Abstract
Introduction
Over the past couple of years, the cost of drug development has sharply increased along with the high rate of clinical trial failures. Such increase in expenses is partially due to the inability of the “one drug – one target” approach to predict drug side effects and toxicities. To tackle this issue, an alternative approach, known as polypharmacology, is being adopted to study small molecule interactions with multiple targets. Apart from developing more potent and effective drugs, this approach allows for studies of off-target activities and the facilitation of drug repositioning. Although exhaustive polypharmacology studies in-vitro or in-vivo are not practical, computational methods of predicting unknown targets or side effects are being developed.
Areas Covered
This article describes various computational approaches that have been developed to study polypharmacology profiles of small molecules. It also provides a brief description of the algorithms used in these state-of-the-art methods.
Expert opinion
Recent success in computational prediction of multi-targeting drugs has established polypharmacology as a promising alternative approach to tackle some of the daunting complications in drug discovery. This will not only help discover more effective agents, but also present tremendous opportunities to study novel target pharmacology and facilitate drug repositioning efforts in the pharmaceutical industry.
Keywords: Drug polypharmacology, Multi-targeting ligands, Drug repurposing, Computer-aided drug design, In silico prediction
1. Introduction
Before adopting a reductionist approach, pharmaceutical companies relied on phenotypic screenings to discover novel drugs. Later, progress in biochemistry and molecular biology allowed researchers to identify key targets in disease mechanisms. The “one drug, one target, one disease” approach has helped the pharmaceutical industry develop drugs that act on specific targets and understand the molecular mechanisms of drug-target interactions. However, the reductionist approach fails to appreciate the complexities of disease pathways and the system wide effects of drugs 1.
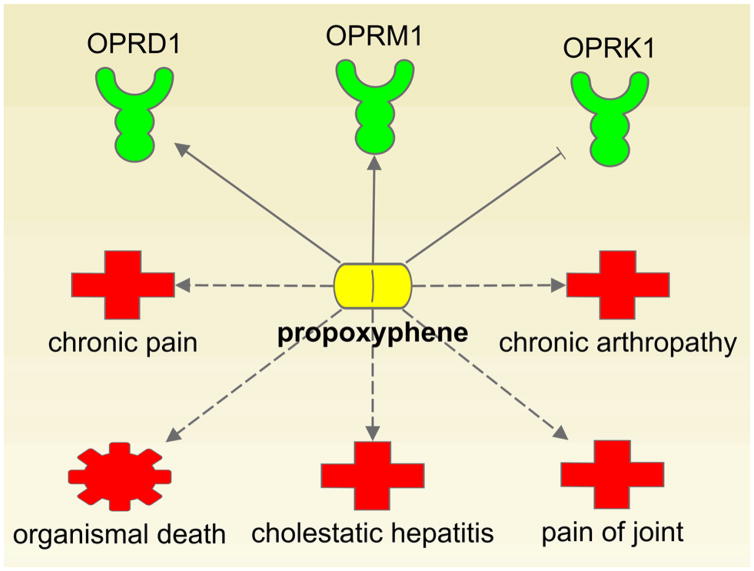
Recently, the number of clinical trial failures has been increasing. Some system wide understanding of small molecule targeting and prediction of side effects well ahead of clinical trials can help the pharmaceutical industry curb drug development costs. To address this issue, many pharmaceutical companies have begun to share their high-throughput small molecule screening data and attempted to understand drug system-wide effects in human. Paolini et al. mapped global pharmacological space by assembling a dataset of 276,122 active compounds and revealed that ~35% of the compounds had activity against more than one receptor 2. This is referred to as polypharmacology, the ability of a single agent to interact with and modulate multiple receptors. Traditionally, such off-target activity has been largely associated with drug side effects. For instance, propoxyphene, used as an analgesic agent, is a mu opioid receptor agonist with noncompetitive anti-α3, β4, and neuronal nicotinic acetylcholine activity (Figure 1). In 2010, the drug was withdrawn from the market owing to some fatal side effects. However, off-target properties can also be beneficial. Recently, the promiscuous nature of some drugs has been identified for novel applications to treat different diseases, known as drug repositioning. Repurposing previous approved drugs significantly decreases the time and cost in drug development compared with developing novel drugs from brand-new scaffolds, because previous clinical trial studies provide precious data of drug pharmacokinetics/pharmacodynamics (PK/PD) and toxicity profiles. A well-known example of drug repositioning is sildenafil (Viagra®), a drug that was originally designed for hypertension but is now marketed to treat penile erection dysfunction.
Figure 1.
Multiple targets of propoxyphene, along with its medical use and side effects.
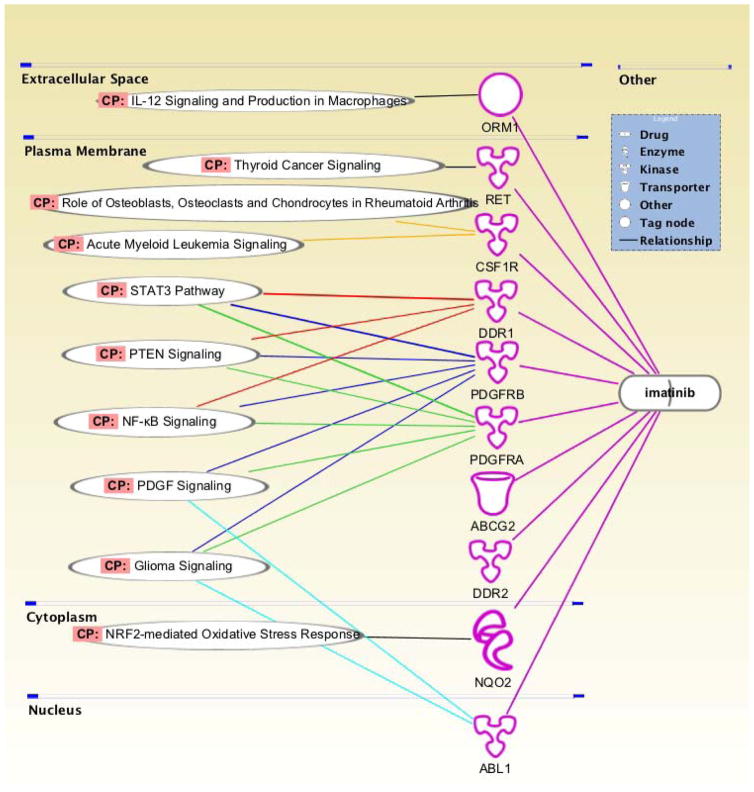
Due to its potential applications and recent successes, polypharmacology has generated great interest in drug discovery 3, 4. Kinase inhibitors are a good demonstration of polypharmacological properties. For instance, imatinib was initially developed to target the BCR-ABL protein, for which it was approved by FDA to treat Chronic Myelogenous Leukemia (CML) 5. Interestingly, it was also found to inhibit CD117, platelet-derived growth factor receptor (PDGFR), and c-KIT in the later subsequent research. It was then approved to use in patients with gastrointestinal stromal tumors 6. Imatinib is now known to inhibit even more targets and exerts various pharmacological effects. The signaling pathways, which have been identified to be targeted by imatinib are shown in Figure 2. Through targeting multiple signaling pathways, such drugs hold some significant advantages such as use in a larger population of patients. Also, some of these compounds have been reported to produce better therapeutic effects compared with selective drugs 7. Even in the field of neurodegenerative medicine, various studies have been reported about the use of multi-target-directed ligands to treat complex diseases such as Alzhemer’s Disease (AD), Parkinson’s Disease (PD), Huntington’s Disease (HD), and Amyotrophic Lateral Sclerosis (ALS) 8–10.
Figure 2.
The polypharmacology profile of imatinib, generated using the QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity).
In polypharmacology, one of the most important goals is to rationally design compounds that act on multiple key targets driving the pathogenesis of a given disease. Therefore targeting multiple proteins simultaneously stands a good chance to increase drug efficacy and decrease the possibility of drug resistance. In order to achieve these goals, it would be necessary to develop state-of-art computational techniques for data curation, model development, and quantitative predictions. In recent years, polypharmacology databases have been developed including STITCH 11, Polypharma 12, SuperTarget 13, IBIS 14, and SIDER 15. Tools such as PyMine bring relevant data from disease to chemical structure together and provide a convenient platform that could be potentially used for polypharmacology studies 16. A variety of methods that could be used to predict unknown targets for small molecules can be classified into three categories: structure-based methods, ligand-based methods and system biology methods.
In this review, we provide a summary of recent computational methods (Table 1) reported to study drug polypharmacology. We expect that a better understanding of the algorithms used in these methods will help improve current tools and develop novel technologies for polypharmacological studies. It is worth mentioning that, due to the length limit, rather than attempting to cover all reported methods, we will briefly discuss a few specific examples in each category. Readers are recommended to refer to the original articles for further understanding of these methods or other tools of their interest.
Table 1.
Methods and algorithms used for polypharmacology studies
| Methods | Algorithms Used | Web Links |
|---|---|---|
| Structure-based Methods | ||
| DOCK17 | Geometric shape matching algorithm, anchor and grow algorithm | http://dock.compbio.ucsf.edu/ |
| INVDOCK 18 | Geometric algorithm | http://bidd.nus.edu.sg/group/softwares/invdock.htm |
| idTarget 19 | Modified DOCK algorithm | http://idtarget.rcas.sinica.edu.tw |
| DRAR-CPI 20 | Connectivity maps with DOCK6 algorithm | http://cpi.bio-x.cn/drar/ |
| TarFisDock 21 | Modified DOCK Algorithm | http://www.dddc.ac.cn/tarfisdock/ |
| Glide 22 | Stochastic Search Algorithm | http://www.schrodinger.com/Glide |
| FRED23 | Stochastic Search Algorithm | https://docs.eyesopen.com/oedocking/fred.html |
| SiteEngines 24 | Geometric hashing method | http://bioinfo3d.cs.tau.ac.il/SiteEngine/ |
| SuMo 25 | Geometric matching of triplets | http://sumo-pbil.ibcp.fr/cgi-bin/sumo-welcome |
| IsoMIF Finder 26 | FLAP algorithm, Tanimoto coefficient | http://bcb.med.usherbrooke.ca/imfi.php |
| BioGPS27 | FLAP algorithm | NA |
| PocketMatch 28 | Greedy alignment algorithm, PM scoring method | http://proline.physics.iisc.ernet.in/pocketmatch/ |
| PARIS 29 | Convolution kernel based method | http://cbio.ensmp.fr/paris/paris.html |
| BSAlign 30 | Subgraph algorithm | http://www.aungz.com/BSAlign/index.htm |
| PharmMapper 31 | Kabsch Algorithm | http://59.78.96.61/pharmmapper/ |
| TRAP 32 | MD simulation, ARDR and PCA | http://trapp.h-its.org/trapp/ |
| GANDI 33 | Genetic algorithm with tabu search | http://www.biochem-caflisch.uzh.ch/download/ |
| AutoT&T 34 | Automatic tailoring and transplanting algorithm | http://www.sioc-ccbg.ac.cn/software/att2/ |
| ReCore 35 | Geometric rank searching algorithm | https://www.biosolveit.de/ReCore/ |
| AutoGrow 36 | Click chemistry assisted evolutionary algorithm | http://autogrow.ucsd.edu |
| Ligand-based Methods | ||
| SEA 37 | Chemical similarity, Kruskal’s algorithm | http://sea.bkslab.org |
| TarPred 38 | Extended-connectivity fingerprint 4 (ECFP4), Tanimoto Coefficient | http://www.dddc.ac.cn/tarpred |
| SuperPred 39 | Extended-connectivity fingerprint 4 (ECFP4), Tanimoto Coefficient | http://prediction.charite.de |
| SwissTarget 40 | Chemical and structure similarity | http://www.swisstargetprediction.ch |
| System Biology Methods | ||
| Cmap 41 | Pattern matching | https://www.broadinstitute.org/cmap/ |
| STITCH11, 42 | Text mining | http://stitch.embl.de/ |
| LINCS Canvas Browser43 | LINCS database browser | http://www.maayanlab.net/LINCS/LCB/ |
| Ingenuity Pathway Analysis ® | Pathway analysis | http://www.ingenuity.com/ |
| cBIOPortal44, 45 | A web portal to visualize genomics and proteomic data | http://www.cbioportal.org/ |
1.1. Structure-based Methods
1.1.1 Inverse Docking
Molecular docking of small molecules into the receptor active sites is one of the most widely used structure-based methods in the field of computer-aided drug discovery. It has been shown that current docking algorithms are able to predict docking poses of small molecules similar to experimental structures. Recent advances in high-throughput protein crystallography, nuclear magnetic resonance (NMR) spectroscopy, and electron microscopy have generated a large number of 3D protein structures in the Protein Data Bank (PDB). Availability of such structural data has allowed development of inverse docking methods where the primary aim is to dock a small molecule into binding sites of multiple targets for hit identification. Unlike traditional docking algorithms where small molecules are scored and ranked, in inverse docking target receptors are ranked according to their scores. In theory, all of the available docking algorithms should be able to carry out inverse docking. However the major challenge is to handle a large amount of structural data of macromolecules and implement scoring functions that could accurately rank the targets instead of small molecules. It has been found that traditional scoring functions used in current docking algorithms are not sufficient to rank targets accurately and robustly 46, 47. For example, Wang et al. observed that the original Glide scoring functions were insufficient in ranking targets 48, but a binding-site specific correction term was included, the results were significantly improved. Kellenberger et al. published another study where they ranked the targets using the Gold fitness score and topological molecular interaction fingerprint (IFP). It was shown that combination of these two scoring functions performed better than any other single docking scoring function 46. These studies suggest that novel target-specific scoring functions are needed to accurately rank targets for polypharmacological predictions.
Therefore in recent years, significant efforts have been put to develop modified scoring functions specific for target ranking such as INVDOCK 18, TarFisDock 21, and idTarget 19. Chen et al. developed the first automated inverse docking method INVDOCK [18]. Briefly for each small molecule and target pair, INVDOCK calculates ligand-protein interaction energy (ΔELP). In order to screen targets, this energy score is then compared with two threshold values: ΔEThreshold calculated from PDB receptor-ligand complex analysis and ΔEcompetitor calculated from the ligand present in the PDB structure of that receptor target. Targets are considered as binders if the calculated ΔELP is less than ΔEThreshold and ΔEcompetitor. The ΔEcompetitor threshold value is used to assess if the given small molecule is a better binder than the one present in the crystal structure. INVDOCK has been used in many studies that predicted multiple targets of given small molecules49–52. For example, Zhao et al. predicted 32 targets for Astragaloside IV (AGS-IV) using INVDOCK method 52. In the follow up studies, they experimentally validated three targets, namely calcineurin (CN), c-Jun N-terminal kinase (JNK), and angiotensin-converting enzyme (ACE).
Another popular inverse docking tool is idTarget, a webserver for target fishing developed by Wang et al. 19. It uses a divide-and-conquer approach where the entire receptor surface is used for docking. The search space is limited by adaptively constructing small overlapping grids to converge docking results. For each grid an affinity profile is generated with Gaussian function and the binding affinity range is used to calculate the width of the function. This profile is then employed to compute the Z-score. The target is considered significant if a large negative Z value is obtained. For a given binding pocket i and ligand j, the Z value can be calculated using the following equations:
| (1) |
| (2) |
| (3) |
where, Eij is the dock energy of ligand j and pocket i; sdi is the width of the affinity profile for pocket i; Ei is the center of the affinity profile for pocket i and Ec is the dock energy from the crystal pose. This idTarget method can be used for hit identification. Recently, Kumar et al. employed idTarget to predict multiple targets of kinetin 53, and they found that 4 out of 7 predicted targets are consistent to experimental reports in the subsequent literature search.
1.1.2 Binding Site Similarity
In addition to inverse docking, binding site similarity-based search is also frequently used for target prediction. It is based on the assumption that structurally similar proteins have similar molecular function and thus it is likely that they bind to structurally similar compounds. This phenomenon can be explained by the “Lock & Key” model that was postulated by Emil Fisher in 1894. Even differing in their overall sequences or structures, proteins may share local similarities in the form of binding sites, enabling them to bind similar ligands. Computational tools implemented by the community to identify similar binding sites are listed in Table 1. A majority of these tools utilize some variations of geometric patterns where the 3D coordinates of proteins are converted into simplified patterns that can be compared. The geometric matching of triplets is the most commonly used strategy, and the triplets are aligned based on the measurement of the distances between each triplet.
The Fingerprints for Ligands And Proteins (FLAP) algorithm was developed recently by combining the GRID Molecular Interaction Fields (GRID-MIFs) and pharmacophoric features 54. In the initial steps, various chemical probes that cover all types of chemical interactions are used to generate MIFs. These MIFs are then converted into pharmacophoric features using the weighted energy-based and space-coverage functions. All possible combinations of pharmacophoric features are used to generate four-point pharmacophore fingerprints, which are used to find ligand-ligand, ligand-receptor, and receptor-receptor similarities. The FLAP algorithm has been implemented in BioGPS, an automated method that calculates binding site similarities 27. This method works in two steps. First, It uses the Fixpdb algorithm for automatic protein structure preparation. In the second step, it employs the FLAPsite algorithm for binding site detection using 16 different GRID probes. As described earlier the FLAP protocol then calculates MIFs for the binding sites. These MIFs can then be superposed against all binding site MIFs in the database. Such binding site similarity tools can be used for drug repurposing and hit identifications. By comparing binding sites, it can be used in lead optimization process as well.
1.1.3 Inverse Pharmacophore Modeling
Traditionally, pharmacophore modeling is used as a ligand-based approach in drug discovery to identify small molecules with similar 3D features of the active molecule. Recently, advanced pharmacophore methods have been developed to map structure-based pharmacophore models of targets with small molecule pharmacophoric features 31, 54. One such tool, PharmMapper is devised by Liu et al. to predict targets of a given small molecule by reversely mapping small molecule conformations to the receptor pharmacophores. Similar to the widely used geometric hashing methods, PharmMapper creates a triangle-hashing table from triplets of feature points. These ligand-based triangles are then matched with a pre-computed library of pharmacophores generated from proteins. The alignment of these set of ligand triplets and protein pharmacophore is then carried out using the Kabsch algorithm, and a distance-dependent score is calculated, which is a weighted sum of point score (Spoint) and vector score (Svector) defined as below:
| (4) |
| (5) |
where, d(p)i is the distance between the ligand point feature p and the matching receptor pharmacophore feature; T(p)i is the matching tolerance value for d(p)i; F(v)i is same as F(p)i with the directionality difference and θ is the angle between the two vectors from the matching pair of vector features in the ligand and pharmacophore. The final fit score is calculated using the following equation:
| (6) |
where, Spenalty is calculated in the same way as Spoint but only between aligned pharmacophore excluded volume features and ligand heavy atoms. Final fit score is normalized to the range of [0,1]. Chen et al. applied this method to predict targets for a series of acenaphtho[1,2]pyrrole derivatives 55, and they identified several tyrosine kinases as the putative targets and experimentally validated the results thereafter.
1.1.4 Molecular Dynamic (MD) Simulations
MD simulations are often used to study thermodynamics and kinetics of the drug-receptor interactions. Cavalli et al. recently reviewed the role of MD simulation and related methods in drug discovery 56. They reviewed the theoretical background of MD and enhanced sampling methods most consistently used to study ligand-receptor interaction. They also assessed how these approaches help improve target affinity and drug residence time for better drug efficacies. In particular they discussed that, with the help of sampling techniques, clustering methods and accelerated MD simulations, some transient or allosteric binding pockets can be discovered using the static X-ray crystallographic structures 57–59.
Also Wade et al. published TRAPP (TRansient Pocket in Proteins), a tool to discover, analyze and visualize transient binding pockets in proteins using conformational ensemble data obtained from MD simulations 32. The authors showed that TRAPP was able to identify transient pockets observed in MD trajectory/conformational ensemble generated from the holo-structures with closed or partially closed binding pockets of HSP90. Another widely used and successful approach is Markov State Models (MSMs) with clustering algorithms and ensemble data generated from MD simulations 57. This technique has been used to identify transient pockets as well as to study activation pathway. Pande et al. performed one such study to identify intermediate states of c-SRC that can be targeted for drug design 60. Obviously, such newly identified transient pockets, although no reports have been published yet, can be exploited for polypharmcological agent design or drug repurposing studies.
1.1.5 Fragment-based Multi-target Drug Design
Fragments are small and relatively simpler chemical entities as compared to drug/lead-like molecules, and they tend to be more promiscuous. A small number of fragments can cover vast chemical search space, and thus fragment-based approaches increase probability of finding hits and help in designing novel compounds. Therefore they can be used for hit identification, hit-to-lead development, and lead optimization. Bottegoni et al. recently published a review article illustrating how fragment-based computational approaches can contribute significantly in a drug discovery pipeline starting from target identification to hit-to-lead optimization 61. In the article, the authors described various biophysical methods for fragment-based screening and how multiple solvent crystal structure (MSCS) can be used for multi-target design. The authors also described various studies indicating promiscuous nature of fragments. Along the same line, Hopkins and coworkers extracted the binding promiscuity information of fragments from Pfizer corporate screening data that included 220 diverse biological targets and 75,000 compounds 62. They revealed that smaller molecular weight fragments with less negative interacting features had more promiscuous nature towards biological targets than their counterparts. Combining such promiscuous fragments of the drugs from multiple drug-receptor complexes seems to be an obvious and clear strategy to develop polypharmacological drugs. Similarly Proschak et al. developed multiSOM, an in-silico approach for fragment-based design of polypharmacological ligands 63. It was applied to screen molecular fragments targeting 5-lipoxygenase (5-LO) and soluble epoxide hydrolase (sEH), and the authors identified fragments that could target both enzymes. Later, they were able to develop one of the fragments to a potent dual inhibitor with high affinity to both targets (IC50 of 0.03 μM toward 5-LO and 0.17 μM toward sEH).
1.2. Ligand-based Methods
The prediction of unknown targets using ligand-based methods is based on the compound properties and activities. One important assumption behind this is that structurally similar compounds bind to similar targets. In the past decades, many similarity-based methods have been developed to predict the targets of small molecules (Table 1). Herein, we take several examples to discuss their methodologies and algorithms.
The Similarity Ensemble Approach (SEA) is a similarity-based method that assesses the possibility of a compound binding to a target based on their ligand topological similarities 37. The statistical method of SEA used to calculate the binding likelihood is similar to that applied in Basic Local Alignment Search Tool (BLAST) 64. The SEA dataset consists of 65,241 ligands, which are annotated into 246 sets of receptors filtered from MDL Drug Data Report (MDDR). Firstly, the two-dimensional topological Daylight fingerprints were calculated for each ligand, and a pair-wise similarity metric was generated for each ligand in each set by calculating the Tanimoto Coefficient (Tc). For each set comparison, Tc between ligand pairs that passed the threshold were summed up and termed raw score (Sraw) for that comparison set. In the next step, z-score is derived using the following formula:
| (7) |
where, μ(Srandom) is the expected random raw score mean for that combination set size and σrandom is the random standard deviation. μ(Srandom) and σrandom values were calculated from the statistical model generated from the filtered subset of the MDD database. The z-score was then converted to the expectation value (E-value) in order to assess the probability that the score was achieved by a random chance:
| (8) |
where Ndb was the number of sets compared in the database and P(z) was the probability that the z-score was achieved by random. Herein, the background z-score has an extreme value distribution. As a result, probability of getting better raw score as compared to random chance P (Z>z) calculated using the following equation:
| (9) |
where and Γ′(1) is Euler Mascheroni constant. A matrix of the E-values was constructed for all pairs of sets in the database, and using the Kruskal’s algorithm, a minimum spanning tree is built as a similarity map. Although the similarity map is calculated based on ligand similarities, it is able to generate clusters of targets that are biologically related. Keiser et al. applied the SEA method to predict targets for methadone, emetine, and loperamide 37. They found that these drugs might act as antagonist of muscarinic M3, α2 adrenergic, and neurokinin NK2 receptors, respectively. The follow-up experimental studies indeed validated their prediction results.
Alternatively Zauhar et al. developed Shape Signature, a novel fragment-based ligand similarity search method65. This program uses smooth surface triangulator (SMART) algorithm to generate the ligand surface. In addition to ligand similarity, it can also compare the ligand shape with the shape of the binding site. Such shape-based similarity function can be further explored for polypharmacological applications during hit identification.
Currently, a variety of machine learning methods such as pairwise kernel method (PKM) 66, Gaussian interaction profile (GIP) 67, Laplacian regularized least squares (LapRLS) 68, kernel regression 69, kernelized Bayesian matrix factorization with twin kernels (KBMF2K) 70, and bipartite local method (BLM) 71 have been developed. In a comparison study of these methods, Ding et al. concluded that KBMF2K and PKM performed better than the other methods 72. Jacob et al. adopted SVM-based PKM method to predict drug-target interactions 66. The similarity of drug–target pairs from the drug-drug similarity score and the target-target similarity score is computed by following formula:
| (10) |
The generated similarity matrix (kernel) of drug-target pairs is then used to train SVM classifier for prediction of drug-target interactions. This algorithm was tested on three classes of targets (GPCRs, enzymes and ion channels) and it was reported that the method did exceptionally well compared to the traditional ligand-based methods, especially when targets do not have any known ligand.
Most recently, a new branch of machine learning known as deep learning is gaining significant attention in a variety of areas including drug discovery. The deep feed-forward neural network is the most basic deep learning algorithm, and can be characterized by cascaded layers of nonlinear processing units for feature extraction and transformation. Based on its architecture, several different neural networks have been proposed, including convolutional neural network, deep coding network and deep Boltzmann Machine (BM) 73. BM consists of a set of binary stochastic visible units used as a training vector in a layer named visible layer. It then successively connects a set of layers filled with binary stochastic hidden units named hidden layer 74.
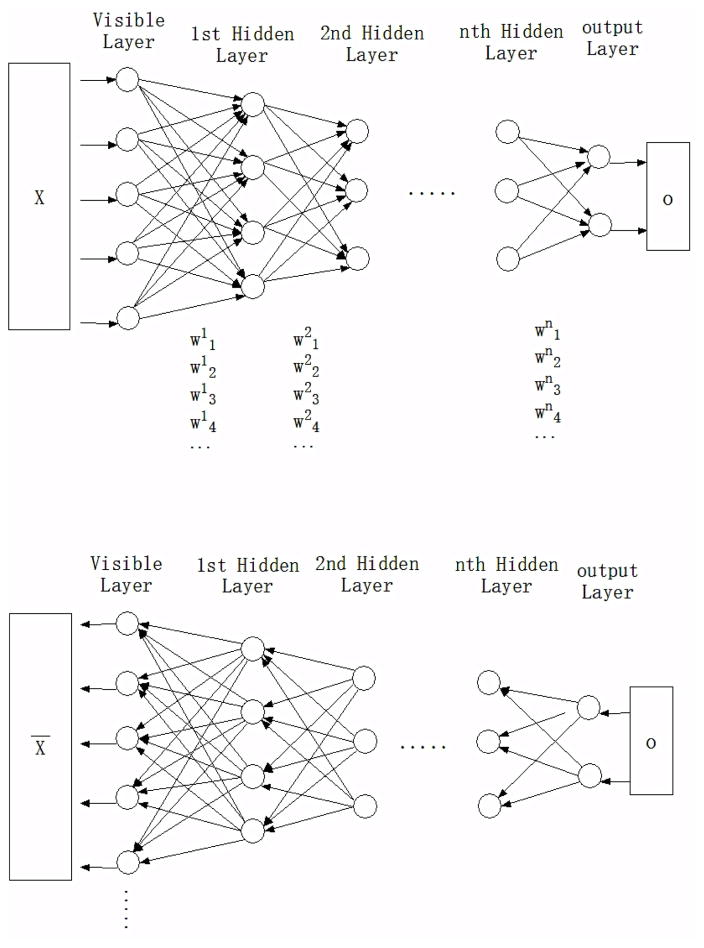
Restricted Boltzmann Machine (RBM) is a modified BM in which the training process can be divided into two phases 75. As illustrated by Figure 3 (Top), in the first phase, RBM visible layer learns the structure of the input data by its activation function and the output will be passed to the first hidden layer. The first hidden layer takes on the role of the visible layer, and its output data passes recursively until to the last layer. It is noted that each node in a hidden layer receives data from nodes of all previous layers, and the weight for each connection of paired nodes is randomly generated. In the second phase, namely reconstruction phase (Figure 3, Bottom), the output layer becomes the visible layer. The previous output data o is treated as the input data, and then passes backward from the output layer to the previous visible layer. In this process, the weights of each connection are the same as in the first phase. The difference between output X̄ and the previous X is then back propagated against the RBM’s weights, in an iterative learning process, until an error minimum is reached. Schneider et al. recently described various deep learning algorithms that can be applied for drug discovery 76.
Figure 3.
Top: in the first phase, input data x is passed through hidden layers to the final output layer. Bottom: in the reconstruction phase, the output results o are passed from output layer to the previous visible layer.
In drug discovery, deep learning algorithms can be used to interpret and understand the features extracted from chemical as well as biological data. For example, Lusci et al. developed a deep learning-based webserver AquaSol that predicts aqueous solubility of drug-like molecules using molecular graphs of compounds 77. They showed that such deep learning-based method outperformed other state-of-the-art solubility prediction tools. Most recently, researchers have used deep learning algorithms to predict drug-target interactions, repurpose drugs, and explore drug novel pharmacological properties using chemogenomics data 78, 79. Many start-up companies have emerged to adopt deep learning algorithms for their business. For example, the core technology behind Berg LLC (www.berghealth.com) is their Berg Interrogative Biology® platform that analyzes patients’ biological and clinical data using a proprietary deep learning algorithm. Currently, the company has two small molecule drug candidates in clinical trials. Atomwise (www.atomwise.com) is another company that developed the first deep convolutional neural network known as AtomNet for structure-based drug design. Recently the company reported that its discovery of new Ebola treatment using this technology.
1.3. Systems Biology Approaches
Understanding of diseases, especially complex ones has never been so thorough with the advent of high-throughput techniques producing gigantic amount of data in fields such as genomics, proteomics, and so on. The efforts such as the Connectivity Map Project 41, the largest collection of gene-expression profiles for diseases, genetic mutations, and drug activities, have significantly facilitated our understanding of disease mechanisms and therapeutic treatment. Hopkins et al. introduced the term “Network Pharmacology” and suggested that integrating chemogenomics data with network biology will help develop new approaches to target disease-causing networks instead of single gene/target 80. Vitali et al. also recently used protein-protein interaction network and implemented a network-based method to retrieve different associations with ligands. This has been used to rank targets for polypharmacological studies 81.
STITCH is useful database to predict protein-compound interactions 82. The authors employed natural language processing text-mining techniques and extracted known protein-ligand interactions from literature. The data was then used to predict interactions of query compounds. A new function was added recently to allow filtration of tissue specific proteins in the network 82 based on the data from TISSUES resources 83 and Expression Atlas 84.
Recently our group has embarked on the development of new methods for drug polypharmacology predictions. Based on more than 56,000 protein structures and 5,000 drugs curated in-house, we built an integrated platform to analyze the relationship between drugs and their receptors 85. We also implemented a web-based application that can predict interactions and visualize the molecular networks based on ligand and receptor annotations, chemical similarity, and molecular connectivity maps. Our group has also developed Polypharma, which includes all drug-like ligands that bind to more than two distinct proteins in PDB 12. It provides a collection of confirmed multi-targeted compounds useful in novel target predictions and can be used to predict new multi-targeting compounds. The analysis of these multi-targeting compounds can highlight the differences between single-targeting and multi-targeting agents.
Most impressively, the current high-throughput methods, such as RNA-Seq, Next Generation Sequencing (NGS), and Reverse Protein Phase Array (RPPA), have generated significant amount of gene expression data. With such genome-wide data from treated and untreated patient samples, one can study the overall functional effects of a small molecule and derive gene expression signatures that could be used to build training datasets for polypharmacology studies. For instance, Sirota et al. devised a drug repositioning method to predict drug effects based on the expression pattern of 100 diseases and 164 drugs 86. In a follow-up experimental validation study, they showed that cimetidine could be used to treat lung adenocarcinoma.
In addition, the NIH Library of Integrated Network-based Cellular Signature Program (LINCS) (http://www.lincsproject.org) is an ambitious community-wide program designed to collect and disseminate the massive data constantly generated and updated worldwide using multiple advanced methodologies. It is also involved in development of analytical tools to predict the response of human cells to interference by drugs, environment, or mutations. The database, which contains more than millions of drug-induced gene expression profiles, can be used to explore the potential drugs for polypharmacology 43. Another software package, Ingenuity® Pathway Analysis, developed by Qiagen, helps analyze and visualize data by mapping various chemicals and novel signaling pathways. Other tools such as Cystoscope, an open source molecular interaction network visualizer, can also be utilized for informatics-based polypharmacology predictions 87.
2. Polypharmacological Drug Design (PDD)
Polypharmacological drug design is the concept of rationally designing small molecules that can act on multiple targets of interest in order to achieve better efficacies, lower toxicities, less resistance, and more convenient administration. Since 2004, Morphy et al. have published several articles describing the opportunities, challenges and strategies for developing such multi-targeted ligands 88–90. One of the major advantages of this method is facilitation of discovery of novel lead compounds with unique structural and chemical properties. Although developing multi-targeted drug is extremely challenging, investigators have started to make attempts to identify/develop such multi-targeted agents.
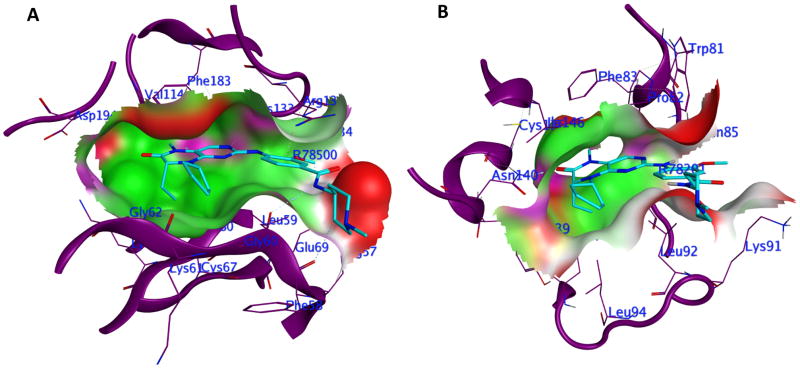
Recently, efforts have been made in developing kinase inhibitors with suitable polypharmacological profile. For instance, Ciceri et al. employed an inter-family strategy based on the fact that the bromodomains play important role in multiple diseases where kinase inhibitors are used as therapeutic drugs 91, 92. Through structural modeling, they provided a rationale for new polypharmacological drug discovery strategies focused on the two complementary target classes. Upon testing 628 kinase inhibitors, they identified PLK1 inhibitor BI2536 as the potent inhibitor of BRD1 receptor. Comparison of co-crystal structures also explained the kinase-bromodomain cross-reactivity (Figure 4) and showed a high degree of pharmacophore feature similarity in the binding sites. This demonstrates proof of principle that it is possible to develop potent and polypharmacological drugs, an idea that is anticipated to add important therapeutic benefits to future healthcare.
Figure 4.
Binding of the small molecule BI2536 in A) the binding pocket of PLK1 receptor (PDB ID: 2RKU); and B) the binding pocket of the first bromodomain of human BRD4 (PDB ID: 4OGI).
Besnard et al. reported about the adaptive optimization algorithm, a novel lead optimization approach based on the automated design of compounds using medicinal chemistry transformation rules 93. In this method, the ranking of the target is based on predefined polypharmacology profile objectives. In the first step, the parent population of chemical structures (a set of compounds that needs to be optimized) and a set of ideal objectives (properties of an ideal target compound) are generated. The second step is to obtain all possible analogues of the parent compounds through medicinal chemistry transformations. For each generated analogue, target activity and molecular descriptors are calculated. Based on these descriptors, each analogue is then scored against Laplacian-corrected naïve Bayesian classifier models built for 784 targets using FCFP6 fingerprints of small molecules. Next, analogues are ranked based on the distance between predicted score of the analogue and ideal achievement objective using the vector scalarization method.
By combining compounds that meet threshold values with random compounds from a previous parent population, a new parent population is generated and the process is iterated until no further improvement occurs. This algorithm was tested using donepezil, an approved acetylcholinesterase inhibitor, as a starting point to develop new compounds with blood brain barrier (BBB) permeability and selectivity against D2 and D4 dopamine receptors 93. The authors were able to identify novel compounds with dual activity against acetylcholinesterase and D2/D4 dopamine receptors with improved BBB permeability. They reported that 75% of the predictions from 800 ligand-receptor interactions studied were correct. It suggests that this method can be very useful in the development of novel leads for suitable polypharmacology profiles.
Alzhemer’s Disease (AD) is a complex diseased caused by multiple factors such as formation of insoluble neurotoxic plaques of Aβ through amyloid aggregation, oxidative stress, hyperphosphorylation of tau protein, calcium imbalance, mitochondrial dysfunction, or deterioration of synaptic transmission. Such multi-factorial etiology is most suitable for multi-targeted drug discovery approach. For instance, Viegas et al. reported recent advances and applications of multi-targeted drug design strategies for AD treatment 94. They described various examples of multi-targeted drugs by hybridizing Tacrine with caffeic acid, pyrines, carvedilol, ferulic acid, and cystamine. They also discussed Memoquin, a dimeric agent with anticholinesterasic and antimuscarinic activity developed by Cavalli and co-workers. It has IC50 of 1.55 nM for AChE and 108nM against BACE-1. Thus by restoring cholinergic deficit and reducing AB expression and accumulation, the agent helps in AD recovery. Recently, Grover et al. published highly predictive group-based QSAR (GQSAR) model for the fragments of 20 1,4-dihydropyridine (DHP) derivatives. A large combinatorial library of DHP analogues was created, the activity of each compound was predicted, and the top compounds were analyzed using refined molecular docking. A detailed interaction analysis was carried out for the top two compounds (EDC and FDC), which showed significant binding affinity for BACE-1 and AChE 95. Cordeiro et al. published a fragment-based QSAR model that showed 94.06% and 92.92% for sensitivity and specificity in training sets respectively for anti-PCa activity 96. Some fragments were extracted from the molecules and their contributions to anti-PCa activity were calculated. They identified several fragments as potential substructural features responsible of anti-PCa activity. They reported new molecular entities designed from fragments with positive contributions as possible anti-PCa agents. Success in such studies shows promising future for polypharmacological drug design.
3. Conclusions
Pharmaceutical companies have successfully carried out their drug discovery programs using a reductionist approach in the last several decades. However, the cost increase in drug development has promoted the pharmaceutical industry to opt for alternative strategies, and recent success in discovering previously unidentified uses of well-studied FDA drugs has provided a great motivation for such efforts. The assumption that similar compounds bind to similar targets remains as the basic principle behind most of the polypharmacology studies. A major challenge is to develop accurate and robust scoring functions that can rank targets instead of small molecules. Currently novel approaches to rational design of multi-targeting small molecules are being explored. Apart from classical structure-based and ligand-based methods, interest in system biology and bioinformatics-based methods, along with community-wide efforts, has increased. These methods are shown to not only predict new targets of small molecules, but also help understand the complex dynamics of diseases and the molecular interaction pathways underneath.
4. Expert Opinion
4.1. Key Findings and Potentials
Polypharmacology has certainly emerged as a new paradigm in drug discovery. It is not only being used to identify toxicities or new uses for approved drugs but also develop novel molecular entities with controlled multiple targeting activities. Recently, we have witnessed rapid growth and wide application of polypharmacology studies. Particularly, the successes in drug repurposing 97 brought a high expectation to this field and such efforts have become international initiatives 98, 99. As polypharmacology can predict drug effects, both on-target and off-target, it could provide a better picture of targeting diseases. Therefore, the rational polypharmacological drug design holds a great promise and potential for the next generation of drug discovery. However, to achieve such ambitious goals and ultimately translate the knowledge to effective patient therapies, we still need to overcome a variety of weaknesses and challenges.
4.2. Weakness and Challenges
One of the biggest challenges in polypharmacology studies is that we only understand the pathways/mechanism of many diseases partially at the molecular level. It is of great issue to derive the polypharmacological network using incomplete data. Even after the complex networks are constructed 100, understanding the convoluted associations is still a very challenging task. Also computational polypharmacology methods rely heavily on experimental data for model building or program learning, and frequently such data is either missing or varies in quality. For instance, structure-based methods need high quality 3D structures of receptors. While this is the case for certain proteins such as kinases, only representative structures or no structures are available for many other protein families. There is also a tendency in the scientific literature that research efforts concentrate on a small number of targets, resulting in the lack of functional annotation of many, sometimes even most, family members. In addition, inverse docking suffers from several issues including difficulty to address target flexibility and limitations in accurately and efficiently scoring targets instead of small molecules. More accurate mining techniques and mapping methodologies are needed to analyze the complex data. Currently it is still difficult to seamlessly integrate and constantly update the enormous data from non-standard, non-synchronized sources.
Although it is now possible to study system-wide functional data with the help of high-throughput screening technologies, integration of such data will provide a better understanding of drug effects on complex diseases and their molecular regulation pathways. Unfortunately, most computational programs follow the “garbage in, garbage out (GIGO)” rule. Hence a close attention needs to be given to the quality of input data and thus a significant amount of effort on data curation is required. In addition, most of the current methods are implemented either as web servers or standalone packages. Since community efforts become more crucial, it will be important to develop portable programming libraries which can be employed by the community developers to modify existing or implement new toolkits. It is also critical to integrate concepts and approaches into this field from other disciplines such as cheminformatics, bioinformatics, and systems biology, as demonstrated by examples such as the SEA 37 and Polypharma 12 methods.
4.3. Current status and future perspective
It is well recognized that, drug polypharmacology goes well beyond target families and drug off-targets are poorly understood or largely unknown, in most cases. However, it is impossible to exhaustively test all of the chemical and biological space with wet lab experiments. Thus more computational approaches are needed, along with increased integration of available data. As described above, numerous methods and databases have been developed, and of course challenges still lie ahead. However it is acknowledged that polypharmacology approaches are showing their promise and will certainly have the potential to transform our next-generation drug discovery and development.
During the next 5–10 years, polypharmacology will grow significantly, with more and more cutting-edge technologies such as deep learning used to extract unexpected relations of biological and chemical entities controlling complex diseases. On one hand, this is important for prediction of possible adverse effects of new drugs in the pipeline of development. On the other hand, it is particularly helpful for drug repurposing where new indications of existing drugs/agents can be identified. To revive the stalling drug discovery engine, both public and private sectors have launched new initiatives such as the NCATS program and the Cures Within Reach mission (http://http://cureswithinreach.org/). Therefore, in the near future we will see wider collaborations between academia and pharmaceutical industry for repositioning FDA approved drugs.
Along the same line, advances in computational sciences for polypharmacology prediction will continue with more widespread applications in drug discovery, especially deep learning, which is essentially to train a computer to take all unstructured big data and look for complex relationships in all that big data using methods like artificial neural networks. This is something the human brain does naturally, with well-known examples such as the IBM cognitive computing platform Watson and the most recent AlphaGo program developed by Google DeepMind. Although the application of deep learning in drug discovery is still limited, with development of new deep learning algorithms, we have been equipped with a potentially game-changing toolbox. Based the success of recent pioneering studies, deep learning will be useful for the coming age of big data analysis in pharmaceutical research and ultimately personalized precision medicine. Down the road in the next few years, we anticipate that more sophisticated, robust, and accurate polypharmacology approaches will boom and rational design of more potent but less toxic multi-targeting agents may become possible, although it remains extremely challenging at the current stage.
Article Highlights.
Polypharmacology involves molecular interactions of drugs and multiple targets implicated in complicated single or multiple disease pathway signaling and treatment. This review article covers computational aspects of polypharmacology studies.
Polypharmacology is alternative to the traditional concept of “one drug –one target”. Computational polypharmacology studies could predict potential drug side effects and disastrous toxicities as well as be used to repurpose existing drugs.
High-level data curation/integration and novel computational approaches have been developed, but more are needed for accurate polypharmacology prediction.
Deep learning is being adopted for retrieving hidden and complex relations between biological targets and chemical entities.
Numerous attempts have been made to develop new molecular entities with optimized polypharmacology profiles.
Significant challenges still lie ahead for polypharmacology studies, in particular for rational design of effective multi-targeting agents.
Acknowledgments
Special thanks to the OpenEye Scientific Software and ChemAxon for their free academic licenses. We also thank the HPC resources from Texas Advanced Computing Center (TACC) and the University of Texas M.D. Anderson Cancer Center.
Funding:
This work is supported by is partially supported by the Cancer Prevention Research Institute of Texas (CPRIT) DP150086, the National Science Foundation (NSF) grant CHE-1411859 and the National Institutes of Health through grants NIH/NIGMS GM070737, and NIH/NCI P30CA016672.
Footnotes
Declaration of Interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
References
- 1.Van Regenmortel MH. Reductionism and complexity in molecular biology. Scientists now have the tools to unravel biological and overcome the limitations of reductionism. EMBO Rep. 2004 Nov;5(11):1016–20. doi: 10.1038/sj.embor.7400284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paolini GV, Shapland RH, van Hoorn WP, et al. Global mapping of pharmacological space. Nat Biotechnol. 2006 Jul;24(7):805–15. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 3**.Anighoro A, Bajorath J, Rastelli G. Polypharmacology: challenges and opportunities in drug discovery. J Med Chem. 2014 Oct 9;57(19):7874–87. doi: 10.1021/jm5006463. Excellent review on polypharmacology that describes advantages and disadvantages between combination therapy, multi-targeted drugs and drug repurposing It also describes applications of polypharmacology in cancer therapy and CNS diseases. [DOI] [PubMed] [Google Scholar]
- 4.Rastelli G, Pinzi L. Computational polypharmacology comes of age. Front Pharmacol. 2015;6:157. doi: 10.3389/fphar.2015.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capdeville R, Buchdunger E, Zimmermann J, et al. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov. 2002 Jul;1(7):493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 6.Joensuu H, Roberts PJ, Sarlomo-Rikala M, et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med. 2001 Apr 5;344(14):1052–6. doi: 10.1056/NEJM200104053441404. [DOI] [PubMed] [Google Scholar]
- 7.Chou WC, Huang SF, Yeh KY, et al. Different responses to gefitinib in lung adenocarcinoma coexpressing mutant- and wild-type epidermal growth factor receptor genes. Jpn J Clin Oncol. 2006 Aug;36(8):523–6. doi: 10.1093/jjco/hyl057. [DOI] [PubMed] [Google Scholar]
- 8.Cavalli A, Bolognesi ML, Minarini A, et al. Multi-target-directed ligands to combat neurodegenerative diseases. J Med Chem. 2008 Feb 14;51(3):347–72. doi: 10.1021/jm7009364. [DOI] [PubMed] [Google Scholar]
- 9.Leon R, Garcia AG, Marco-Contelles J. Recent advances in the multitarget-directed ligands approach for the treatment of Alzheimer’s disease. Med Res Rev. 2013 Jan;33(1):139–89. doi: 10.1002/med.20248. [DOI] [PubMed] [Google Scholar]
- 10.Ambure P, Roy K. CADD modeling of multi-target drugs against Alzheimer’s disease. Curr Drug Targets. 2015 Sep 6; doi: 10.2174/1389450116666150907104855. [DOI] [PubMed] [Google Scholar]
- 11.Kuhn M, von Mering C, Campillos M, et al. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008 Jan;36(Database issue):D684–8. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Reddy AS, Tan Z, Zhang S. Curation and analysis of multitargeting agents for polypharmacological modeling. J Chem Inf Model. 2014 Sep 22;54(9):2536–43. doi: 10.1021/ci500092j. Description of Polypharma, a web-based comprehensive database of high-quality co-crystalized compounds having polypharmacology profiles that can be useful for computational polypharmacology modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunther S, Kuhn M, Dunkel M, et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008 Jan;36(Database issue):D919–22. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoemaker Ba, Zhang D, Tyagi M, et al. IBIS (Inferred Biomolecular Interaction Server) reports, predicts and integrates multiple types of conserved interactions for proteins. Nucleic acids research. 2012;40:D834–40. doi: 10.1093/nar/gkr997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn M, Letunic I, Jensen LJ, Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016 Jan 4;44(D1):D1075–9. doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhari R, Li Z. PyMine: a PyMOL plugin to integrate and visualize data for drug discovery. BMC Res Notes. 2015;8:517. doi: 10.1186/s13104-015-1483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang PT, Brozell SR, Mukherjee S, et al. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA. 2009 Jun;15(6):1219–30. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YZ, Ung CY. Computer automated prediction of potential therapeutic and toxicity protein targets of bioactive compounds from Chinese medicinal plants. Am J Chin Med. 2002;30(1):139–54. doi: 10.1142/S0192415X02000156. [DOI] [PubMed] [Google Scholar]
- 19.Wang JC, Chu PY, Chen CM, et al. idTarget: a web server for identifying protein targets of small chemical molecules with robust scoring functions and a divide-and-conquer docking approach. Nucleic Acids Res. 2012 Jul;40(Web Server issue):W393–9. doi: 10.1093/nar/gks496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo H, Chen J, Shi L, et al. DRAR-CPI: a server for identifying drug repositioning potential and adverse drug reactions via the chemical-protein interactome. Nucleic Acids Res. 2011 Jul;39(Web Server issue):W492–8. doi: 10.1093/nar/gkr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Gao Z, Kang L, et al. TarFisDock: a web server for identifying drug targets with docking approach. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W219–24. doi: 10.1093/nar/gkl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friesner Ra, Banks JL, Murphy RB, et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. Journal of Medicinal Chemistry. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 23.McGann M. FRED pose prediction and virtual screening accuracy. J Chem Inf Model. 2011 Mar 28;51(3):578–96. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- 24.Shulman-Peleg A, Nussinov R, Wolfson HJ. SiteEngines: recognition and comparison of binding sites and protein-protein interfaces. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W337–41. doi: 10.1093/nar/gki482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jambon M, Imberty A, Deleage G, et al. A new bioinformatic approach to detect common 3D sites in protein structures. Proteins. 2003 Aug 1;52(2):137–45. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- 26.Chartier M, Adriansen E, Najmanovich R. IsoMIF Finder: online detection of binding site molecular interaction field similarities. Bioinformatics. 2016 Feb 15;32(4):621–3. doi: 10.1093/bioinformatics/btv616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siragusa L, Cross S, Baroni M, Goracci L, et al. BioGPS: navigating biological space to predict polypharmacology, off-targeting, and selectivity. Proteins. 2015 Mar;83(3):517–32. doi: 10.1002/prot.24753. [DOI] [PubMed] [Google Scholar]
- 28.Yeturu K, Chandra N. PocketMatch: a new algorithm to compare binding sites in protein structures. BMC Bioinformatics. 2008;9:543. doi: 10.1186/1471-2105-9-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann B, Zaslavskiy M, Vert JP, et al. A new protein binding pocket similarity measure based on comparison of clouds of atoms in 3D: application to ligand prediction. BMC Bioinformatics. 2010;11:99. doi: 10.1186/1471-2105-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aung Z, Tong JC. BSAlign: a rapid graph-based algorithm for detecting ligand-binding sites in protein structures. Genome Inform. 2008;21:65–76. [PubMed] [Google Scholar]
- 31.Liu X, Ouyang S, Yu B, et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010 Jul;38(Web Server issue):W609–14. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokh DB, Richter S, Henrich S, et al. TRAPP: a tool for analysis of transient binding pockets in proteins. J Chem Inf Model. 2013 May 24;53(5):1235–52. doi: 10.1021/ci4000294. [DOI] [PubMed] [Google Scholar]
- 33.Dey F, Caflisch A. Fragment-based de novo ligand design by multiobjective evolutionary optimization. J Chem Inf Model. 2008 Mar;48(3):679–90. doi: 10.1021/ci700424b. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Zhao Z, Liu Z, et al. AutoT&T v.2: An Efficient and Versatile Tool for Lead Structure Generation and Optimization. J Chem Inf Model. 2016 Feb 22;56(2):435–53. doi: 10.1021/acs.jcim.5b00691. [DOI] [PubMed] [Google Scholar]
- 35.Maass P, Schulz-Gasch T, Stahl, et al. Recore: a fast and versatile method for scaffold hopping based on small molecule crystal structure conformations. J Chem Inf Model. 2007 Mar-Apr;47(2):390–9. doi: 10.1021/ci060094h. [DOI] [PubMed] [Google Scholar]
- 36.Durrant JD, Lindert S, McCammon JA. AutoGrow 3.0: an improved algorithm for chemically tractable, semi-automated protein inhibitor design. J Mol Graph Model. 2013 Jul;44:104–12. doi: 10.1016/j.jmgm.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37**.Keiser MJ, Roth BL, Armbruster BN, et al. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007 Feb;25(2):197–206. doi: 10.1038/nbt1284. Description of most widely used Similarity Ensemble Approach (SEA) method that relate proteins based on their ligand similarity. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Gao Y, Peng J, et al. TarPred: a web application for predicting therapeutic and side effect targets of chemical compounds. Bioinformatics. 2015 Jun 15;31(12):2049–51. doi: 10.1093/bioinformatics/btv099. [DOI] [PubMed] [Google Scholar]
- 39.Nickel J, Gohlke BO, Erehman J, et al. SuperPred: update on drug classification and target prediction. Nucleic Acids Res. 2014 Jul;42(Web Server issue):W26–31. doi: 10.1093/nar/gku477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gfeller D, Grosdidier A, Wirth M, et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014 Jul;42(Web Server issue):W32–8. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006 Sep 29;313(5795):1929–35. doi: 10.1126/science.1132939. Very useful resource for studying connectivity between diseases, drug actions and genetic mutations. Article describes the database, and analysis on their web server with examples. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn M, Szklarczyk D, Pletscher-Frankild S, et al. STITCH 4: integration of protein-chemical interactions with user data. Nucleic Acids Res. 2014 Jan;42(Database issue):D401–7. doi: 10.1093/nar/gkt1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Duan Q, Flynn C, Niepel M, et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 2014 Jul;42(Web Server issue):W449–60. doi: 10.1093/nar/gku476. Description of LINCS Canvas Browser that can be used in systems biology and systems pharmacology studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013 Apr 2;6(269) doi: 10.1126/scisignal.2004088. pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012 May;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kellenberger E, Rodrigo J, Muller P, et al. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins: Structure, Function and Genetics. 2004;57:225–42. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- 47.Schomburg KT, Bietz S, Briem H, et al. Facing the challenges of structure-based target prediction by inverse virtual screening. J Chem Inf Model. 2014 Jun 23;54(6):1676–86. doi: 10.1021/ci500130e. [DOI] [PubMed] [Google Scholar]
- 48.Wang W, Zhou X, He W, et al. The interprotein scoring noises in glide docking scores. Proteins. 2012 Jan;80(1):169–83. doi: 10.1002/prot.23173. [DOI] [PubMed] [Google Scholar]
- 49.Yang L, Wang K, Chen J, et al. Exploring off-targets and off-systems for adverse drug reactions via chemical-protein interactome--clozapine-induced agranulocytosis as a case study. PLoS Comput Biol. 2011 Mar;7(3):e1002016. doi: 10.1371/journal.pcbi.1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yadav DK, Mudgal V, Agrawal J, et al. Molecular docking and ADME studies of natural compounds of Agarwood oil for topical anti-inflammatory activity. Curr Comput Aided Drug Des. 2013 Sep;9(3):360–70. doi: 10.2174/1573409911309030012. [DOI] [PubMed] [Google Scholar]
- 51.Zhao S, Iyengar R. Systems pharmacology: network analysis to identify multiscale mechanisms of drug action. Annu Rev Pharmacol Toxicol. 2012;52:505–21. doi: 10.1146/annurev-pharmtox-010611-134520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu X, Guo Q, Xiong W, et al. Therapeutic effect of astragaloside-IV on bradycardia is involved in up-regulating klotho expression. Life Sci. 2016 Jan 1;144:94–102. doi: 10.1016/j.lfs.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 53.Kumar SP, Parmar VR, Jasrai YT, et al. Prediction of protein targets of kinetin using in silico and in vitro methods: a case study on spinach seed germination mechanism. J Chem Biol. 2015 Jul;8(3):95–105. doi: 10.1007/s12154-015-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cross S, Baroni M, Carosati E, et al. FLAP: GRID molecular interaction fields in virtual screening. validation using the DUD data set. J Chem Inf Model. 2010 Aug 23;50(8):1442–50. doi: 10.1021/ci100221g. [DOI] [PubMed] [Google Scholar]
- 55.Chen Z, Wang X, Zhu W, et al. Acenaphtho[1,2-b]pyrrole-based selective fibroblast growth factor receptors 1 (FGFR1) inhibitors: design, synthesis, and biological activity. J Med Chem. 2011 Jun 9;54(11):3732–45. doi: 10.1021/jm200258t. [DOI] [PubMed] [Google Scholar]
- 56*.De Vivo M, Masetti M, Bottegoni G, et al. Role of Molecular Dynamics and Related Methods in Drug Discovery. J Med Chem. 2016 May 12;59(9):4035–61. doi: 10.1021/acs.jmedchem.5b01684. Recent comprehensive review of theory and applications of MD simulations in drug discovery. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Dong Z. Effect of Clustering Algorithm on Establishing Markov State Model for Molecular Dynamics Simulations. J Chem Inf Model. 2016 Jun 27;56(6):1205–15. doi: 10.1021/acs.jcim.6b00181. [DOI] [PubMed] [Google Scholar]
- 58.Shenkin PS, Mcdonald DQ. Cluster-Analysis of Molecular-Conformations. Journal of Computational Chemistry. 1994 Aug;15(8):899–916. [Google Scholar]
- 59.Bottegoni G, Cavalli A, Recanatini M. A comparative study on the application of hierarchical-agglomerative clustering approaches to organize outputs of reiterated docking runs. J Chem Inf Model. 2006 Mar-Apr;46(2):852–62. doi: 10.1021/ci050141q. [DOI] [PubMed] [Google Scholar]
- 60.Shukla D, Meng Y, Roux B, et al. Activation pathway of Src kinase reveals intermediate states as targets for drug design. Nat Commun. 2014 Mar 03;5:3397. doi: 10.1038/ncomms4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61**.Bottegoni G, Favia AD, Recanatini M, et al. The role of fragment-based and computational methods in polypharmacology. Drug Discovery Today. 2012:23–34. doi: 10.1016/j.drudis.2011.08.002. Provides indepth description of importance of fragment-based methods in drug design and possible appoaches for the application of polypharmacology. [DOI] [PubMed] [Google Scholar]
- 62.Hopkins AL, Mason JS, Overington JP. Can we rationally design promiscuous drugs? Curr Opin Struct Biol. 2006 Feb;16(1):127–36. doi: 10.1016/j.sbi.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 63.Achenbach J, Klingler FM, Blocher R, et al. Exploring the chemical space of multitarget ligands using aligned self-organizing maps. ACS Med Chem Lett. 2013 Dec 12;4(12):1169–72. doi: 10.1021/ml4002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 65.Zauhar RJ, Gianti E, Welsh WJ. Fragment-based Shape Signatures: a new tool for virtual screening and drug discovery. Journal of Computer-Aided Molecular Design. 2013;27(12):1009–36. doi: 10.1007/s10822-013-9698-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacob L, Vert JP. Protein-ligand interaction prediction: an improved chemogenomics approach. Bioinformatics. 2008 Oct 1;24(19):2149–56. doi: 10.1093/bioinformatics/btn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Laarhoven T, Nabuurs SB, Marchiori E. Gaussian interaction profile kernels for predicting drug-target interaction. Bioinformatics. 2011 Nov 1;27(21):3036–43. doi: 10.1093/bioinformatics/btr500. [DOI] [PubMed] [Google Scholar]
- 68.Xia Z, Wu LY, Zhou X, et al. Semi-supervised drug-protein interaction prediction from heterogeneous biological spaces. BMC Syst Biol. 2010;4( Suppl 2):S6. doi: 10.1186/1752-0509-4-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamanishi Y, Araki M, Gutteridge A, et al. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics. 2008 Jul 1;24(13):i232–40. doi: 10.1093/bioinformatics/btn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonen M. Predicting drug-target interactions from chemical and genomic kernels using Bayesian matrix factorization. Bioinformatics. 2012 Sep 15;28(18):2304–10. doi: 10.1093/bioinformatics/bts360. [DOI] [PubMed] [Google Scholar]
- 71.Bleakley K, Yamanishi Y. Supervised prediction of drug-target interactions using bipartite local models. Bioinformatics. 2009 Sep 15;25(18):2397–403. doi: 10.1093/bioinformatics/btp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding H, Takigawa I, Mamitsuka H, et al. Similarity-based machine learning methods for predicting drug-target interactions: a brief review. Brief Bioinform. 2014 Sep;15(5):734–47. doi: 10.1093/bib/bbt056. [DOI] [PubMed] [Google Scholar]
- 73.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015 Jan;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Sejnowski D. A Learning Algorithm for Boltzmann Machines. Cognitive Science. 1985;9(1):147–69. [Google Scholar]
- 75.Smolensky P. Information Processing in Dynamical Systems: Foundations of Harmony Theory. Parallel Distributed Processing: Exploration in Microstructure of Cognition. 1986;1:194–281. [Google Scholar]
- 76**.Gawehn E, Hiss JA, Schneider G. Deep Learning in Drug Discovery. Mol Inform. 2016 Jan;35(1):3–14. doi: 10.1002/minf.201501008. Excellent paper that describes simple deep neural netwrok, restricted boltzmann machine and convolutional neural network and application of deep learning in drug discovery. [DOI] [PubMed] [Google Scholar]
- 77.Lusci A, Pollastri G, Baldi P. Deep architectures and deep learning in chemoinformatics: the prediction of aqueous solubility for drug-like molecules. J Chem Inf Model. 2013 Jul 22;53(7):1563–75. doi: 10.1021/ci400187y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hamanaka M, Taneishi K, Iwata H, et al. CGBVS-DNN: Prediction of Compound-protein Interactions Based on Deep Learning. Mol Inform. 2016 Aug 12; doi: 10.1002/minf.201600045. [DOI] [PubMed] [Google Scholar]
- 79.Tian K, Shao M, Wang Y, et al. Boosting compound-protein interaction prediction by deep learning. Methods. 2016 Jul 1; doi: 10.1016/j.ymeth.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 80.Hopkins AL. Network pharmacology. Nat Biotechnol. 2007 Oct;25(10):1110–1. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 81.Vitali F, Mulas F, Marini P, et al. Network-based target ranking for polypharmacological therapies. J Biomed Inform. 2013 Oct;46(5):876–81. doi: 10.1016/j.jbi.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 82.Szklarczyk D, Santos A, von Mering CM, et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016 Jan 4;44(D1):D380–4. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santos A, Tsafou K, Stolte C, et al. Comprehensive comparison of large-scale tissue expression datasets. PeerJ. 2015;3:e1054. doi: 10.7717/peerj.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Petryszak R, Keays M, Tang YA, et al. Expression Atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016 Jan 4;44(D1):D746–52. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tian L, Zhang S. Mapping drug-target interaction networks. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:2336–9. doi: 10.1109/IEMBS.2009.5335053. [DOI] [PubMed] [Google Scholar]
- 86.Sirota M, Dudley JT, Kim J, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011 Aug 17;3(96):96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88**.Morphy R, Rankovic Z. Designed multiple ligands. An emerging drug discovery paradigm. J Med Chem. 2005 Oct 20;48(21):6523–43. doi: 10.1021/jm058225d. One of the earliest detailed perspective on strategied for designing multiple ligands/multi-targeted drugs and their application in various diseases and conditions such as depression, schizophrenia, Allergies, hypertension and metabolic diseases. [DOI] [PubMed] [Google Scholar]
- 89.Morphy R, Rankovic Z. The physicochemical challenges of designing multiple ligands. J Med Chem. 2006 Aug 10;49(16):4961–70. doi: 10.1021/jm0603015. [DOI] [PubMed] [Google Scholar]
- 90.Morphy R, Rankovic Z. Fragments, network biology and designing multiple ligands. Drug Discov Today. 2007 Feb;12(3–4):156–60. doi: 10.1016/j.drudis.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 91.Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011 Oct 4;108(40):16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wyce A, Degenhardt Y, Bai Y, et al. Inhibition of BET bromodomain proteins as a therapeutic approach in prostate cancer. Oncotarget. 2013 Dec;4(12):2419–29. doi: 10.18632/oncotarget.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93*.Besnard J, Ruda GF, Setola V, et al. Automated design of ligands to polypharmacological profiles. Nature. 2012 Dec 13;492(7428):215–20. doi: 10.1038/nature11691. Description of automated design of multi-targeted compounds with application. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dias KS, Viegas C., Jr Multi-Target Directed Drugs: A Modern Approach for Design of New Drugs for the treatment of Alzheimer’s Disease. Curr Neuropharmacol. 2014 May;12(3):239–55. doi: 10.2174/1570159X1203140511153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goyal M, Dhanjal JK, Goyal S, et al. Development of dual inhibitors against Alzheimer’s disease using fragment-based QSAR and molecular docking. Biomed Res Int. 2014;2014:979606. doi: 10.1155/2014/979606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Speck-Planche A, Kleandrova VV, Luan F, et al. Multi-target inhibitors for proteins associated with Alzheimer: in silico discovery using fragment-based descriptors. Curr Alzheimer Res. 2013 Feb;10(2):117–24. doi: 10.2174/1567205011310020001. [DOI] [PubMed] [Google Scholar]
- 97.Issa NT, Kruger J, Byers SW, Dakshanamurthy S. Drug repurposing a reality: from computers to the clinic. Expert Rev Clin Pharmacol. 2013 Mar;6(2):95–7. doi: 10.1586/ecp.12.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98*.Allison M. NCATS launches drug repurposing program. Nat Biotechnol. 2012 Jul;30(7):571–2. doi: 10.1038/nbt0712-571a. Description of NCATS drug repuposing program. [DOI] [PubMed] [Google Scholar]
- 99.Power A, Berger AC, Ginsburg GS. Genomics-enabled drug repositioning and repurposing: insights from an IOM Roundtable activity. JAMA. 2014 May;311(20):2063–4. doi: 10.1001/jama.2014.3002. [DOI] [PubMed] [Google Scholar]
- 100.Wren JD, Bekeredjian R, Stewart JA, et al. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004 Feb 12;20(3):389–98. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]