,13]. In this study, we also found that loss of rsp can significantly reduce haemolytic activity in S. aureus NCTC8325. The function of Rsp in haemolysis was further determined by introducing the pLIrsp and pRMCrsp plasmids in which rsp was highly overexpressed into the WT strain. Many secreted toxins involved in pathogenesis of S. aureus infections can influence the haemolysis. The results of qRT-PCR, β-galactosidase activity assay, western blot, and EMSA evidenced that Rsp can upregulate the expression of hla, psmα, and psmβ by directly binding to their promoter regions.
It has been reported that the expression of hla and psm is tightly regulated by the agr locus [21,36], and that Rsp can regulate the expression of virulence genes via an agr-dependent manner [12]. However, there were no dramatic differences in the expression of agrA and RNAIII between the WT and rsp mutant strains in our study, suggesting that Rsp might regulate the expression of virulence genes in an agr-independent manner in S. aureus NCTC8325. We examined the haemolysis of the agr-mutant strains, and found that Rsp can still promote the haemolysis of S. aureus in the absence of agr. We also checked the transcript levels of hla and psm and the production level of Hla in the agr-mutant strains, and the results showed the same tendency. These results suggested that Rsp can modulate the haemolytic activity of S. aureus and the transcription of virulence genes independent of agr. Meanwhile, we constructed the rsp mutant in HA S. aureus strains N315 and Newman to further investigate whether the agr-independent Rsp regulatory pathway is HA S. aureus-specific. The results of haemolysis, qRT-PCR, and western blot corroborated the regulatory effects of Rsp on the transcription of virulence genes. Mutation of rsp in S. aureus N315 displayed decreased transcription of virulence genes independent of agr. However, in S. aureus Newman, mutation of rsp was correlated with the decreased expression of agr, suggesting that Rsp regulates the transcription of virulence genes in this strain might through an agr-dependent manner. These results suggested that the regulatory effects of Rsp on the transcription of virulence genes are variable in different S. aureus strains. A previous report has shown that mutation of rsp decreased the haemolytic activity and the expression of virulence genes in S. aureus USA300 LAC, but had no impact on the expression of agr [13], further indicating that neither the agr-dependent nor agr-independent regulatory pathways mediated by Rsp are strain specific.
Since our data indicated that Rsp contributes to the haemolysis, a hallmark of S. aureus virulence, and that Rsp positively regulates the expression of Hla and PSMs, which are essential for S. aureus to cause diseases in vivo [28,29,60], we further investigated the impact of rsp on virulence using mouse subcutaneous abscess model. Unlike systemic bacteraemia model, which evaluates the ability of bacteria to survive, disseminate and proliferate, the subcutaneous abscess infection does not require dissemination, in which bacterial growth is restricted by the infiltration of polymorphonuclear leukocytes and nutrient limitations [61]. The rsp mutant strain displayed decreased virulence compared with the WT strain, as characterized by a defect in abscess formation and a blunt in bacterial recovery from the abscesses. This finding is also supported by the previous studies of S. aureus infections [12,13]. Furthermore, due to the abscess formation ability of the agr mutant was almost abolished, we failed to observe significant difference in virulence between the agr mutant and the agr rsp double mutant strains. Whether Rsp modulates the virulence of S. aureus in the absence of agr remains to be investigated by other mouse infection models.
In addition to the virulence regulation, Rsp also has been reported to play an important role in the regulation of biofilm formation in S. aureus [11,12]. In this study, we found that the rsp mutants resulted in increased biofilm formation ability compared with those of the WT strains (Figure S2(A–C)). Furthermore, compared with the agr mutant strain, the agr rsp double mutant strain displayed increased biofilm formation both in the NCTC8325 and N315 backgrounds (Figure S2(A,B)), indicating that Rsp can modulate biofilm formation in an agr-independent manner. Considering the significance of PSMs in biofilm formation [62], we assumed that the increased biofilm formation in the rsp mutant strain might partially associated with the decreased expression of psm.
In conclusion, this study has revealed that Rsp can directly bind to its own promoter region for autoregulation, and that Rsp can positively regulate the expression of virulence genes by directly binding to their promoter regions in an agr-independent manner. The contribution of Rsp to the pathogenicity of S. aureus was confirmed by using a mouse subcutaneous abscess model. These findings provide new insights into the regulatory mechanisms of virulence gene expression and S. aureus pathogenesis.
Supplementary Material
Acknowledgments
The authors thank Wen Wen and Guorong Zhang for technical assistance. The authors thank the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for providing the bacterial strains.
Funding Statement
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences [grant number XDB29020000].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 2.Tong SY, Davis JS, Eichenberger E, et al. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freer JH, Arbuthnott JP.. Toxins of Staphylococcus aureus. Pharmacol Ther. 1982;19:55–106. doi: 10.1016/0163-7258(82)90042-0 [DOI] [PubMed] [Google Scholar]
- 4.Foster TJ, Hook M.. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/S0966-842X(98)01400-0 [DOI] [PubMed] [Google Scholar]
- 5.Cheung AL, Bayer AS, Zhang G, et al. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40:1–9. doi: 10.1016/S0928-8244(03)00309-2 [DOI] [PubMed] [Google Scholar]
- 6.Gallegos MT, Schleif R, Bairoch A, et al. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/.61.4.393-410.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibarra JA, Perez-Rueda E, Carroll RK, et al. Global analysis of transcriptional regulators in Staphylococcus aureus. BMC Genomics. 2013;14:126. doi: 10.1186/1471-2164-14-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim Y, Jana M, Luong TT, et al. Control of glucose-and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J Bacteriol. 2004;186:722–729. doi: 10.1128/JB.186.3.722-729.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cue D, Lei MG, Luong TT, et al. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol. 2009;191:6363–6373. doi: 10.1128/JB.00913-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cue D, Lei MG, Lee CY.. Activation of sarX by Rbf is required for biofilm formation and icaADBC expression in Staphylococcus aureus. J Bacteriol. 2013;195:1515–1524. doi: 10.1128/JB.00012-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lei MG, Cue D, Roux CM, et al. Rsp Inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J Bacteriol. 2011;193:5231–5241. doi: 10.1128/JB.05454-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, He L, Song Y, et al. AraC-type regulator Rsp adapts Staphylococcus aureus gene expression to acute infection. Infect Immun. 2015;84:723–734. doi: 10.1128/IAI.01088-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasa S, Lindemann C, Young BC, et al. Natural mutations in a Staphylococcus aureus virulence regulator attenuate cytotoxicity but permit bacteremia and abscess formation. Proc Natl Acad Sci USA. 2016;113:E3101–E3110. doi: 10.1073/pnas.1520255113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chua KYL, Monk IR, Lin YH, et al. Hyperexpression of alpha-hemolysin explains enhanced virulence of sequence type 93 community-associated methicillin-resistant Staphylococcus aureus. BMC Microbiol. 2014;14:31. doi: 10.1186/1471-2180-14-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JY, Kim JW, Moon BY, et al. Characterization of a novel two-component regulatory system, HptRS, the regulator for the hexose phosphate transport system in Staphylococcus aureus. Infect Immun. 2015;83:1620–1628. doi: 10.1128/IAI.03109-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Sun H, Liu X, et al. Regulatory mechanism of the three-component system HptRSA in glucose-6-phosphate uptake in Staphylococcus aureus. Med Microbiol Immunol. 2016;205:241–253. doi: 10.1007/s00430-015-0446-6 [DOI] [PubMed] [Google Scholar]
- 17.Novick RP, Projan SJ, Kornblum J, et al. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645 [DOI] [PubMed] [Google Scholar]
- 18.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x [DOI] [PubMed] [Google Scholar]
- 19.George EA, Muir TW.. Molecular mechanisms of agr quorum sensing in virulent staphylococci. Chembiochem. 2007;8:847–855. doi: 10.1002/cbic.200700023 [DOI] [PubMed] [Google Scholar]
- 20.Boisset S, Geissmann T, Huntzinger E, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Queck SY, Jameson-Lee M, Villaruz AE, et al. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhakdi S, Tranum-Jensen J.. Alpha-toxin of Staphylococcus aureus. Microbiol Rev. 1991;55:733–751. doi: 10.1128/MMBR.55.4.733-751.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilke GA, Bubeck Wardenburg J.. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus alpha-hemolysin-mediated cellular injury. Proc Natl Acad Sci USA. 2010;107:13473–13478. doi: 10.1073/pnas.1001815107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wardenburg JB, Bae T, Otto M, et al. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–1406. doi: 10.1038/nm1207-1405 [DOI] [PubMed] [Google Scholar]
- 25.Wardenburg JB, Schneewind O.. Vaccine protection against Staphylococcus aureus pneumonia. J Exp Med. 2008;205:287–294. doi: 10.1084/jem.20072208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powers ME, Becker REN, Sailer A, et al. Synergistic action of Staphylococcus aureus alpha-toxin on platelets and myeloid lineage cells contributes to lethal sepsis. Cell Host Microbe. 2015;17:775–787. doi: 10.1016/j.chom.2015.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonsson P, Lindberg M, Haraldsson I, et al. Virulence of Staphylococcus aureus in a mouse mastitis model: studies of alpha hemolysin, coagulase, and protein A as possible virulence determinants with protoplast fusion and gene cloning. Infect Immun. 1985;49:765–769. doi: 10.1128/IAI.49.3.765-769.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy AD, Bubeck Wardenburg J, Gardner DJ, et al. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J Infect Dis. 2010;202:1050–1058. doi: 10.1086/656043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R, Braughton KR, Kretschmer D, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656 [DOI] [PubMed] [Google Scholar]
- 30.Janzon L, Lofdahl S, Arvidson S.. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol Gen Genet. 1989;219:480–485. doi: 10.1007/BF00259623 [DOI] [PubMed] [Google Scholar]
- 31.Kretschmer D, Gleske AK, Rautenberg M, et al. Human formyl peptide receptor 2 senses highly pathogenic Staphylococcus aureus. Cell Host Microbe. 2010;7:463–473. doi: 10.1016/j.chom.2010.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otto M. Staphylococcus aureus toxins. Curr Opin Microbiol. 2014;17:32–37. doi: 10.1016/j.mib.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi SD, Malachowa N, Whitney AR, et al. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis. 2011;204:937–941. doi: 10.1093/infdis/jir441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rautenberg M, Joo HS, Otto M, et al. Neutrophil responses to staphylococcal pathogens and commensals via the formyl peptide receptor 2 relates to phenol-soluble modulin release and virulence. FASEB J. 2011;25:1254–1263. doi: 10.1096/fj.10-175208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Diep BA, Villaruz AE, et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morfeldt E, Taylor D, von Gabain A, et al. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan PF, Foster SJ.. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/JB.180.23.6232-6241.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giraudo AT, Cheung AL, Nagel R.. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch Microbiol. 1997;168:53–58. doi: 10.1007/s002030050469 [DOI] [PubMed] [Google Scholar]
- 39.McNamara PJ, Milligan-Monroe KC, Khalili S, et al. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol. 2000;182:3197–3203. doi: 10.1128/JB.182.11.3197-3203.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt KA, Manna AC, Gill S, et al. Sart, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect Immun. 2001;69:4749–4758. doi: 10.1128/IAI.69.8.4749-4758.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duthie ES, Lorenz LL.. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. [DOI] [PubMed] [Google Scholar]
- 42.Dassy B, Hogan T, Foster TJ, et al. Involvement of the accessory gene regulator (agr) in expression of type 5 capsular polysaccharide by Staphylococcus aureus. J Gen Microbiol. 1993;139:1301–1306. doi: 10.1099/00221287-139-6-1301 [DOI] [PubMed] [Google Scholar]
- 43.Hu J, Zhang X, Liu X, et al. Mechanism of reduced vancomycin susceptibility conferred by walK mutation in community-acquired methicillin-resistant Staphylococcus aureus strain MW2. Antimicrob Agents Chemother. 2015;59:1352–1355. doi: 10.1128/AAC.04290-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CY, Buranen SL, Ye ZH.. Construction of single-copy integration vectors for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-V [DOI] [PubMed] [Google Scholar]
- 45.Corrigan RM, Foster TJ.. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid. 2009;61:126–129. doi: 10.1016/j.plasmid.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 46.Schneewind O, Mihaylova-Petkov D, Model P.. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen W, Liu B, Xue L, et al. Autoregulation and virulence control by the toxin-antitoxin system SavRS in Staphylococcus aureus. Infect Immun. 2018;86:e00032-18. doi: 10.1128/IAI.00032-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bae T, Schneewind O.. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 49.Valihrach L, Demnerova K.. Impact of normalization method on experimental outcome using RT-qPCR in Staphylococcus aureus. J Microbiol Methods. 2012;90:214–216. doi: 10.1016/j.mimet.2012.05.008 [DOI] [PubMed] [Google Scholar]
- 50.Jiang Q, Jin Z, Sun B.. Mgra negatively regulates biofilm formation and detachment by repressing the expression of psm operons in Staphylococcus aureus. Appl Environ Microbiol. 2018;84:e01008-18. doi: 10.1128/AEM.01008-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu GY, Essex A, Buchanan JT, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–215. doi: 10.1084/jem.20050846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yarwood JM, Bartels DJ, Volper EM, et al. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemotte PK, Walker GC.. Induction and autoregulation of ada, a positively acting element regulating the response of Escherichia coli K-12 to methylating agents. J Bacteriol. 1985;161:888–895. doi: 10.1128/JB.161.3.888-895.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Froehlich B, Husmann L, Caron J, et al. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J Bacteriol. 1994;176:5385–5392. doi: 10.1128/JB.176.17.5385-5392.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato JY, Ohnishi Y, Horinouchi S.. Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. J Mol Biol. 2005;350:12–26. doi: 10.1016/j.jmb.2005.04.058 [DOI] [PubMed] [Google Scholar]
- 56.Ma R, Qiu S, Jiang Q, et al. AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int J Med Microbiol. 2017;307:257–267. doi: 10.1016/j.ijmm.2017.03.003 [DOI] [PubMed] [Google Scholar]
- 57.Montgomery CP, Boyle-Vavra S, Daum RS.. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PloS One. 2010;5:e15177. doi: 10.1371/journal.pone.0015177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bronner S, Monteil H, Prevost G.. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev. 2004;28:183–200. doi: 10.1016/j.femsre.2003.09.003 [DOI] [PubMed] [Google Scholar]
- 59.Bronesky D, Wu ZF, Marzi S, et al. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu Rev Microbiol. 2016;70:299–316. doi: 10.1146/annurev-micro-102215-095708 [DOI] [PubMed] [Google Scholar]
- 60.Rauch S, DeDent AC, Kim HK, et al. Abscess formation and alpha-hemolysin induced toxicity in a mouse model of Staphylococcus aureus peritoneal infection. Infect Immun. 2012;80:3721–3732. doi: 10.1128/IAI.00442-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coulter SN, Schwan WR, Ng EY, et al. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x [DOI] [PubMed] [Google Scholar]
- 62.Periasamy S, Joo HS, Duong AC, et al. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci USA. 2012;109:1281–1286. doi: 10.1073/pnas.1115006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.