Abstract
We describe the isolation and characterization of Fusarium volatile from a bronchoalveolar lavage (BAL) sample of a female patient living in French Guiana with underlying pulmonary infections. Phylogenetic analysis of fragments of the calmodulin (cmdA), translation elongation factor (tef1), RNA polymerase second largest subunit (rpb2), and β-tubulin (tub) loci revealed that strain CBS 143874 was closely related to isolate NRRL 25615, a known but undescribed phylogenetic species belonging to the African clade of the Fusarium fujikuroi species complex. The fungus differed phylogenetically and morphologically from related known species, and is therefore described as the new taxon Fusarium volatile. Antifungal susceptibility testing suggested that the new species is resistant to echinocandins, fluconazole, itraconazole with lower MICs against amphotericin B, voriconazole and posaconazole.
Keywords: clinical samples, FFSC, French Guiana, fungal taxonomy, phylogeny
INTRODUCTION
Species of the fujikuroi species complex (FFSC) of the genus Fusarium have been extensively studied in view of their ability to cause infections in plants and to produce toxins that may lead to food poisoning (Chilaka et al. 2017). Moreover, some members of the FFSC have repeatedly been reported from opportunistic infections in humans, which may be mild or local in the case of onychomycosis and keratitis, or invasive and severe in individuals with extended burn wounds and bone marrow transplant recipients. In addition, systemic and disseminated infections occur in severely immunocompromised patients (Guarro 2013, Al-Hatmi et al. 2016b, de Hoog et al. 2019).
The FFSC is one of the larger groups within the genus Fusarium and contains species with diverse ecologies (Nirenberg & O’Donnell 1998, O’Donnell et al. 2000). Species of the FFSC are characterised by forming yellow, orange or purple colonies on potato dextrose agar (PDA); globose, oval, napiform or clavate microconidia are borne in chains and false heads on mono- and polyphialides, while different combinations of microconidial morphologies and phialide types can coexist; the macroconidia are thin-walled, almost straight to slightly curved, with a well-developed pedicellate basal cell. Chlamydospores are rarely formed in the FFSC, although they can be present in some species, formed mostly intercalarily on the hyphae, grouped in chains or clusters (Nirenberg & O’Donnell 1998, Leslie & Summerell 2006).
Molecular studies suggested that at least 50 phylogenetic lineages may be recognized within FFSC. Three major clades with limited biogeographic distribution can be distinguished, and these are termed the African, American and Asian clades (O’Donnell et al. 1998). Thus far 16 species of the FFSC have been reported to cause human infections, namely F. acutatum, F. ananatum, F. andiyazi, F. anthophilum, F. fujikuroi, F. guttiforme, F. musae, F. napiforme, F. nygamai, F. proliferatum, F. ramigenum, F. sacchari, F. subglutinans, F. temperatum, F. thapsinum and F. verticillioides (Al-Hatmi et al. 2016a, de Hoog et al. 2019).
Identification to species level in FFSC is often difficult because of the high morphological diversity and intraspecific variation of microscopic features (O’Donnell et al. 2015). Currently known species of the FFSC are morphologically and genetically very similar and often can only be reliably separated using multilocus molecular analyses (Geiser et al. 2004). However, considering that some members of the FFSC are relevant human opportunistic pathogens or important toxin producers, their correct identification is crucial. The present paper describes a species of FFSC, based on isolate CBS 143874 collected from a human patient specimen in Cayenne, French Guiana, and characterised by morphological and phylogenetic methods. A morphological identification key is provided to identify the novel species and additional FFSC species known from human clinical specimens.
CASE REPORT
The patient was a 22-yr-old woman, native of Brazil, but has lived in French Guiana for 8 yr. She presented at the Internal Medicine Department of the Andrée Rosemon hospital centre in Cayenne (French Guiana) with a 4-mo history of chronic asthenia at the moment of clinical examination. A right cervical, sub-angulomaxillary adenophlegmon had appeared 1 mo earlier. She did not have any other clinical signs and had no cough or weight loss and denied night sweats. She had antecedents of disseminated lupus erythematous diagnosed a year before and was treated with an association of hydroxychloroquine and corticoids. Corticotherapy had been interrupted 4 mo earlier.
Chest computed tomography revealed the presence of a left apical pulmonary cavern with alveolar opacity. Computed tomography of the head and neck highlighted the presence of several cervical necrotic adenomegalies on the right side associated with an abscessed subcutaneous collection. Inflammatory syndrome was mild (C-reactive protein = 7 mg/L). A chest radiography suggested the presence of an aspergilloma, with an image compatible with an Aspergillus ball into the pulmonary cavern.
Presence of bacteria and fungi was assessed by culturing a broncho-alveolar fluid (BAL) sample. Cultures were negative for bacterial growth including mycobacteria. Cultures for fungi were positive for a fusarium-like fungus. Although there was no evidence of bacterial infection, the patient was diagnosed as having a tuberculosis adenitis based on the presence of a pulmonary cavern at radiography. She was treated with the following antituberculosis quadritherapy (Ethambutol, Isoniazide, Pyrazinamide and Rifampicine). Fusarium growth was not taken into consideration due to the absence of pulmonary and systemic clinical signs. Patient evolution under treatment was spectacular with a rapid regression of adenitis and a diminution of the asthenia. At 6 mo, lesions had completely disappeared at computed tomography. The Fusarium isolate was sent to the Westerdijk Fungal Biodiversity Institute (WI), Utrecht, The Netherlands, for further characterization.
MATERIALS AND METHODS
Strains
A Fusarium sp. strain was isolated from BAL fluid specimen of the patient in French Guiana and submitted to the WI, under accession number CBS 143874. Ex-type and reference strains spanning the known diversity of the FFSC were selected based on phylogenetic and morphological similarity to the strain under study (Table 1). The clinical isolate was grown on PDA slants for 7 d at 25 °C, and was maintained as working culture or stored in 20 % (v/v) glycerol at −80 °C for prolonged use.
Table 1.
GenBank accession numbers of Fusarium species included in this study.
| Species | Collection1 | Source | Country |
GenBank/ENA accession number2 |
|||
|---|---|---|---|---|---|---|---|
| cmdA | rpb2 | tef1 | tub | ||||
| F. agapanthi | NRRL 54463T | Agapanthus praecox | Australia | KU900611 | KU900625 | KU900630 | KU900635 |
| NRRL 54464 | Agapanthus praecox | Australia | KU900613 | KU900627 | KU900632 | KU900637 | |
| F. ananatum | CBS 118516T | Ananas comosus fruit | South Africa | LT996175 | LT996137 | LT996091 | LT996112 |
| CBS 118517 | Ananas comosus fruit | South Africa | KU603990 | KU604273 | KU604480 | KU603895 | |
| F. andiyazi | CBS 119857T | Sorghum bicolor soil debris | South Africa | LT996176 | LT996138 | LT996092 | LT996113 |
| CBS 134430 | Human | Turkey | KU603956 | KU604232 | KU604450 | KU603867 | |
| F. anthophilum | CBS 737.97 | Hippeastrum sp. | Germany | LT996177 | LT996139 | LT996093 | LT996114 |
| CBS 119859 | Cymbidium sp. leaf spot | New Zealand | KU603988 | KU604279 | KU604427 | KU603931 | |
| F. bactridioides | NRRL 20476 | Cronartium conigenum | USA | AF158343 | - | AF160290 | U34434 |
| F. begoniae | CBS 403.97 | Begonia elatior hybrid | Germany | AF158346 | LT996140 | AF160293 | U61543 |
| NRRL 31848 | Begonia elatior hybrid | USA | - | - | AY329035 | AY329044 | |
| F. bulbicola | CBS 220.76T | Nerine bowdenii | Germany | KF466327 | KF466404 | KF466415 | KF466437 |
| F. circinatum | CBS 405.97T | Pinus radiata | USA | KM231393 | HM068354 | KM231943 | KM232080 |
| F. coicis | NRRL 66233T | Coix gasteenii | Australia | LT996178 | KP083274 | KP083251 | LT996115 |
| F. ficicrescens | CBS 125178T | Ficus carica fruit | Iran | KU603958 | KT154002 | KU604452 | KP662896 |
| CBS 125181 | Ficus carica fruit | Iran | KU603959 | KT154003 | KU604453 | KP662897 | |
| F. lactis | CBS 411.97NT | Ficus carica | USA | AF158325 | LT996149 | AF160272 | U61551 |
| NRRL 31630 | Capsicum annuum | Belgium | FR870301 | FR870313 | FR870289 | FR870325 | |
| F. mexicanum | NRRL 47473 | Mangifera indica infloresence | Mexico | GU737389 | Not public | GU737416 | GU737308 |
| NRRL 53580 | Mangifera indica infloresence | Mexico | GU737394 | - | GU737421 | GU737367 | |
| F. nygamai | NRRL 13448T | Necrotic Sorghum root | Australia | AF158326 | EF470114 | AF160273 | U34426 |
| NRRL 26421 | Human | Egypt | KU603949 | EF470127 | HM347121 | KU603865 | |
| F. oxysporum | CBS 716.74 | Vicia faba | Germany | AF158366 | JX171583 | AF008479 | U34435 |
| CBS 744.97 | Pseudotsuga menziesii | USA | AF158365 | LT575065 | AF160312 | U34424 | |
| F. phyllophilum | CBS 216. 76T | Dracaena deremensis leaf | Italy | KF466333 | KF466410 | KF466421 | KF466443 |
| F. pseudocircinatum | CBS 449.97T | Solanum sp. | Ghana | AF158324 | LT996151 | AF160271 | U34427 |
| F. pseudonygamai | CBS 417.97T | Pennisetum typhoides | Nigeria | AF158316 | LT996152 | AF160263 | U34421 |
| F. ramigenum | CBS 418.98T | Ficus carica | USA | KF466335 | KF466412 | KF466423 | KF466445 |
| Fusarium sp. | NRRL 25346 | Pine pitch canker | USA | AF158349 | - | AF160296 | U61642 |
| NRRL 26756 | Plant leaf litter | Cuba | AF158360 | Not public | AF160307 | AF160336 | |
| NRRL 26757 | Pine pitch canker | USA | AF158361 | - | AF160308 | AF160351 | |
| F. subglutinans | CBS 747.97 | Zea mays | USA | AF158342 | JX171599 | AF160289 | U34417 |
| F. sudanense | CBS 454.97T | Striga hermonthica | Sudan | LT996185 | LT996155 | KU711697 | KU603909 |
| F. temperatum | NRRL 25622 | Zea mays | South Africa | AF158354 | Not public | AF16030 | AF160317 |
| F. terricola | CBS 483.94T | Soil | Australia | KU603951 | LT996156 | KU711698 | KU603908 |
| F. thapsinum | CBS 733.97 | Sorghum bicolor | South Africa | LT996186 | JX171600 | AF160270 | U34418 |
| F. tjaetaba | NRRL 66243T | Sorghum interjectum | Australia | LT996187 | KP083275 | KP083263 | GU737296 |
| F. udum | NRRL 22949 | Lactarius pubescens | Germany | AF158328 | LT996172 | AF160275 | U34433 |
| F. verticillioides | CBS 734.97 | Zea mays | Germany | AF158315 | EF470122 | AF160262 | U34413 |
| CBS 115135 | Human | Sweden | KU603944 | KU604217 | KU604384 | KU603861 | |
| F. volatile | CBS 143874T | Human bronchoalveolar lavage fluid | French Guiana | MK984595 | LR596006 | LR596007 | LR596008 |
| NRRL 25615 | Oryza sativa seed | Nigeria | AF158357 | - | AF160304 | AF160348 | |
| F. werrikimbe | CBS 125535T | Sorghum leiocladum | Australia | - | - | EF107131 | EF107133 |
| F19361 | Sorghum leiocladum | Australia | - | - | EF107132 | EF107134 | |
| F. xylarioides | NRRL 25486T | Coffea trunk | Ivory Coast | - | HM068355 | AY707136 | AY707118 |
1 CBS: Westerdijk Fungal Biodiverity Institute, Utrecht, The Netherlands. F: University of Sydney, Sydney, New South Wales, Australia. NRRL: Agricultural Research Service Culture Collection, National Center for Agricultural Utilization Research, USDA, Peoria, IL, USA. NT: ex-neotype. T: ex-type.
2 ENA: European Nucleotide Archive. cmdA: calmodulin. rpb2: RNA polymerase second largest subunit. tef1: translation elongation factor–alpha. tub: β-tubulin. Sequences marked as Not public are available on the sequence datasets published by Edwards et al. (2016).
Morphology
CBS 143874 was characterised morphologically following procedures described elsewhere (Aoki et al. 2013, Leslie & Summerell 2006, Sandoval-Denis et al. 2018). Colony growth rates and production of diffusible pigments were evaluated on PDA, colony features were also recorded on malt extract agar (MEA) and oatmeal agar (OA). Colour notations followed those of Rayner (1970). Micro-morphological features were studied from cultures grown for 7–10 d at 24 °C, using a 12 h light/dark cycle with near-UV and white fluorescent light. Features of the aerial and sporodochial conidiophores, conidia and production of chlamydospores were assessed on synthetic nutrient-poor agar (SNA; Nirenberg 1976) and on carnation leaf agar (CLA; Fisher et al. 1982). Measurements and photomicrographs were recorded using sterile water as mounting medium and a Nikon Eclipse 80i (Nikon, Tokyo, Japan) microscope with Differential Interference Contrast (DIC) optics and a Nikon AZ100 dissecting microscope, both equipped with a Nikon DS-Ri2 or a Nikon DS-5 high definition colour digital camera and the Nikon software NIS-elements D software v. 4.30.
Growth rates
Cardinal growth temperatures were determined on MEA and PDA plates incubated in the dark for 2 wk at temperatures of 18–40 °C at intervals of 3 °C; with two replicates for each isolate. Average growth rates per species were calculated and expressed as diametric growth per 24 h.
DNA extraction, amplification and sequencing
The Wizard® Genomic DNA purification Kit (Promega, Madison, WI, USA) was used to extract total genomic DNA from fresh mycelium scraped from the surface of 7-d-old cultures on MEA at 24 °C. Partial fragments of four loci were PCR-amplified following previously published protocols using the following primer pairs: BT-2a/BT-2b for the β-tubulin gene (tub) (Glass & Donaldson 1995), CL1/CL2 for the calmodulin gene (cmdA) (O’Donnell et al. 2009), EF-1/EF-2 for the translation elongation factor-alpha gene (tef1) (O’Donnell et al. 1998), and RPB2-5f2/7cr plus RPB2-7cf/11ar for two non-contiguous fragments of the RNA polymerase second largest subunit (rpb2) (Liu et al. 1999, Sung et al. 2007). Sequencing was done in both directions using the respective PCR primers on an Applied Biosystems 3730xl DNA Analyzer (Life Technologies, Carlsbad, CA, USA). The DNA sequences were analysed and consensus sequences were assembled using SeqMan Pro v. 13 (DNAStar, Madison, WI, USA).
Phylogenetic analyses
Phylogenetic analyses of single loci and the combined dataset were carried out using three independent algorithms: Maximum-likelihood (ML), Maximum Parsimony (MP) and Bayesian inference (BI). Both ML and BI were run on the CIPRES Science Gateway portal (Miller et al. 2012). Evolutionary models were calculated with MrModelTest v. 2.3 using the Akaike information criterion (Nylander 2004). For ML, RAxML-HPC2 v. 8.2.10 on XSEDE was used (Stamatakis 2014), with a bootstrap analysis (BS) based on default parameters. The BI analyses were run using MrBayes v. 3.2.6 on XSEDE (Ronquist & Huelsenbeck 2003) using four incrementally heated MCMC chains for 5 M generations and a sample frequency of every 1 000 trees. The 50 % consensus trees and posterior probabilities (PP) values were calculated after discarding the first 25 % of samples as burn-in. Maximum-parsimony analyses were run using PAUP v. 4.0b10 (Swofford 2003). Heuristic searches included 1 000 random stepwise addition replicates, with tree bisection and reconstruction (TBR) branch swapping; all characters were equally weighted and gaps treated as missing data. Branches of zero length were collapsed and all multiple, equally parsimonious trees were saved. Tree statistics [tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency index (RC)] were calculated. Clade stability was evaluated using a bootstrap analysis (BS) of 1 000 replicates.
Antifungal susceptibility
Antifungal susceptibility testing of CBS 143874 was performed by the CLSI broth microdilution as described in the CLSI document M38-A2 (Clinical and Laboratory Standards Institute 2008) with modifications according to Al-Hatmi et al. (2015). The following drugs were used: amphotericin B (Sigma-Aldrich), fluconazole (Pfizer, Groton, CT, USA), itraconazole (Janssen Pharmaceutica, Tilburg, The Netherlands), voriconazole (Pfizer), posaconazole (Merck), isavuconazole (Basilea Pharmaceutica, Basel, Switzerland), micafungin (Astellas, Ibaraki, Japan), and anidulafungin (Pfizer). Three reference strains (Paecilomyces variotii ATCC 22319, Candida krusei ATCC 6258, and Candida parapsilosis ATCC 22019) were included as quality controls.
RESULTS
Molecular analyses
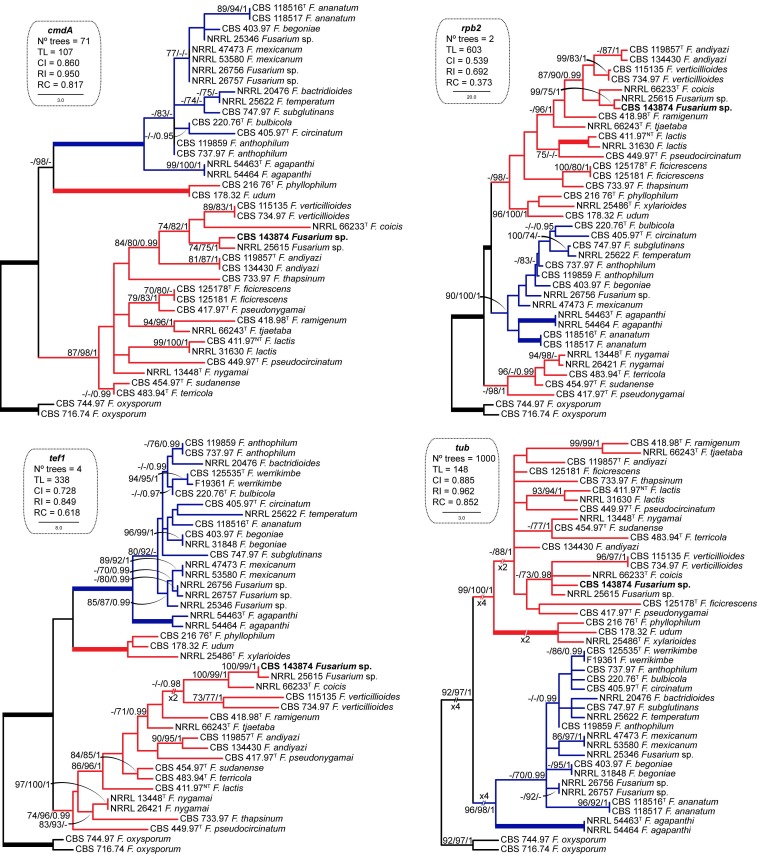
Topologies obtained from the analyses of the majority of individual gene datasets were congruent, differing only in the positions of unsupported nodes. Individual analyses of the cmdA, rpb2, tef1 and tub loci consistently resolved Fusarium sp. CBS 143874 as a member of the African clade sensu O’Donnell et al. (1998) (Fig. 1). Phylogenies based on cmdA, rpb2 and tef sequences showed that strain CBS 143874, together with an unidentified isolate (NRRL 25615) formed a genetically exclusive lineage, phylogenetically related to F. coicis and F. verticillioides. In contrast, the tub phylogeny failed to unambiguously identify most of the Fusarium species included in this study, showing mostly marginal statistical support values that did not allow unequivocal separation of lineages. Nevertheless, this locus confirmed the phylogenetic position of CBS 143874 and NRRL 25615 as close relatives of F. coicis. With the exception of tub, single locus phylogenies did not support the monophyly of the African clade of FFSC, which resolved as polyphyletic.
Fig. 1.
Maximum Parsimony (MP) trees obtained from the individual phylogenetic analyses of the cmdA, rpb2, tef1 and tub datasets of representative isolates of the Fusarium fujikuroi species complex. Numbers on the nodes are MP and Maximum-Likelihood (ML) bootstrap values (BS) above 70 % and Bayesian posterior probability values (PP) above 0.95. Thickened branches indicate full statistical support (MP-BS, ML-BS = 100 % and PP = 1). Coloured branches indicate the African (red) and American (blue) clades according to O’Donnell et al. (1998). The clinical isolate is highlighted in bold. Ex-type and ex-neotype strains are indicated with T and NT, respectively. The trees are rooted with Fusarium oxysporum CBS 744.97 and CBS 716.74. TL = tree length, CI = consistency index, RI = retention index, RC = rescaled consistency index.
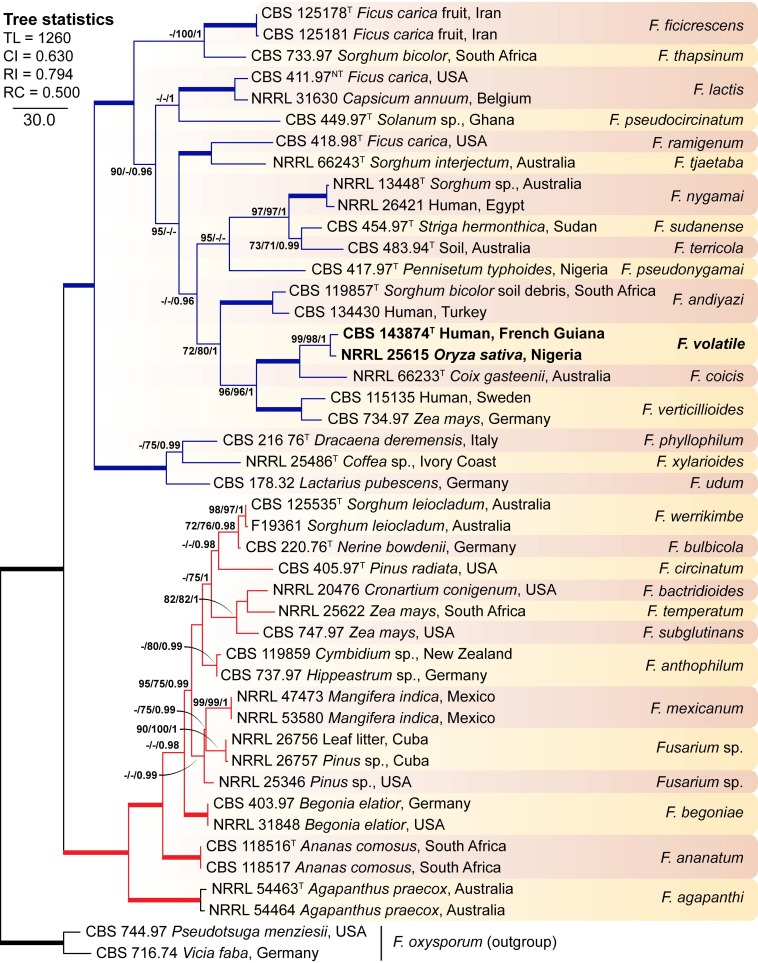
The final, combined alignment encompassed a total of 3 305 positions (cmdA 545, rpb2 1 591, tef1 680, tub 489), of which 696 sites were variable (cmdA 86, rpb2 295, tef1 194, tub 121) and 452 were phylogenetically informative (cmdA 61, rpb2 204, tef1 119, tub 68). The combined phylogeny confirmed the results of the individual phylogenetic analyses, Fusarium sp. strains CBS 143874 and NRRL 25615 formed a highly-supported, genetically exclusive group, closely related to but different from F. coicis and F. verticillioides (Fig. 2); consequently, the above-mentioned phylogenetic clade is proposed here as the new species Fusarium volatile.
Fig. 2.
The first of 16 most parsimonious trees obtained from the combined cmdA, rpb2, tef1 and tub sequences of 43 strains belonging to the Fusarium fujikuroi species complex (FFSC). Numbers on the nodes are MP and Maximum-Likelihood (ML) bootstrap values (BS) above 70 % and Bayesian posterior probability values (PP) above 0.95. Thickened branches indicate full statistical support (MP-BS, ML-BS = 100 % and PP = 1). Coloured branches indicate the African (red) and American (blue) clades according to O’Donnell et al. (1998). Isolates and name of the new species are highlighted in bold. Ex-type and ex-neotype strains are indicated with T and NT, respectively. The tree is rooted with Fusarium oxysporum CBS 744.97 and CBS 716.74. TL = tree length, CI = consistency index, RI = retention index, RC = rescaled consistency index.
Antifungal susceptibility testing
Antifungal susceptibility testing according to CLSI M38A (Clinical and Laboratory Standards Institute 2008) demonstrated that Fusarium volatile had a low MIC of 1 μg/mL against amphotericin B, voriconazole (1 μg/mL), and posaconazole (0.5 μg/mL), whereas the fungus had high MICs for fluconazole (>64 μg/mL), itraconazole (>16 μg/mL), isavuconazole (4 μg/mL), anidulafungin (>8 μg/mL), and micafungin (>8 μg/mL) (Table 2).
Table 2.
MIC values of clinical isolate CBS 143874 (μg/mL).
| Strain | AMB | FLC | ITC | VOR | POS | ISA | ANI | MICA |
|---|---|---|---|---|---|---|---|---|
| CBS 143874 | 1 | >64 | >16 | 1 | 0.5 | 4 | >8 | >8 |
AMB: amphotericin B. FLC: fluconazole. ITC: itraconazole. VOR: voriconazole. POS: posaconazole. ISA: isavuconazole. ANI: anidulafungin. MICA: micafungin.
Taxonomy
Fusariumvolatile Al-Hatmi, Sand.-Den., S.A. Ahmed & de Hoog, sp. nov. MycoBank MB831243. Figs 3, 4.
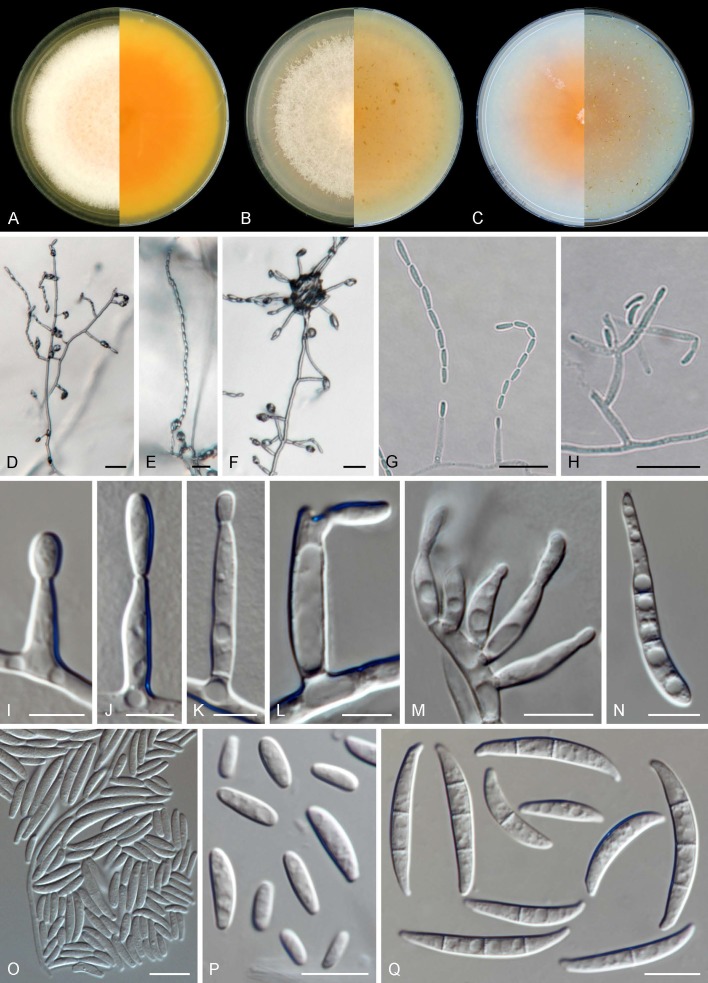
Fig. 3.
Fusarium volatile (ex-type CBS 143874). A–C. Colonies (left obverse, right reverse) on MEA, PDA and OA, respectively, after 14 d at 24 °C. D–H. Aerial conidiophores and chains of conidia. I–L. Aerial phialides. M. Sporodochial phialides. N. Aerial conidia showing microcyclic conidiation. O, P. Aerial conidia. Q. Sporodochial conidia. Scale bars: D–H = 20 μm, I–L = 5 μm, all others = 10 μm.
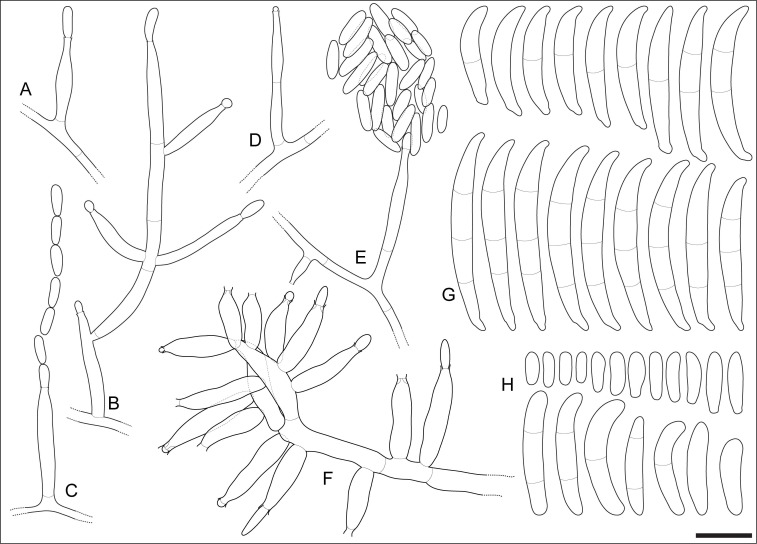
Fig. 4.
Fusarium volatile (ex-type CBS 143874). A–E. Aerial conidiophores. F. Sporodochial conidiophore. G. Sporodochial conidia. H. Aerial conidia. Scale bar = 10 μm.
Etymology: Named after its supposedly airborne entry in the human patient.
Typus: French Guiana, Cayenne, bronchoalveolar lavage (BAL) effusion of patient with lung infection, 2017, M. Demar (holotype CBS H-24004, culture ex-type CBS 143874).
Mycelium consisting of hyaline, smooth, branched, 1.5–5.5 μm diam hyphae. Aerial conidiophores erect, often reduced to conidiogenous cells, borne laterally on hyphae, less commonly irregularly or verticillately branched, up to 80 μm long, rarely proliferating, bearing terminal single mono- and polyphialides; aerial phialides subulate to subcylindrical, smooth- and thin-walled, (7–)11.5–21(–29) × 2–4.5 μm (av. 16.2 × 3.3 μm), commonly bearing a single conidiogenous locus, rarely polyphialidic, lacking noticeable periclinal thickening or collarettes; aerial conidia ellipsoid, allantoid, club-shaped to somewhat cylindrical, often with a flattened base, straight or gently curved, smooth- and thin-walled, 0–2(–3)-septate, (4.5–)5.5–19(–30) × (1.5–)2–4(–5.5) μm (av. 12.4 × 3.1 μm), grouped in moderately long, straight or flexuous chains, quickly collapsing to form discrete conidial heads; microcyclic conidiation present. Sporodochia produced infrequently in the aerial mycelium, rarely on the surface of carnation leaves, tan to pale orange; sporodochial conidiophores simple or sparingly irregularly branched, 16–21(–22) × (3–)3.5–4.5 μm (av. 18.6 × 3.8 μm), bearing terminal and lateral single monophialides or terminal whorls of up to 4 monophialides; sporodochial phialides doliiform to subcylindrical, smooth- and thin-walled, (9.5–)11–15(–19) × (2.5–)3–4(–4.5) μm (av. 12.9 × 3.6 μm), sporodochial conidia falcate, straight or dorsiventrally curved, curvature often more distinctly pronounced on the dorsal line, tapering toward the basal part; apical cell conical and slightly hooked; basal cell foot-shaped, (0–)1–3-septate, hyaline, thin- and smooth-walled. Aseptate conidia (14–)15–20.5 × 3–4 μm (av. 17.8 × 3.5 μm), 1-septate conidia (17–)19–24.5(–27) × (2–)3–4 μm (av. 21.7 × 3.5 μm), 2-septate conidia (26–)27–30(–31) × 3.5–4 μm (av. 28.4 × 3.9 μm), 3-septate conidia (27–)29–34.5(–36.5) × (3–)3.5–4(–4.5) μm (av. 31.7 × 4 μm); overall (14–)21–32.5(–36.5) × (2–)3–4.5 μm (av. 26.6 × 3.8 μm). Chlamydospores not observed.
Colonies growing in the dark after 7 d at 24 °C. On MEA reaching 50–68 mm diam, white, salmon to peach; colony surface raised to slightly umbonate, velvety felty; margin undulate to filiform; reverse orange to luteous with pale luteous periphery. On OA reaching 65–74 mm diam, saffron to rosy buff, turning pale vinaceous toward the periphery, membranous; margin entire with abundant submerged mycelium; reverse saffron to salmon, pale orange at the centre. On PDA with an average radial growth rate of 4.7–5.7 mm/d, reaching 60–80 mm diam, salmon, saffron to pale ochraceous with peach centre; flat with slightly raised centre, felty to cottony; margin irregular, undulate to lobate; reverse pale orange to ochraceous.
Cardinal growth temperatures: optimal development at 27–33 °C, minimum 18 °C, maximum 37 °C. The species was still able to grow at 37 °C, but not at 40 °C.
Notes: Fusarium volatile is phylogenetically closely related to F. coicis and F. verticillioides. The three mentioned species share the common morphological features attributed to the FFSC, such as the lack of chlamydospores, formation of oval to clavate microconidia and presence of monophialides, while sporodochia are not commonly produced. However, F. coicis and F. volatile differ significantly from F. verticillioides by having up to 3-septate microconidia (usually aseptate in F. verticillioides) and the additional presence, although rare, of polyphialides and microconidia formed on false heads and chains (strictly monophialidic, forming chains of conidia in F. verticillioides; Leslie & Summerell 2006). Fusarium volatile differs from F. coicis by its much shorter and less septate, curved macroconidia (up to 123 μm long, 4–10-septate and almost straight in F. coicis; Laurence et al. 2015).
Key to species of the Fusarium fujikuroi species complex known from human clinical specimens
1. Polyphialides present ................................................................................................................................................................ 2
1 Polyphialides absent .................................................................................................................................................................. 6
2. Microconidial chains present .................................................................................................................................................... 3
2. Microconidial chains absent ...................................................................................................................................................... 8
3. Sporodochia orange ................................................................................................................................................................... 4
3. Sporodochia tan to pale orange ............................................................................................................................................... 10
4. Chlamydospores present ............................................................................................................................................ F. nygamai
4. Chlamydospores absent ............................................................................................................................................................. 5
5. Pyriform conidia present ............................................................................................................................................. F. fujikuroi
5. Pyriform conidia absent ......................................................................................................................................... F. ramigenum
6. Napiform conidia present .......................................................................................................................................................... 7
6. Napiform conidia absent .......................................................................................................................................................... 11
7. Chlamydospores present ........................................................................................................................................ F. napiforme
7. Chlamydospores absent ......................................................................................................................................... F. thapsinum
8. Conidiophores mainly prostrate, rarely branched ..................................................................................................................... 9
8. Conidiophores mainly erect, branched .................................................................................................................................... 13
9. Chlamydospores present, polyphialides rare ............................................................................................................ F. acutatum
9. Chlamydospores absent, polyphialides abundant and proliferating extensively ........................................................ F. sacchari
10. Microconidia 0-septate; macroconidia 3–5-septate, straight or almost so .......................................................... F. proliferatum
10. Microconidia 0–2(–3)-septate; macroconidia 0-3-septate, gently curved .................................................................... F. volatile
11. Microconidia in chains only ............................................................................................................................... F. verticillioides
11. Microconidia in head and chains ............................................................................................................................................. 12
12. Macroconidia present, 3–6-septate ............................................................................................................................ F. andiyazi
12. Macroconidia absent .................................................................................................................................................... F. musae
13. Globose microconidia present ............................................................................................................................. F. anthophilum
13. Globose microconidia absent .................................................................................................................................................. 14
14. Colonies on PDA orange ........................................................................................................................................................... 15
14. Colonies on PDA purple or violet ............................................................................................................................................ 16
15. Macroconidia formed only on aerial mycelium, sporodochia not produced ........................................................... F. ananatum
15. Macroconidia formed on sporodochia ................................................................................................................. F. temperatum
16. Microconidia 0-septate, macroconidia rare or absent ............................................................................................ F. guttiforme
16. Microconidia 0–1-septate, macroconidia abundant ............................................................................................ F. subglutinans
DISCUSSION
Fusariosis is usually acquired by inhalation of conidia or after trauma, skin burns, or sometimes through central venous access, or at lower incidence the gastrointestinal tract after consumption of contaminated food (Carneiro et al. 2011, Muhammed et al. 2011). Fusarium species can affect humans either by infection (Al-Hatmi et al. 2016b) or by mycotoxicosis (Marasas et al. 1984). Clinically, a disseminated fusariosis is characterised by persistent sepsis despite broad-spectrum antibiotic therapy. Although all organs may be concerned, cutaneous involvement is predominant, followed by pulmonary infection. Given that they are widespread in natural and human-made environments, Fusarium species may contaminate laboratory specimens and yield false-positive responses. The interpretation of Fusarium growth from clinical materials strongly depends on the clinical context (Nucci & Anaissie 2007). However, repeated isolation of the fungus and culture from sinus aspirate or deep respiratory secretions in severely immunocompromised hosts should always be considered as diagnostic of fusariosis (Nucci & Anaissie 2007).
In our study, we isolated a novel Fusarium species from broncho-alveolar lavage (BAL) aspirate. We cannot definitively differentiate between a fungal colonisation or an environmental contamination as the novel species was isolated in a single BAL sample and as the patient did not have any pulmonary clinical symptoms and evolved positively without antifungal treatment. Nevertheless, the chest radiography suggested the presence of an aspergilloma, given a suspicious image compatible with a Monod sign into the pulmonary cavern that could have been attributed to the presence of Fusarium, as deep infections by Fusarium and Aspergillus spp. may be confused. These opportunists present similar radiologic results, share comparable histologic appearances with hyaline, septate, branched hyphae and can cause similar clinical syndromes (Hayden et al. 2003). It is, therefore, important to make the correct diagnosis to optimise treatment and improve prognosis. Similarly, the correct species-level identification for Fusarium infections is crucial for a positive outcome since different infectious species may present marked differences in their antifungal susceptibility patterns, at least in vitro (Al-Hatmi et al. 2015).
Our phylogenetic analysis based on a four gene dataset (cmdA, rpb2, tef and tub) showed that F. volatile belongs to the FFSC, where it formed a genetically exclusive, strongly supported monophyletic clade, phylogenetically related to F. coicis and F. verticillioides. The phylogenetic clade representing F. volatile had already been recognised as a distinct species in FFSC, however, it was not formally described as such (O’Donnell et al. 2000). The FFSC is a species-rich group, currently comprising more than 50 taxa, including opportunists on humans, economically relevant plant pathogens, and mycotoxin producers. Most of these species have been recognised based on phylogenetic analyses (Kvas et al. 2009). A large number of cryptic, unnamed phylogenetic species remain to be formally described. Based on phylogenetic relationships, O’Donnell et al. (1998) organised the FFSC into three main clades (African, American and Asian clades), each encompassing numerous species with similar biogeographic patterns. Our results showed that, despite being isolated in South America, F. volatile clusters in the African clade, which matches with the African origin of a genetically identical strain (NRRL 25615) from Nigeria. Interestingly, our individual phylogenies do not support the current delimitation of the African clade of FFSC, which was found to be polyphyletic using cmdA, rpb2 and tef1 markers. The phylogenies support the results previously reported by other authors using the same and additional phylogenetic markers (Kvas et al. 2009, Walsh et al. 2010, O’Donnell et al. 2013, Laurence et al. 2015, Sandoval-Denis et al. 2018), which shows that the biogeographic clade distribution of the FFSC needs further re-valuation.
The voucher strain CBS 143874 of F. volatile clusters as a sister clade to F. coicis and F. verticillioides, the latter species being known to be capable of causing human infection (Al-Hatmi et al. 2016b). Fusarium verticillioides is an important producer of mycotoxins, including fumonisins (Leslie & Summerell 2006, Rosa Junior et al. 2019); these toxins are highly detrimental to animals and are suspected to be responsible for acute and chronic human diseases (Leslie & Summerell 2006). In contrast, F. coicis is known only from Coix gasteenii (Poaceae), a rare Australian grass species, while no human infection or toxin production has been reported. These clinical data suggest that the new species F. volatile is unlikely a primary human pathogen but rather an opportunist that takes advantage of the host’s compromised immune response. Production of mycotoxins, however, was not tested in F. volatile and remains to be studied.
Among the FFSC species known to occur on humans (de Hoog et al. 2019), F. volatile is morphologically similar to F. fujikuroi, F. nygamai, F. proliferatum and F. ramigenum. All the latter species are characterised by obovoid to clavate microconidia formed in false heads and chains from mono- and polyphialides, and by, except F. nygamai, absence of chlamydospores. Fusarium volatile can be recognised by its ellipsoid to cylindrical microconidia (vs. the pyriform microconidia of F. fujikuroi and F. proliferatum, and the obovoid microconidia of F. ramigenum), its less-septate macroconidia (up to 3-septate vs. up to 5-septate in all the species listed above), and its tan to pale orange sporodochia (orange in F. fujikuroi and F. nygamai) (Leslie & Summerell 2006). Two additional opportunistic species on humans, F. acutatum and F. sacchari, produce microconidia only on false heads, but might also be confused with F. volatile because of their similar morphology and septation of macroconidia. Fusarium volatile differs from F. acutatum by its 0–3-septate microconidia (0-septate in the latter species) and its less curved macroconidia. It can be separated from F. sacchari by the microconidial shape (oval in F. sacchari), while polyphialides are rarely seen in F. volatile (common in F. sacchari).
Antifungal susceptibility testing demonstrated that F. volatile had reduced MICs for posaconazole (0.5 μg/mL), followed by amphotericin B and voriconazole (1 μg/mL), whereas MIC values for fluconazole, isavuconazole, itraconazole and the echinocandins were elevated (Table 2). In the present case, no antifungal therapy was given. In general, Fusarium spp. show a remarkably high degree of intrinsic resistance to a wide spectrum of clinically available antifungal drugs. Prior to the voriconazole era, the initial approach of treating invasive fusariosis consisted on the administration of high-dose (>5 mg/kg/d) liposomal amphotericin B (Al-Hatmi et al. 2017). After FDA approval in 2002, voriconazole has become the first-line treatment because of its lower toxicity and higher clinical efficacy against fusariosis (Walsh et al. 1998, Stempel et al. 2015). The European Fungal Infection Study Group and the European Confederation of Medical Mycology recommended a lipid formulation of amphotericin B or voriconazole for treating invasive fusariosis (Tortorano et al. 2014).
In conclusion, we described the new species F. volatile, belonging to the FFSC. This new taxon was found in a BAL sample from a patient with non-haematological predisposing conditions. Further studies are required to determine natural ecology, transmission routes and the potential pathogenic role of this new species.
REFERENCES
- Al-Hatmi AM, Bonifaz A, Ranque R, et al. (2017). Current antifungal treatment of fusariosis. International Journal of Antimicrobial 51: 326–332. [DOI] [PubMed] [Google Scholar]
- Al-Hatmi AM, van Diepeningen AD, Curfs-Breuker I, et al. (2015). Specific antifungal susceptibility profiles of opportunists in the Fusarium fujikuroi complex. Journal of Antimicrobial Chemotherapy 70: 1068–1071. [DOI] [PubMed] [Google Scholar]
- Al-Hatmi AMS, Hagen F, Menken SBJ, et al. (2016a). Global molecular epidemiology and genetic diversity of Fusarium, a significant emerging human opportunist from 1958-2015. Emerging Microbes and Infections 5: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hatmi AMS, Meis JF, de Hoog GS. (2016b). Fusarium: molecular diversity and intrinsic drug resistance. PLoS Pathogens 12: e1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T, Smith JA, Mount LL, et al. (2013). Fusarium torreyae sp. nov., a pathogen causing canker disease of Florida torreya (Torreya taxifolia), a critically endangered conifer restricted to northern Florida and southwestern Georgia. Mycologia 105: 312–319. [DOI] [PubMed] [Google Scholar]
- Carneiro HA, Coleman JJ, Restrepo A, et al. (2011). Fusarium infection in lung transplant patients: Report of 6 cases and review of the literature. Medicine 90: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilaka CA, De Boevre M, Atanda OO, et al. (2017). The status of Fusarium mycotoxins in sub-Saharan Africa: A review of emerging trends and post-harvest mitigation strategies towards food control. Toxins 9: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Guarro J, Gené J, et al. (2019). Atlas of Clinical Fungi, 4th ed. Westerdijk Institute / Universitat Rovira i Virgili, Utrecht / Reus. [Google Scholar]
- Edwards J, Auer D, de Alwis SK, et al. (2016). Fusarium agapanthi sp. nov., a novel bikaverin and fusarubin-producing leaf and stem spot pathogen of Agapanthus praecox (African lily) from Australia and Italy. Mycologia 108: 981–992. [DOI] [PubMed] [Google Scholar]
- Fisher NL, Burguess LW, Toussoun TA, et al. (1982). Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 72: 151–153. [Google Scholar]
- Geiser DM, Jiménez-Gasco M, Kang S. (2004). FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. European Journal of Plant Pathology 110: 473–479. [Google Scholar]
- Glass NL, Donaldson GC. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarro J. (2013). Fusariosis, a complex infection caused by a high diversity of fungal species refractory to treatment. European Journal of Clinical Microbiology & Infectious Diseases 32: 1491–1500. [DOI] [PubMed] [Google Scholar]
- Hayden RT, Isotalo PA, Parrett T, et al. (2003). In situ hybridization for the differentiation of Aspergillus, Fusarium, and Pseudallescheria species in tissue section. Diagnostic Molecular Pathology 12: 21–26. [DOI] [PubMed] [Google Scholar]
- Kvas M, Marasas WFO, Wingfield BD, et al. (2009). Diversity and evolution of Fusarium species in the Gibberella fujikuroi complex. Fungal Diversity 34: 1–21. [Google Scholar]
- Laurence MH, Walsh JL, Shuttleworth LA, et al. (2015). Six novel species of Fusarium from natural ecosystems in Australia. Fungal Diversity 77: 349–366. [Google Scholar]
- Leslie JF, Summerell BA. (2006). The Fusarium laboratory manual. Blackwell Publishing, Ames. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Marasas WFO, Nelson PE, Toussoun TA. (1984). Toxigenic Fusarium species. Identity and mycotoxicology. The Pennsylvania State University Press, USA. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012). The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the extreme to the campus and beyond: 1–8. Association for Computing Machinery, USA. [Google Scholar]
- Muhammed M, Coleman JJ, Carneiro HA, et al. (2011). The challenge of managing fusariosis. Virulence 2: 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg HI. (1976). Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. [Google Scholar]
- Nirenberg HI, O’Donnell K. (1998). New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 90: 434–458. [Google Scholar]
- Nucci M, Anaissie E. (2007). Fusarium infections in immunocompromised patients. Clinical Microbiology Reviews 20: 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. (2004). MrModeltest v2. Program distributed by the author. [Google Scholar]
- O’Donnell K, Cigelnik E, Nirenberg HI. (1998). Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90: 465–493. [Google Scholar]
- O’Donnell K, Nirenberg HI, Aoki T, et al. (2000). A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. [Google Scholar]
- O’Donnell K, Rooney AP, Proctor RH, et al. (2013). Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genetics and Biology 52: 20–31. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Ward TJ, Robert VARG, et al. (2015). DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica 43: 583–595. [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, et al. (2009b). Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum - F. equiseti and F. chlamydosporum species complexes within the United States. Journal of Clinical Microbiology 47: 3851–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner RW. (1970). A mycological colour chart. CMI and British Mycological Society, Kew, Surrey. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rosa Junior OF, Dalcin MS, Nascimento VL, et al. (2019). Fumonisin production by Fusarium verticillioides in maize genotypes cultivated in different environments. Toxins 11: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Guarnaccia V, Polizzi G, et al. (2018). Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 40: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Denis M, Swart WJ, Crous PW. (2018). New Fusarium species from the Kruger National Park, South Africa. MycoKeys 34: 63–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stempel JM, Hammond SP, Sutton DA, et al (2015). Invasive fusariosis in the voriconazole era: Single-center 13-year experience. Open Forum Infectious Diseases 2: ofv099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung GH, Sung JM, Hywel-Jones NL, et al. (2007). A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution 44: 1204–1223. [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts, USA. [Google Scholar]
- Tortorano AM, Richardson M, Roilides E, et al. (2014). ESCMID and ECMM joint guidelines on diagnosis and management of hyalohyphomycosis: Fusarium spp., Scedosporium spp. and others. Clinical Microbiology and Infection 3: 27–46. [DOI] [PubMed] [Google Scholar]
- Walsh J, Laurence M, Liew E, et al. (2010). Fusarium: two endophytic novel species from tropical grasses of northern Australia. Fungal Diversity 44: 149–159. [Google Scholar]
- Walsh TJ, Hiemenz JW, Seibel NL, et al. (1998). Amphotericin B lipid complex for invasive fungal infections: analysis of safety and efficacy in 556 cases. Clinical Infectious Diseases 26: 1383–1396. [DOI] [PubMed] [Google Scholar]
- Wingfield MJ, de Beer ZW, Slippers B, et al. (2012). One fungus, one name promotes progressive plant pathology. Molecular Plant Pathology 13: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]