Abstract
Epidemiologic and genomic studies have progressively improved our understanding of the causation of hypertension and the complex relationship with diet and environment. The majority of Mendelian forms of syndromic hypotension and hypertension (HTN) have all been linked to mutations in genes whose encoded proteins regulate salt–water balance in the kidney, supporting the primacy of the kidneys in blood pressure regulation. There are more than 1,477 single nucleotide polymorphisms associated with blood pressure and hypertension and the challenge is establishing a causal role for these variants. Hypertension is a complex multifactorial phenotype and it is likely to be influenced by multiple factors including interactions between diet and lifestyle factors, microbiome, and epigenetics. Given the finite genetic variability that is possible in humans, it is likely that incremental gains from single marker analyses have now plateaued and a greater leap in our understanding of the genetic basis of disease will come from integration of other omics and the interacting environmental factors. In this review, we focus on emerging results from the microbiome and metabolomics and discuss how leveraging these findings may facilitate a deeper understanding of the interrelationships between genomics, diet, and microbial ecology in humans in the causation of essential hypertension.
Keywords: blood pressure, diet, genomics, hypertension, metabolomics, metagenomics, microbiome, salt
The major risk factor underpinning cardiovascular (CV) diseases is hypertension which directly accounts for up to 10.5 million of the 18 million CV deaths that occur annually or 12% of total global deaths.1 By 2025, hypertension is projected to affect more than 1.5 billion people globally and modeling indicates that effective control of hypertension through improving treatment rates and lifestyle measures could save more lives than any other clinical intervention.2 Epidemiologic and genomic studies have progressively improved our understanding of the causation of hypertension and the complex relationship with diet and environment. Family studies have consistently demonstrated a genetic component influencing blood pressure (BP) and hypertension (HTN). The Montreal adoption study3 demonstrated correlation coefficients of 0.38 and 0.16 between biological and adoptive sibs, respectively, indicating that 61% of the population correlation for systolic BP was due to shared genes and 39% to environment shared by both parents and children. The heritability of clinic systolic BP and diastolic BP is around 15–40% and 15–30%, respectively, whereas for ambulatory night-time systolic and diastolic BP the heritabilities are, respectively, 32–70% and 32–50%.4,5
The identification of rare mutations in genes causing monogenic forms of HTN come from linkage analysis of pedigrees exhibiting a Mendelian pattern of inheritance of the BP phenotype. The study of monogenic syndromes has expanded our understanding of some of the pathways that regulate blood pressure. Table 1 summarizes the different forms of monogenic hypertension, their key features, and causal genes and they are detailed in recent authoritative reviews.6–8 The majority of Mendelian forms of syndromic hypotension and HTN have all been linked to mutations in genes whose encoded proteins regulate salt–water balance in the kidney, supporting the primacy of the kidneys in BP regulation.8 While these monogenic mutations are germline mutations which are inherited, there is now recognition of somatic mutations that cause aldosteronism and hypertension. A gain-of-function somatic mutation in a K+ channel, KCNJ5, which results in membrane depolarization and enhanced aldosterone production, is a common genetic defect noted for ~40% of aldosterone-producing adenomas (APAs).6 Mutations in three other genes, encoding the α-subunit of Na+-K+-ATPase (ATP1A1); ATP2B3, a plasma membrane Ca2+-ATPase homologous to the sarcoplasmic endoplasmic reticulum Ca2+-ATPases (SERCA); and CACNA1D, encoding an L-type Ca2+ channel CaV1.3, are observed in ~7% of the cases.6
Table 1.
Monogenic forms of hypertension and hypotension
| Syndrome | Gene | BP | Renin | Aldosterone | Serum K+ | Catecholamines | Treatment |
|---|---|---|---|---|---|---|---|
| Liddle syndrome MIM 177200 | SCNN1B - SCNN1G | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Amiloride or triamterene |
| Gitelman syndrome MIM 263800 | SLC12A3 | ↓↓ | ↑↑ | - | ↓↓ | — | Oral potassium and magnesium supplementation with adequate salt and water |
| Bartter syndrome MIM 601678, 241200, 602522, 613090, 300971 | SLC12A1 KCNJ1 CLCNKB BSND CLCNKA CLCNKB MAGED2, MAGED, BARTS5 | ↓↓ | ↑↑ | ↑↑ | ↓↓ | — | Potassium supplementation and use of cyclooxygenase inhibitors, angiotensin converting enzyme (ACE)- inhibitors and potassium sparing diuretics |
| Familial hyperaldosteronism (FH) MIM 103900, 605635, 613677 “glucocorticoid remediable aldosteronism” | CYP11B1 KCNJ5 | ↑↑ | ↓↓ | ↑↑ | ↓ | — | Dexamethasone |
| Apparent mineralocorticoid excess (AME) MIM 218030 | HSD11B2 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Low sodium diet and spironolactone |
| Pseudohypoaldosteronism (PHA) MIM 177735, 614491, 614492, 614495, 614496 “Gordon syndrome” | NR3C2 WNK4 WNK1 KLHL3 CUL3 | ↓↓ | ↑↑ | ↑↑ | ↑↑ | — | Thiazide diuretics, prostaglandin inhibitors, alkalizing agents, and potassium-binding resins |
| ↑↑ | ↓↓ | ↑ | ↑ | — | |||
| Sporadic aldosterone- producing adenoma (APA), or primary aldosteronism | KCNJ5 ATP1A1 CACNA1D ATP2B3 | ↑↑ | ↓↓ | ↑↑ | ↓ | — | Surgery, aldosterone antagonists |
| Hypertension exacerbation in pregnancy MIM 605115 | NR3C2 | ↑↑ | ↓↓ | ↓↓ | ↓ | — | Spironolactone contraindicated; sodium chloride treatment |
| 11β-hydroxylase MIM 202010 | CYP11B1 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Glucocorticoid therapy |
| 3β-hydroxysteroid dehydrogenase OMIM 613890 | HSD3B2 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Glucocorticoid therapy |
| 17α-hydroxylase deficiency MIM 202110 | CYP17A1 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Glucocorticoid therapy, potassium sparing diuretics |
| 21-Hydroxylase deficiency MIM 201910 | CYP21A2 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Glucocorticoid therapy |
| Hypertension and brachydactyly syndrome MIM 112410 “Bilginturan syndrome” | PDE3A | ↑↑ | — | — | — | — | Possible role for PDE3 inhibition |
| Paragangliomas (PGL1-5) MIM 168000, 601650, 605373, 115310, 614165 | SDHD SDHAF2 SDHC SDHB SDHA | ↑↑ | — | — | — | ↑↑ | Surgery, adrenergic blockers (alpha blockade followed by beta-blockade) |
| von Hippel–Lindau syndrome MIM 193300 | VHL | ↑↑ | — | — | — | ↑↑ | |
| Multiple endocrine neoplasia, type IIA MIM 171400 | RET | ↑↑ | — | — | — | ↑↑ | |
| NOS3-pregnancy-induced hypertension MIM +163729 | NOS3 | ↑↑ | — | — | — | ↑↑ |
The era of genome-wide association studies (GWAS) commenced in 2007, resulting in 4,346 publications and 166,103 associations for a wide range of polygenic traits including hypertension and BP (https://www.ebi.ac.uk/gwas/home). The hypothesis underlying GWAS is that common variations (single nucleotide polymorphisms (SNPs)) can have significant impact on common traits and thence human health (common disease/common variant hypothesis). There are more than 1,477 SNPs associated with blood pressure, but these SNPs account for about a third of the estimated 30–50% heritability of blood pressure and explain just about 5.7% of the population phenotypic variance of SBP.8,9 Multiple reasons have been invoked to explain the missing heritability of blood pressure including gene–gene interactions, gene–environment interactions, diet and lifestyle factors, microbiome, and epigenetics. Furthermore, establishing a causal role for common SNPs from GWAS in essential hypertension has been challenging. Hypertension is a complex multifactorial phenotype and it is likely to be influenced by multiple factors including interactions between these factors. Genome-wide association studies which only look at single marker associations are blind to this complex aspect of hypertension pathogenesis. Given the finite genetic variability that is possible in humans, it is likely that incremental gains from single marker analyses have plateaued and a greater leap in our understanding of the genetic basis of disease will come from integration of other omics and the interacting environmental factors.10
In this review, we focus on emerging results from the microbiome and metabolomics and discuss how leveraging these findings may facilitate a deeper understanding of the interrelationships between genomics, diet and microbial ecology in humans in the causation of essential hypertension and identify new interventions to manage hypertension.
INSIGHTS FROM GENOMICS AND DIET ON HYPERTENSION
There is consistent epidemiologic evidence between dietary sodium intake and blood pressure.11,12 The majority of the Mendelian forms of hypertension and hypotension exert their effect by perturbing renal sodium handling pathways (Table 1).8 A classic example is Gitelman syndrome (GS), which is a salt-losing tubulopathy characterized by hypokalemic metabolic alkalosis with hypomagnesemia and hypocalciuria.13 With a prevalence at 1–10 per 40,000, and potentially higher in Asia, GS is arguably the most frequent inherited tubulopathy. GS is caused by biallelic inactivating mutations in the SLC12A3 gene encoding the thiazide-sensitive sodium chloride cotransporter (NCC), is usually detected in adolescence or adulthood and exhibits high phenotypic variability.13 Affected individuals have low blood pressure and may present with salt craving (i.e., preference for salty food or a salted treat during childhood) or in many cases diagnosed incidentally in asymptomatic adults. The first GWAS of gene–salt interaction was conducted in 1,876 Chinese participants of the Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study with dietary salt intake imputed from overnight urine collections.14 Single SNP and gene-based analyses identified signals in the following genes—UST, CLGN, MKNK1, C2orf80, EPHA6, SCOC-AS1, SCOC, MGAT4D, ARHGAP42, CASP4, and LINC01478 for gene–sodium intake interaction. A cross-sectional study of 2,728 male Japanese adults where dietary salt consumption was estimated using electronically collected meal purchase data from cafeteria showed a nominally significant association between salt consumption and the rs5063 SNP in the NPPA gene.15
The most compelling example of gene–salt interaction on blood pressure comes from a GWAS study that identified a 5′-promoter SNP, rs13333226 near the Uromodulin gene (UMOD) which is almost exclusively expressed in the thick ascending limb of the loop of Henle in the kidney.16 The minor G allele of this SNP, rs13333226, was associated with a lower risk of hypertension and reduced urinary UMOD excretion. The main sodium transporter in thick ascending limb is NKCC2 which is blocked by the commonly used loop-diuretic furosemide. Trudu et al.17 showed furosemide treatment significantly enhanced natriuresis and reduced BP levels both in the transgenic mice and in the hypertensive individuals homozygous for the UMOD increasing allele. This has identified a novel pathway of blood pressure and renal function regulation through possible interaction with the sodium transporter NKCC2 in the thick ascending limb of the loop of Henle. This is now the basis of a clinical trial (www.clinicaltrials.gov, NCT03354897) to reposition a loop diuretic in the hypertension care pathway.
There are only a few studies that looked at diet–gene interactions. A study of 723 obese adults showed the risk allele (rs16147-C) in neuropeptide Y (NPY) gene was associated with a greater reduction of BP phenotypes in response to low-fat diet, whereas an opposite genetic effect was observed in response to high-fat diet.18
THE MICROBIOME AND HYPERTENSION
The human body is colonized by hundreds of trillions of microbes, which collectively possess hundreds of times as many genes as coded in the human genome and is collectively referred to as the “microbiome.” 19 The microbiota is involved in energy harvest and storage, as well as in a variety of metabolic functions such as fermenting and absorbing undigested carbohydrates and interaction with the immune system, providing signals for the normal development of immune functions. Advances in high-throughput sequencing technology has allowed for the identification of human-associated microorganisms without the need for culturing.20 Under physiological conditions, there is a balance between the intestinal bacteria and the host. Disruption of this intricate system (dysbiosis) has been implicated in many human diseases, including cardiometabolic diseases and hypertension.21–28 The gut microbiota can produce a range of bioactive metabolites, such as enzymes, peptides, antibiotics, amino acids, hormones, and vitamins that can mediate host receptor activation, signaling, and immunomodulatory effects and several of these metabolites have been linked with CV diseases and drug response.26,29–31 There is compelling evidence that the gut microbiome is modified by diet and this has implications on both health and precision medicine strategies of this is of relevance in hypertension.32,33
Metabolomics studies have so far identified numerous circulating metabolites (Supplementary Table) associated with BP and HTN in cohorts of different ethnicities, gender, and age range. The metabolites identified fall into these broad classes: 39% lipids, 27% amino acids, 19% xenobiotic, 7% carbohydrates, 2% energy, 2% nucleotide, 2% peptide, and 2% cofactors and vitamins26,27,34–44. These include 24 metabolites produced by the gut microbiome and released into the blood stream. We now describe the most promising metabolites.
HEXADECANEDIOATE
The dicarboxylic acid hexadecanedioate was significantly associated both with BP and mortality indicating a sustained detrimental effect of higher levels of the metabolite through increased BP.26 Evidence for a causal role was obtained by feeding this compound to rats resulting in significant increases in BP, indicating that it is not a by-product, but a cause of high blood pressure. Higher hexadecanedioate are associated with stroke in the Women’s Health Initiative cohort45 and the Atherosclerosis Risk In Communities study.46 GWAS showed hexadecanedioate levels to be associated with genetic variants in ADH1B and SLC01B1,47,48In vitro studies confirmed hexadecanedioate was a novel substrate of OATP1B1 (encoded by SLCO1B1) as well as OAT1 and OAT3 indicating that is an endogenous biomarker of OATP1B1 function.49 A single copy of a loss-of-function variant in SLC01B1 increased serum hexadecanedioate levels and resulted in a 29% increased risk of heart failure incidence in an African American cohort.50 A potential pharmacogenetic application of hexadecanedioate is suggested by correlations observed between BP reduction after amlodipine administration with reductions in plasma hexadecanedioate levels.51 Hexadecanedioic acid is a long chain dicarboxylic acid which is generated during fatty acid ω-oxidation and thence metabolized by β-oxidation in peroxisomes. ω-oxidation is a minor metabolic pathway that occurs in the smooth endoplasmic reticulum and also contributes to 5–10% of total fatty acids metabolism in the liver. Human ω-hydroxylases are all members of the cytochrome P450 family specifically CYP4A and CYP4F. Following addition of the ω-hydroxyl group, the fatty acid becomes a substrate for alcohol dehydrogenase (ADH) which generates an oxo-fatty acid, followed by generation of the corresponding dicarboxylic acid via the action of aldehyde dehydrogenases (ALDH). Furthermore, gene expression of ADH1A, ADH1B and CYP4 involved in ω-oxidation pathways were strongly correlated to hexadecanedioate levels indicating a endogenous biomarker for alcohol’s effect on BP.50,52,53 Tang et al.54 recently associated hexadecanedioate with Lachnospira, a specific microbial taxon, and linked the metabolite with vitamin E, folate, lutein, zeaxanthin, cheese, and tomato intakes.
SHORT-CHAIN FATTY ACIDS
Short-chain fatty acids (SCFA) are microbially mediated metabolic by-product of dietary fiber fermentation in the colon subsequently absorbed into the bloodstream of the host.55 SCFAs can bind to and activate host receptors (Gpr41, Gpr43, Gpr109a, and Olfr78), thereby acting as a route of “communication” between gut microbial metabolism and host physiology. The most common SCFA are acetate, propionate, and butyrate, which account for ~80% of all SCFAs and are of significant biological interest, as they have very tangible roles in modulating direct and indirect BP pathways.56 Studies in animal models with interventions that manipulated levels of SCFAs showed that an increase in SCFA was associated with lower BP.24,57 Decreases in bacterial taxa thought to produce short chain fatty acids have been reported in two different rat models of hypertension to increase BP,28 while another study found that rats which received a microbial transplant which increased blood pressure had higher levels of plasma acetate.58 Chronic acetate intake in animal models was shown to reduce both SBP and DBP by 21 mm Hg, and improved cardiac function in line with the mitigation of inflammatory disorders.24 Propionate has been shown to increase BP in animal models by stimulating renin secretion through Olfr78 and Gpr41.57 When wild-type and Olfr78 KO mice were treated with a mixture of antibiotics with blood pressure measurements, wild-type mice had a mild increase in blood pressure on antibiotics, Olfr78 KO mice had a much more dramatic increase indicating Olfr78 and Gpr41 normally act in physiologically opposite roles and balance out wide swings in BP.57 Additionally, there is evidence for GPR109A and GPR43 activation by SCFAs in controlling inflammation and promoting epithelial repair in the colon with implications for BP regulation through inflammatory pathways.59 Thus, the role of SCFAs in blood pressure regulation is multi-faceted, involves at least two different SCFA receptors, multiple species of bacteria, and multiple host tissues.60 There is data showing variants in the host genome can influence the composition of the gut microbiota61 and causal role for SCFA in metabolic disease was evaluated using Mendelian randomization that showed host genetic-driven increase in gut production of the SCFA butyrate is associated with improved insulin response following an oral glucose test.62 This opens possibilities for studies to assess the causal relationship between SCFA and blood pressure in the future.
TRIMETHYLAMINE N-OXIDE
Similar to SCFAs, trimethylamine N-oxide (TMAO) is generated by gut microbes from choline, betaine, and carnitine through dietary phosphatidylcholine oxidation. The plasma level of TMAO is determined by several factors including diet, gut microbial flora, drug administration and liver flavin monooxygenase activity.63 There is accruing evidence that TMAO is associated with inflammation and atherosclerosis, though it is unclear whether it is proatherogenic or just a biomarker of increased cardiovascular disease.64,67 In Sprague-Dawley rats, infusion of TMAO did not affect BP in normotensive animals, but it prolonged the hypertensive effect of Ang II.68 Genomic studies indicate that TMAO levels are not determined by genetic variation, rather they reflect the influence diet and gut microbiota.69
4-HYDROXYHIPPURATE
A microbial metabolite of benzoate, 4-hydroxyhippurate, is the most widely detected urinary metabolite of host-microbial origin in humans and its urinary concentrations are modulated by diet, stress, disease, and microbial presence or activity.34 Zheng et al.35 showed 4-hydroxyhippurate was associated with incident hypertension in a small study of 896 black normotensive subjects.
INDOLES
Indoles are products of gut bacterial metabolism of tryptophan and have been shown to impact homeostatic processes including inflammatory pathways,70 gut barrier permeability,71 and impact arterial blood pressure pathways via the inhibition of serotonin receptor blockers.72
PHENYLACETYLGLUTAMINE
Phenylacetylglutamine is involved in amino acid metabolism and has been associated with CV disorders including diastolic blood pressure, incidence of heart failure,73 carotid femoral pulse wave velocity,74 and overall mortality in chronic kidney disease patients.75 However, findings appear to be dependent on kidney status and glomerular filtration rate.
Gut microbiota and blood pressure
Studies in animal models of HTN have been instrumental in understanding the role of gut microbiota and metabolites and its association with hypertension (Figure 1).24,76 High salt intake affects the gut microbiome in mice, particularly by depleting Lactobacillus murinus and treatment of mice with L. murinus prevented salt-sensitive hypertension by modulating TH17 cells.77 Rat models of HTN showed a reduction in the proportion of Bacteroidetes and an increase in Firmicutes in the gut with a corresponding shift of the metabolic profile.28 In humans, a greater percentage of Prevotella and reduced abundance of Bacteroidetes were observed in patients with HTN and prehypertension compared with healthy controls along with a different metabolic profile.27 In particular, the metabolites 3,4,5-trimethoxycinnamic and S-carboxymethyl-l-cysteine were lower in prehypertension and HTN compared with controls. 3,4,5-Trimethoxycinnamic acid suppresses cell adhesion molecules in vascular endothelium protecting against dysregulated inflammatory disorders. S-carboxymethyl-l-cysteine, on the other hand, has been shown to mitigate inflammatory damage.27,78 In the Coronary Artery Risk Development in Young Adults (CARDIA) study, gut microbial diversity was inversely associated with both hypertension and systolic BP.79 Finally, antibiotic therapy in the Dahl salt-sensitive (S) rat and the spontaneously hypertensive rat (SHR) appeared to increase systolic BP in the former but decrease in the latter accompanied by significant alterations in gut microbiota.80 This highlights potential interactions between host genome and interventions that modify the gut microbiome and blood pressure. These results illustrate the interconnection between potential hypertensive pathways and microbial metabolites that merit further validation and elucidation of underpinning mechanisms.
Figure 1.
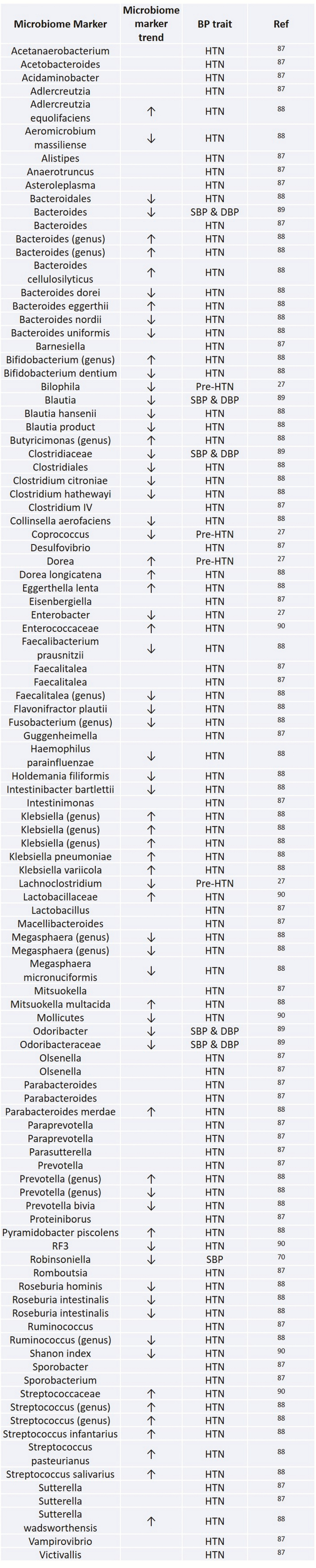
Gut microbiome and blood pressure traits.
Dietary interventions to modify the microbiome and blood pressure
Gut microbiota composition and function is shaped from infancy and persists into adulthood, but it retains some degree of flexibility that allows modulation through exposure to a variety of environmental factors, the most important of which is diet.81,82
The Mediterranean diet is understood to exert a cardioprotective effect, including mitigating blood pressure. This beneficial effect may be partially dependent on changes to the gut microbiome, two recent studies suggest that adhering to a Mediterranean diet is associated with a more favorable gut microbiome composition and increased SCFA generation potential (increased Faecalibacterium prausnitzii and Clostridium cluster XIVa).83
The OmniHeart study was a randomized crossover study of three dietary patterns—OmniCarb diet (58% kcal from carbohydrates, 15% from protein, and 27% from fat), OmniProt (10% of the kcal from carbohydrates replaced by mainly vegetable sourced protein), or OmniMFA (10% of the kcal from carbohydrates replaced by unsaturated fat).36 An inverse association was observed for proline–betaine with SBP and DBP in OmniCarb and OmniMFA diets. Carnitine was directly associated with SBP in OmniProt, hippurate with both SBP and DBP in OmniCarb. Comparisons between OmniProt and OmniMFA diets identified inverse associations for BP with 4-cresyl sulfate and with phenylacetylglutamine, metabolites of tyrosine, and phenylalanine. A metabolite of tryptophan-NAD, N-methyl-2-pyridone-5-carboxamide was also inversely associated in the OmniCarb diet compared with baseline.36
Derkach et al.84 randomly assigned 119 participants a 12-week DASH diet or a 12-week typical American diet (control). Each participant was randomly assigned high-, medium-, or low-versions of their respective diets, crossing over after 30 days. They identified six metabolic pathways associated with higher sodium intake. The strongest associations were generated from fatty acid, benzoate, methionine, and tryptophan pathways. Moreover, switching from high- to low-sodium intake had a greater effect on metabolites in comparison to switching from high- to medium-sodium, this effect was particularly apparent for metabolites involved in the γ-glutamyl amino acid pathway.84 The enzyme responsible for γ-glutamyl metabolite formation γ-glutamyl transferase has previously been positively associated with prehypertension.85 The key finding from the study does not relate to BP and comes from the strongest associated metabolite 4-ethylphenylsulfate, which increased with sodium restriction. This metabolite is generated by gut bacteria and related to numerous disorders, highlighting this gut–diet-health interaction.
Lee et al.86 investigated serum metabolites associated with incident HTN within a Korean cohort (n = 1,529), grouping participants into tertiles of MUFA intake. They reported an inverse association between the highest MUFA intake group and risk of HTN when compared with the lowest MUFA intake. Moreover, researchers associated MUFA intake with metabolite concentration, higher MUFA intake was associated with an increase in the metabolite phosphatidylcholine-diacyl (PC aa) C 38:1.
CONCLUSIONS
Advances in high throughput genomics, metabolomics and other omics have vastly increased our understanding of the complex architecture of BP and hypertension. The burgeoning list of genomic variants associated with BP and hypertension provides a realistic basis for refining the molecular taxonomy of hypertension, but this requires incorporation of other characteristics including lifestyle and environmental influences in addition to molecular and genomic information. This step is critical in realizing the potential of genomics and other omics in precision hypertension prevention and therapy.
Supplementary Material
ACKNOWLEDGMENTS
C.M. is funded by the MRC Aim-Hy project grant and Chronic Disease Research Foundation. P.L. is funded by the Chronic Disease Research Foundation. S.P. is funded by the Medical Research Council (MRC) (MR/M016560/1; Ancestry and biological Informative Markers for stratification of Hypertension [AIM-HY] Study), the British Heart Foundation (BHF) (CS/16/1/31878) and BHF Centre of Excellence (RE/18/6/34217).
DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1. GBDRF Collaborators. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017; 390:1345–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Whelton PK, Carey RM, Aronow WS, Casey DE Jr, Collins KJ, Dennison Himmelfarb C, DePalma SM, Gidding S, Jamerson KA, Jones DW, MacLaughlin EJ, Muntner P, Ovbiagele B, Smith SC Jr, Spencer CC, Stafford RS, Taler SJ, Thomas RJ, Williams KA Sr, Williamson JD, Wright JT Jr. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines. Circulation 2018; 138:e426–e483. [DOI] [PubMed] [Google Scholar]
- 3. Mongeau JG, Biron P, Sing CF. The influence of genetics and household environment upon the variability of normal blood pressure: the Montreal Adoption Survey. Clin Exp Hypertens A 1986; 8:653–660. [DOI] [PubMed] [Google Scholar]
- 4. Fava C, Burri P, Almgren P, Groop L, Hulthén UL, Melander O. Heritability of ambulatory and office blood pressure phenotypes in Swedish families. J Hypertens 2004; 22:1717–1721. [DOI] [PubMed] [Google Scholar]
- 5. Hottenga JJ, Boomsma DI, Kupper N, Posthuma D, Snieder H, Willemsen G, de Geus EJ. Heritability and stability of resting blood pressure. Twin Res Hum Genet 2005; 8:499–508. [DOI] [PubMed] [Google Scholar]
- 6. Funder JW. Primary aldosteronism. Hypertension 2019; 74:458–466. [DOI] [PubMed] [Google Scholar]
- 7. Levanovich PE, Diaczok A, Rossi NF. Clinical and molecular perspectives of monogenic hypertension. Curr Hypertens Rev 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Padmanabhan S, Joe B. Towards precision medicine for hypertension: a review of genomic, epigenomic, and microbiomic effects on blood pressure in experimental rat models and humans. Physiol Rev 2017; 97:1469–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, Ng FL, Evangelou M, Witkowska K, Tzanis E, Hellwege JN, Giri A, Velez Edwards DR, Sun YV, Cho K, Gaziano JM, Wilson PWF, Tsao PS, Kovesdy CP, Esko T, Mägi R, Milani L, Almgren P, Boutin T, Debette S, Ding J, Giulianini F, Holliday EG, Jackson AU, Li-Gao R, Lin WY, Luan J, Mangino M, Oldmeadow C, Prins BP, Qian Y, Sargurupremraj M, Shah N, Surendran P, Thériault S, Verweij N, Willems SM, Zhao JH, Amouyel P, Connell J, de Mutsert R, Doney ASF, Farrall M, Menni C, Morris AD, Noordam R, Paré G, Poulter NR, Shields DC, Stanton A, Thom S, Abecasis G, Amin N, Arking DE, Ayers KL, Barbieri CM, Batini C, Bis JC, Blake T, Bochud M, Boehnke M, Boerwinkle E, Boomsma DI, Bottinger EP, Braund PS, Brumat M, Campbell A, Campbell H, Chakravarti A, Chambers JC, Chauhan G, Ciullo M, Cocca M, Collins F, Cordell HJ, Davies G, de Borst MH, de Geus EJ, Deary IJ, Deelen J, Del Greco M F, Demirkale CY, Dörr M, Ehret GB, Elosua R, Enroth S, Erzurumluoglu AM, Ferreira T, Frånberg M, Franco OH, Gandin I, Gasparini P, Giedraitis V, Gieger C, Girotto G, Goel A, Gow AJ, Gudnason V, Guo X, Gyllensten U, Hamsten A, Harris TB, Harris SE, Hartman CA, Havulinna AS, Hicks AA, Hofer E, Hofman A, Hottenga JJ, Huffman JE, Hwang SJ, Ingelsson E, James A, Jansen R, Jarvelin MR, Joehanes R, Johansson Å, Johnson AD, Joshi PK, Jousilahti P, Jukema JW, Jula A, Kähönen M, Kathiresan S, Keavney BD, Khaw KT, Knekt P, Knight J, Kolcic I, Kooner JS, Koskinen S, Kristiansson K, Kutalik Z, Laan M, Larson M, Launer LJ, Lehne B, Lehtimäki T, Liewald DCM, Lin L, Lind L, Lindgren CM, Liu Y, Loos RJF, Lopez LM, Lu Y, Lyytikäinen LP, Mahajan A, Mamasoula C, Marrugat J, Marten J, Milaneschi Y, Morgan A, Morris AP, Morrison AC, Munson PJ, Nalls MA, Nandakumar P, Nelson CP, Niiranen T, Nolte IM, Nutile T, Oldehinkel AJ, Oostra BA, O’Reilly PF, Org E, Padmanabhan S, Palmas W, Palotie A, Pattie A, Penninx BWJH, Perola M, Peters A, Polasek O, Pramstaller PP, Nguyen QT, Raitakari OT, Ren M, Rettig R, Rice K, Ridker PM, Ried JS, Riese H, Ripatti S, Robino A, Rose LM, Rotter JI, Rudan I, Ruggiero D, Saba Y, Sala CF, Salomaa V, Samani NJ, Sarin AP, Schmidt R, Schmidt H, Shrine N, Siscovick D, Smith AV, Snieder H, Sõber S, Sorice R, Starr JM, Stott DJ, Strachan DP, Strawbridge RJ, Sundström J, Swertz MA, Taylor KD, Teumer A, Tobin MD, Tomaszewski M, Toniolo D, Traglia M, Trompet S, Tuomilehto J, Tzourio C, Uitterlinden AG, Vaez A, van der Most PJ, van Duijn CM, Vergnaud AC, Verwoert GC, Vitart V, Völker U, Vollenweider P, Vuckovic D, Watkins H, Wild SH, Willemsen G, Wilson JF, Wright AF, Yao J, Zemunik T, Zhang W, Attia JR, Butterworth AS, Chasman DI, Conen D, Cucca F, Danesh J, Hayward C, Howson JMM, Laakso M, Lakatta EG, Langenberg C, Melander O, Mook-Kanamori DO, Palmer CNA, Risch L, Scott RA, Scott RJ, Sever P, Spector TD, van der Harst P, Wareham NJ, Zeggini E, Levy D, Munroe PB, Newton-Cheh C, Brown MJ, Metspalu A, Hung AM, O’Donnell CJ, Edwards TL, Psaty BM, Tzoulaki I, Barnes MR, Wain LV, Elliott P, Caulfield MJ; Million Veteran Program . Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet 2018; 50:1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabási AL. Disease networks. Uncovering disease–disease relationships through the incomplete interactome. Science 2015; 347:1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Intersalt: an international study of electrolyte excretion and blood pressure. Results for 24 hour urinary sodium and potassium excretion. Intersalt Cooperative Research Group. BMJ 1988; 297:319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sacks FM, Svetkey LP, Vollmer WM, Appel LJ, Bray GA, Harsha D, Obarzanek E, Conlin PR, Miller ER III, Simons-Morton DG, Karanja N, Lin PH; DASH-Sodium Collaborative Research Group . Effects on blood pressure of reduced dietary sodium and the Dietary Approaches to Stop Hypertension (DASH) diet. DASH-Sodium Collaborative Research Group. N Engl J Med 2001; 344:3–10. [DOI] [PubMed] [Google Scholar]
- 13. Blanchard A, Bockenhauer D, Bolignano D, Calò LA, Cosyns E, Devuyst O, Ellison DH, Karet Frankl FE, Knoers NV, Konrad M, Lin SH, Vargas-Poussou R. Gitelman syndrome: consensus and guidance from a Kidney Disease: improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int 2017; 91:24–33. [DOI] [PubMed] [Google Scholar]
- 14. Li C, He J, Chen J, Zhao J, Gu D, Hixson JE, Rao DC, Jaquish CE, Gu CC, Chen J, Huang J, Chen S, Kelly TN. Genome-wide gene–sodium interaction analyses on blood pressure: the genetic epidemiology network of salt-sensitivity study. Hypertension 2016; 68:348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Imaizumi T, Ando M, Nakatochi M, Maruyama S, Yasuda Y, Honda H, Kuwatsuka Y, Kato S, Kondo T, Iwata M, Nakashima T, Yasui H, Takamatsu H, Okajima H, Yoshida Y, Matsuo S. Association of interactions between dietary salt consumption and hypertension-susceptibility genetic polymorphisms with blood pressure among Japanese male workers. Clin Exp Nephrol 2017; 21:457–464. [DOI] [PubMed] [Google Scholar]
- 16. Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie CE, Menni C, Monti MC, Delles C, Laing S, Corso B, Navis G, Kwakernaak AJ, van der Harst P, Bochud M, Maillard M, Burnier M, Hedner T, Kjeldsen S, Wahlstrand B, Sjögren M, Fava C, Montagnana M, Danese E, Torffvit O, Hedblad B, Snieder H, Connell JM, Brown M, Samani NJ, Farrall M, Cesana G, Mancia G, Signorini S, Grassi G, Eyheramendy S, Wichmann HE, Laan M, Strachan DP, Sever P, Shields DC, Stanton A, Vollenweider P, Teumer A, Völzke H, Rettig R, Newton-Cheh C, Arora P, Zhang F, Soranzo N, Spector TD, Lucas G, Kathiresan S, Siscovick DS, Luan J, Loos RJ, Wareham NJ, Penninx BW, Nolte IM, McBride M, Miller WH, Nicklin SA, Baker AH, Graham D, McDonald RA, Pell JP, Sattar N, Welsh P, Munroe P, Caulfield MJ, Zanchetti A, Dominiczak AF; Global BPgen Consortium . Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet 2010; 6:e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, Citterio L, Demaretz S, Trevisani F, Ristagno G, Glaudemans B, Laghmani K, Dell’Antonio G, Loffing J, Rastaldi MP, Manunta P, Devuyst O, Rampoldi L; SKIPOGH team . Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med 2013; 19:1655–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang X, Qi Q, Liang J, Hu FB, Sacks FM, Qi L. Neuropeptide Y promoter polymorphism modifies effects of a weight-loss diet on 2-year changes of blood pressure: the preventing overweight using novel dietary strategies trial. Hypertension 2012; 60:1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science 2005; 307:1915–1920. [DOI] [PubMed] [Google Scholar]
- 20. Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun 2016; 469:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell 2012; 148:1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Bäckhed F, Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 2012; 3:1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, Bäckhed F. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA 2011; 108(Suppl 1):4592–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marques FZ, Mackay CR, Kaye DM. Beyond gut feelings: how the gut microbiota regulates blood pressure. Nat Rev Cardiol 2018; 15:20–32. [DOI] [PubMed] [Google Scholar]
- 25. Menni C, Lin C, Cecelja M, Mangino M, Matey-Hernandez ML, Keehn L, Mohney RP, Steves CJ, Spector TD, Kuo CF, Chowienczyk P, Valdes AM. Gut microbial diversity is associated with lower arterial stiffness in women. Eur Heart J 2018; 39:2390–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menni C, Graham D, Kastenmüller G, Alharbi NH, Alsanosi SM, McBride M, Mangino M, Titcombe P, Shin SY, Psatha M, Geisendorfer T, Huber A, Peters A, Wang-Sattler R, Xu T, Brosnan MJ, Trimmer J, Reichel C, Mohney RP, Soranzo N, Edwards MH, Cooper C, Church AC, Suhre K, Gieger C, Dominiczak AF, Spector TD, Padmanabhan S, Valdes AM. Metabolomic identification of a novel pathway of blood pressure regulation involving hexadecanedioate. Hypertension 2015; 66:422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, Wu S, Liu W, Cui Q, Geng B, Zhang W, Weldon R, Auguste K, Yang L, Liu X, Chen L, Yang X, Zhu B, Cai J. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017; 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, Zadeh M, Gong M, Qi Y, Zubcevic J, Sahay B, Pepine CJ, Raizada MK, Mohamadzadeh M. Gut dysbiosis is linked to hypertension. Hypertension 2015; 65:1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Donia MS, Fischbach MA. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science 2015; 349:1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA 2009; 106:3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tuteja S, Ferguson JF. Gut microbiome and response to cardiovascular drugs. Circ Genom Precis Med 2019; 12:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W. Influence of diet on the gut microbiome and implications for human health. J Transl Med 2017; 15:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C, Zhao G, Chen Y, Zhao L. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 2010; 4:232–241. [DOI] [PubMed] [Google Scholar]
- 34.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008; 453:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Y, Yu B, Alexander D, Mosley TH, Heiss G, Nettleton JA, Boerwinkle E. Metabolomics and incident hypertension among blacks: the atherosclerosis risk in communities study. Hypertension 2013; 62:398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loo RL, Zou X, Appel LJ, Nicholson JK, Holmes E. Characterization of metabolic responses to healthy diets and association with blood pressure: application to the Optimal Macronutrient Intake Trial for Heart Health (OmniHeart), a randomized controlled study. Am J Clin Nutr 2018; 107:323–334. [DOI] [PubMed] [Google Scholar]
- 37.Ke C, Zhu X, Zhang Y, Shen Y. Metabolomic characterization of hypertension and dyslipidemia. Metabolomics 2018; 14:117. [DOI] [PubMed] [Google Scholar]
- 38.Shahin MH, Gong Y, McDonough CW, Rotroff DM, Beitelshees AL, Garrett TJ, Gums JG, Motsinger-Reif A, Chapman AB, Turner ST, Boerwinkle E, Frye RF, Fiehn O, Cooper-DeHoff RM, Kaddurah- Daouk R, Johnson JA. A genetic response score for hydrochlorothiazide use: insights from genomics and metabolomics integration. Hypertension 2016; 68:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao Y, Wang Y, Xi L, Li G, Zhao F, Qi Y, Liu J, Zhao D. A nested case–control study of association between metabolome and Hypertension risk. Biomed Res Int 2016; 2016:7646979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakraborty S, Galla S, Cheng X, Yeo JY, Mell B, Singh V, Yeoh B, Saha P, Mathew AV, Vijay-Kumar M, Joe B. Salt-responsive metabolite, β-hydroxybutyrate, attenuates hypertension. Cell Rep 2018; 25:677–689.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai Q, Peng B, Wu X, Cao Y, Sun X, Hong M, Na R, Liu B, Li Q, Li Z, Fang W, Zhu N, Zong C, Yu Q. Metabolomic study for essential hypertension patients based on dried blood spot mass spectrometry approach. IUBMB Life 2018; 70:777–785. [DOI] [PubMed] [Google Scholar]
- 42.de la Cuesta-Zuluaga J, Mueller NT, Alvarez-Quintero R, Velasquez- Mejia EP, Sierra JA, Corrales-Agudelo V, Carmona JA, Abad JM, Escobar JS. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients 2018; 11:pii: E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Hou E, Wang L, Wang Y, Yang L, Zheng X, Xie G, Sun Q, Liang M, Tian Z. Reconstruction and analysis of correlation networks based on GC-MS metabolomics data for young hypertensive men. Anal Chim Acta 2015; 854:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulkarni H, Meikle PJ, Mamtani M, Weir JM, Barlow CK, Jowett JB, Bellis C, Dyer TD, Johnson MP, Rainwater DL, Almasy L, Mahaney MC, Comuzzie AG, Blangero J, Curran JE. Plasma lipidomic profile signature of hypertension in Mexican American families: specific role of diacylglycerols. Hypertension 2013; 62:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Balasubramanian R, Paynter NP, Giulianini F, Manson JE, Zhao Y, Chen JC, Vitolins MZ, Albert CA, Clish C, Rexrode KM. Metabolomic profiles associated with all-cause mortality in the Women’s Health Initiative. Int J Epidemiol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun D, Tiedt S, Yu B, Jian X, Gottesman RF, Mosley TH, Boerwinkle E, Dichgans M, Fornage M. A prospective study of serum metabolites and risk of ischemic stroke. Neurology 2019; 92:e1890–e1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suhre K, Gieger C. Genetic variation in metabolic phenotypes: study designs and applications. Nat Rev Genet 2012; 13:759–769. [DOI] [PubMed] [Google Scholar]
- 48. Yokoyama A, Mizukami T, Matsui T, Yokoyama T, Kimura M, Matsushita S, Higuchi S, Maruyama K. Genetic polymorphisms of alcohol dehydrogenase-1B and aldehyde dehydrogenase-2 and liver cirrhosis, chronic calcific pancreatitis, diabetes mellitus, and hypertension among Japanese alcoholic men. Alcohol Clin Exp Res 2013; 37:1391–1401. [DOI] [PubMed] [Google Scholar]
- 49. Müller F, Sharma A, König J, Fromm MF. Biomarkers for in vivo assessment of transporter function. Pharmacol Rev 2018; 70:246–277. [DOI] [PubMed] [Google Scholar]
- 50. Yu B, Li AH, Metcalf GA, Muzny DM, Morrison AC, White S, Mosley TH, Gibbs RA, Boerwinkle E. Loss-of-function variants influence the human serum metabolome. Sci Adv 2016; 2:e1600800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hiltunen TP, Rimpelä JM, Mohney RP, Stirdivant SM, Kontula KK. Effects of four different antihypertensive drugs on plasma metabolomic profiles in patients with essential hypertension. PLoS One 2017; 12:e0187729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao Y, Peng J, Lu C, Hsin M, Mura M, Wu L, Chu L, Zamel R, Machuca T, Waddell T, Liu M, Keshavjee S, Granton J, de Perrot M. Metabolomic heterogeneity of pulmonary arterial hypertension. PLoS One 2014; 9:e88727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Menni C, Metrustry SJ, Ehret G, Dominiczak AF, Chowienczyk P, Spector TD, Padmanabhan S, Valdes AM. Molecular pathways associated with blood pressure and hexadecanedioate levels. PLoS One 2017; 12:e0175479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tang ZZ, Chen G, Hong Q, Huang S, Smith HM, Shah RD, Scholz M, Ferguson JF. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Front Genet 2019; 10:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006; 124:837–848. [DOI] [PubMed] [Google Scholar]
- 56. Jama HA, Beale A, Shihata WA, Marques FZ. The effect of diet on hypertensive pathology: is there a link via gut microbiota-driven immunometabolism? Cardiovasc Res 2019; 115:1435–1447. [DOI] [PubMed] [Google Scholar]
- 57. Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA 2013; 110:4410–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang Y, Haribabu B, Vijay-Kumar M, Pennathur S, Joe B. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics 2015; 47:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jobin C. GPR109a: the missing link between microbiome and good health? Immunity 2014; 40:8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chambers ES, Preston T, Frost G, Morrison DJ. Role of Gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutr Rep 2018; 7:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, Wang J, Imhann F, Brandsma E, Jankipersadsing SA, Joossens M, Cenit MC, Deelen P, Swertz MA, Weersma RK, Feskens EJ, Netea MG, Gevers D, Jonkers D, Franke L, Aulchenko YS, Huttenhower C, Raes J, Hofker MH, Xavier RJ, Wijmenga C, Fu J; LifeLines cohort study . Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016; 352:565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, Mujagic Z, Masclee AAM, Jonkers DMAE, Oosting M, Joosten LAB, Netea MG, Franke L, Zhernakova A, Fu J, Wijmenga C, McCarthy MI. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 2019; 51:600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Janeiro MH, Ramirez MJ, Milagro FI, Martinez JA, Solas M. Implication of trimethylamine N-oxide (TMAO) in disease: potential biomarker or new therapeutic target. Nutrients 2018; 10:pii: E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gruppen EG, Garcia E, Connelly MA, Jeyarajah EJ, Otvos JD, Bakker SJL, Dullaart RPF. TMAO is associated with mortality: impact of modestly impaired renal function. Sci Rep 2017; 7:13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mafune A, Iwamoto T, Tsutsumi Y, Nakashima A, Yamamoto I, Yokoyama K, Yokoo T, Urashima M. Associations among serum trimethylamine-N-oxide (TMAO) levels, kidney function and infarcted coronary artery number in patients undergoing cardiovascular surgery: a cross-sectional study. Clin Exp Nephrol 2016; 20:731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Senthong V, Wang Z, Li XS, Fan Y, Wu Y, Tang WH, Hazen SL. Intestinal microbiota-generated metabolite trimethylamine-N-oxide and 5-year mortality risk in stable coronary artery disease: the contributory role of intestinal microbiota in a COURAGE-like patient cohort. J Am Heart Assoc 2016; 5:pii: e002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tang WH, Wang Z, Fan Y, Levison B, Hazen JE, Donahue LM, Wu Y, Hazen SL. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: refining the gut hypothesis. J Am Coll Cardiol 2014; 64:1908–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ufnal M, Jazwiec R, Dadlez M, Drapala A, Sikora M, Skrzypecki J. Trimethylamine-N-oxide: a carnitine-derived metabolite that prolongs the hypertensive effect of angiotensin II in rats. Can J Cardiol 2014; 30:1700–1705. [DOI] [PubMed] [Google Scholar]
- 69. Hartiala J, Bennett BJ, Tang WH, Wang Z, Stewart AF, Roberts R, McPherson R, Lusis AJ, Hazen SL, Allayee H; CARDIoGRAM Consortium . Comparative genome-wide association studies in mice and humans for trimethylamine N-oxide, a proatherogenic metabolite of choline and L-carnitine. Arterioscler Thromb Vasc Biol 2014; 34:1307–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007; 56:1761–1772. [DOI] [PubMed] [Google Scholar]
- 71. Abdul Rahim MBH, Chilloux J, Martinez-Gili L, Neves AL, Myridakis A, Gooderham N, Dumas ME. Diet-induced metabolic changes of the human gut microbiome: importance of short-chain fatty acids, methylamines and indoles. Acta Diabetol 2019; 56:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Huć T, Nowinski A, Drapala A, Konopelski P, Ufnal M. Indole and indoxyl sulfate, gut bacteria metabolites of tryptophan, change arterial blood pressure via peripheral and central mechanisms in rats. Pharmacol Res 2018; 130:172–179. [DOI] [PubMed] [Google Scholar]
- 73. Zheng Y, Yu B, Alexander D, Manolio TA, Aguilar D, Coresh J, Heiss G, Boerwinkle E, Nettleton JA. Associations between metabolomic compounds and incident heart failure among African Americans: the ARIC Study. Am J Epidemiol 2013; 178:534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Menni C, Mangino M, Cecelja M, Psatha M, Brosnan MJ, Trimmer J, Mohney RP, Chowienczyk P, Padmanabhan S, Spector TD, Valdes AM. Metabolomic study of carotid-femoral pulse-wave velocity in women. J Hypertens 2015; 33:791–796; discussion 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Poesen R, Claes K, Evenepoel P, de Loor H, Augustijns P, Kuypers D, Meijers B. Microbiota-derived phenylacetylglutamine associates with overall mortality and cardiovascular disease in patients with CKD. J Am Soc Nephrol 2016; 27:3479–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kolodziejczyk AA, Zheng D, Elinav E. Diet–microbiota interactions and personalized nutrition. Nat Rev Microbiol 2019; 17:742–753. [DOI] [PubMed] [Google Scholar]
- 77. Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mähler A, Balogh A, Markó L, Vvedenskaya O, Kleiner FH, Tsvetkov D, Klug L, Costea PI, Sunagawa S, Maier L, Rakova N, Schatz V, Neubert P, Frätzer C, Krannich A, Gollasch M, Grohme DA, Côrte-Real BF, Gerlach RG, Basic M, Typas A, Wu C, Titze JM, Jantsch J, Boschmann M, Dechend R, Kleinewietfeld M, Kempa S, Bork P, Linker RA, Alm EJ, Müller DN. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017; 551:585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang W, Zheng JP, Zhu SX, Guan WJ, Chen M, Zhong NS. Carbocisteine attenuates hydrogen peroxide-induced inflammatory injury in A549 cells via NF-κB and ERK1/2 MAPK pathways. Int Immunopharmacol 2015; 24:306–313. [DOI] [PubMed] [Google Scholar]
- 79. Sun S, Lulla A, Sioda M, Winglee K, Wu MC, Jacobs DR Jr, Shikany JM, Lloyd-Jones DM, Launer LJ, Fodor AA, Meyer KA. Gut microbiota composition and blood pressure. Hypertension 2019; 73:998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Galla S, Chakraborty S, Cheng X, Yeo J, Mell B, Zhang H, Mathew AV, Vijay-Kumar M, Joe B. Disparate effects of antibiotics on hypertension. Physiol Genomics 2018; 50:837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010; 107:14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gutiérrez-Díaz I, Fernández-Navarro T, Salazar N, Bartolomé B, Moreno-Arribas MV, de Andres-Galiana EJ, Fernández-Martínez JL, de Los Reyes-Gavilán CG, Gueimonde M, González S. Adherence to a mediterranean diet influences the fecal metabolic profile of microbial-derived phenolics in a spanish cohort of middle-age and older people. J Agric Food Chem 2017; 65:586–595. [DOI] [PubMed] [Google Scholar]
- 84. Derkach A, Sampson J, Joseph J, Playdon MC, Stolzenberg-Solomon RZ. Effects of dietary sodium on metabolites: the dietary approaches to stop hypertension (DASH)-Sodium Feeding Study. Am J Clin Nutr 2017; 106:1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shankar A, Li J. Association between serum gamma-glutamyltransferase level and prehypertension among US adults. Circ J 2007; 71:1567–1572. [DOI] [PubMed] [Google Scholar]
- 86. Lee H, Jang HB, Yoo MG, Chung KS, Lee HJ. Protective effects of dietary MUFAs mediating metabolites against hypertension risk in the Korean genome and epidemiology study. Nutrients 2019; 11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dan X, Mushi Z, Baili W, Han L, Enqi W, Huanhu Z, Shuchun L. Differential analysis of hypertension-associated intestinal microbiota. Int J Med Sci 2019; 16:872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yan Q, Gu Y, Li X, Yang W, Jia L, Chen C, Han X, Huang Y, Zhao L, Li P, Fang Z, Zhou J, Guan X, Ding Y, Wang S, Khan M, Xin Y, Li S, Ma Y. Alterations of the gut microbiome in hypertension. Front Cell Infect Microbiol 2017; 7:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Dekker Nitert M; SPRING Trial Group . Increased systolic and diastolic blood pressure is associated with altered gut microbiota composition and butyrate production in early pregnancy. Hypertension 2016; 68:974–981. [DOI] [PubMed] [Google Scholar]
- 90. Jackson MA, Verdi S, Maxan ME, Shin CM, Zierer J, Bowyer RCE, Martin T, Williams FMK, Menni C, Bell JT, Spector TD, Steves CJ. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun 2018; 9:2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


