Abstract
Cas1 integrase associates with Cas2 to insert short DNA fragments into a CRISPR array, establishing nucleic acid memory in prokaryotes. Here we applied single-molecule FRET methods to the Enterococcus faecalis (Efa) Cas1–Cas2 system to establish a kinetic framework describing target-searching, integration, and post-synapsis events. EfaCas1–Cas2 on its own is not able to find the CRISPR repeat in the CRISPR array; it only does so after prespacer loading. The leader sequence adjacent to the repeat further stabilizes EfaCas1–Cas2 contacts, enabling leader-side integration and subsequent spacer-side integration. The resulting post-synaptic complex has a surprisingly short mean lifetime. Remarkably, transcription efficiently resolves the postsynaptic complex and we predict that this is a conserved mechanism that ensures efficient and directional spacer integration in many CRISPR systems. Overall, our study provides a complete model of spacer acquisition, which can be harnessed for DNA-based information storage and cell lineage tracing technologies.
Prokaryotes and vertebrates utilize transposon-derived recombinases and integrases to establish adaptive immunity. In prokaryotes, this involves the integrase-mediated insertion of short foreign DNA-derived spacers into the CRISPR array, updating the molecular memory at the nucleic acid level1–4. Whereas the subsequent RNA-guided CRISPR interference mechanism varies significantly among various CRISPR-Cas systems1,5–9, the immunity acquisition mechanism is essentially identical10–12. The universally conserved Cas1 and Cas2 proteins form an integrase complex3,4,13, capture a short double-stranded (ds) DNA (prespacer) that was excised from foreign DNA (protospacers), and insert it into the CRISPR locus as a new spacer11. Early studies of the Escherichia coli Type I-E CRISPR system revealed the architecture of the Cas1–Cas2 complex3,4, its preference for 3’ overhang-containing DNA duplexes as prespacers4,14,15 and the leader-proximal CRISPR repeat as the integration target1–3,16, and its host-factor dependence on RecBCD17 and IHF18. In Type II-A CRISPR-Cas systems, spacer integration reaction solely relies on Cas1–Cas219, although prespacer biogenesis requires Cas9 and Csn220–23. Structural studies from these two systems further provided high-resolution explanations for prespacer integration24,25. However, prespacer biogenesis, processing, and resolution of the integration complex are less well understood17,23,26–30.
Results
Resolving EfaCas1–Cas2-mediated binding and stepwise integration using smFRET
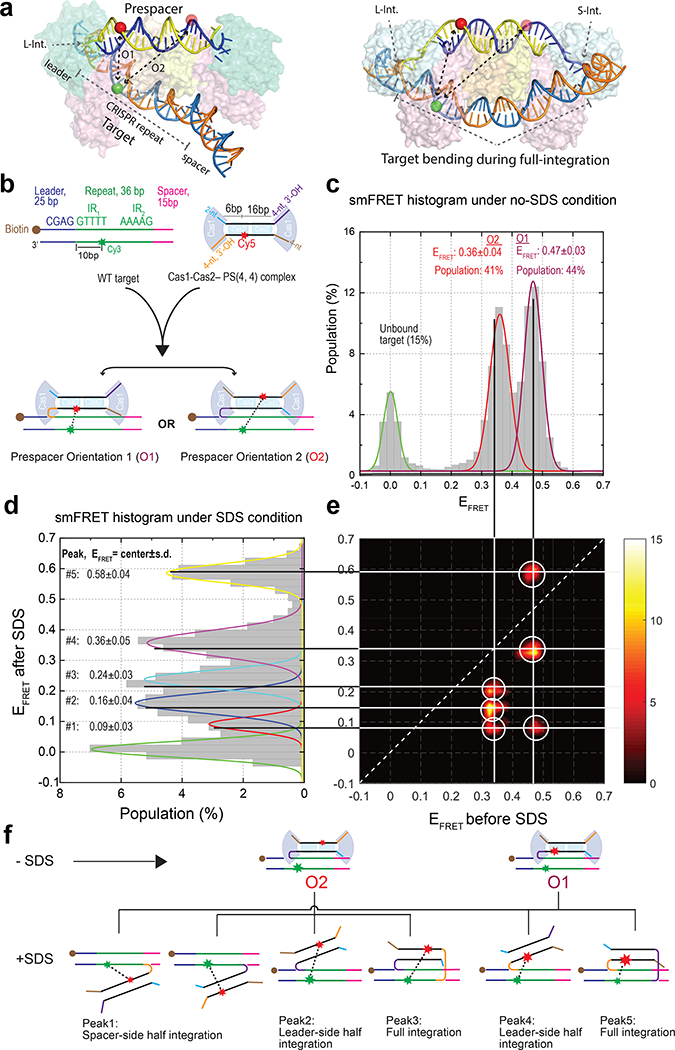
Because the spacer integration reaction involves multiple intermediates, it is inherently challenging to dissect using bulk biochemistry. We therefore carried out single-molecule Förster Resonance Energy Transfer (smFRET) experiments to establish the kinetic framework for the Cas1–Cas2-mediated prespacer integration. Focusing on the Type II-A Cas1–Cas2 from Enterococcus faecalis, for which crystal structures of key functional states have been determined25, we introduced a donor fluorophore (Cy3) into the leader-side integration target and an acceptor fluorophore (Cy5) into an asymmetric position in the integration-ready prespacer [PS(4, 4)] (Fig. 1a). Based on the structures, these modifications were not expected to interfere with EfaCas1–Cas2-catalyzed integration reaction, which was confirmed in bulk assays (Extended Data Fig. 1). This labeling scheme is particularly sensitive in detecting leader-proximal binding or integration; however, it does not further distinguish the conformational differences in the target DNA during the half-to-full integration transition25 (Fig. 1a). When premixed EfaCas1–Cas2 and Cy5-labeled prespacer were introduced together into the flow cell, the FRET-induced Cy5 signals were detected from the immobilized Cy3-targets within seconds, suggesting that target capture was fast and efficient. Representative single-molecule traces from five-minute movie recordings revealed a mixture of transient and long-lasting FRET events, presumably corresponding to cycles of target binding or dissociation and integration or disintegration, respectively (Extended Data Fig. 2a–d). Assuming bulk biochemistry observations hold, that Cas1–Cas2 preferentially integrates the prespacer to the leader-proximal side of the target3,19,24,25, we would expect two equally populated native FRET states corresponding to Cas1–Cas2 integrating prespacer in two orientations to the leader-side target. This was indeed the case. The two almost equally populated states in the FRET histogram agreed extremely well with the measured distances in the half-integration structure25 (Fig. 1a–c).
Fig. 1. Reconstitution of the integration reaction at the single-molecule level.
a, Location of the fluorophores on half and full integration crystal structures (PDB accession code: 5XVO and 5XVP). Full integration requires DNA bending, but this does not affect the Cy3-Cy5 distance in the leader-side labeling scheme. b, Schematic of the target, Cas1–Cas2–PS(4,4), and two orientations in which Cas1–Cas2 can bind/integrate the prespacer to the target. c, d, EFRET histogram under native (no-SDS) and denaturing (SDS) conditions, respectively. EFRET peaks are reported as mean ± s.d (n= 7500 and 7000 single-molecules traces for c and d, respectively). e, Transition density plot showing the transition from native peaks to denatured peaks. Each orientation, O1 and O2, is split into corresponding leader-side, spacer-side or full-integration configuration. Transitions to FRET zero were removed to augment the weaker transition peaks. f, Schematic showing transition from the native conformation(O1 and O2) to various SDS-denatured integration configurations.
Because binding and integration were not easily distinguished under the native condition without prior knowledge of their single molecule behavior, a 2% SDS wash step was introduced towards the end of the recording to denature EfaCas1–Cas2. Only integrated prespacers would survive the SDS wash due to its covalent linkage to the surface-anchored target DNA; unintegrated prespacers would be removed. On average ~76% of the FRET pairs survived the SDS wash, suggesting that prespacer integration by EfaCas1–Cas2 was quite efficient. Transition density plot (TDP) analysis (Fig. 1e) further resolved the correlation between native and SDS-denatured states (Fig. 1c, d). Each native state was found to partition into three denaturing states. Further analysis (Supplemental Information) revealed that they corresponded to all six possible integration scenarios (two scenarios overlapped under denaturing FRET peak 1) – the leader-half, spacer-half, and full-integrations, each in two prespacer orientations (Fig. 1d, f, Extended Data Fig. 2e–i). According to the full-integration crystal structure25, each half-site is in dynamic equilibrium between EfaCas1–Cas2-catalyzed disintegration and reintegration in the post-synaptic complex (PSC). This rationalizes the spacer-side-only peaks.
We also explored spacer-side target-labeling schemes in an effort to distinguish half- and full-integration states based on the conformational difference in target DNA, as revealed in crystal structures25. Four native FRET states were evident (Extended Data Fig. 3c). The half-integration state was indeed distinct from the corresponding full-integration state (Extended Data Fig. 3a), consistent with the observation that the target DNA is bent at the central dyad during the half-to-full integration transition24,25. However, the spacer-side labeling schemes decreased the full-integration rate despite several structure-guided labeling schemes to avoid steric hindrance. We therefore carried out the rest of the analysis using the leader-side labeling scheme.
Preference for leader-proximal integration
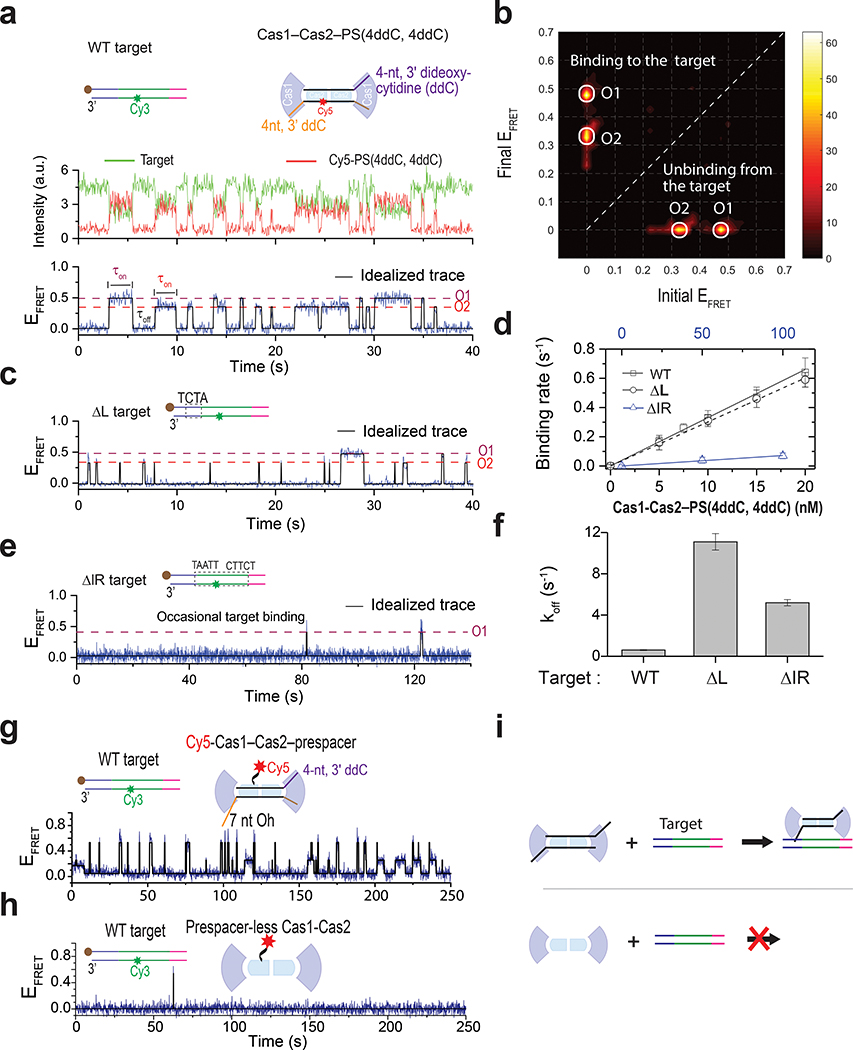
Next, we performed a focused analysis of the target-searching process by preventing the integration chemistry from taking place using a prespacer containing dideoxy-termini [PS(4ddC, 4ddC]. Long-dwelling states (Extended Data Fig. 2) disappeared consequently, and the smFRET traces were dominated by fast on and off binding events (Fig. 2a, b). Dwell time analysis revealed that Cas1–Cas2–PS(4ddC, 4ddC) on average spends 1.65 seconds on target (τon) before dissociation, and another 3.03 seconds to rediscover a target (τoff) at 10 nM concentration (Fig. 2d). The derived microscopic Kd (=koff/kon) of 18 nM at 25 ˚C is consistent with the value derived from the time-resolved single-molecule counting experiments (Extended Data Fig. 4). The pair of 5-bp inverted repeats (IR) within the CRISPR repeat and the flanking 4-bp leader sequence are critical cis-elements 3,19,25 (Fig. 2a). We evaluated their roles in guiding target discovery using smFRET experiments. Interestingly, EfaCas1–Cas2 was equally efficient at discovering the leader-less CRISPR repeat (ΔL); τoff for leader-flanking and leader-less targets were comparable (Fig. 2d). However, each binding event was much shorter (τon was 18-fold shorter, Fig. 2c, f), the average dwell time was so transient that the integration chemistry was not expected to take place (koff 11-fold faster than the later measured khalf; Extended Data Fig. 5a–c). In contrast, when the pair of IR was removed from the CRISPR target (ΔIR1+2), contacts by EfaCas1–Cas2– PS(4ddC, 4ddC) dropped to the background level (kon 46-fold slower, Fig. 2d, e). Taken together, our data suggest that the spacer-loaded EfaCas1–Cas2 is capable of making frequent contacts to every CRISPR repeat in the CRISPR array; however, only the leader-flanking CRISPR repeat-binding events lead to productive integration, because the leader sequence enables EfaCas1–Cas2 to dwell much longer, enabling half-site integration (koff 1.5-fold slower than khalf). This rationalizes the observation that Cas1–Cas2 mediated spacer integration occurs overwhelmingly at the first CRISPR repeat flanked by the leader sequence1–3,16,31.
Fig. 2. Mechanism of target searching by EfaCas1–Cas2.
a, (Top) Schematic of the target and Cas1–Cas2–PS(4ddC,4ddC). The prespacer lacks 3’-OH at both ends. (Bottom) A representative smFRET trace showing binding and unbinding events with 20 nM Cas1–Cas2–PS(4ddC,4ddC). Two FRET levels, O1 and O2, were observed consistent the data in Fig. 1c, representing target binding in two prespacer orientations. Dwell times in the on-state and off-state are denoted on the trace. b, Transition density plot created from 843 binding-unbinding transitions (51 traces). c, e, Representative traces from ΔL and ΔIR targets under the same concentration used in a. d, Plot of the binding rate (1/τoff) vs Cas1–Cas2–PS(4ddC,4ddC) concentration for different targets. The slope provides the binding rate constant (kon), which is reported as mean ± s.e. The values for off-state dwell times used to derivethe binding rate are provided in source data. f, Plot of 1/τon to derive the dissociation constant (koff). koff was determined from four concentrations for WT and ΔL target and two concentrations for ΔIR target. koff is reported as mean ± s.e. The values for on-state dwell times used to derive dissociation rate are provided in source data. g, A representative trace showing frequent binding and unbinding events obtained for 15 nM Cy5-labeled Cas1–Cas2 and unlabeled prespacer. h, A representative trace showing rare binding and unbinding events in the absence of a prespacer. i, Schematic illustrating target binding in the presence (top) and absence (bottom) of prespacer. In the presence of prespacer, Cas1–Cas2 can successfully find the target and integrates the prespacer, but it fails to identify a target in the absence of a prespacer. Source data for panels d and f are available online.
Only prespacer-loaded Cas1–Cas2 is capable of target-searching
Given that only one or a handful of integration targets are present in the prokaryotic genome, and that typically the biogenesis of prespacers is limiting32,33, the vast majority of Cas1–Cas2s exist in the apo form, which may compete with prespacer-loaded Cas1–Cas2 for integration targets. We evaluated such possibility by comparing the target-binding behavior of EfaCas1–Cas2, with or without prespacers present. Cas2 with an A19C mutation was Cy5-labeled via maleimide-thiol chemistry; ~70% of the assembled EfaCas14-Cas22 contained a single Cy5 label with our protocol. The prespacer-bound EfaCas1–Cas2-Cy5 behaved similarly to the EfaCas1–Cas2–Cy5-prespacer in target-searching, suggesting that the mutagenesis and Cy5- Cas2 labeling did not alter EfaCas1–Cas2 behavior (Fig. 2g, Extended Data Fig. 5d). Strikingly, under the same condition but without prespacer present, apo Cas1–Cas2 failed to discover the target, therefore would not interfere with spacer integration (Fig. 2h, Extended Data Fig. 5e). It appears that prespacer-binding configures Cas1–Cas2 into the target-searching mode, presumably by inducing a conformational change in Cas1–Cas23,14,15. The target searching by apo and prespacer-loaded EfaCas1-Cas2 is illustrated in Fig. 5i.
Establishing a complete kinetic framework for two-step integration
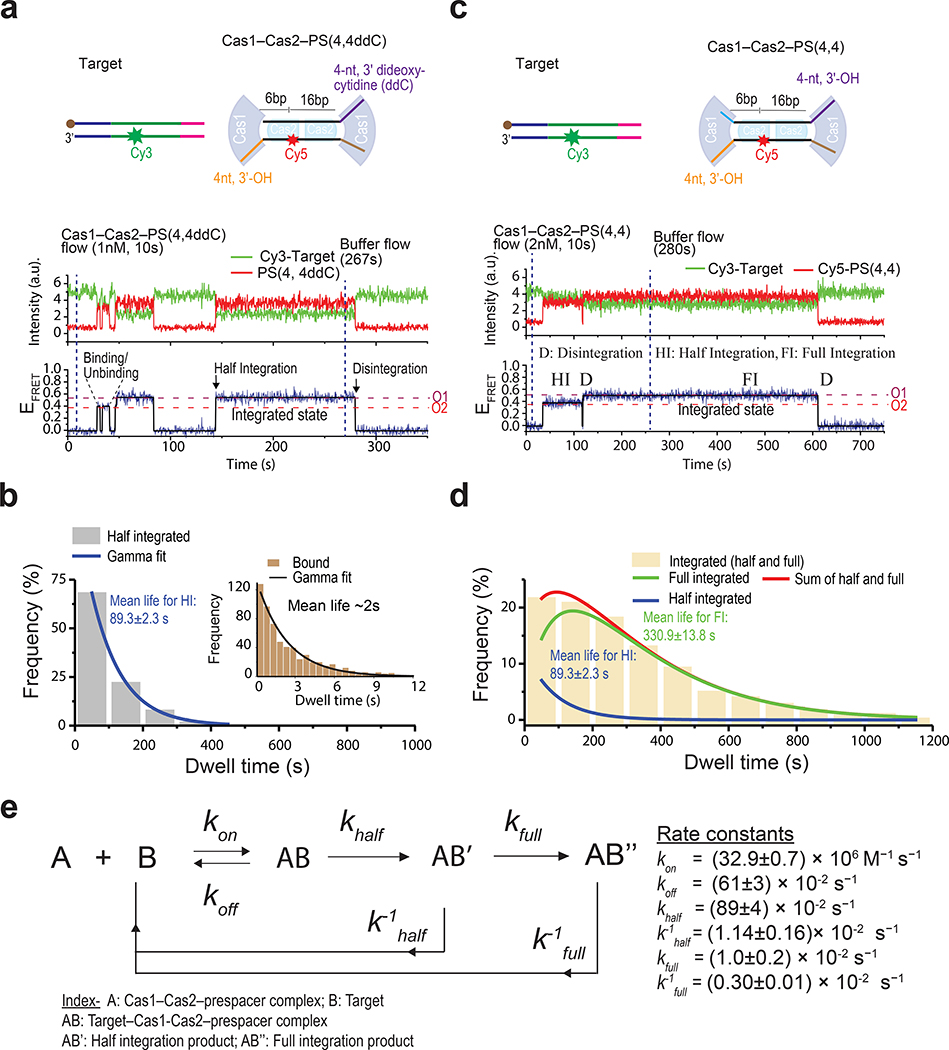
Next, we focused on the half-integration process by programming Cas1–Cas2 with a single di-deoxy-containing prespacer [PS(4, 4ddC)]. When EfaCas1–Cas2 oriented the di-deoxy end of prespacer towards the leader-side target, evident by its FRET state, only target-searching behaviors were observed (Fig. 3a, Extended Data Fig. 6a). In contrast, when the 3’-OH end of prespacer was brought to the leader-side target by EfaCas1–Cas2, significantly longer contacts were observed (Fig. 3a, Extended Data Fig. 6a), which were confirmed by SDS wash to represent the leader-side half-integration events (Extended Data Fig. 6a). The transition from native O1 FRET level to its corresponding denatured FRET level is shown in Extended Data Fig. 6b with schematic illustration. Next, we swapped -ddC and -OH groups on PS(4, 4ddC), which should result in a reversion of the dwell times on smFRET traces. Indeed, the short dwell time became long and vice versa, confirming that long contacts are due to half integration from the 3’-OH group of the prespacer (Extended Data Fig. 6c). We derived a half-integration reaction rate (khalf) of ~0.9 s−1 from single-molecule stop-flow experiments, by applying SDS-quenching at different time point and quantifying the accumulation of the half-integrated molecules (Extended Data Fig. 5b, c). After integration reached equilibrium, excess EfaCas1–Cas2 was washed out, and the system was continuously monitored for another 15 minutes to capture disintegration events; such long-exposure did not bleach fluorophores significantly in our experimental setup (Extended Data Fig. 7b, c). The half-integrated prespacer disintegrated rather fast, with a mean survival time (mean lifetime) of ~ 1.5 minutes (k−1half=1.14±0.16×10−2s−1). Being able to quickly disintegrate products that failed to proceed to full-integration in a timely fashion has been proposed as a mechanism to protect genome integrity19.
Fig. 3. Stability of half and full integration complexes.
a, Schematic of target and Cas1–Cas2–PS(4,4ddC) used in the experiment (top) and a representative smFRET trace (bottom). The dashed lines show prespacer orientations (O1 and O2). O2 corresponds to binding events whereas O1 corresponds to half-integration (HI) and was assigned as such based on mean lifetime and post-SDS denaturation. b, Dwell times of binding (inset) and half-integration events were collected from traces like the one shown in panel a and plotted on the histograms. Mean lifetimes for binding and half-integration events were obtained using gamma fit on the histogram data. 358 traces were included in the analysis (each trace contributes one dwell time data point). c, Schematic of target and Cas1–Cas2–PS(4,4) used in the experiment (top) and a representative smFRET trace (bottom). Important events, such as binding-unbinding, integration (half and full) and disintegration, are marked. Since both orientations O1 and O2 correspond to integration in this case, the potential half and full integration events were assigned based on mean lifetime. d, A total of 606 smFRET traces, which showed wide dwell time distribution, were analyzed to collect dwell times for integration events, which were plotted in the histogram. A sum of two gamma functions fitted the histogram, providing two mean lifetimes: one for half and one for full integration. e, Kinetic parameters of Cas1–Cas2–prespacer and target interaction. Source data for graphs in b and d are available online.
The rate of half-to-full conversion and the stability of the full-integration products were measured from 20-minute recordings using the full-integration-competent prespacer PS(4,4). A portion of the FRET events were significantly longer than the half-integration events (Fig. 3c). These were confirmed by SDS-wash to be full-integration events (Extended Data Fig. 7d–f). In a single molecule stop-flow experiment similar to that in Extended Data Fig. 5, one can derive the half-to-full conversion rate by either quantifying the depletion of the half-integration species or the accumulation of the full-integration species; both methods yielded essentially the same rate constant (kfull=0.010±0.02 s−1; Extended Data Fig. 8). The stability of the full-integration product was measured in a similar procedure as in Fig. 3a, by monitoring EfaCas1–Cas2–PS(4,4) dissociation from the long-lasting traces after removing excess Cas1–Cas2, which eliminated new integration events. Dwell time analysis revealed that the full-integration product has a mean lifetime of ~5.5 minutes (Fig. 3d). With this, we established a complete kinetic framework for EfaCas1–Cas2-mediated prespacer integration (Fig. 3e).
Stepwise and in situ nucleolytic processing of prespacer leads to directional integration
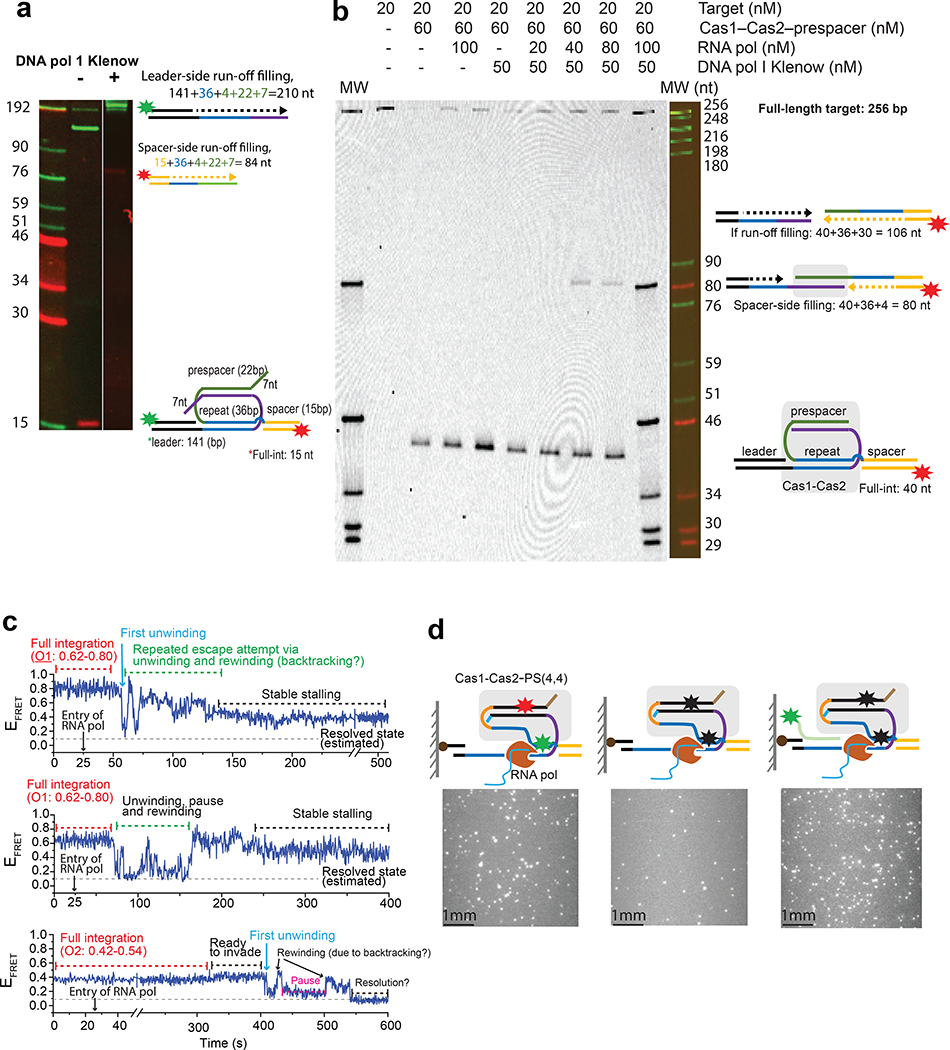
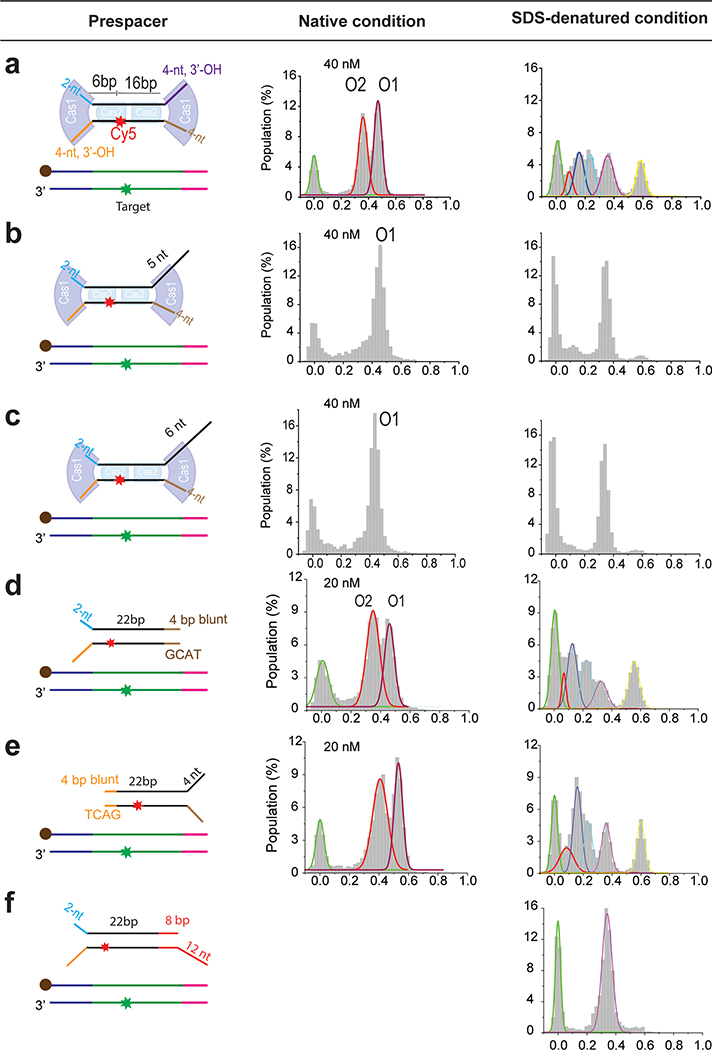
We next explored the less understood prespacer biogenesis mechanism. New spacers are integrated with fixed directionality relative to the protospacer adjacent motif (PAM)34,35. This ensures that the transcribed CRISPR RNA can guide interference in a PAM-dependent fashion. Furthermore, each CRISPR system acquires spacers of defined length. The length specification varies among different CRISPR systems. It is unlikely that each prokaryote has evolved a dedicated process to custom-feed the preferred prespacers for Cas1–Cas2 to integrate. We hypothesized that Cas1–Cas2 itself defines the prespacer specification by recruiting and protecting a portion of the prespacer precursor from host or Cas nuclease trimming, then integrating the protected portion as a mature prespacer. In our hands, when the prespacer already contained a 22-bp mid-duplex preferred by EfaCas1–Cas2, the integration outcome depended critically on the 3’-overhang length. The end containing an optimal 4-nt 3’-overhang was integrated very efficiently, whereas the opposite overhang that was merely 1–2 nucleotides longer strongly inhibited integration (Fig. 4a, Extended Data Fig. 9b, c). We next explored whether the longer overhangs could be trimmed by a host nuclease to allow integration. Indeed, incubating with the 3’-to-5’ single-stranded nuclease E. coli ExoI (SbcB) enabled EfaCas1–Cas2 to also integrate from the 26-nt overhang side of the prespacer, which refracted integration without nuclease treatment (Fig. 4a). Because a prespacer precursor is unlikely to contain a perfect 22-bp duplexed region, we systematically tested the integration behavior of EfaCas1–Cas2 on prespacers containing a longer duplexed region. When the entire duplex was 26-bp in length, EfaCas1–Cas2 apparently was able to specify a 22-bp mid-duplex and integrate the frayed 4-bp terminus (Extended Data Fig. 9d–e); another 4-bp or 16-bp extra completely inhibited integration from the duplexed end (Extended Data Fig. 9f, Fig. 4b). Importantly, integration from the protruding duplexed end in a 42-bp prespacer precursor could be enabled by E. coli ExoIII treatment, which exonucleolytically trims the 3’-strand from a DNA duplex (Fig. 4b). The accumulating data hinted that an ordered prespacer processing scheme could lead to directional integration. Indeed, when a long duplex-containing prespacer was synchronized to the half-integration state from the 4-nt overhang end, ExoIII rescued the stalled half-to-full transition, presumably by processing the unprotected duplex into an optimal overhang for full integration. Importantly, the full-integration outcome was unidirectional because the leader-side integration had already taken place (Fig. 4c). While the tested nucleases may or may not be solely responsible for prespacer processing in vivo, we think the principle to establish directional integration likely holds true for all CRISPR systems, that unidirectional integration involves Cas1–Cas2 integrating partially processed prespacers and allowing further nucleolytic trimming in between the two integration steps, and that the tug of war between Cas1–Cas2 protection and nucleolytic trimming define the idiosyncratic prespacer length. Our data did not address the PAM-dependent prespacer biogenesis, which necessarily involves Cas9 and Csn2 in Type II-A CRISPR20–23 and Cas4 in many other systems27–29,36. We envision that the PAM-dependent prespacer trimming fits into this mechanistic framework, and that the idiosyncratic timing of the PAM-dependent overhang trimming explains the observed orientation differences between Type I and II CRISPR systems.
Fig. 4. Prespacer processing and unidirectional integration.
a, Left, Pre ExoI treatment. Schematic showing assembly of precursor prespacer with Cas1–Cas2 (boxed) and integration. The prespacer has 22bp mid-duplex and two 3’ overhangs of 4 and 26 nt. The prespacer can only be half-integrated from the 4-nt end as shown in histograms presented below the schematic. Both native and SDS condition histograms are presented. Right, Post-ExoI treatment. Schematic showing processing by host nucleases (boxed), such as ExoI, which can act on the longer overhang of the prespacer and trim it to 4nt, the correct length for integration. The resulting prespacer can then be integrated, which is shown in histogram below the schematic. Since the prespacer is integrated in both O1 and O2 orientations, integration is called bidirectional. b, Left, Pre- ExoIII treatment. Schematic showing assembly of Cas1-Cas2 on a precursor prespacer with mid-duplex extended by 20bp (boxed). The prespacer has a 4-nt overhang on one end. Right, Post-ExoIII treatment. ExoIII can act on the duplex end, trimming it to 4bp.The end can then be frayed by Cas1–Cas2 itself. The prespacer is ready for integration and can be integrated in either orientation. The histogram shows peaks corresponding to half and full integration in both O1 and O2 orientations. c, (Left) Scheme of the experimental approach to test the model for unidirectional integration. In this model, precursor prespacer is first integrated from the 4nt overhang end in one orientation (O1 in this case). The other end is then processed in-situ without dissociation from the target. Once the duplex end trimmed to the proper length, Cas1–Cas2 integrates the end to the spacer side, which ensures unidirectional full integration. (Right) Histogram showing unidirectional integration. The peak with EFRET ~0.35 represents half integration and the peak with EFRET ~0.55 represents full integration in O1 orientation, consistent with the proposed model.
Transcription and unscheduled DNA synthesis converts PSC to a new spacer
Both structural and smFRET evidences suggest that upon full-integration, Cas1–Cas2 is caged inside the post-synaptic complex (PSC) by the covalently connected CRISPR repeat and prespacer. Importantly, smFRET reveals that the PSC is only stable for 5.5 minutes on average (Fig. 3d). The disintegrated prespacer may reintegrate with compromised directionality, leading to the incorporation of useless spacers. Consistent with this idea, high-throughput analysis revealed that ~ 2% of the E. coli spacers were derived from the same set of PAM-flanking protospacers but inserted in the opposite orientation into the CRISPR array37. For this reason, DNA replication is unlikely the main mechanism for PSC resolution because the frequency of a replication fork passing through the CRISPR locus is highly dependent on chromosome size and the growth rate of the cell at the time of prespacer integration38. While possible in fast-growing bacteria, such as E. coli, DNA replication would be an unreliable mechanism to resolve the PSC in slower-growing bacteria and archaea. Moreover, resolving the sophisticated topology in the PSC may also require strong force and tight regulation. If the CRISPR repeat and the prespacer are simultaneously unwound and replicated, it would result in double-strand break (DSB) formation and consequently, genome instability. Interestingly, when we assembled the PSC and then washed away Cas1–Cas2 using SDS, the Klenow fragment of E. coli DNA polymerase I, the main polymerase for unscheduled DNA synthesis, could recognize the nicks and efficiently unwound the naked PSC through DNA polymerization. However, this action replicated both CRISPR repeat and spacer, resulting in DSB formation (Fig. 5a). Importantly, DNA pol I was not able to polymerize from an intact PSC (Fig. 5b), presumably because the nicks were inaccessible due to EfaCas1–Cas2 protection. If the DNA replication is unlikely the default process to resolve a PSC, what other molecular processes could be involved? Based on the reasoning that it has to be a frequent and fundamentally conserved process common to both bacteria and archaea, we hypothesized that RNA transcription, which happens frequently at every CRISPR locus, may stall at the PSC and trigger transcription-coupled DNA repair39,40 to resolve it. To test this possibility, we immobilized a promoter-containing integration target in the TIRF experiment, introduced EfaCas1–Cas2 to reach integration equilibrium, and then allowed E. coli RNA polymerase to transcribe and clash into the PSC. Initially, we speculated that additional transcription-coupled repair proteins such as Mfd may be required to destabilize the PSC39,40. To our surprise, smFRET traces indicated that transcription alone triggered partial resolution of the PSC (Fig. 5c). To detect whether the CRISPR repeat was unwound by the transcribing RNA polymerase, we introduced a Cy3-labeled ssDNA complementary to the first 12-nt of the CRISPR repeat. Many probes annealed to the immobilized target when transcription was initiated, suggesting that the CRISPR repeat was at least partially unwound by the transcribing RNA polymerase (Fig. 5d); the same probes did not anneal to targets when transcription was omitted (Extended Data Fig. 10). We reasoned that this created an opportunity for the DNA polymerase to follow up and irreversibly resolve the PSC. Indeed, when we carried out the bulk version of the transcription-towards-PSC experiment described in Fig. 5c and subsequently introduced E. coli DNA pol I Klenow and dNTP, it was found that the DNA polymerase captured the exposed DNA lesion and replicated the entire CRISPR repeat, as monitored from the spacer side (Fig. 5b). This gap-filling polymerization was not observed when only Klenow or RNA polymerase was present (Fig. 5b). Importantly, in this scenario DNA replication stopped at the CRISPR repeat-spacer boundary rather than traversing through the spacer region, as it did on a naked PSC (Fig. 5a). This strongly suggests that during PSC resolution EfaCas1–Cas2 protects the duplex region of the prespacer from strand-displacement DNA synthesis, which would lead to DSB formation. Collectively, our data revealed a novel transcription-assisted DNA repair mechanism for PSC resolution, and that Cas1–Cas2 safeguards this process by defining the replication boundary and preventing DSB formation.
Fig. 5. Resolution of post-synaptic complex (PSC).
a, Urea gel resolving DNA Pol I mediated extension of leader-side and spacer-side fragments from the naked PSC, without Cas1–Cas2 protection. b, Urea gel showing the extension of the spacer-side fragment, indicating duplication of CRISPR repeat. Extension occurs when both RNA polymerase and DNA polymerase are present simultaneously in the reaction, but when only RNA polymerase or DNA polymerase is present, extension is not observed. c, Representative smFRET traces showing FRET transitions due to transcription of a promoter upstream of leader. The traces are annotated with presumptive events such as unwinding of the repeat, pausing, rewinding, and stalling. Prior to transcription (first 25 seconds), integration of the prespacer is achieved by flowing Cas1–Cas2–PS(4,4). Target Cy3 label is on the coding (top) strand to prevent RNA polymerase blockade, which changes the FRET level of O1 and O2. d, Experimental set-up to show that RNA polymerase can unwind the CRISPR repeat. Left, Cy3 spots from the target. After integration was confirmed through the presence of Cy5 spots in the acceptor channel, transcription was initiated. Middle,Cy3 fluorophores were photobleached (although some Cy3 survived). Right, The Cy3-IR1 DNA probe is added to the channel. It should anneal with the template (bottom) strand of the repeat, if the template strand is exposed by RNA polymerase-mediated unwinding. New Cy3 spots in the photobleached area indicate annealing of the Cy3 probe. Control experiments without transcription are shown in Extended Data Fig. 10a–c. Uncropped gel images for a and b are available online.
Transcription at the CRISPR locus promotes timely new spacer incorporation in vivo
The effect of transcription on new spacer acquisition was further investigated in an in vivo setting. The E. faecalis spacer acquisition cis-elements (CRISPR leader and a single repeat) were grafted into the E. coli chromosome. E. faecalis Cas1 and Cas2 expression were induced and optimal prespacers were electroporated into E. coli cells to bypass the prespacer biogenesis bottleneck. Possible run-through transcription was insulated by upstream and downstream transcriptional terminators, and a native E. coli promoter was either included in or omitted from the upstream of the CRISPR leader (Fig. 6a). Results showed that the spacer acquisition efficiency was reproducibly higher (~2.8-fold) when a promoter was present upstream of CRISPR leader. An even stronger stimulatory effect was observed (~3.5-fold) when DNA replication was stalled by Nalidixic acid (Fig. 6b). We attempted to further distinguish the influence of transcription from that of replication by examining the timing of new spacer acquisition using a PCR detection scheme that specifically amplified the fully incorporated new spacers (Fig. 6a). Because the replication fork passes through the CRISPR locus much less frequently than a transcribing RNA polymerase, new spacers should only be detected after a lag if the acquisition is coupled with DNA replication, whereas a transcription-coupled acquisition process should incorporate new spacers much earlier. Our results were consistent with the latter scheme. New spacer incorporation could be detected as early as five minutes after electroporation when transcription was enabled, and the process was not negatively affected by Nalidixic acid, whereas when the promoter was not present, new spacers were detected much later and to a lesser extent (Fig. 6c). Furthermore, we found the integration of multiple (up to 5) spacers at later time points (for example, 80 minutes) when transcription of the array is occurring, regardless of whether cells are dividing or not; however, as expected, these events occurred at a lower frequency than that of a single integration (Extended Data Fig. 10d). The background level of new spacer incorporation could be due to cryptic transcription, residual replication, or the possible existence of additional PSC resolution pathways. Nonetheless, our in vivo data corroborates the in vitro reconstitution results in suggesting that transcription at the CRISPR locus actively promotes new spacer acquisition in the E. faecalis Type II-A CRISPR-Cas system.
Fig. 6. In vivo evidence that transcription from the CRISPR locus promotes new spacer incorporation.
a, In vivo spacer acquisition assay. E. coli expressing Cas1 and Cas2 (bacteria depicted as green circle) were electroporated with spacers (lightning bolt) and the of CRISPR array expansion was analyzed by PCR at specified time intervals. Primer set 1 (orange) amplifies both expanded and unexpanded CRISPR arrays from genomic DNA, whereas primer set 2 (green) selectively amplifies new spacers incorporated in one orientation. b, Quantification of new spacer acquisition (top), with or without transcription from the CRISPR locus, under conditions of replication arrest, which was achieved by adding nalidixic acid in the growth medium. The bars represent mean of 3 independent experiments; p-value: * < 0.05; ** <0.005 (two-tailed t-test; t-scores were converted to corresponding p-values). c, Image of agarose gel showing PCR products using primer set 2 from time-course experiments in the above four conditions. Uncropped gel images for panel b and c and source data for the graph in b are available online.
Discussion
Our work provides the temporal resolution to dissect the kinetic framework governing prespacer biogenesis, integration, and incorporation processes. It establishes the foundation to further understand spacer acquisitions processes that involve specialized processing factors, such as Cas3 and Cas4 nucleases27–29,36,41–43 and reverse transcriptase44–47. The most important conceptual advance of this study is the realization that an efficient mechanism to resolve the post-synaptic complex is essential to maintain robust CRISPR-Cas immunity. A persistent PSC would prevent new CRISPR RNA production and potentially weaken or halt CRISPR surveillance, whereas premature disintegration would lead to the loss of preestablished prespacer directionality. Both scenarios are detrimental to CRISPR immunity. Here we reveal that transcription from the CRISPR leader, in combination with unscheduled DNA synthesis and the continuous spacer protection by Cas1–Cas2 efficiently and precisely resolves the post-synaptic complex, allowing the completion of new spacer incorporation; the step-by-step events of spacer incorporation are displayed in Fig. 7. The efficiency of such mechanism may help rationalize the interesting observation that the S. pyogenes Type II-A CRISPR system is able to acquire new spacers almost instantaneously while a phage is injecting its DNA genome, and the updated crRNA guides Cas9 to protect the host from any subsequent infection from the same phage48. Exceptions to this theme may exist. For example, spacer acquisition in the E. coli Type I-E system requires the integration host factor protein to bend the leader, which sequesters the promoter and most likely shuts off transcription18. An integral component of the DNA replisome, DnaJ, is further required to nucleolytically process the PAM-side of the prespacer to complete full-integration49. The PSC resolution in this subset of the CRISPR systems may indeed be replication-coupled; however, the coupling does not always take place in a timely fashion, as a small percentage of the E. coli spacers appear to have been acquired from disintegrated prespacer reintegrated in the wrong orientation37. Given the potential fitness cost of halting CRISPR RNA production during integration, we think the majority of the slower-growing prokaryotes would likely rely on the transcription-coupled mechanism to resolve the PSC in a timely fashion. Lastly, despite its great potential in cell lineage tracing and information storage applications32,33, Cas1–Cas2-mediated spacer acquisition has been difficult to reconstitute in eukaryotic cells. Our study clearly defines the bottlenecks in the spacer integration process. We hope this would lead to renewed effort to harness the power of the Cas1–Cas2 integrase.
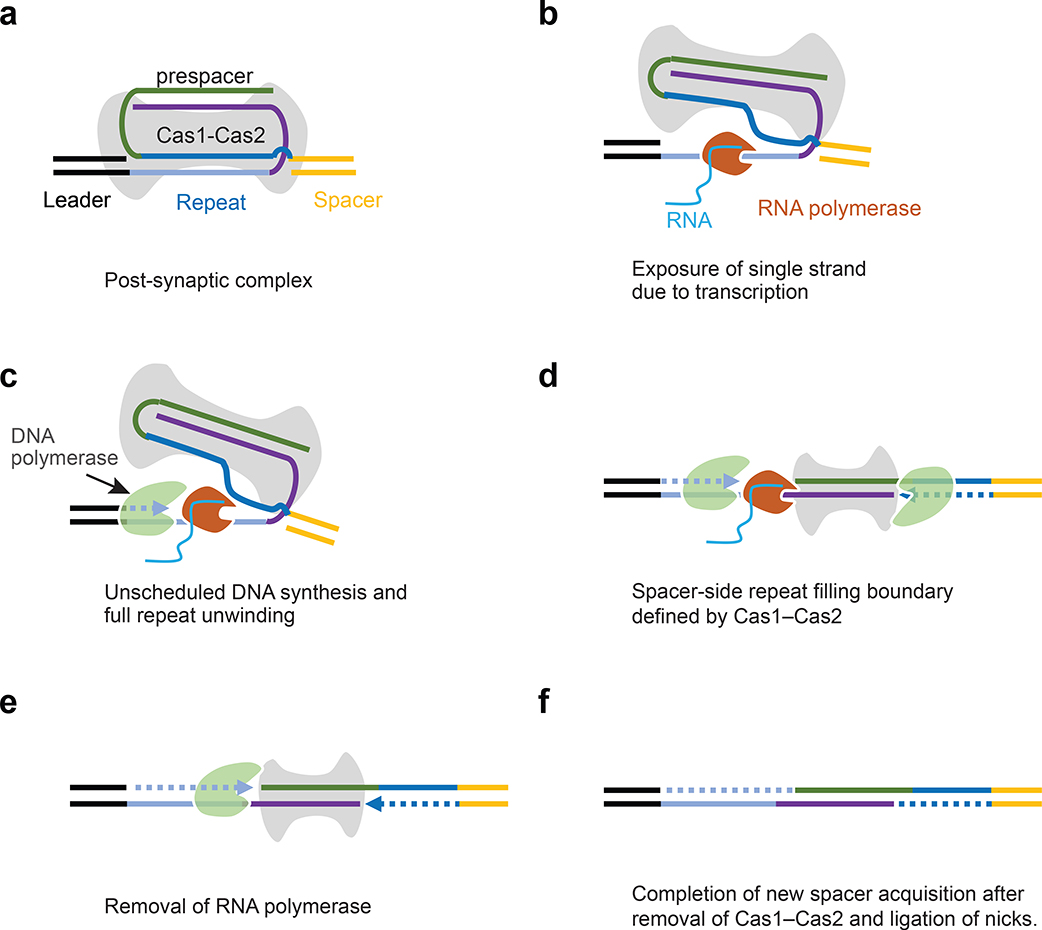
Fig. 7:
Model explaining how transcription-coupled repair resolves PSC and allows the final spacer incorporation into a CRISPR array. a, Integration of the prespacer by Cas1–Cas2 and formation of post-synaptic complex (PSC). b, RNA polymerase invades into the PSC upon transcription of the CRISPR locus and exposes single strands of the CRISPR repeat. c, Exposed repeat is filled by DNA polymerase. d, Cas1–Cas2 remain bound with new spacer and defines the spacer-side filling boundary. It also prevents double-strand DNA breaks. e, RNA polymerase is removed, and the leader-side is filled completely. f, Cas1–Cas2 is removed and the nicks are ligated, completing the incorporation of the new spacer.
Methods
Protein expression and purification
The expression and purification of EfaCas1–Cas2 were done by following the protocol published in our previous work25. Briefly, the His6-Sumo-tagged Cas1 and Cas2 were expressed and purified separately from E. coli BL21(DE3) cells following the same protocol. Briefly, the expression cells were grown separately at 37C until OD reached ~0.8. The culture was cooled to 18 ˚C, and protein expression was induced with 1 mM final concentration of IPTG. Cells were harvested from overnight cell culture by centrifuging at 4000g for 20 minutes, resuspended in the lysis buffer (25 mM HEPES, pH 7.5, 500 mM NaCl, 20 mM Imidazole), and lysed by sonication. After centrifugation at 15000 rpm for 40 minutes, the soluble fraction was loaded onto a Ni-NTA column pre-equilibrated in the lysis buffer. The unbound proteins were washed from the column by three rounds of five column-volume lysis buffer, and the bound proteins were eluted with the lysis buffer supplemented with 300mM imidazole. The eluted Cas1 and Cas2 proteins were concentrated and mixed at 2:1 molar ratio to form the apo Cas1–Cas2 complex. The sumo protease was then added to cleave the His6-Sumo-tag from both Cas1 and Cas2. The apo Cas1–Cas2 complex with the correct stoichiometry was separated from the individual components and other impurities using size exclusion chromatography (Superdex 200, GE Healthcare).
Introducing Cy5-label via thio-chemistry to EfaCas1–Cas2 complex
Because EfaCas2 lacks cysteine, we performed structure-guided mutagenesis and introduced an Ala19Cys substitution into EfaCas2; this residue is not conserved, and the sidechain change is not expected to interfere with EfaCas1–Cas2 function. Cas2_A19C was expressed and purified following the same procedure as for EfaCas2_WT, with the addition of a Cy5 maleimide labeling step while the protein was bound on the Ni-NTA column50. The unreacted Cy5 was removed by applying the lysis buffer without imidazole. The bound Cas2 was then eluted from the column, pooled, and concentrated. Cy5-labeled Cas2 was then complexed with Cas1 and further purified as previously described. Based on the histogram of the single-molecule fluorescence intensity, over 70% of the EfaCas1–Cas2 molecules from this labeling and purification approach were singly labeled.
Bulk integration reaction procedure for evaluating smFRET compatibility of Cy3-target and Cy5-prespacer
Bulk biochemistry was carried out to evaluate whether the attachment of Cy3 and Cy5 (both backboned labeled) on the target and prespacer affected integration reaction. The prespacer, PS(4,4), was mixed with Cas1–Cas2 in 1:1 molar ratio in a binding buffer (100 mM NaCl, 50 mM HEPES, pH 7.5). The pre-formed complex was then reacted with 10 nM Cy3-labeled target in the molar ratio of 4:1 at 10 mM MgCl2 final concentration. 50 μl of the reaction was removed at a different time point and mixed with Tris-equilibrated phenol-chloroform, pH 8 to stop the reaction. Each mixture was vortexed for 10s and spun at 13.6k rpm to separate the phases. The top layer was removed and mixed with an equal volume of 95% formamide-EDTA solution for gel analysis. The samples were separated on 10% urea-PAGE gel. The gel was scanned using appropriate lasers on Typhoon imager.
Construction of Total Internal Reflection (TIR) based imaging system
A prism-type total internal reflection fluorescence imaging system was built around an IX73 inverted microscope (Olympus). A green laser (OBIS 532 nm LS 100 mW, Coherent) and a red laser (OBIS 640 nm LX 40 mW) were installed on the optical table, and the incident beams were guided along the same optical path to excite donor (Cy3) and acceptor (Cy5) fluorophores, respectively. The Cy3 and Cy5 fluorescence signals were collected using a 60X water objective (UplanSApo, 60x/ 1.2 w, Olympus), filtered through a long-pass filter (BLP01–532R-25, Semrock) when the green laser was used or a notch filter (ZET635NF, Chroma) when the red laser was used. The filtered signals were then partitioned by a dichroic mirror (ZT633rdc-UF1, Chroma), and then projected onto two separate areas on an EMCCD camera (iXon, Andor), creating donor and acceptor channels for Cy3 and Cy5 signal visualization.
smFRET experimental setup
The slide cleaning, passivation and flow chamber preparation were performed by using the protocol, as described51. The microfluidic chamber was first filled with neutravidin to bind surface-biotin, and the excess unbound neutravidin was removed after 10 minutes of incubation. The biotinylated target DNA (10–20 pM) diluted in the imaging buffer (50 mM HEPES, pH 7.5, 2 mM Trolox, 0.8 mg/ml glucose, 0.1 mg/ml BSA, 0.1 mg/ml glucose oxidase, 0.02 mg/ml catalase, 10 mM MgCl2 and 100 mM NaCl) was introduced into the channel. The gradual appearance of bright spots on the donor channel indicated target immobilization. Once the desired target number reached 300−400, the channel was flushed with imaging buffer to remove unbound DNA. For the actual smFRET experiments, Cas1–Cas2 and prespacer were pre-assembled in 1:1 molar ratio at 200–500 nM concentration for 20 minutes to assemble the binary complex, in a buffer containing 50 mM HEPES (pH 7.5) and 100 mM NaCl.
smFRET recording of prespacer integration under native conditions
The pre-assembled Cas1–Cas2–PS(4,4) was diluted in the desired concentration (1–50 nM) using imaging buffer just prior to use in an experiment. The diluted solution was then loaded to the buffer reservoir mounted on one end of the flow cell. We recorded movies of varying lengths for different purposes. For example, to capture the initial interaction of Cas1–Cas2–PS(4,4) upon target encounter (pre-steady state), we recorded a relatively long movie lasting for about 5 minutes. The movie recording started 10 seconds prior to the introduction of 50–80 μLCas1–Cas2–PS(4,4), drawn from the reservoir into the chamber by applying negative pressure from the opposite opening. The movie recording continued until photobleaching of Cy3. This way we were able to capture multiple cycles and/or different modes of interactions. After system attained a steady-state, up to four additional 2–3-minute movies were also recorded to capture the steady state behavior. At last, 25 short movies (12 frames each) were also recorded for capturing the distribution of molecules in different FRET states. All movies were analyzed by using an analysis software package downloaded from the CPLC webpage <https://cplc.illinois.edu/software>. Long movies were converted to FRET trajectories for single molecules using the movie analysis program. The first 10 frames of each trajectory were combined to build FRET efficiency histogram. We refer to this histogram as ‘native-state histogram’ as Cas1–Cas2 is still interacting with the prespacer and target. MatLab graphical user interface was created (which can be made available upon request) to further process smFRET traces for vbFRET, dwell time and TDP plot analyses.
SDS denaturation procedure for the purpose of distinguishing binding from integration
2% SDS in the imaging solution (50 mM Tris-HCl, pH 6.8, 2 mM Trolox, 0.8 mg/ml glucose, 0.1 mg/ml glucose oxidase, 0.02 mg/ml catalase, NaCl 100 mM, 2% SDS added at last) was introduced during movie recording to denature and remove Cas1–Cas2. Unreacted prespacer DNA was removed by SDS whereas integrated prespacer remained. This washing step was typically applied towards the end of the native-state movie recording. The transition from the native to denaturing FRET state allowed us to define the integration status of a given single molecule. Twenty-five short movies with SDS-wash step incorporated were recorded at different areas and combined to generate the denaturing-state histogram.
Transition density plot (TDP) analysis
smFRET traces containing binding and unbinding transitions (Fig. 2a, c, e) or native to SDS-denatured transitions (Extended Data Fig. 6a, 7b, c) were hand-picked and fed to vbFRET software52 for finding hidden FRET states. The resulting ‘idealized traces’ were analyzed by in-house Matlab script to generate TDP.
Dwell time analysis to determine association rate constant (kon), dissociation rate constant (koff), reverse half-integration rate (k−1half) and reverse full-integration rate (k−1full)
Before carrying out kon and koff analyses, smFRET traces were subjected to vbFRET hidden state analyzer. Once the idealized traces were generated by vbFRET, rest of the procedure was semi-automated via custom-made Matlab graphical user interface. To determine kon (which is based on off-time or τoff), off-state dwell times were collected from at least 100 idealized traces (Fig. 2a, c, e) programmatically and plotted on a histogram. By fitting the gamma function, average τoff was determined, the inverse of which gives binding rate. The binding rate for each concentration of Cas1–Cas2–PS(4ddC, 4ddC) was determined from the program and plotted against the corresponding concentration. The binding rate vs concentration was linearly fitted, the slope of which gives kon (Fig. 2d). To find koff, on-state dwell times (τon) were collected. Even though Cas1–Cas2–PS(4ddC, 4ddC) binding occurs in two orientations, we took both as on-state in our analysis of koff. The rest of the procedure for finding τon was similar to that of τoff. The inverse of τon gives koff (Fig. 2f). The koff was found from multiple concentrations and reported as mean±s.e.
For the determination of k−1half and k−1full, experiments were carried out at low laser power of 3–5 mW. To compensate for the signal loss due to low laser power, movies were recorded at 2.5–3 Hz speed. The challenge with these experiments was to record a long movie for about 20 minutes, the doubling time for E. coli. Therefore, the stability of Cy3 and Cy5 were tested beforehand (Extended Data Fig. 7), which showed remarkable durability against photobleaching and blinking. The procedure in both types of the experiment (k−1half or k−1full) is to introduce Cas1–Cas2–PS(4, 4ddC) or Cas1–Cas2–PS(4,4), in the target-containing channel and allow integration for 4–5 minutes. While integration is undergoing, excess Cas1–Cas2–prespacer was washed out by gentle imaging buffer flow (30–40 μl/min). Once 30–40 μl of the buffer has flowed through the channel (volume of the channel is ~10 μl), the buffer flow was stopped. The surviving molecules on the imaging area after the flow are in an integrated state. The time it takes for them to dissociate from the target determines the rate of disintegration. The movie recording began 10s prior to Cas1–Cas2–prespacer flow and continued for next 20–25 minutes under continuous laser exposure.
To determine k−1half, Cas1–Cas2–PS(4, 4ddC) was flowed through the channel and the above procedure was followed to collect a movie. Then the movie was analyzed to generate the smFRET trace. The trace was marked at the time when imaging buffer flowed in, for example 267s on Fig. 3a. The time point at which the Cy5 signal disappear was noted (time2), which is due to detachment of Cas1–Cas2–PS(4, 4ddC) from the target. Tracing back along the FRET efficiency curve, the point of integration (time1) was also noted. The difference between these two time-points is half-integration dwell time. As many as five independent experiments were performed to acquire 300–400 dwell time data points for half integration. The dwell times were plotted on the histogram and fitted with gamma function () to determine the mean half-integration dwell time (= k * θ), the inverse of which gives k−1half.
To determine k−1full, Cas1–Cas2–PS(4,4) was used. The experimental procedure, movie recording, and trace generation are identical to previously described for k−1half. With PS(4,4) integration can occur from either end of prespacer giving rise to O1 and O2. In each orientation, a molecule can remain in half or full integration state, but FRET value of the orientation does not change due to leader-side Cy3 labeling on the target. One way to distinguish between half and full integration is to examine dwell time. Presumably, full integration dwell time is longer than that of half-integration dwell time. In this situation where half is mixed with full, the half-integration dwell time can be used to extract dwell time of full integration. All dwell time data points from the experiments were plotted in a histogram. The data were fitted with a sum of two gamma function (). As k and θ are known from the half-integration reaction, those parameters were fixed in the equation, and the subsequent equation was fitted which returned l and η. The inverse of (l*η) gives k−1full.
Time-resolved SDS-quenching experiment to determine integration rates khalf and kfull
In order to determine reaction rates for half and full integration, the target was immobilized on the surface and reacted with Cas1–Cas2–PS(4,4) for time points specified on ;;a and Extended Data Fig. 8 b–f. Once the reaction time was over, the 2% SDS solution was flowed at 250μl/min (time delay for SDS to reach across the channel was adjusted, which is about 1 second in our setup) to quench the reaction. After five minutes of SDS incubation, the channel was thoroughly washed with 500 μl of buffer (100 mM NaCl, 50 mM HEPES, pH 7.5) to remove SDS completely. Then, 100 μl of imaging buffer was passed through the channel and incubated for another five minutes before acquiring 25 short movies at different imaging areas. The movies were analyzed as described above, and histograms were generated for each reaction time points. To find the magnitude of integration, histogram data were fitted with Gaussian function (ORIGIN Pro 2018b) and area under the Gaussian peaks were obtained, which represents the amount of integration. Half integration peaks were added to get a total half integration population and the same procedure followed for full integration peaks. The half and full integration population were plotted against respective reaction time (Extended Data Fig. 5c, 8f) and the resulting plot was fitted with a single-phase exponential equation. The rate constants of the exponential equations provided the rate of reaction.
Fluorescence (Cy5) spot counting
As Cas1–Cas2–PS(4,4) (PS(4,4) has Cy5) flows through the surface (channel) pre-bound with Cy3-target, FRET-induced Cy5 signal starts to appear due to binding or integration of Cas1–Cas2 with the target and number gradually increases. In our experiments, the specified concentration of Cas1–Cas2–PS(4,4) (Extended Data Fig. 4b) flowed through the channel at 150 μl/min flow rate. The flow of Cas1–Cas2–PS(4,4) and movie recording started simultaneously and the movie was recorded continuously at 10Hz while visualizing molecular interactions. Within a minute of flow, movie recording stopped and Cy5 spots were counted in every 3–4 frames from the recorded movie. The Cy5 count was plotted against time and fitted with a single-phase exponential growth equation to obtain the observed rate (kobs). This process was repeated for four concentrations and corresponding kobs were obtained from the fit (Extended Data Fig. 4b). Then the plot of kobs vs Cas1–Cas2–PS(4,4) concentration was created and the resulting plot was fitted with the non-linear equation because kobs deviated from the line, indicating the presence of other processes apart from pure binding-unbinding (integration reaction in this case). The line was well fit by equation. The fit provides reaction rate constant (k2) as well as Kd (Extended Data Fig. 4c). The Kd derived this way does not involve kon and koff but has identical meaning to that derived as the ratio of .
Experimental procedure related to PSC resolution
Cas1–Cas2-less naked constructs (half or full) for testing repeat duplication were generated by reacting 192-bp target and Cas1–Cas2–PS(4,4) (unlabeled PS(4,4)). The 5’ ends of the 192-bp target were labeled with Cy3 and Cy5 to the leader and spacer side, respectively (Fig. 5a). The complex of Cas1–Cas2 and PS(4,4) was assembled the same way described above. The target was mixed with Cas1–Cas2–PS(4,4) at a 1:3 ratio and reacted for 5 minutes. Then the reaction was quenched with 0.5% SDS (the amount of SDS needed to successfully quench the reaction was titrated beforehand) in the final concentration. To remove SDS from the reaction, DNA was ethanol-precipitated and dissolved in the binding buffer. The concentration of resuspended constructs was estimated to be about 200 nM by comparing its fluorescence against known standards. The constructs were analyzed on 12% UREA page before using in extension experiments. Finally, the unprotected constructs were tested in a reaction containing 20 nM constructs, 50 nM DNA polymerase (NEB, Catalogue # M0212S), 200 μM dNTPs, and 50 mM (pH 7.5) HEPES. The reaction tube was placed in 37 ˚C water bath for 10 minutes, quenched with an equal volume of formamide-EDTA (formamide 90%, EDTA 50 mM, pH 8.0), and reaction products were analyzed on 12% urea page.
To test whether RNA pol facilitates repeat duplication, experiments were done in the order of integration, transcription, and extension. The length of target was increased from 192 bp to 256 bp to provide more separation between promoter and leader to avoid steric clash between RNA pol and Cas1–Cas2. The amount of protein and DNA used in the experiments are written in Fig. 5b. First, the integration reaction was carried out for 10 minutes at 37C at 1:3 ratio for the target to Cas1–Cas2–PS(4,4). While keeping the integration reaction at 37C, transcription mix (apart from E. coli RNA pol (NEB, Catalogue # M0551S), transcription mix also has ApU dinucleotide and rNTPs, 200 μM each) was added on one delivery. After a minute of transcription, extension mix (DNA Polymerase I Klenow fragment and 200 μM dNTPs) were also added, and the triple-reaction continued for 10 more minutes. The reaction was stopped by 100% phenol and routed through the phenol extraction procedure for DNA extraction. The products were analyzed on 9% UREA gel.
The single-molecule version of PSC resolution experiments (Fig. 5c, d) were done by immobilizing Cy3-labeled 200 bp targets containing promoter, leader, repeat, and spacer (see Supplementary Table 1 for the sequence). 10 nM Cas1–Cas2–PS(4,4) complex was flowed through the immobilized targets and reacted for five minutes. Then transcription mixture containing 17 nM E. coli RNA pol, 200 μM ApU, 1 μM rNTPs was introduced to the channel at a low flow rate (20–30 μl/minute). Following this, a Cy3-labeled 16nt oligo complementary to the first 12-nt of the CRISPR repeat was added to detect unwinding of repeat sequence. Imaging buffer without gloxy was added while illuminating the imaging area with the laser at relatively high power to photobleach Cy3. For this set of experiments, microscope stage was warmed up by locally heating the stage plate (custom built), which set temperate of slide to about 37C at the center. The temperature of room was also increase to 30C during the experiment to minimize heat loss from the slide. After recording a long movie (traces shown in Fig. 6c), Cy3 on the targets was photobleached. The control experiments (Extended Data Fig. 10a–c) were also performed the same way, but addition of transcription components was omitted from the experiments.
Prespacer trimming by ExoI or ExoIII
ExoI (NEB #M0293S), a processive 3’−5’ single-stranded nuclease, was used to trim the single-stranded 3’-overhang not protected by EfaCas1–Cas2 (Fig. 4a). EfaCas1–Cas2–PS(4, 26) complex were pre-assembled by incubating 550 nM of EfaCas1–Cas2 and 500 nM PS(4,26) in ice for 20 minutes in a buffer containing 50 mM HEPES (pH 7.5) and 100 mM NaCl. 50 μl of the reaction was transferred to 37 ˚C water bath for 5 minutes, and 20 units of ExoI (1 μl) was subsequently added. After a 30-minute incubation, the ExoI-treated EfaCas1–Cas2– PS(4, 26) complex was diluted to 50 nM and introduced into the quartz slide to react with the immobilized integration target. The remaining procedure has been described previously.
When a long-duplex containing prespacer precursor (i.e. PS(4,20-bp duplex)) is bound by EfaCas1–Cas2, the protruding dsDNA not protected by Cas1–Cas2 would be vulnerable to digestion by a dsDNase. Whereas ExoI has little activity for dsDNA, ExoIII (NEB #M0206S) is capable of selectively degrading the unprotected dsDNA from 3’ end. The pre-treatment procedure by ExoIII was similar to that by ExoI, except that only 10 units of ExoIII were used in the 50 μl reaction (Fig. 4b). Due to the high activity of ExoIII, the reaction was carried out at room temperature (~22 ˚C). For in situ prespacer processing (Fig. 4c), 1 μl of 10 unit/μl ExoIII was mixed with 50 μl reaction buffer (50 mM HEPES (pH 7.5) and 100 mM NaCl, 10 mM MgCl2) and introduced into the flow cell after allowing integration reaction to occur first. To prevent target DNA degradation by ExoIII, the G-quadruplex sequence (5’-TTGGGTGGGTGGGTGGG) was introduced to the 3’-end of the target (not shown in the schematics). After 5 minutes of treatment, the reaction was quenched by 2% SDS wash, and the integration pattern were imaged as described before.
Strains, Plasmids, and Reagents related to in vivo spacer acquisition assays
In vivo spacer acquisition was performed in strains derived from E. coli BL21-AI cells. In strain BL21-AI_Efa_pTrc, CRISPR Array I (NC_012947.1 position 1002802–1004320) was replaced with a synthetic Enterococcus faecalis array. This synthetic array consisted of 3 transcriptional terminators53, an E. coli RNA polymerase promoter (pTrc), 99bp of randomized sequence, a truncated (26-bp) E. faecalis leader, an E. faecalis repeat sequence, an E. faecalis spacer, a randomized region of 50bp, and a strong terminator in the reverse orientation (ECK120029600 from 54). Strain BL21-AI_Efa was identical to BL21-AI_Efa_pTrc except the pTrc promoter was removed from the synthetic array. pCas1,2 has a pRSF DUET 1 backbone and the E. faecalis Cas1 and Cas2 genes in MCS 1. pCas (addgene: 62225) and pTarget (addgene: 62226) were used for genome editing55. PCR site-directed mutagenesis was performed to change the gRNA sequence in pTarget (primers: F_pTarget_gRNA, R_pTarget_gRNA). The synthetic Efa array was flanked by ~300bp of homology to the regions flanking the BL21-AI CRISPR Array I, purchased as a gblock sequence (Integrated DNA Technologies), and cloned into pTarget to make pTarget_Efa. The pTrc promoter was added to pTarget_Efa using site directed mutagenesis to make pTarget_Efa_pTrc (primers: F_pTarget_pTrc, R_pTarget_pTrc). Primer and gblock sequences are documented in Supplementary Table 1. Plasmids were constructed using FastDigest restriction enzymes (Thermoscientific) according to the manufacturer’s instructions. All Cells were grown in LB (Luria–Bertani) media from Teknova. Reagents used were Kanamycin (50 μg/mL), Spectinomycin (50 μg/mL), Arabinose (0.2% w/v), IPTG (1 mM), and Nalidixic Acid (10 μg/mL).
E. coli Chromosome Editing
Chromosomal editing in BL21-AI was performed using a Cas9/Lambda Red system as described55. Briefly, the temperature-sensitive pCas (addgene: #62225) harboring Cas9 and Lambda Red components was transformed into BL21-AI and grown at 30˚C. An overnight culture of BL21-AI pCas was diluted and grown to OD A600 ~0.6 and lambda red genes were induced with 10 mM final concentration of Arabinose for 15 minutes. pTarget_Efa or pTarget_Efa_pTrc, which contained the gRNA sequence and the repair template, was electroporated into induced cells to allow for genome editing. The Cas9 gRNA (TTAAGTACTCTTTAACATAAAGG) targeted the leader region of the native BL21_AI CRISPR array. Cells were recovered in LB at 30˚C for 45 minutes and plated on LB+Kanamycin+Spectinomycin plates. Colony PCR (primers: F_genome_check and R_genome_check) was performed to detect successful genome editing and PCR products were Sanger sequenced to ensure successful editing. pTarget plasmids were cured after overnight growth in LB+IPTG (0.5mM)+Kanamycin and pCas was cured after overnight growth in LB at 37˚C. Successful removal of pCas and pTarget plasmids was ensured by the inhibition of growth on LB with the corresponding antibiotic.
Spacer Acquisition Assay
Top_Efa_prespacer and Bottom_Efa_prespacer were annealed in duplex annealing buffer (100 mM Potassium acetate, 30 mM HEPES, pH=7.5) to form prespacers used in the spacer acquisition assay. Salts were removed from prespacers by EtOH precipitation and resuspended in sterile MilliQ water to a concentration of 3.125 μM. The spacer acquisition was based on the published method32,33. Overnight cultures of BL21-AI_Efa_pTrc pCas1,2 and BL21-AI_Efa pCas1,2 were grown overnight at 37 ˚C. 100 μL of culture was diluted to 3 mL LB with induction components (1mM IPTG, 0.2% Arabinose, 50 μg/mL Kanamycin) and placed at 37 ˚C for 1 hour. Cultures were induced at 37˚C with shaking (180 rpm) for 2 hours. Where stated, nalidixic acid (10μg/mL) was added to the culture 1 hour and 40 minutes after the start of shaking. After induction, cultures were placed on ice and 1 mL of culture was centrifuged at 13,000x g for 1 minute. The supernatant was discarded, and the cell pellet was resuspended in 1 mL of chilled MilliQ water. The suspension was centrifuged at 13,000x g for 1 minute and washed with 1 mL of chilled MilliQ water; this wash step was repeated 3 times. The cell pellet was resuspended in 50uL of 3.125 μM prespacer in MilliQ water. This mixture was added to a 1mm gap cuvette (Laboratory Product Sales Inc.) and electroporated at 1.8kV, 25μF, and 200Ω using a Bio Rad Gene Pulser™. Cells were immediately recovered in 3 mL of cold LB or LB + Nalidixic acid (10 μg/mL) where noted. Cells remained on ice for 6 minutes and were then grown at 37 ˚C for 2 hours. 50 μL samples were taken at multiple time points (Time 0 minutes was taken before electroporation) and immediately heated to 95˚C for 5 minutes and stored at −20 ˚C.
Detection of Spacer Acquisition
Spacer integration was detected using PCR. 1 μL of the sample was used as the template for PCR. For Primer set 1 the PCR conditions were as follows: initial denaturation 3 minutes 95˚C, denaturation 10 seconds 98 ˚C, annealing 15 seconds 62.5 ˚C, and extension 20 seconds 72 ˚C for 25 cycles, and final extension 5 minutes 72 ˚C. PCR products were run on a 3% agarose TAE gel. Percent spacer integration was measured by quantifying the band intensity of the expanded array (350 bp) compared to the unexpanded array (284 bp) as an internal control in GelQuantNET v1.8.2. For the spacer integration time-course, a PCR of each sample using Primer set 1 (same conditions as above) was performed. The PCR product was purified using a GeneJET PCR Purification Kit (Thermo Scientific). 2.5 ng of DNA was used as a template for the Primer set 2 PCR reactions, with conditions as follows: of each sample was used as template initial denaturation 1 minutes 95˚C, denaturation 10 seconds 98˚C, annealing 15 seconds 67.5˚C, and extension 15 seconds 72˚C for 34 cycles, and final extension 5 minutes 72˚C.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
Source data for figure Fig. 2d and f, Fig. 3b and d, and Fig. 6b are available with the paper online. Information extracted from single-molecule movies is presented in Fig. 1–6 and Extended Data Fig. 1–10 in the manuscript. Raw movies data are available upon reasonable request.
Extended Data
Extended Data Fig. 1. Bulk biochemistry showing that the structure-guided fluorescent labeling scheme did not alter the integration activity of EfaCas1–Cas2.
a, Location of the Cy3 (green) and Cy5 (red) fluorophores and the six possible integration schemes (leader-half, spacer-half, and full-integration in two prespacer orientations); b, The expected length of the fluorophore-containing products from each integration scheme on a. c, Product of EfaCas1–Cas2 catalyzed integration over time, resolved on Urea-PAGE. Green band: Cy3-containing products; red band: Cy5-containing products; yellow: products containing both Cy3 and Cy5 fluorophores; leftmost lane: 5’-Cy3-labeled ssDNA size ladder. Uncropped gel images for panel c are shown in the Source Data.
Extended Data Fig. 2. Efficient target capture by EfaCas1–Cas2–PS(4,4) and interpretation of denaturing FRET states after SDS wash.
a, Histogram (native condition) collected from 25 short movies each having 325 FRET pairs on average after 10 min of 10 nM Cas1–Cas2–PS(4,4) incubation. Only two peaks were observed in steady state representing two orientation of prespacer, but it is not clear whether prespacer is integrated into the leader-side/spacer side, or, in half or full integration state. EFRET= center ± s.d. b-d, Representative smFRET traces showing potential binding, half-integration or full-integration events. Within five minutes of recording, more than 90% of traces recorded Cas1–Cas2 activities in the form of binding-unbinding or integration-disintegration. e, f, Oligonucleotide annealing scheme to mimic the leader-side half integration in two prespacer orientations. g, h, FRET histogram from single-molecule constructs depicted in e and f, respectively. i, smFRET histogram (denatured condition) after EfaCas1–Cas2 catalyzed integration from half-integration-only prespacers [i.e. PS(4, 4ddC)]. Integration only took place from the non-dideoxy end of the prespacer. Leader-side integration was strongly preferred. Spacer-side integration peak was only present after extended incubation.
Extended Data Fig. 3. Spacer-side labeling scheme revealed DNA bending and four native FRET levels for half and full integration.
a, The Crystal structure of half and full integration shown in both prespacer orientations; half to full conversion bends the target DNA and changes the FRET states, positions of donor and acceptor fluorophores are as indicated on DNA. b, Schematic of half and full integration in native states; six integration possibilities are shown. c, Steady-state FRET efficiency histogram showing binding-integration of Cas1–Cas2–PS(4,4) in the native state. Only four peaks were observed, two for half integration (unbent target) and two for the bent state in each orientation. Mostly, bent state corresponded to full integration (as detected by SDS wash), but a small fraction of bent population also showed half integration (both leader and spacer side) due to integration-disintegration phenomenon (see TDP); EFRET= center ± s.d. d, EFRET transition from the native to denatured state tabulated. e, Schematics of six integration configurations in the protein-denatured state.
Extended Data Fig. 4. Kinetic measurement of Kd by counting Cy5 spots.
a, A schematic of target and Cas1–Cas2–PS(4,4) used in experiments. b, Plot of bound or integrated single molecule population (measured via Cy5 signal on target) after the introduction of EfaCas1–Cas2–PS(4,4) at different concentration into the flow cell. Fitting the data with single exponential equation yields rate constant kobs for each concentration, which when plotted against concentration (b) gives equilibrium constant Kd and a reaction rate constant, k2.
Extended Data Fig. 5. Measurement of leader side reaction rate khalf.
a, A schematic of target and Cas1–Cas2–PS(4,4) used in experiments b, FRET histogram collected following SDS denaturation at varied reaction times, showing the changing free and integrated population (leader side) at different reaction times. c, Plot of leader-side half integrated population vs reaction time. Data fitting gives the rate of formation (khalf) of leader-side half integration. The khalf is comparable to k2 (Extended Data Fig. 3) and represents a lower limit of reaction rate because the integration reaction was difficult to perform reliably by hand for a reaction time of 1s or less.
Extended Data Fig. 6. Capturing binding-unbinding and integration-disintegration events using one-ended ddC prespacer.
a, A representative smFRET trace from a 15-min long movie with prespacer PS(4, 4ddC). O1 has longer dwell time than O2 because of integration from 3’-OH. The trace captures binding and unbinding, integration and disintegration, and FRET transition from native to the denatured state upon SDS treatment on a single shot. Two orientations of prespacer are shown by dashed lines. b, Plot of transitions from two native peaks, O1 and O2, to corresponding denatured peaks representing leader and spacer side integration, respectively. The plot was generated for PS(4,4ddC). c, Two smFRET traces for prespacer PS(4ddC, 4) after swapping -OH group and -ddC from PS(4, 4ddC). The dwell time for O1 and O2 is reversed due to swap.
Extended Data Fig. 7. Long movie trace showing finite stability of half and full integration.
a, Schematics of construct used in photo-stability test with 532 nm excitation, Cy3 emission, and FRET-induced Cy5 emissions. b, Representative smFRET traces showing eventual photobleaching of Cy3 (top) and Cy5 (bottom) after long-time excitation. c, Percentage of live molecules vs survival time for Cy3 (green) and Cy5 (red). d, A schematic of target and Cas1–Cas2–PS(4,4) used in experiments. e, f, Representative smFRET traces from 20-min long recording. As the one Cas1–Cas2–PS(4,4) molecule integrates and later disintegrates, another comes and interacts with the target as the excess molecules were not washed out. After 18 minutes of recording, SDS solution flowed through the channel to identify the fate of PS(4,4) prespacer if it was integrated at the time of flow. The last part of trace was used to create TDP as it contains both native and denatured state FRET levels.
Extended Data Fig. 8. Kinetic measurement of full-integration reaction.
a, A schematic of target and Cas1–Cas2–PS(4,4) used in experiments. b-e, Denaturing FRET histogram of the integration reaction quenched at different time point using SDS wash. Prespacer PS(4,4) was used in the measurement to allow full integration. Histogram for each time point was constructed from 25 short movies, each with about 300 FRET pairs. f, Histograms in b-e were quantified and the percentage of half- (black) and full-integration (red) products were plotted against reaction time, which shows the depletion of half-integration and the compensatory accumulation of full-integration population. The rate of formation full-integration (kfull) was derived from fitting the single-exponential equation, y = y0 + A * exp(−kfull * x), where y is population, x is reaction time.
Extended Data Fig. 9. Integration of various prespacer precursors.
a, Both overhangs 4 nt for positive control; both native and SDS histogram are shown. b, c, One side overhang 4 nt, another side overhang 5 and 6 nt, respectively. Histograms under native condition show integration from only one orientation, i.e. O1, and histograms under SDS treatment indicates that only one side of prespacer is attached as half integration. d, e, EfaCas1–Cas2 can spontaneously unwind 4 bp duplex blunt end. As a result, both orientations O1 and O2 appear under the native condition, and SDS wash resolves native peaks into several new peaks as seen before (Fig.1 d). f, With duplex length extended to 30 bp from its optimal 22 bp length, only one side with 4 nt overhang is integrated. Data were collected only for the denaturing condition.
Extended Data Fig. 10. Probe does not bind to repeat without transcription.
a, Cy3 spots from the 200bp target duplex five minutes after the flow of Cas1-Cas2-PS(4,4). The integration was detected by the appearance of Cy5 spots in the acceptor channel (but Cy5 spots not shown). b, The Cy3 spots were photobleached quickly by introducing imaging solution without gloxy under regular illumination of green laser (~25 mW). c, Image collected after the flow of Cy3-IR1 probe. The lack of Cy3 spots suggests that repeat is not exposed where the probe is expected to bind. A slight increase in spot number compared to ‘b’ (second image) may be due to non-specific binding of probe on surface-adsorbed Cas1–Cas2 that did not have prespacer or reappearance of some dark Cy3 (which appeared photobleached in ‘b’). d, Gel image showing multiple integrated spacers (bands) after 80 minutes under the different condition of replication and transcription.
Supplementary Material
Acknowledgements
This work is supported by National Institutes of Health (NIH) grants GM118174 and GM102543 to A.K. The authors declare no competing financial interests.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Yosef I, Goren MG & Qimron U Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic acids research 40, 5569–5576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nuñez JK et al. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nature structural & molecular biology 21, 528–534 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nuñez JK, Lee AS, Engelman A & Doudna JA Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519, 193–198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova KS, Grishin NV, Shabalina SA, Wolf YI & Koonin EV A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1, 7, doi: 10.1186/1745-6150-1-7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotin A, Quinquis B, Sorokin A & Ehrlich SD Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Mojica FJ, García-Martínez J & Soria E Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of molecular evolution 60, 174–182 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Pourcel C, Salvignol G & Vergnaud G CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663, doi: 10.1099/mic.0.27437-0 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Marraffini LA & Sontheimer EJ CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845, doi: 10.1126/science.1165771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amitai G & Sorek R CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol 14, 67–76, doi: 10.1038/nrmicro.2015.14 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Jackson SA et al. CRISPR-Cas: Adapting to change. Science 356, eaal5056 (2017). [DOI] [PubMed] [Google Scholar]
- 12.McGinn J & Marraffini LA Molecular mechanisms of CRISPR-Cas spacer acquisition. Nat Rev Microbiol 17, 7–12, doi: 10.1038/s41579-018-0071-7 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Wiedenheft B et al. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 17, 904–912, doi: 10.1016/j.str.2009.03.019 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Wang J et al. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163, 840–853 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Nuñez JK, Harrington LB, Kranzusch PJ, Engelman AN & Doudna JA Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature 527, 535–538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Díez-Villaseñor C, Guzmán NM, Almendros C, García-Martínez J & Mojica FJ CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas IE variants of Escherichia coli. RNA biology 10, 792–802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goren MG et al. Repeat size determination by two molecular rulers in the type IE CRISPR array. Cell reports 16, 2811–2818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuñez JK, Bai L, Harrington LB, Hinder TL & Doudna JA CRISPR immunological memory requires a host factor for specificity. Molecular cell 62, 824–833 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Wright AV & Doudna JA Protecting genome integrity during CRISPR immune adaptation. Nature Structural & Molecular Biology (2016). [DOI] [PubMed] [Google Scholar]
- 20.Heler R et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519, 199–202, doi: 10.1038/nature14245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Y, Terns RM & Terns MP Cas9 function and host genome sampling in Type II-A CRISPR–Cas adaptation. Genes & Development 29, 356–361, doi: 10.1101/gad.257550.114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ka D et al. Crystal Structure of Streptococcus pyogenes Cas1 and Its Interaction with Csn2 in the Type II CRISPR-Cas System. Structure 24, 70–79, doi: 10.1016/j.str.2015.10.019 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson M et al. Structure of the DNA-Bound Spacer Capture Complex of a Type II CRISPR-Cas System. Mol Cell 75, 90–101 e105, doi: 10.1016/j.molcel.2019.04.020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright AV et al. Structures of the CRISPR genome integration complex. Science 357, 1113–1118, doi: 10.1126/science.aao0679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Y, Ng S, Nam KH & Ke A How type II CRISPR-Cas establish immunity through Cas1–Cas2-mediated spacer integration. Nature 550, 137–141, doi: 10.1038/nature24020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunne T et al. Cas3-Derived Target DNA Degradation Fragments Fuel Primed CRISPR Adaptation. Mol Cell 63, 852–864, doi: 10.1016/j.molcel.2016.07.011 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Kieper SN et al. Cas4 Facilitates PAM-Compatible Spacer Selection during CRISPR Adaptation. Cell Rep 22, 3377–3384, doi: 10.1016/j.celrep.2018.02.103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, Zhou Y, Taylor DW & Sashital DG Cas4-Dependent Prespacer Processing Ensures High-Fidelity Programming of CRISPR Arrays. Mol Cell 70, 48–59 e45, doi: 10.1016/j.molcel.2018.03.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiimori M, Garrett SC, Graveley BR & Terns MP Cas4 Nucleases Define the PAM, Length, and Orientation of DNA Fragments Integrated at CRISPR Loci. Mol Cell 70, 814–824 e816, doi: 10.1016/j.molcel.2018.05.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drabavicius G et al. DnaQ exonuclease-like domain of Cas2 promotes spacer integration in a type I-E CRISPR-Cas system. EMBO Rep 19, doi: 10.15252/embr.201745543 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rollie C, Schneider S, Brinkmann AS, Bolt EL & White MF Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. Elife 4, e08716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shipman SL, Nivala J, Macklis JD & Church GM Molecular recordings by directed CRISPR spacer acquisition. Science 353, aaf1175, doi: 10.1126/science.aaf1175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shipman SL, Nivala J, Macklis JD & Church GM CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature 547, 345–349, doi: 10.1038/nature23017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J & Almendros C Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740, doi: 10.1099/mic.0.023960-0 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Marraffini LA & Sontheimer EJ Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571, doi: 10.1038/nature08703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Kasciukovic T & White MF The CRISPR associated protein Cas4 Is a 5’ to 3’ DNA exonuclease with an iron-sulfur cluster. PLoS One 7, e47232, doi: 10.1371/journal.pone.0047232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shmakov S et al. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res 42, 5907–5916, doi: 10.1093/nar/gku226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fossum S, Crooke E & Skarstad K Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli. EMBO J 26, 4514–4522, doi: 10.1038/sj.emboj.7601871 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adebali O, Chiou YY, Hu J, Sancar A & Selby CP Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase. Proc Natl Acad Sci U S A 114, E2116–E2125, doi: 10.1073/pnas.1700230114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park JS, Marr MT & Roberts JWE coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109, 757–767 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Lee H, Dhingra Y & Sashital DG The Cas4-Cas1–Cas2 complex mediates precise prespacer processing during CRISPR adaptation. Elife 8, doi: 10.7554/eLife.44248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dillard KE et al. Assembly and Translocation of a CRISPR-Cas Primed Acquisition Complex. Cell 175, 934–946 e915, doi: 10.1016/j.cell.2018.09.039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redding S et al. Surveillance and Processing of Foreign DNA by the Escherichia coli CRISPR-Cas System. Cell 163, 854–865, doi: 10.1016/j.cell.2015.10.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silas S et al. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science 351, aad4234, doi: 10.1126/science.aad4234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silas S et al. On the Origin of Reverse Transcriptase-Using CRISPR-Cas Systems and Their Hyperdiverse, Enigmatic Spacer Repertoires. MBio 8, doi: 10.1128/mBio.00897-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez-Delgado A, Mestre MR, Martinez-Abarca F & Toro N Spacer acquisition from RNA mediated by a natural reverse transcriptase-Cas1 fusion protein associated with a type III-D CRISPR-Cas system in Vibrio vulnificus. Nucleic Acids Res, doi: 10.1093/nar/gkz746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohr G et al. A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition. Mol Cell 72, 700–714 e708, doi: 10.1016/j.molcel.2018.09.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modell JW, Jiang W & Marraffini LA CRISPR-Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 544, 101–104, doi: 10.1038/nature21719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim S, Loeff L, Colombo S, Brouns SJJ & Joo C Selective Prespacer Processing Ensures Precise CRISPR-Cas Adaptation. bioRxiv, 608976, doi: 10.1101/608976 (2019). [DOI] [Google Scholar]
References for the Methods and Supplemental text sections
- 50.Joo C & Ha T Labeling Proteins for Single-Molecule FRET. Cold Spring Harbor Protocols 2012, pdb.prot071035 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Roy R, Hohng S & Ha T A practical guide to single-molecule FRET. Nat Methods 5, 507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bronson JE, Fei J, Hofman JM, Gonzalez RL & Wiggins CH Learning Rates and States from Biophysical Time Series: A Bayesian Approach to Model Selection and Single-Molecule FRET Data. Biophysical Journal 97, 3196–3205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mairhofer J, Wittwer A, Cserjan-Puschmann M & Striedner G Preventing T7 RNA polymerase read-through transcription-A synthetic termination signal capable of improving bioprocess stability. ACS Synth Biol 4, 265–273, doi: 10.1021/sb5000115 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Chen YJ et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods 10, 659–+, doi: 10.1038/Nmeth.2515 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Jiang Y et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81, 2506–2514, doi: 10.1128/AEM.04023-14 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for figure Fig. 2d and f, Fig. 3b and d, and Fig. 6b are available with the paper online. Information extracted from single-molecule movies is presented in Fig. 1–6 and Extended Data Fig. 1–10 in the manuscript. Raw movies data are available upon reasonable request.