Summary
In Sub-Saharan Africa most individuals are homozygous for the DARC promoter mutation, -46T>C, abrogating DARC expression on erythrocytes. It was reported that -46C/C is associated with 40% increase in HIV-1 acquisition in a USA population and may account for 11% of HIV-1 burden in Africa. We determined -46T>C genotypes for HIV-1 infected and uninfected African Americans. We found no evidence that the DARC negative phenotype caused by -46C/C affects HIV infection or rates of progression to AIDS in people of African ancestry. The earlier result is likely due to population substructure that led to inflated associations with DARC -46C/C.
Editors
Recently He et al. (He et al., 2008) reported that persons of Sub-Saharan African ancestry, the majority of whom do not express DARC on their red blood cells (RBC), have a genetic vulnerability to HIV infection. They estimated that the DARC negative phenotype conferred by the -46CC genotype increases the odds of HIV infection by 40% and extrapolated that as much as 11% of the HIV burden in Africa may be due to -46C homozygosity. He et al. also reported that the DARC negative genotype was associated with delayed progression to AIDS-associated dementia, delayed CD4+ T cell loss and longer survival. DARC expression on RBC is abolished by the -46T→C transition in the DARC promoter region that disrupts the erythrocyte-specific transcription binding site (Tournamille et al., 1995). The -46T allele is fixed outside of Africa while the -46C allele occurs only in Africa and in persons with African ancestry. It is hypothesized that the -46C allele arose to near-fixation in Africa through positive selection by Plasmodium vivax—DARC negative individuals lacking the Duffy receptor used for cell entry by P. vivax for RBC entry are highly resistant to P. vivax malaria (Horuk et al., 1993). DARC is a plausible candidate gene for HIV infection and pathogenesis—it binds proinflammatory chemokines, one of which, CCL5 (RANTES), also binds to CCR5, required by R5 HIV strains for cell entry (Alkhatib et al., 1996; Gardner et al., 2004). CCL5 is a HIV-1 suppression factor that inhibits replication by both competitively binding to and down-regulating surface expression of CCR5(Alkhatib et al., 1997). It has also reported that DARC binds HIV to the surface of red blood cells (Lachgar et al., 1998; He et al., 2008). The DARC negative phenotype is coincident with the world’s highest HIV prevalence rates; therefore, the reported influence of DARC -46T→C polymorphism on the HIV epidemic warrants replication and confirmation.
We determined the extent of population substructure for 454 HIV infected and 425 HIV uninfected persons enrolled in the ALIVE cohort in Baltimore, Maryland, all of whom self-reported to be injecting drug users (IDU)(Vlahov et al., 1998a). Population substructure is a major confounding factor since DARC -46C is itself a marker for African ancestry. A principle components analysis (PCA) using the EIGENSOFT program with 70 independent, ancestry-informative markers (AIMs) revealed that there is minimal population substructure stratification between the ALIVE HIV+ and HIV− groups (p=0.2) (Supplemental Fig 1; Supplemental Table 1) (Kopp et al., 2008; Price et al., 2006). The first principal component (the component of the first eigenvector in the principle component transformation) captured the majority of the variation, and more relevantly, captured the variation differentiating African from European ancestry (the eigenvalues of the first 5 components were 206, 16, 15, 15 and 14, respectively).
Genotypes were determined for DARC -46T→C using theTaqMan Assays on Demand for rs2814778 (ABI, Foster City, CA). The genotype frequencies for -46C/C, T/C, and C/C did not violate Hardy-Weinberg expectations in either the HIV+ or HIV− groups. Most importantly, the frequencies of the -46C/C genotypes in the HIV+ (66.5%) and HIV− (66.4%) groups were nearly identical indicating no association between -46C/C and HIV acquisition, either in the unadjusted model (OR=0.96, p=0.896) or with the first principal component for each individual included as a covariate to control for population stratification (OR=1.00, p=0.98). The He et al group reported an OR≈1.5; given our population size of 879 individuals and an allele frequency of 66%, we had 75% power to detect an association of this strength at the 0.05 level. On the other hand, assuming an actual OR value of 1.5 as reported by He et al., the probability of observing OR <= 1 in a sample of our size is 0.01 (calculated assuming a normal approximation of the log odds ratio) indicating that our results differ significantly from the those reported by He et al. (2008).
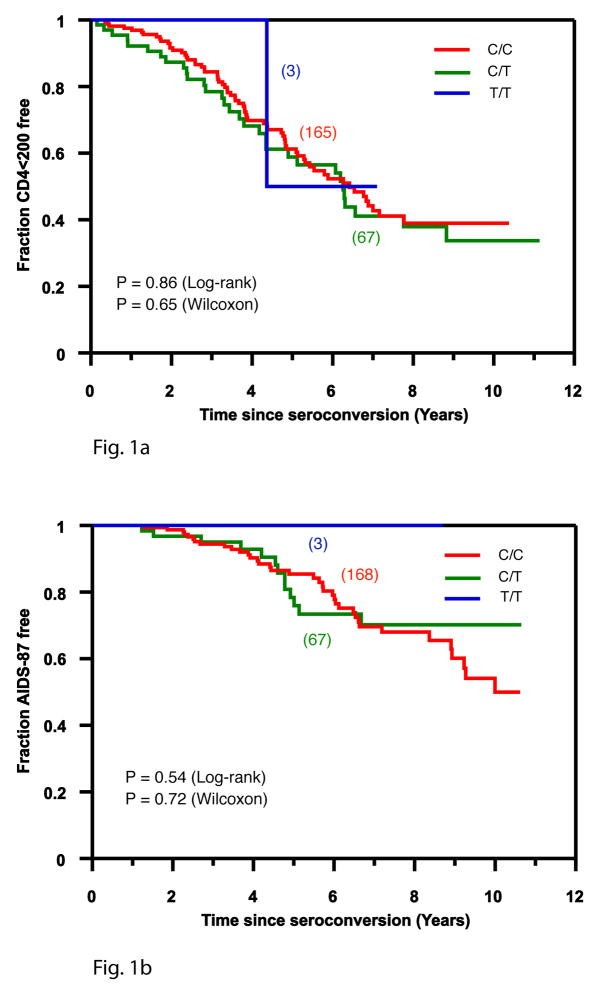
We also tested the role of DARC -46C/C on two HIV outcomes, CD4+ T cell <200 and AIDS-defining conditions (CDC 1987 case definition) using Kaplan-Meier statistics (Figure 1) and the Cox proportional hazards model (Table 1). To avoid potential confounding by anti-retroviral therapy, the censoring date was the earliest of the date of the last recorded visit, or July 31, 1997 (Celentano et al., 2001). We did not attempt to replicate the He et al. finding of favorable outcomes for AIDS-associated dementia or death because reliable data was not available to us for the former and we had too few events to evaluate for AIDS-related death. As shown in Table 1 and Figure 1, we did not observe significant associations with time to CD4<200 or AIDS 1987 using a well-characterized group of seroconverters. In the simple Cox model, stratified by age at seroconversion, the RH was 0.85 and 1.24 for CD4<200 and AIDS 1987 (p≥0.47). The results for CD4<200 and AIDS 1987 were similar after adjusting for the effects of HLA, CCR5, and RANTES genetic factors (RH=0.84 (95% CI: 0.53–1.31), P=0.43, RH=1.03 (95% CI: 0.53–2.0) or after including the eigenvalue scores for the first principal component as a covariate to control for population stratification (RH=0.99, 1.27, p≥0.48 respectively). Inspection of the Kaplan-Meier survival curves and Kaplan-Meier statistics (Fig. 1) suggests no differences in rates of progression to CD4<200 or to AIDS 1987 due to -46 T→C genotypes. These data do not support a role for DARC -46 T→C in HIV progression to clinical AIDS.
Figure 1.
Kaplan Meier plots showing progression to two AIDS outcomes: < 200 CD4 T cells/μl and clinical AIDS symptoms (AIDS-87) for subjects from the ALIVE cohort, for three DARC -46 genotypes: C/C (DARC null phenotype), red; C/T, green; and T/T, blue. Kaplan-Meier statistics for the log-rank and Wilcoxon test, for a recessive model (DARC -46 CC vs. CT and TT) are shown.
Table 1.
Impact of DARC -46CC conferring the DARC negative phenotype on HIV-1 infection and disease progression.
| DARC -46 CC versus CT and TT | ||||||
|---|---|---|---|---|---|---|
| Unadjusted1 |
Adjusted for Ancestry2 |
|||||
| HIV+ N (%) | HIV− N (%) | OR, RH (95% CI) | P | OR, RH (95% CI) | P | |
| HIV-1 Infection | ||||||
|
| ||||||
| Genotype | n = 454 | n = 425 | ||||
| C/C | 302 (66.5) | 282 (66.4) | 0.96 (0.75–1.31) | 0.896 | 1.00 (0.69–1.47) | 0.98 |
| T/C | 140 (30.8) | 131 (30.8) | ||||
| T/T | 12 (2.6) | 12 (2.8) | ||||
| HWE P3 | 0.37 | 0.49 | ||||
|
| ||||||
| AIDS Progression4 | ||||||
|
| ||||||
| Endpoint | N HIV+ SC)5 | Events | RH (95% CI) | P | RH (95% CI) | P |
|
|
|
|||||
| CD4 200 | 235 | 99 | 0.85 (0.56–1.31) | 0.47 | 0.99 (0.63–1.57) | 0.97 |
| AIDS 87 | 238 | 49 | 1.24 (0.66–2.35) | 0.51 | 1.27 (0.66–2.46 | 0.48 |
Odds ratio (OR) and 95% Confidence interval (CI) were obtained from a conditional logistic regression analysis comparing HIV-1 infected (+) to HIV-1 uninfected (−) individuals;
Adjusted for African ancestry considering the first eigenvector as a covariate in the analysis;
Hardy-Weinberg Equilibrium test for observed versus expected genotypes;
Relative hazards (RH) and 95% confidence interval (CI) were from Cox proportional hazards model. All models were stratified by age group at seroconversion;
HIV-1 seroconverters (SC).
The results of the He et al. (2008) study and this one differ markedly for association of HIV infection susceptibility with the DARC negative phenotype conferred by -46C/C. The ALIVE cohort shows very little population stratification by the results of the PCA. Our finding challenges the conclusion of the He et al. (2008) that DARC -46C/C may contribute to the HIV burden observed in Africa. We suggest that the infection signal in the He et al. (2008) study is the result of inadequate correction for population stratification due to substructure between the cases and controls that lead to the false positive association of the DARC -46C/C genotype with HIV infection. The He et al. (2008) used 11 informative markers to determine the probability of either African or European ancestry and considered this probability as a covariate to correct for stratification due to ancestry substructure between cases and controls; however, it is the degree of African or European ancestry that is relevant—particularly since the DARC -46 T→C polymorphism is among the most informative markers of African versus non-African ancestry. In summary, we find no evidence that the absence of DARC on red blood cells due abrogation of gene expression by the -46C/C genotype is associated with greater risk for HIV infection or delayed progression to AIDS in African Americans IVD either in the unadjusted analysis or after adjusting for ancestry using the results of the PCA.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contracts N01-CO-12400. Additional funding was received from NIH contract N02-CP-55504. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Alkhatib G, Locati M, Kennedy PE, Murphy PM, Berger EA. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- Celentano DD, Galai N, Sethi AK, Shah NG, Strathdee SA, Vlahov D, Gallant JE. Time to initiating highly active antiretroviral therapy among HIV-infected injection drug users. Aids. 2001;15:1707–1715. doi: 10.1097/00002030-200109070-00015. [DOI] [PubMed] [Google Scholar]
- Gardner L, Patterson AM, Ashton BA, Stone MA, Middleton J. The human Duffy antigen binds selected inflammatory but not homeostatic chemokines. Biochem Biophys Res Commun. 2004;321:306–312. doi: 10.1016/j.bbrc.2004.06.146. [DOI] [PubMed] [Google Scholar]
- He W, Neil S, Kulkarni H, Wright E, Agan BK, Marconi VC, Dolan MJ, Weiss RA, Ahuja SK. Duffy antigen receptor for chemokines mediates trans-infection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility. Cell Host Microbe. 2008;4:52–62. doi: 10.1016/j.chom.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horuk R, Chitnis CE, Darbonne WC, Colby TJ, Rybicki A, Hadley TJ, Miller LH. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261:1182–1184. doi: 10.1126/science.7689250. [DOI] [PubMed] [Google Scholar]
- Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachgar A, Jaureguiberry G, Le Buenac H, Bizzini B, Zagury JF, Rappaport J, Zagury D. Binding of HIV-1 to RBCs involves the Duffy antigen receptors for chemokines (DARC) Biomed Pharmacother. 1998;52:436–439. doi: 10.1016/s0753-3322(99)80021-3. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.