Aetiological and clinical heterogeneity in Alzheimer's disease complicates diagnosis, treatment and drug trials. Badhwar et al. review three omics data reduction techniques that capture disease-relevant heterogeneity, and present a roadmap for assembling a multiomics diagnostic tool.
Keywords: Alzheimer’s disease, multiomics biomarkers, neuroimaging subtype, metabolite panel, polygenic risk score
Abstract
Aetiological and clinical heterogeneity is increasingly recognized as a common characteristic of Alzheimer’s disease and related dementias. This heterogeneity complicates diagnosis, treatment, and the design and testing of new drugs. An important line of research is discovery of multimodal biomarkers that will facilitate the targeting of subpopulations with homogeneous pathophysiological signatures. High-throughput ‘omics’ are unbiased data-driven techniques that probe the complex aetiology of Alzheimer’s disease from multiple levels (e.g. network, cellular, and molecular) and thereby account for pathophysiological heterogeneity in clinical populations. This review focuses on data reduction analyses that identify complementary disease-relevant perturbations for three omics techniques: neuroimaging-based subtypes, metabolomics-derived metabolite panels, and genomics-related polygenic risk scores. Neuroimaging can track accrued neurodegeneration and other sources of network impairments, metabolomics provides a global small-molecule snapshot that is sensitive to ongoing pathological processes, and genomics characterizes relatively invariant genetic risk factors representing key pathways associated with Alzheimer’s disease. Following this focused review, we present a roadmap for assembling these multiomics measurements into a diagnostic tool highly predictive of individual clinical trajectories, to further the goal of personalized medicine in Alzheimer’s disease.
Introduction
Alzheimer’s disease is a complex, multifactorial pathology that manifests itself along a continuum of conditions, ranging from asymptomatic, to mild cognitive impairment (MCI), to dementia (specifically Alzheimer’s disease dementia). Trials of disease-modifying therapies remain unsuccessful, and these persistent failures have been attributed to (i) intervention late in the disease process (i.e. symptomatic stage), by which time extensive irreversible damage has accrued; and (ii) lack of precision intervention targets in a multifactorial condition. Accordingly, an important line of current research is directed at the discovery of multimodal biomarkers that will help facilitate the detection of Alzheimer’s disease in asymptomatic populations, and the adaptation of intervention regimens to different target subpopulations in prevention trials (Anstey et al., 2015; Olanrewaju et al., 2015). This work reviews recent data-driven approaches to biomarker discovery, in three omics fields that capture complementary aspects of neurodegeneration and Alzheimer’s disease risk factors. We further propose a roadmap for integrating these multiomics biomarkers to advance our understanding of heterogeneity in Alzheimer’s disease and other age-related dementias, and promote efficacy in intervention trials.
Established Alzheimer’s disease biomarkers currently capture three facets of the disease pathophysiology: amyloidosis (A), tauopathy (T), and specific aspects of neurodegeneration (N) or A/T/N (Jack et al., 2018). Although these biomarkers have been usefully applied to the crucial goal of early Alzheimer’s disease detection (Sperling et al., 2011), they fall short in explaining the heterogeneity of individual clinical trajectories, and their ability to predict differential cognitive decline is modest (Dumurgier et al., 2017). Predicting progression to dementia is challenging, as patients diagnosed with probable Alzheimer’s disease dementia show considerable heterogeneity in the cognitive domains impaired (Scheltens et al., 2016), and the presence or severity of established Alzheimer’s disease biomarkers. For example, amyloidosis-and-tauopathy-defined ‘pure Alzheimer’s disease neuropathology’ is observed in only 30–50% of patients with probable Alzheimer’s disease dementia (Beach et al., 2012; Robinson et al., 2018). The remaining cases show co-occurrence of multiple brain pathologies that overlap with other neurodegenerative diseases (NDDs) of ageing, such as cerebral small vessel disease, and Lewy body dementia. Minimum to above-threshold levels of Alzheimer’s disease pathology are also observed in a considerable proportion (39%) of dementia patients not clinically diagnosed as probable Alzheimer’s disease (Beach et al., 2012). Alzheimer’s disease pathology has also been demonstrated in post-mortem studies of cognitively normal older adults (Bennett et al., 2006), and it remains unclear whether such individuals would have developed Alzheimer’s disease symptoms with time, should they have lived longer (Jagust, 2013). Overall, ‘top-down’ clinical labels, based primarily on cognitive symptoms, imperfectly align with biomarkers of neurodegeneration. Additional biomarkers are thus urgently needed to characterize the clinicopathological heterogeneity of Alzheimer’s disease, and to disambiguate it from other age-related NDDs and normal ageing (Jack et al., 2018).
A radically different paradigm to NDDs is to move away from ‘top-down’ clinical labels, and concentrate on pathological signatures built ‘bottom up’ using unsupervised machine learning algorithms and high-throughput ‘omics’ metrics that screen global facets of an organism. ‘Omics’ refers to several areas of study in biology, all of which end in the suffix -omics, and implies a comprehensive (or global) assessment of the subject using high-throughput technologies. For example, the study of an organism’s entire collection of genes (the genome) versus a single or a few genes is termed genomics (Hasin et al., 2017). Data-driven approaches on ‘omics’ technologies-generated data provide new opportunities to probe the complex aetiology of Alzheimer’s disease from multiple levels (e.g. network, cellular, and molecular), and to identify biomarker signatures with high diagnostic/prognostic value. This review focuses on the following omics approaches: brain connectomics, metabolomics, and genomics. These omics data capture complementary information on Alzheimer’s disease emergence and progression: brain connectomics (and morphometry) can track accrued neurodegeneration and other sources of network impairments, metabolomics provides a global small molecule snapshot that is sensitive to ongoing pathological processes, and genomics characterizes relatively invariant genetic risk factors representing key pathways associated with Alzheimer’s disease (Jack et al., 2018). The high-dimensional nature of omics ‘big data’ can prove challenging to process, manipulate, and visualize, even when a single modality is involved. Multiple redundancies are often present in these measures, and not all data points provide independent information as they tend to co-vary because of shared biological processes. We focus this review on three omics data reduction techniques that capture disease-relevant population heterogeneity with a limited number of indicators: neuroimaging-based subtypes, metabolite panels, and polygenic risk score (PRS). Neuroimaging subtypes are based on data-driven algorithms that identify patient subgroups with homogeneous brain imaging features. Metabolite panels are developed via data-driven algorithms applied to thousands of small molecules representing global biochemical events and distinguishing clinical phenotypes. PRS and other empirically derived representations of interactive or multi-gene risk may represent key domains of mechanisms and pathways to Alzheimer’s disease. Following this focused review, we discuss the rationale and challenges for assembling multiomics diagnostic tools highly predictive of individual clinical trajectories in the context of Alzheimer’s disease, and in particular, the importance of pathophysiological heterogeneity in research clinical cohorts, with the intent to extend these findings to the discrimination of Alzheimer’s disease, as well as other dementia related subtypes.
Materials and methods
We conducted parallel focused reviews of PubMed articles published between January 2011 to June 2018 in three omics domains: brain connectomics (and morphometry), metabolomics, and genomics. It should be noted that for brain connectomics (and morphometry), we only focused on MRI-based studies, and did not include studies using PET imaging methods, as data-driven subtype literature is sparse in the latter. We included studies investigating Alzheimer’s disease in humans and published in English (termed ‘common' inclusion criteria). Additional articles (that met criteria) were identified by scanning the reference lists of selected PubMed articles. Described in the following three sections are search characteristics specific to each omics domain. Characteristics of the 40 domain-specific omics studies included are provided in Supplementary Tables 1, 2 and 5.
Brain morphology and connectomics
Search term combinations used for brain morphology and connectomics in neuroimaging were: (i) Alzheimer’s disease OR Alzheimer pathology AND subtype; (ii) mild cognitive impairment OR amnestic mild cognitive impairment OR MCI OR amnestic MCI AND hierarchical clustering; resting-state AND functional MRI AND Alzheimer AND clustering; (iii) Alzheimer’s disease OR Alzheimer pathology AND structural subtype; (iv) Alzheimer’s disease OR Alzheimer pathology AND structural MRI AND clustering; (v) resting-state AND functional MRI AND Alzheimer AND hierarchical clustering; (vi) diffusion MRI AND Alzheimer AND clustering; and (vii) diffusion MRI AND Alzheimer AND hierarchical clustering; diffusion MRI AND hierarchical clustering. To these we applied the ‘common’ inclusion criteria. Thereafter, only studies reporting Alzheimer’s disease spectrum subgroups identified using data-driven methods were included. It should be noted that neuroimaging-based subtyping is an emergent field and there is still a lack of consistent terminology. Therefore, to avoid missing relevant studies due to stringent use of terminology, search combinations (i) and (ii), using relaxed and inclusive keywords, captured all of the morphometry-based literature. In total, 12 papers met our criteria, and were reviewed.
Metabolomics
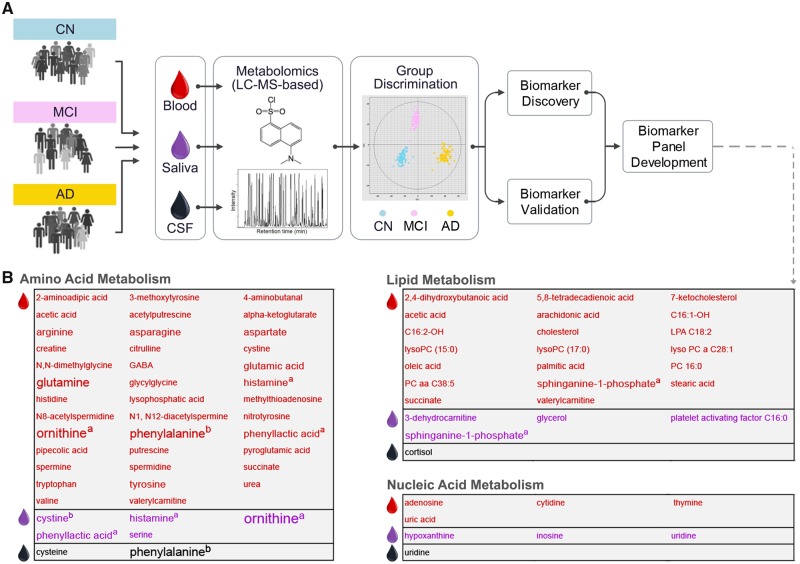
Search term combinations for metabolomics were: (i) Alzheimer’s disease AND metabolomics panels; (ii) Alzheimer’s disease AND metabolomics profiles; (iii) Alzheimer’s disease AND metabolomics networks; (iv) Alzheimer’s disease AND metabolomics pathways; and (v) Alzheimer’s disease AND metabolomics biomarkers. To these we applied the common inclusion criteria. We further excluded: reviews and technical reports; articles not relevant to Alzheimer’s disease metabolomic panels, pathways, or networks; and studies using a targeted metabolomics approach. Five papers were identified from reference list scans. In total, 11 studies were reviewed. Details on key metabolites highlighted in the 11 studies were compiled from three different databases: Human Metabolome Database (HMDB, http://www.hmdb.ca/), Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/pathway.html) Pathway Database, and PubMed. These details were used to generate Fig. 2B and are provided in Supplementary Table 3.
Figure 2.
A typical Alzheimer’s disease metabolomics biomarker discovery pipeline. Metabolomics is a relatively recent addition to the systems biology toolkit for the study of NDDs of ageing (Wilkins and Trushina, 2017). It encompasses the global study of small molecules (50–1500 Da in mass) that are substrates and products of metabolism. Together, these metabolites (e.g. amino acids, antioxidants, vitamins) represent the overall physiological status of the organism. An individual’s metabolic activity is influenced by an individual’s genotype and environment (Kaddurah-Daouk et al., 2011). Analysis of the metabolome, therefore, provides an opportunity to study the dynamic molecular phenotype of an individual. Untargeted metabolomics approaches are increasingly used to compare two or more groups (e.g. Alzheimer’s disease dementia and cognitively normal participants) and identify metabolite profiles associated with a disease. These profiles provide insight into underlying disease mechanisms, as well as constitute candidates for biomarker discovery and drug development. In the field of Alzheimer’s disease research, metabolomics studies (targeted and untargeted) over the past decade have examined several biofluids and tissues, including serum, plasma, CSF, saliva, urine, and brain tissue (Wilkins and Trushina, 2017). Technologies include NMR (nuclear magnetic resonance) spectroscopy and mass spectrometry. (A) A typical Alzheimer’s disease metabolomics biomarker discovery pipeline using mass spectrometry (MS)-liquid chromatography (LC) is depicted. Subsequent to metabolite extraction, identification, and quantification, most studies apply multivariate statistical methods to the metabolome data to identify the top discriminant metabolites. These can be further combined into metabolite panels to increase discriminative power (i.e. sensitivity and specificity) in Alzheimer’s disease prediction and progression (Liang et al., 2015, 2016; Huan et al., 2018). Significant discriminative power is commonly tested with the receiver operating characteristic curve analysis (AUC values). Discriminant metabolite panels are then validated in independent samples. Following discriminant metabolite(s) discovery, researchers conduct pathway and network analyses, which provide crucial mechanistic insights into the sequences of processes leading to the heterogeneous phenotypes of neurodegeneration. Pathway analysies focus on identifying sequences of processes that lead to the presence of a discriminant metabolite. Network analyses examine how discriminant metabolites are connected to each other within Alzheimer’s disease and related dementias. (B) The three main metabolism pathways (namely, amino acid, lipid and nucleic acid) that 90 Alzheimer’s disease-associated metabolites in our review (n = 11 publications, Supplementary Table 2) were found to belong. The text colour indicates the biofluid metabolome each metabolite was identified in: red = serum or plasma, purple = saliva, black = CSF. A larger font size indicates that the metabolite was identified in more than one study (Supplementary Table 3) The maximum number of studies a metabolite was detected in our review was four. aPresence in plasma or serum and saliva. bPresence in plasma or serum and CSF. AD = Alzheimer’s disease; CN = cognitively normal.
Genomics
Search term combinations used for the genomics were: (Polygenic Risk Scale OR Polygenic Risk Index OR Genetic Risk Score OR Genetic Risk Scale OR Genetic Risk Index) AND (Alzheimer Disease OR Alzheimers Disease OR Alzheimer’s Disease), which identified 264 potential papers. To these we applied the ‘common’ inclusion criteria, followed by exclusion of single-gene studies. In total, 17 studies were reviewed. References used for the compilation of the genes in Supplementary Table 4 are provided in the Supplementary material.
Brain subtypes
Anatomical subtypes
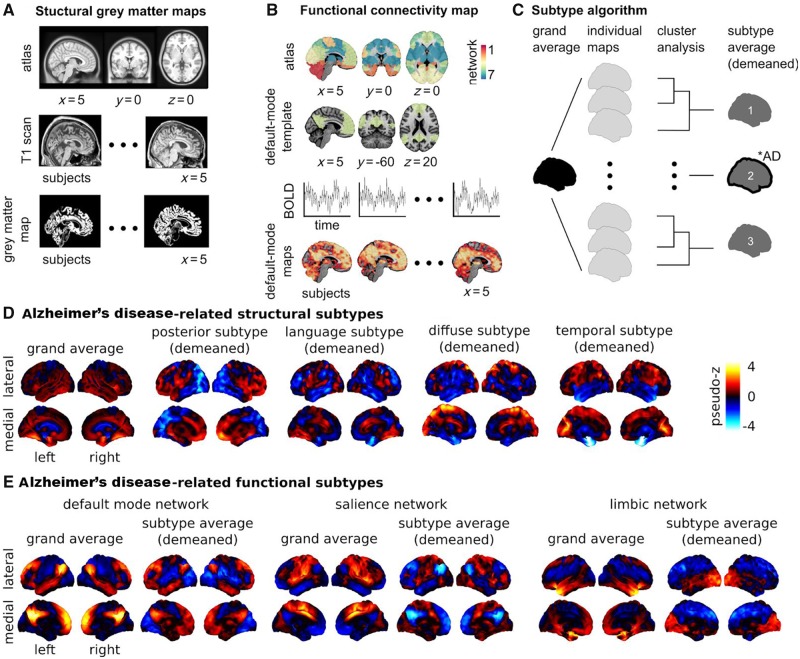
The spatial distribution of brain atrophy on structural MRI is highly heterogeneous in MCI, and Alzheimer’s disease dementia patients (Nettiksimmons et al., 2014; Poulakis et al., 2018). Using data-driven clustering algorithms, 11 studies have attempted to subtype and characterize this inherent heterogeneity (Supplementary Table 1). Seven studies reported at least three distinct atrophy subtypes in Alzheimer’s disease dementia (Noh et al., 2014; Hwang et al., 2016; Park et al., 2017; Poulakis et al., 2018; ten Kate et al., 2018), or mixed (Alzheimer’s disease dementia and cognitively normal) cohorts (Varol et al., 2017; Tam et al., 2019). Subtypes were generally consistent across studies, and can be described as diffuse, medial temporal-predominant (temporal), and temporo-occipito-parietal-predominant (posterior) (Fig. 1D). They were generated by applying (i) hierarchical agglomerative clustering using Ward’s clustering linkage (Noh et al., 2014; Hwang et al., 2016; Tam et al., 2019), Louvain clustering (Park et al., 2017), random forest clustering (Poulakis et al., 2018), or non-negative matrix factorization (ten Kate et al., 2018) on cortical thickness or grey matter density maps; or (ii) clustering on grey matter density maps using a novel approach called HYDRA (Varol et al., 2017). Good agreement across studies may, in part, reflect usage of the same data sample [Alzheimer’s Disease Neuroimaging Initiative (ADNI)] for subtype identification in four studies (Hwang et al., 2016; Varol et al., 2017; Poulakis et al., 2018; Tam et al., 2019). Some studies report two-subtype decomposition (Dong et al., 2016; Malpas, 2016), but these lack interstudy consensus. Using model-based clustering on regional cortical thickness measures from ADNI, Malpas (2016) reported normal and atrophic-entorhinal subtypes in a sample including Alzheimer’s disease dementia and cognitively normal individuals. The atrophic-entorhinal subtype demonstrated considerable heterogeneity in entorhinal thickness, suggesting the presence of additional subtypes (Malpas, 2016). Dong et al. (2016) reported limbic-insular and parietal-occipital atrophy subtypes using CHIMERA clustering on brain volume data from Alzheimer’s disease dementia and cognitively normal ADNI participants. In a separate study, the same group reported four atrophy subtypes using CHIMERA: normal, temporal, and two diffuse subtypes—one with predominant temporal involvement (diffuse-temporal), and one without (diffuse) (Dong et al., 2017). Visually, the diffuse subtype from CHIMERA shared some overlap with the posterior subtype described previously. In general, the reported subtypes (Dong et al., 2017) fit better with the three subtype solution, considering that, unlike previous studies, the CHIMERA study included cognitively normal individuals. Finally, Tam et al. (2019) identified a fourth atrophy subtype involving several language-related areas.
Figure 1.
Brain morphology and connectomics Alzheimer’s disease-related subtypes. Neuroimaging provides insight into the effect of neurodegeneration on brain health. There exist different tools that can capture distinct, yet complementary, aspects of brain structure and function. The most established neuroimaging marker of neurodegeneration is grey matter atrophy, measured by structural MRI. Structural MRI is a non-invasive technique widely used in both research and clinical practice. To generate structural maps, individual structural MRI scans are first spatially aligned to a reference template or atlas (A). Then for each individual and each voxel (smallest volume element in MRI data), a metric characterizing the local structure of the grey matter is generated, such as (A) grey matter volume, cortical thickness or surface area. Using these approaches, it is possible to monitor the thinning of grey matter, which likely reflects the death of neuronal cell bodies at advanced stages of neurodegeneration. Synaptic disruption is an early event in Alzheimer’s disease (Sperling et al., 2011), and functional networks may have the ability to compensate the impact of neurodegeneration on cognitive symptoms (Franzmeier et al., 2017). For these reasons, intrinsic functional connectivity from resting state functional MRI is an emerging Alzheimer’s disease biomarker that holds promise for early diagnosis (Sperling et al., 2011; Badhwar et al., 2017). To analyse resting state functional MRI, select regions in canonical brain networks previously established in the literature are generally considered (B). An individual resting state functional MRI connectivity map can be generated for different networks, with the default mode, limbic, and salience networks being the key components affected by Alzheimer’s disease (Badhwar et al., 2017) (B). Structural and functional brain maps can enter a subtyping procedure, which identifies groups of individuals with homogeneous brain maps (C).The number of subtypes are defined a priori or through various metrics for model selection (Seghier, 2018), for example n = 3 in C. A subtype map is generated by averaging the maps within each subgroup and subtracting the grand average (i.e. demeaned) to emphasize the features of the subtype. Chi square statistics are applied to identify groups that include a greater number of Alzheimer’s disease patients than expected by chance (illustrated by a ‘*AD’ annotation for subtype 2 in C). (D) The subtyping procedure was applied on maps of grey matter density from cognitively normal and Alzheimer’s disease dementia individuals in the ADNI database (n = 377). Four of seven subtypes were identified as Alzheimer’s disease dementia-related (results adapted from Tam et al., 2019). Three subtypes were consistent with previous reports: posterior (or temporo-occipito-parietal-predominant), diffuse, and temporal (or medial temporal-predominant) atrophy subtypes. A novel language atrophy subtype was also identified. (E) The subtype procedure was applied to resting state functional MRI data collected on cognitively normal, MCI, and Alzheimer’s disease dementia individuals in a dataset pooling ADNI2 with several independent samples (n = 130). Three subtypes were extracted for three resting state networks known to be impacted by Alzheimer’s disease: default mode, salience, and limbic (Badhwar et al., 2017). One Alzheimer’s disease dementia/MCI-related subtype was found for each network. The salience and default mode followed similar patterns: increased within-network connectivity, and a lower (negative) connectivity between networks. The limbic subtypes showed lower connectivity with frontal regions, and increased connectivity with occipital regions. Results adapted from Orban et al. (2017b). The section on ‘Brain subtypes’ compares results from the abovementioned and other studies with similar approaches and objectives. Supplementary Table 1 provides detailed characteristics of the 12 neuroimaging subtyping studies (structural MRI and resting state functional MRI) that met our search criteria. BOLD = blood oxygenation level-dependent signal.
The choice of the number of subtypes is, to some degree, arbitrary. Two studies showed that their three subtypes could be decomposed into six (Noh et al., 2014), or more (Tam et al., 2019) homogeneous groups. Finally, an additional study looking at heterogeneity with a linear mixture model, instead of a discrete cluster analysis, showed that most individuals tend to express varying levels of multiple subtypes (Zhang et al., 2016). Continuous measures of subtype similarity are thus more advisable than discrete assignment (Zhang et al., 2016; Tam et al., 2019).
We now highlight various associations between Alzheimer’s disease markers/risk factors and the three atrophy subtypes consistently reported. In three studies, Alzheimer’s disease dementia patients with the posterior subtype were reported to be the youngest, and had the earliest age at onset (Noh et al., 2014; Hwang et al., 2016; Park et al., 2017). They also demonstrated greater PET-detectable amyloidosis (Hwang et al., 2016), and pathological levels of CSF amyloid-β42 and tau (Noh et al., 2014; Varol et al., 2017; ten Kate et al., 2018). Differences across subtypes were reported with fluorodeoxyglucose-PET-detectable glucose hypometabolism (Hwang et al., 2016), and white matter hyperintensities (ten Kate et al., 2018). Subtype-specific associations with Alzheimer’s disease-related genes were also observed, specifically, apolipoprotein E (APOE) (Noh et al., 2014; Varol et al., 2017), CD2AP (CD2-associated protein) (Varol et al., 2017), SPON1 (Spondin-1) (Varol et al., 2017), LOC390956 or PPIAP59 (peptidyl-prolyl cis-trans isomerase A pseudogene) (Varol et al., 2017), though the association with APOE was not consistently found (Hwang et al., 2016). Associations of subtypes with cognition were observed for global (Varol et al., 2017; Tam et al., 2019) and domain-specific (e.g. episodic memory) (Noh et al., 2014; Park et al., 2017; Poulakis et al., 2018; ten Kate et al., 2018) measures, but not by all studies (Hwang et al., 2016). Associations between subtypes and sex were found to be significant in two (Noh et al., 2014; Varol et al., 2017) of four (Noh et al., 2014; Hwang et al., 2016; Varol et al., 2017; Tam et al., 2019) studies.
Functional subtypes
By coupling cluster analysis and resting state functional MRI, a preprint report by Orban et al. (2017b) investigated connectivity subtypes in cognitively normal, MCI, and Alzheimer’s disease dementia patients (Supplementary Table 1). They noted associations between functional connectivity subtypes and cognitive symptoms in the default mode, limbic, and salience networks in MCI, and Alzheimer’s disease dementia patients (Fig. 1E). Limbic subtypes were also associated with Alzheimer’s disease biomarkers (CSF amyloid-β42 levels, APOE4 genotype) in an independent cohort at increased risk for familial Alzheimer’s disease, suggesting that functional connectivity subtypes may be sensitive to the presence and progression of preclinical disease (Orban et al., 2017b).
Summary
Our review found convergent evidence of distinct brain atrophy subtypes in Alzheimer’s disease dementia patients, including at least three data-driven atrophy subtypes: diffuse, temporal, and posterior. These structural subtypes seem to associate with established biomarkers, risk factors, and clinical symptoms of Alzheimer’s disease, as well as cognitive subtypes: e.g. temporal subtype with memory impairment, and diffuse subtype with impaired executive function (Zhang et al., 2016). While Alzheimer’s disease associated resting state functional subtypes in the default mode, limbic, and salience networks were only reported by one study (Orban et al., 2017b), the finding is in line with a recent meta-analysis (Badhwar et al., 2017) reporting consistent alterations in connectivity in the same three networks in patients with Alzheimer’s disease dementia, MCI, or in both groups. The picture emerging from functional MRI data is one of aberrant between-network connectivity initiating in the mesolimbic network at the preclinical stage and propagating to the salience and default mode network with Alzheimer’s disease progression (Orban et al., 2017b).
Metabolomics panels
Metabolite panels
We reviewed six Alzheimer’s disease studies that constructed metabolite panels from top discriminant metabolites in biofluids: two plasma, one serum, two saliva, and one CSF (Supplementary Table 2). Platforms used for metabolomics analysis, performed alone or in combination, were as follows: ultra-performance liquid chromatography mass spectrometry (MS) and gas MS (Wang et al., 2014), liquid chromatography MS (Mapstone et al., 2017), faster ultra-performance liquid chromatography MS (Liang et al., 2015, 2016), liquid chromatography-tandem (also with solid phase extraction) and gas MS (Czech et al., 2012), and nuclear magnetic resonance spectroscopy (Figueira et al., 2016) (Supplementary Table 2).
Using plasma metabolome data, Wang et al. (2014) constructed a six-metabolite panel to discriminate Alzheimer’s disease dementia from cognitively normal, and a five-metabolite panel to discriminate amnestic MCI from cognitively normal. Arachidonic acid, N,N-dimethylglycine, and thymine were present in both panels. Association of these three panel metabolites with lipid, amino acid, and nucleic acid metabolism, respectively, suggested specific metabolic deregulations early in Alzheimer’s disease, resulting largely in increased inflammation and oxidative stress. Arachidonic acid (a polyunsaturated fatty acid) is a known modulator of neuroinflammation (Wang et al., 2014), while perturbations in N,N-dimethylglycine, and thymine levels can lead to oxidative DNA damage (Wang et al., 2014). Mapstone et al. (2017) used a 12 plasma metabolite panel to discriminate the following cohorts from cognitively normal: older adults with superior memory; amnestic MCI + Alzheimer’s disease dementia patients; and participants who converted to amnestic MCI or Alzheimer’s disease dementia in ∼2 years. Similar to Wang et al. (2014), several panel metabolites were associated with lipid or amino acid metabolism. Panel metabolites were also found to be constituents of pathways regulating oxidative stress, inflammation, and nitric oxide bioavailability. Using serum metabolome data, Liang et al. (2016) identified a panel of two upregulated lipid metabolites (7-ketocholesterol, sphinganine-1-phosphate) that discriminated MCI from Alzheimer’s disease dementia. 7-Ketocholesterol, a major oxidation product of cholesterol, has been reported to increase with Alzheimer’s disease development, with potential role in the inflammatory responses (Testa et al., 2016), and amyloid-β formation/accumulation and induced neurotoxicity (Phan et al., 2013). Sphinganine-1-phosphate is an intermediate of sphingolipid metabolism, and sphingolipids have also been suggested to modulate the inflammation-associated pathogenesis of Alzheimer’s disease, amyloid-β levels, and oxidative stress-driven neuronal apoptosis (Jazvinšćak Jembrek et al., 2015; Mizuno et al., 2016; Lin et al., 2017). Sphinganine-1-phosphate was also present in a three-metabolite panel constructed from the salivary metabolome, and discriminated patients with Alzheimer’s disease dementia from cognitively normal individuals (Liang et al., 2015). The other two panel members, ornithine and phenyllactic acid, were amino acid metabolites (Ogata et al., 1999) with links to the same oxidative stress pathway reported by Mapstone et al. (2017). A separate study using saliva reported a seven-metabolite panel that discriminated pre-dementia (i.e. 5 years prior to dementia onset) from cognitively normal (Figueira et al., 2016). Of these seven metabolites, the inflammatory modulator histamine was also included as a panel metabolite by Mapstone et al. (2017), as mentioned before. Overall, the metabolites included in the panel were associated with amino acid, lipid, or energy metabolism. Czech et al. (2012) assessed multiple combinations of 16 CSF metabolites to discriminate Alzheimer’s disease dementia from cognitively normal. Highest discrimination was obtained with a five metabolite panel consisting of amino acids and cortisol (Czech et al., 2012). Our focused review of Alzheimer’s disease-associated metabolite panels highlight that the majority of discriminant molecules detected in biofluids are involved in amino acid, lipid, or nucleic acid metabolism (Fig. 2B).
Metabolomics pathways and networks
We reviewed five Alzheimer’s disease studies that followed-up non-targeted metabolomics research in biofluids with pathway or network analyses: two plasma, one plasma plus CSF, and two serum (Supplementary Table 2). Platforms used for metabolomics analysis were as follows: ultra-performance liquid chromatography-tandem MS (de Leeuw et al., 2017), MS (Graham et al., 2015), liquid chromatography MS (Trushina et al., 2013), gas MS (González-Domínguez et al., 2015), and ultra-performance liquid chromatography and gas MS (Orešič et al., 2011) (Supplementary Table 2).
In plasma, de Leeuw et al. (2017) identified 26 metabolites composed of mainly amino acids and lipids with significantly altered levels in Alzheimer’s disease dementia patients. Network analyses suggested a shift in Alzheimer’s disease towards amine and oxidative stress compounds, known to cause imbalances in neurotransmitter production, amyloid-β generation, inflammation, and neurovascular health. Perturbations in amino acid metabolism (interlinked polyamine and l-arginine pathways) were also demonstrated in the plasma metabolome of MCI to Alzheimer’s disease dementia converters (Graham et al., 2015). Changes in polyamine and l-arginine metabolism have been linked to neurotoxicity, and deregulations in genesis and/or death of neural cells and neurotransmitter production (Graham et al., 2015). Other metabolic pathways notably impacted were prostaglandin, glucose and cholesterol (Graham et al., 2015). As inflammation promoting cyclo-oxygenase enzymes are involved in prostaglandin synthesis, altered prostaglandin biosynthesis in converters suggests an underlying inflammatory response (Graham et al., 2015). Deregulated glucose metabolism in converters may be due to brain insulin resistance and microvascular disease (Graham et al., 2015). Moreover, perturbations in cholesterol metabolism have been linked to amyloid-β deposition and tau hyperphosphorylation (Gamba et al., 2012; Graham et al., 2015). Cholesterol metabolism (specifically cholesterol and sphingolipids transport) was also found to be abnormal in both plasma and CSF from patients with Alzheimer’s disease dementia (Trushina et al., 2013). In serum, metabolism of amino acids dominated the top pathways altered in patients with Alzheimer’s disease dementia in one study (González-Domínguez et al., 2015), a finding in line with plasma metabolome data (de Leeuw et al., 2017). A second study in serum reported a three-metabolite panel predictive of progression from MCI to Alzheimer’s disease dementia (within 27 ± 18 months), with major contribution from upregulated 2,4-dihydroxybutanoic acid, a metabolite potentially overproduced during hypoperfusion-related hypoxia (Orešič et al., 2011). Upregulation of the pentose phosphate pathway in progressors further supported the involvement of secondary hypoxia in Alzheimer’s disease pathogenesis. More glucose is metabolized via the pentose phosphate pathway in the brain under hypoxic conditions.
Summary
Overall, metabolite panels, and metabolomics pathway and network analyses provide the following insights: (i) discriminant Alzheimer’s disease-associated metabolites may be narrowly or broadly interconnected (Wilkins and Trushina, 2017); (ii) metabolomes of different biofluids provide convergent and biofluid-related mechanistic insights into Alzheimer’s disease pathology (Trushina et al., 2013); (iii) genotype-associated (e.g. APOE status) differences in preclinical and clinical groups suggest different routes to Alzheimer’s disease (de Leeuw et al., 2017); and (iv) neurodegenerative disease subtypes can be characterized by metabolomics analyses (de Leeuw et al., 2017).
Genomics-derived polygenic risk scores
Polygenic risk score approach
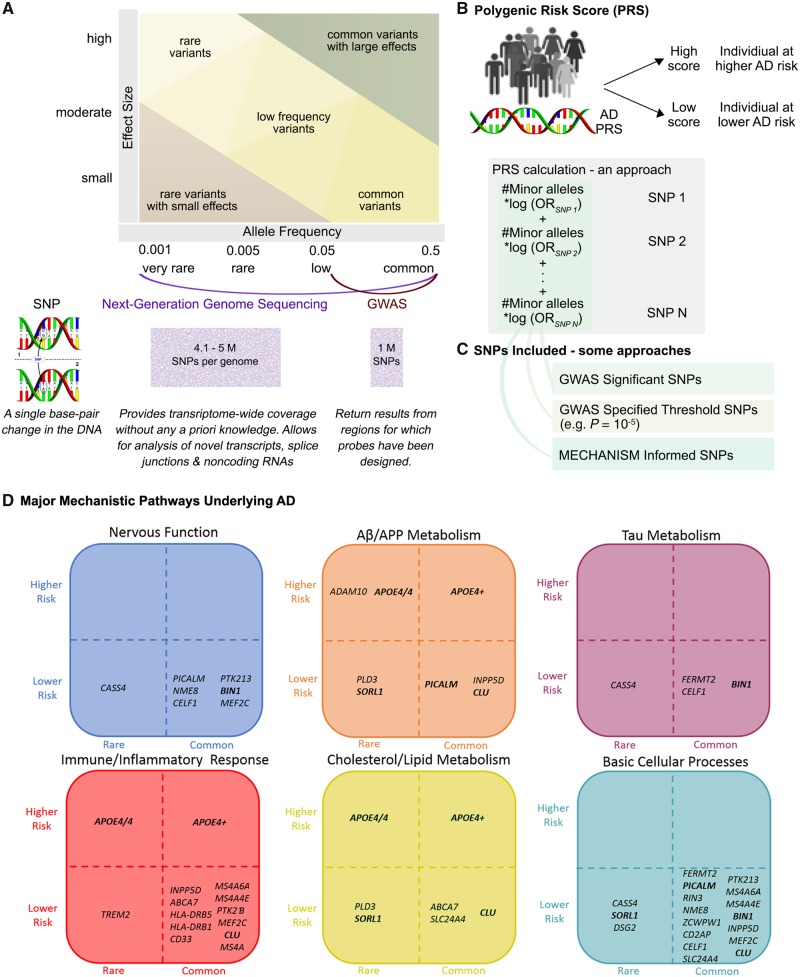
Constructed of multiple single nucleotide polymorphisms (SNPs) that implicate one or more biological mechanisms, PRSs (Fig. 3) are better at discriminating Alzheimer’s disease from cognitively normal than single-gene analysis (Escott-Price et al., 2017b; Torkamani et al., 2018). We reviewed 11 Alzheimer’s disease PRS studies (Supplementary Tables 4 and 5), the majority of which comprised genome-wide association studies (GWAS)-detected SNPs. To identify genetic risk beyond that of APOE alone, several studies assessed PRS with (APOE-PRS) and without (non-APOE-PRS) APOE. Desikan et al. (2017) found that an APOE-PRS associated with age-at-onset of Alzheimer’s disease symptoms, decreased amyloid-β and increased tau in CSF, and increased atrophy, tau, and amyloid-β load in brain. APOE-PRS also associated with plasma inflammatory markers in Alzheimer’s disease patients (Morgan et al., 2017). An APOE-PRS including a rare TREM2 (triggering receptor expressed on myeloid cells 2) variant discriminated Alzheimer’s disease dementia and cognitively normal, with increasing scores associating with decreasing age at onset, and CSF amyloid-β42 (Sleegers et al., 2015). Discriminative power of APOE-PRS was found to improve with diagnostic accuracy, as demonstrated using a pathologically confirmed Alzheimer’s disease cohort (Escott-Price et al., 2017a). In four separate studies, a non-APOE-PRS was reported to discriminate between Alzheimer’s disease dementia and cognitively normal (Xiao et al., 2015), as well as associate with MCI (Adams et al., 2015), increased risk of Alzheimer’s disease dementia (Adams et al., 2015; Chouraki et al., 2016; Tosto et al., 2017), and earlier Alzheimer’s disease onset (Tosto et al., 2017). Inclusion of APOE either resulted in a modest increase in discriminative power (Xiao et al., 2015), stronger clinical or biomarker associations (Adams et al., 2015; Chouraki et al., 2016), or had no additional effect (Tosto et al., 2017). In another non-APOE-PRS study in Alzheimer’s disease patients, PRS scores correlated negatively with CSF amyloid-β42 levels, and positively with temporal cortex amyloid-β pathology, and γ-secretase activity (Martiskainen et al., 2015). Naj et al. (2014) found that although APOE contributed to 3.7% of age at onset variability in Alzheimer’s disease dementia patients, adding a non-APOE-PRS accounted for an additional 2.2%. Overall, Alzheimer’s disease heritability has a large polygenic contribution beyond APOE, which makes PRS approaches pivotal for Alzheimer’s disease risk prediction (Escott-Price et al., 2015).
Figure 3.
Polygenic risk scores. High-throughput genotyping technologies have revolutionized studies in diseases with complex genetics by enabling detection of common genetic variants with low effect sizes, and rarer variants with relatively higher effect sizes (A). In Alzheimer’s disease, the prevalent late-onset variant is genetically complex and demonstrates high heritability (up to 80%) (Gatz et al., 2006), whereas the early-onset familial variant is deterministically driven by single gene mutation(s) in PSEN1 (presenilin 1), PSEN2 (presenilin 2) or APP (amyloid precursor protein) (Guerreiro et al., 2013). The genetics of late-onset Alzheimer’s disease has been predominantly investigated using GWAS. Designed to rapidly scan for statistical links between a set of known SNPs and a phenotype of interest, GWAS can identify common variants with minor allele frequency >5% (Torkamani et al., 2018) (A). Up to 24 key Alzheimer’s disease-risk genes have been identified using GWAS (Supplementary Table 4). The major limitation (and strength) of GWAS is the data-driven, hypothesis-free approach in which multiple genes are identified, though the majority of significant SNPs are (i) located in non-coding or gene-rich areas of the genome making it difficult to identify which gene is being modified by the SNP; and (ii) in high linkage disequilibrium with many SNPs making it difficult to identify which functional variant is responsible for modifying Alzheimer’s disease risk (Karch et al., 2016). Identification of rarer Alzheimer’s disease-associated SNPs (minor allele frequency >0.5% and <5%), that often escape detection with GWAS, is being enabled by next-generation genome sequencing (NGS) technologies, such as whole-exome sequencing and targeted resequencing of disease-associated genes (Bras et al., 2012; Masellis et al., 2013) (Supplementary Table 4). NGS technologies provide transcriptome-wide coverage without requiring any a priori knowledge of SNPs (A). To date, Alzheimer’s disease prediction using individual, high-throughput genotyping technologies identified, risk genes have been predominantly non-significant, with the exception of APOE, which accounts for up to 30% of the genetic risk (Daw et al., 2000). Therefore, the search for risk genes beyond APOE now include PRS (also referred to as genetic risk scores, risk indexes or scales) approaches (B). A PRS is a calculation (e.g. weighted sum) based on the number of risk alleles carried by an individual, where the risk alleles and their weights are defined by GWAS-detected loci and their measured effects (Torkamani et al., 2018). In the most common scenario, only SNPs reaching conventional GWAS significance (P < 5 × 10−8) are included (C). A threshold lower than the genome-wide statistical significance (e.g. P = 10−5) can also be used to improve or estimate total predictability (Torkamani et al., 2018) (C). SNPs representing multiple hits among Alzheimer’s disease risk genes from one or more major mechanistic pathways can also be included into a PRS (C). Displayed are six main mechanistic clusters, each populated by genetic variants thought to represent the cluster (D). Genetic variants have been placed within the cluster according to population frequency (horizontal axis) and level of estimated risk (vertical axis). For example, an amyloid-β/APP metabolism cluster is made up of rare ADAM10 (a disintegrin and metalloproteinase domain-containing protein 10) and common APOE4+ higher risk genes, and rare PLD3 (phospholipase D family member 3) and common PICALM (phosphatidylinositol binding clathrin assembly protein) lower risk genes. Some genes are involved with multiple mechanisms as can be seen for PICALM’s involvement in nervous function, basic cellular processes, and amyloid-β/APP metabolism. As implied in the figure, when creating PRS, it may be very useful to select genes within mechanistic groups, and select groups depending on the purpose of the research. In sum, PRS reflects a large number of SNPs and a complex set of biological mechanisms related to Alzheimer’s disease pathogenesis, and can improve the precision of early Alzheimer’s disease risk or diagnosis (Desikan et al., 2017; Escott-Price et al., 2017b; Morgan et al., 2017).
Mechanism-based interaction and network approaches
Alzheimer’s disease risk genes can be clustered into functional/mechanistic pathways (Fig. 3D), and the information gained used to improve Alzheimer’s disease discrimination and/or risk prediction (Gaiteri et al., 2016; Hu et al., 2017). We reviewed six mechanism-based Alzheimer’s disease studies (Supplementary Table 5). Functional variants of Alzheimer’s disease GWAS-significant SNPs (e.g. CELF1 or CUGBP Elav-like family member 1) was reported to associate with human brain expression quantitative trait loci, and preferentially expressed in specific cell types (e.g. microglia) (Karch et al., 2016). Rosenthal et al. (2014) highlighted the potential regulatory functions of non-coding Alzheimer’s disease GWAS SNPs. Protein-protein interaction network analyses highlighted that Alzheimer’s disease risk genes, whose protein products interact physically, may be under positive evolutionary selection (e.g. PICALM or phosphatidylinositol binding clathrin assembly protein, BIN1 or bridging integrator 1, CD2AP or CD2 associated protein, EPHA1 or EPH receptor A1) (Raj et al., 2012). Ebbert et al. (2014) reported that while an APOE-PRS did not improve discrimination of Alzheimer’s disease from cognitively normal over APOE, a model allowing for epistatic interactions between SNPs increased discriminative power. Patel et al. (2016) applied a stratified false discovery rate approach, used to increase GWAS power by adjusting significance levels to the amount of overall signal present in data, to identify gene networks and provide links with structural MRI phenotypes: e.g. linking genes involved in transport [e.g. SLC4A10 or (solute carrier family 4 member 10), KCNH7 (potassium voltage-gated channel subfamily H member 7)] with hippocampal volume. Huang et al. (2018) integrated Alzheimer’s disease GWAS genes with human brain-specific gene network using machine learning to identify additional Alzheimer’s disease-risk genes.
Summary
In summary, PRSs may contribute substantially to accounting for the genetic variability that distinguishes Alzheimer’s disease from MCI and cognitively normal groups. They may also be used to probe genetic underpinnings of Alzheimer’s disease subtypes as well as related and disparate NDDs. Thus far, research reporting PRSs in relation to conversion rates of cognitively normal or MCI to Alzheimer’s disease dementia have been mixed (Adams et al., 2015; Lacour et al., 2017), however, early PRS prediction of cognitive trajectories and clinical outcomes have also been reported (Desikan et al., 2017; Sapkota and Dixon, 2018).
Discussion
Complementarity of omics biomarkers
Individuals clinically diagnosed with an NDD of ageing (e.g. Alzheimer’s disease) exhibit varying loads of neurodegenerative markers (e.g. amyloid-β, tau, α-synuclein, brain atrophy, vascular abnormalities) (Beach et al., 2012; Robinson et al., 2018). Single-domain omics biomarkers can, to an extent, characterize this heterogeneity in vivo. Some data-driven brain atrophy subtypes parallel established clinical diagnoses. For example, the posterior atrophy subtype is evocative of the posterior cortical atrophy Alzheimer’s disease variant, and the language atrophy subtype of the logopenic progressive aphasia variant (Ossenkoppele et al., 2015). An active area of research is to determine to what degree the ‘bottom-up’, fully automated and data-driven subtypes match with established ‘top-down’ clinical assessments, which usually start with cognitive symptoms and then incorporate specific neuroimaging characteristics, such as left temporoparietal atrophy in logopenic progressive aphasia (Ossenkoppele et al., 2015). The fact that reviewed studies included participants with typical late-onset Alzheimer’s disease dementia, and not atypical variants such as posterior cortical atrophy, suggest that specific brain atrophy phenotypes comprise a spectrum of involvement that may overlap a clinical label, but not associate uniquely with one. It is unclear how functional connectivity subtypes tie in with atrophy subtypes, although they both associate with clinical diagnoses and biomarkers and risk factors of Alzheimer’s disease (Zhang et al., 2016; Orban et al., 2017b). Although the propagation of functional dysconnectivity parallels the Braak staging of Alzheimer’s disease, the mix of connectivity increases and decreases observed in patients may reflect transient compensatory mechanisms as well as neurodegeneration (Badhwar et al., 2017).
To date, we are unaware of studies that have associated data-driven Alzheimer’s disease brain subtypes with PRS and/or metabolite panels. However, brain subtypes could be targeted and validated with data-driven metabolomics analyses, with genomics analyses likely contributing precision information to these complementary omics approaches. Our review strongly supports such coordinated multiomics approaches. For example, an Alzheimer’s disease PRS composed of genes linked to lipid metabolism and inflammatory response may associate with panels composed of metabolites involved in corresponding pathways. One could further speculate whether the resulting inflammation may cause the diffuse atrophy subtype, and trigger specific functional compensatory mechanisms. Testing these hypotheses will require a cohort that is both deeply phenotyped and captures the entire spectrum of age-related dementias.
Complementarity of analytical approaches
There is potential complementary in the information that can be gathered across different analytical approaches, even within a single omics modality.
Different flavours of subtyping methods were used in the imaging papers we reviewed. Some variants, e.g. hierarchical agglomerative clustering using Ward’s clustering linkage (Noh et al., 2014; Hwang et al., 2016; Tam et al., 2019) and Louvain clustering (Park et al., 2017), are just different algorithms that produce similar types of cluster representations. These algorithms may still differ by their quantitative performance, but this is hard to establish in the absence of ground truth, in a purely data-driven analysis. A more qualitative difference in some techniques is the use of continuous (rather than discrete) subtypes, with multiple subtype factors contributing to explain the brain map of any particular individual, such as Bayesian latent factor analysis (Zhang et al., 2016). Zhang et al. argued that capturing interindividual variations as a continuous spectrum is important, and that there are no clearly separated, discrete biological clusters. We note that even using traditional, discrete, clustering techniques, it is possible to compute continuous indices of similarity between a subtype map and a given individual, and some groups that applied discrete cluster analyses actually performed statistical analyses using such continuous proximity measures (Tam et al., 2019). Another important analytical variation is to use clinical labels to build discriminant subtypes between clinical cohorts, such as CHIMERA clustering (Dong et al., 2016, 2017). Such an approach will by construction generate subtypes that associate more strongly with clinical variables than unsupervised subtypes based solely on the similarity of brain maps across all subjects, including controls. It can be noted, however, that some studies apply a second stage analysis to combine multiple unsupervised subtype map into a single multivariate signature with high clinical predictive value (Tam et al., 2019). So the distinction between unsupervised versus supervised subtype generation may not be as fundamental as it superficially appears.
Regarding metabolomics studies, five of six of the studies in the section ‘Metabolite panels’ implemented an orthogonal projection to latent structures-discriminant analysis (OPLS-DA) (Supplementary Table 2). This technique shares similarities with some of the subtyping approaches used in imaging. First, OPLS-DA generates continuous loadings for each factor across subjects, which means that metabolomic data from a particular individual are a combination of multiple latent variables, rather than being associated with one discrete subtype. Second, this technique uses the clinical labels (typically three clinical groups, cognitively normal, MCI and Alzheimer’s disease dementia) in its decomposition, attempting to separate clinically relevant factors from (orthogonal) sources of within-group variance. Metabolomic studies typically proceed by further selecting a small panel of metabolites and assess its predictive power, and this panel can be further linked to larger metabolomic pathways, parallel to network analysis in genetics (refer to the ‘Metabolomics pathways and networks’ section). Emerging analytics in the metabolomic field are thus mixing ideas applied separately in the imaging connectomics and genomics (discussed below) sections of our review.
Regarding the genomics section, the majority of the studies reviewed used PRS derived from omics-based GWAS using both data-driven and hypothesis-driven approaches (Supplementary Tables 4 and 5). These PRSs can be constructed in several ways: (i) all significant SNPs identified by GWAS; (ii) all nominally associated variants based on a particular significance threshold; (iii) combinations of mechanism-related SNPs; and (iv) a variety of interaction analyses. Identifying Alzheimer’s disease-associated markers (i.e. genotype-phenotype models) has been an important first step, but, like any single omics approach, GWAS analyses do not directly consider the multigenic and multifactorial mechanisms underlying Alzheimer’s disease. Other limitations are low effect size of risk variants and the possible heterogeneity of disease risk variants across populations, with the implication that some rare risk alleles may be under-identified. There is also a need to investigate gene-gene or gene-environment interactions further (Ertekin-Taner, 2011; Bras et al., 2012). Notably, recent machine learning technologies are moving towards analyses of multiple genes, multiple phenotypes, and interactions between them (Gaiteri et al., 2016). Genetic interaction models include epistasis (synergistic or dys-synergistic gene-gene interactions), edgetic (genetic variations of protein-protein interactions), and directed network analysis such as biologically constrained multiscale models that provide information about the genetic variants associated with molecular networks and brain networks. Six studies testing network or multiscale models were reviewed (Supplementary Table 5).
Reproducibility of subtypes, panels and polygenic risk score
An important methodological consideration is the reproducibility of subtypes, panels and PRSs. A rigorous evaluation of reproducibility would consist of a series of experiments deriving these subtypes, panels and PRSs in either two independent group samples, or the same sample with different methods. We are not aware of studies conducting such systematic reproducibility experiments. Within the scope of this review, we could only qualitatively comment on the convergence across studies, but it was not possible for us to quantify this convergence (in part because of the limited number of studies) or to identify the factors that influence reproducibility, such as methodological variability, variations due to small sample size, or biological heterogeneity across different recruitment strategies. As these multivariate approaches get more mature and possibly get translated to clinical practice, such systematic reproducibility analyses will be an important area of future work.
Validity of predictive power
Another critical methodological consideration in this review is validation of the predictive value of a biomarker. In the machine learning community, the best practice is to estimate the parameters of a predictive model on a training set, and then evaluate the performance of the trained model on data that were not touched during training, called the test set. This principle should in theory guarantee an unbiased estimate of model performance, yet in practice many caveats exist and call for cautious interpretation of some published results. A model with many degrees of freedom can achieve very high accuracy on any training set, but this performance will not generalize to the test set, a phenomenon called overfitting. For a given model complexity, it is however easier to over-fit a small sample size than a large one. If a research group does try many models on the test set and only reports results for the best model, this also leads to overfitting. A recent review of functional MRI biomarkers demonstrated a strong trend across many brain disorders (including Alzheimer’s disease), where studies with small sample size tend to report markedly higher accuracy scores than studies with large sample sizes (Fig. 4 in Varoquaux, 2018). This strongly suggests that over-fitting in small samples is pervasive in the neuroimaging literature. However, this is not particular to neuroimaging: some of the metabolomics paper reviewed lacked out of sample validation (Wang et al., 2014; Liang et al., 2015) and reported very high area under the curve (AUC) (>0.99), while papers implementing out of sample cross-validation reported more modest effect sizes (Figueira et al., 2016). It is also in general true that accuracy estimates with small sample sizes (n < 100) have a very wide confidence interval, which means that prediction scores reported in studies with a small sample size should be interpreted with caution as the true performance may be very different from the values reported (Fig. 1 in Varoquaux, 2018). Both neuroimaging subtypes and metabolomics panels are relatively young, emerging technologies, and some papers reviewed have a relatively small sample size per clinical group, e.g. ∼50 (Wang et al., 2014; Figueira et al., 2016). Genomics, by contrast, is a much more mature field, and some studies reviewed tested predictive powers across very large samples (Desikan et al., 2017).
Another important consideration is that many types of generalization and test datasets can be implemented, with radically different interpretations. One can test generalization on a different group of subjects, but with data collected using similar methods to the training set and at the same location. This is an important validation step, but remains only a proof-of-concept. Only by testing data collected at different institutions as well as different locations throughout the world, and possibly different data acquisition protocols, can the true predictive performance expected in a ‘real world’ clinical setting be assessed. Almost none of the imaging or metabolomic studies implemented such large-scale generalization experiments.
A roadmap for parsing heterogeneity in neurodegeneration
A data-driven characterization of heterogeneity across the NDDs of ageing will require cohorts representative of the spectrum of neurodegeneration.
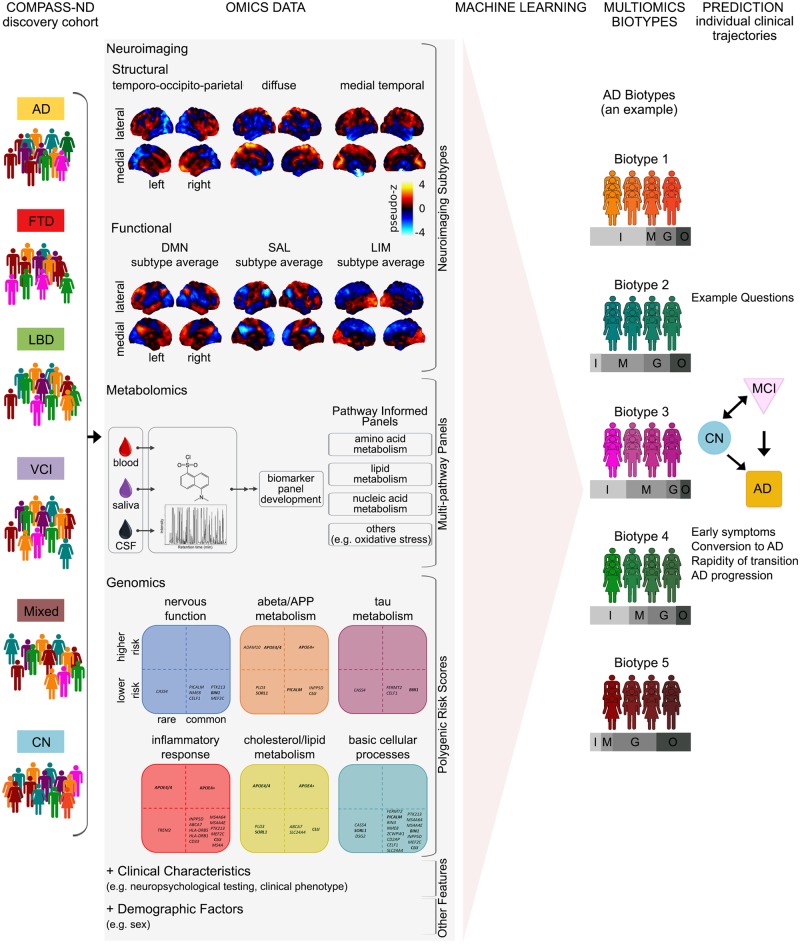
The cohort assembled by the Canadian Consortium on Neurodegeneration in Aging (CCNA, http://ccna-ccnv.ca/) provides a new opportunity to study the full spectrum of age-related dementia. By 2020, the cohort will include 2310 individuals (aged 50–90) featuring the following cognitive conditions: Alzheimer’s disease, vascular, Lewy body, Parkinson’s, frontotemporal, and mixed aetiology dementias, as well as subjective cognitive impairment, MCI, vascular MCI, and cognitively normal (Fig. 4, COMPASS-ND column). The cohort composition ensures that age-related dementias are more or less equally represented, even for less prevalent dementia types (e.g. frontotemporal dementia). Participants will be deeply phenotyped with extensive clinical, neuropsychological, neuroimaging, biospecimen, and neuropathological assessments.
Figure 4.
Proposed roadmap to discovering multiomics Alzheimer’s disease biomarkers. COMPASS-ND: The COMPASS-ND cohort is composed of individuals with various types of dementia or cognitive complaints, as well as healthy, cognitively normal individuals. Omics data: Performing dimension reduction for omics data. Featured as examples are some of the results of our review of the Alzheimer’s disease literature as presented in the paper. Machine learning, Multiomics biotypes and Prediction: These panels demonstrate how signatures of neurodegeneration derived from the integration of multiomics data using machine learning techniques will better identify individuals on an Alzheimer’s disease spectrum trajectory. While our proposed roadmap addresses multiomics biomarkers for Alzheimer’s disease, a similar approach can be used for other neurodegenerative diseases of ageing. AD = Alzheimer's disease; CN = cognitively normal; DMN = default mode network; FTD = frontotemporal dementia; G = genomics features; I = imaging features; LBD = Lewy body disease; LIM = limbic network; M = metabolic features; Mixed = mixed aetiology dementia; O = demographic features; SAL = salience network; SCI = subjective cognitive impairment; VCI = vascular cognitive impairment.
In Fig. 4 we present a roadmap for a multiomics approach to heterogeneity in NDD. We begin with a heterogeneous clinical cohort design (Fig. 4, COMPASS-ND column) that enables the discovery of subgroups sharing a common signature across multiple omics domains (biotypes) that are highly predictive of the clinical status and evolution of individual patients. Multiomics biotypes will be complemented by other important variables such as sex, presence of amyloid-β and tau deposits, and vascular abnormalities. Machine learning tools will be applied to identify an optimal combination of different biotypes and explanatory variables that either discriminate different clinical cohorts, or are predictive of future progression of specific symptoms (Fig. 4, Machine learning column).
We have three complementary lines of reasoning for including a heterogeneous clinical cohort design (i.e. diverse dementia aetiologies) in our proposed roadmap. First, if we were to just consider the data-driven multiomics biotypes generated using an Alzheimer’s disease population (Fig. 4, Multiomics biotypes column), having access to diverse dementia aetiologies will allow us to evaluate the uniqueness of each biotype to Alzheimer’s disease. Second, as heterogeneity is a feature of Alzheimer’s disease as well as other NDDs of ageing (Robinson et al., 2018), our proposed roadmap can be applied to generate multiomics biotypes in other dementia aetiologies. Similar to our review, this will initially require identification of disease-specific indicators from single omic modalities (Fig. 4, Omics data column). We envision that for any given NDD of ageing, multiomics biotypes identified will range from pure (but rare) disease-specific biotypes, to biotypes featuring mixed pathologies. Having access to multiomics biotypes from the spectrum of NDDs of ageing will allow better delineation of biotypes, namely, those that are unique to a specific NDD of ageing, and those that show overlap with other NDDs of ageing. Third, recent work has shown that by training machine learning models on large and heterogeneous data it is possible to generalize better to new studies relying on different methodologies and run on slightly different populations (Abraham et al., 2017; Orban et al., 2017a). Such generalizability is critical for a successful translation in clinical practice.
Towards highly predictive multiomics signatures for prognosis
Because of the emergent nature of the three omics techniques (neuroimaging-based subtypes, metabolite panels, and polygenic risk score), our review was largely composed of proof-of-concept cross-sectional comparisons of cognitively normal older adults with individuals classified with prodromal or diagnosed Alzheimer’s disease, as opposed to the preclinical population. However, the publication of the A/T/N criteria (Jack et al., 2016), along with increasing availability of CSF and PET biomarker data (e.g. amyloid, tau), and longitudinal cohorts provide fertile grounds for additional (and much warranted) research addressing heterogeneity in preclinical and/or at-risk cohorts. Specifically, longitudinal trajectory studies with data-driven neuroimaging subtypes differentially transitioning from cognitively normal to preclinical to prodromal or dementia stages of Alzheimer’s disease are needed. In these designs, metabolomics and genomics (PRS or APOE) would probably serve as a variable to add precision to a biomarkers-based prognosis.
Current biomarkers of Alzheimer’s disease dementia demonstrate limited predictive power for prognosis in the prodromal phase (Rathore et al., 2017). The best models include a combination of cognitive, structural MRI, fluorodeoxyglucose-PET, and/or amyloid-PET measures (Rathore et al., 2017). A substantial proportion of patients identified as progressors, even by the best model, will remain stable over time. Multiomics signatures will hopefully improve the precision of early prognosis. They will also capture a range of information, ranging from brain networks targeted by the disease, metabolic abnormalities in specific pathways, and distinct genetic backgrounds. The multiomics signature may thus also help elucidate the specific pathophysiological pathways involved, and help refine the A/T/N model. Overall, multiomics biomarkers have the potential to reshape clinical diagnosis, and define new ‘bottom-up’ cohorts based on markers of underlying pathologies to design and evaluate drugs. Based on this focused but substantial review, we recommend that additional multiomics analyses be performed.
Supplementary Material
Acknowledgements
We would like to thank Dr Sridar Narayanan for helpful comments on the manuscript.
Funding
This review was performed by members of the Biomarkers Team of the Canadian Consortium on Neurodegeneration in Aging (CCNA; Team Leads: R.A.D. and P.B.) which is funded by the Canadian Institutes of Health Research (CIHR) and partners. Additional sources of support are as follows. A.B. is currently supported by a CIHR Postdoctoral Fellowship (funding reference number #152548) and the Courtois Foundation. At the start of the project A.B. was supported by the Alzheimer Society of Canada Postdoctoral Fellowship. P.B. is a Research Scholar from the Fonds de recherche du Québec, and is also supported by the Courtois Foundation. S.S. is supported by the Alzheimer Society of Canada Postdoctoral Fellowship. R.A.D. and G.P.M. are also supported by the U.S. National Institutes of Health (National Institute on Aging, R01 AG008235). S.D. is a Research Scholar from the Fonds de recherche du Québec – Santé [grant number 30801] and also receives funding from the Consortium d’identification précoce de la maladie d’Alzheimer – Québec is financed through the Fonds de recherche du Québec – Santé / Pfizer Canada Innovation Fund (grant number 25262).
Competing interests
The authors report no competing interests.
Glossary
Abbreviations
- ADNI =
Alzheimer’s Disease Neuroimaging Initiative
- GWAS =
genome-wide association studies
- MCI =
mild cognitive impairment
- NDD =
neurodegenerative disease
- PRS =
polygenic risk score
- SNP =
single nucleotide polymorphism
References
- Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B, et al. Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example. Neuroimage 2017; 147: 736–45. [DOI] [PubMed] [Google Scholar]
- Adams HHH, de Bruijn RFAG, Hofman A, Uitterlinden AG, van Duijn CM, Vernooij MW, et al. Genetic risk of neurodegenerative diseases is associated with mild cognitive impairment and conversion to dementia. Alzheimers Dement 2015; 11: 1277–85. [DOI] [PubMed] [Google Scholar]
- Anstey KJ, Eramudugolla R, Hosking DE, Lautenschlager NT, Dixon RA. Bridging the translation gap: from Dementia risk assessment to advice on risk reduction. J Prev Alzheimers Dis 2015; 2: 189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badhwar A, Tam A, Dansereau C, Orban P, Hoffstaedter F, Bellec P. Resting-state network dysfunction in Alzheimer’s disease: a systematic review and meta-analysis. Alzheimers Dement 2017; 8: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach TG, Monsell SE, Phillips LE, Kukull W. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005-2010. J Neuropathol Exp Neurol 2012; 71: 266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Arvanitakis Z, Kelly JF, Aggarwal NT, Shah RC, et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology 2006; 66: 1837–44. [DOI] [PubMed] [Google Scholar]
- Bras J, Guerreiro R, Hardy J. Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nat Rev Neurosci 2012; 13: 453–64. [DOI] [PubMed] [Google Scholar]
- Chouraki V, Reitz C, Maury F, Bis JC, Bellenguez C, Yu L, et al. Evaluation of a genetic risk score to improve risk prediction for Alzheimer’s disease. J Alzheimers Dis 2016; 53: 921–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech C, Berndt P, Busch K, Schmitz O, Wiemer J, Most V, et al. Metabolite profiling of Alzheimer’s disease cerebrospinal fluid. PLoS One 2012; 7: e31501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, et al. The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet 2000; 66: 196–204.10631151 [Google Scholar]
- Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples LA, et al. Genetic assessment of age-associated Alzheimer disease risk: development and validation of a polygenic hazard score. PLoS Med 2017; 14: e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw FA, Peeters CFW, Kester MI, Harms AC, Struys EA, Hankemeier T, et al. Blood-based metabolic signatures in Alzheimer’s disease. Alzheimers Dement 2017; 8: 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A, Honnorat N, Gaonkar B, Davatzikos C. CHIMERA: Clustering of heterogeneous disease effects via distribution matching of imaging patterns. IEEE Trans Med Imaging 2016; 35: 612–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A, Toledo JB, Honnorat N, Doshi J, Varol E, Sotiras A, et al. Heterogeneity of neuroanatomical patterns in prodromal Alzheimer’s disease: links to cognition, progression and biomarkers. Brain 2017; 140: 735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumurgier J, Hanseeuw BJ, Hatling FB, Judge KA, Schultz AP, Chhatwal JP, et al. Alzheimer’s disease biomarkers and future decline in cognitive normal older adults. J Alzheimers Dis 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbert MTW, Ridge PG, Wilson AR, Sharp AR, Bailey M, Norton MC, et al. Population-based analysis of Alzheimer’s disease risk alleles implicates genetic interactions. Biol Psychiatry 2014; 75: 732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertekin-Taner N. Gene expression endophenotypes: a novel approach for gene discovery in Alzheimer’s disease. Mol Neurodegener 2011; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escott-Price V, Myers AJ, Huentelman M, Hardy J. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann Neurol 2017a; 82: 311–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escott-Price V, Shoai M, Pither R, Williams J, Hardy J. Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiol Aging 2017b; 49: 214.e7–e11. [DOI] [PubMed] [Google Scholar]
- Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 2015; 138: 3673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira J, Jonsson P, Nordin Adolfsson A, Adolfsson R, Nyberg L, Öhman A. NMR analysis of the human saliva metabolome distinguishes dementia patients from matched controls. Mol Biosyst 2016; 12: 2562–71. [DOI] [PubMed] [Google Scholar]
- Franzmeier N, Duering M, Weiner M, Dichgans M, Ewers M, Alzheimer’s Disease Neuroimaging Initiative (ADNI). Left frontal cortex connectivity underlies cognitive reserve in prodromal Alzheimer disease. Neurology 2017; 88: 1054–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Mostafavi S, Honey CJ, De Jager PL, Bennett DA. Genetic variants in Alzheimer disease-molecular and brain network approaches. Nat Rev Neurol 2016; 12: 413–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba P, Testa G, Sottero B, Gargiulo S, Poli G, Leonarduzzi G. The link between altered cholesterol metabolism and Alzheimer’s disease. Ann N Y Acad Sci 2012; 1259: 54–64. [DOI] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 2006; 63: 168–74. [DOI] [PubMed] [Google Scholar]
- González-Domínguez R, García-Barrera T, Gómez-Ariza JL. Metabolite profiling for the identification of altered metabolic pathways in Alzheimer’s disease. J Pharm Biomed Anal 2015; 107: 75–81. [DOI] [PubMed] [Google Scholar]
- Graham SF, Chevallier OP, Elliott CT, Hölscher C, Johnston J, McGuinness B, et al. Untargeted metabolomic analysis of human plasma indicates differentially affected polyamine and L-arginine metabolism in mild cognitive impairment subjects converting to Alzheimer’s disease. PLoS One 2015; 10: e0119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Brás J, Hardy J. SnapShot: Genetics of Alzheimer’s Disease. Cell 2013; 155: 968–968.e1. [DOI] [PubMed] [Google Scholar]
- Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol 2017; 18: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu H, Li X, Guan L, Li J, Tellier LCAM, et al. Revealing Alzheimer’s disease genes spectrum in the whole-genome by machine learning. BMC Neurol 2018; 18: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan T, Tran T, Zheng J, Sapkota S, MacDonald SW, Camicioli R, et al. Metabolomics analyses of saliva detect novel biomarkers of Alzheimer’s disease. J Alzheimers Dis 2018; 65: 1401–16. [DOI] [PubMed] [Google Scholar]
- Hu Y-S, Xin J, Hu Y, Zhang L, Wang J. Analyzing the genes related to Alzheimer’s disease via a network and pathway-based approach. Alzheimers Res Ther 2017; 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Kim CM, Jeon S, Lee JM, Hong YJ, Roh JH, et al. Prediction of Alzheimer’s disease pathophysiology based on cortical thickness patterns. Alzheimers Dement 2016; 2: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA Research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement 2018; 14: 535–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Feldman HH, Frisoni GB, et al. A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology 2016; 87: 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust W. Vulnerable neural systems and the borderland of brain aging and neurodegeneration. Neuron 2013; 77: 219–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazvinšćak Jembrek M, Hof PR, Šimić G. Ceramides in Alzheimer’s disease: key mediators of neuronal apoptosis induced by oxidative stress and Aβ accumulation. Oxid Med Cell Longev 2015; 2015: 346783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Rozen S, Matson W, Han X, Hulette CM, Burke JR, et al. Metabolomic changes in autopsy-confirmed Alzheimer’s disease. Alzheimers Dement 2011; 7: 309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Ezerskiy LA, Bertelsen SAlzheimer’s Disease Genetics Consortium (ADGC)Goate AM. Alzheimer’s disease risk polymorphisms regulate gene expression in the ZCWPW1 and the CELF1 Loci. PLoS One 2016; 11: e0148717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacour A, Espinosa A, Louwersheimer E, Heilmann S, Hernández I, Wolfsgruber S, et al. Genome-wide significant risk factors for Alzheimer’s disease: role in progression to dementia due to Alzheimer's disease among subjects with mild cognitive impairment. Mol Psychiatry 2017; 22: 153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Liu H, Zhang T, Jiang Y, Xing H, Zhang A-H. Metabolomics-based screening of salivary biomarkers for early diagnosis of Alzheimer’s disease. RSC Adv 2015; 5: 96074–9. [Google Scholar]
- Liang Q, Liu H, Zhang T, Jiang Y, Xing H, Zhang A-H. Discovery of serum metabolites for diagnosis of progression of mild cognitive impairment to Alzheimer’s disease using an optimized metabolomics method. RSC Adv 2016; 6: 3586–91. [Google Scholar]
- Lin W, Zhang J, Liu Y, Wu R, Yang H, Hu X, et al. Studies on diagnostic biomarkers and therapeutic mechanism of Alzheimer’s disease through metabolomics and hippocampal proteomics. Eur J Pharm Sci 2017; 105: 119–26. [DOI] [PubMed] [Google Scholar]
- Malpas CB. Structural neuroimaging correlates of cognitive status in older adults: a person-oriented approach. J Clin Neurosci 2016; 30: 77–82. [DOI] [PubMed] [Google Scholar]
- Mapstone M, Lin F, Nalls MA, Cheema AK, Singleton AB, Fiandaca MS, et al. What success can teach us about failure: the plasma metabolome of older adults with superior memory and lessons for Alzheimer’s disease. Neurobiol Aging 2017; 51: 148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiskainen H, Helisalmi S, Viswanathan J, Kurki M, Hall A, Herukka S-K, et al. Effects of Alzheimer’s disease-associated risk loci on cerebrospinal fluid biomarkers and disease progression: a polygenic risk score approach. J Alzheimers Dis 2015; 43: 565–73. [DOI] [PubMed] [Google Scholar]
- Masellis M, Sherborn K, Neto PR, Sadovnick DA, Hsiung G-YR, Black SE, et al. Early-onset dementias: diagnostic and etiological considerations. Alzheimers Res Ther 2013; 5: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S, Ogishima S, Kitatani K, Kikuchi M, Tanaka H, Yaegashi N, et al. Network analysis of a comprehensive knowledge repository reveals a dual role for ceramide in Alzheimer’s disease. PLoS One 2016; 11: e0148431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AR, Touchard S, O’Hagan C, Sims R, Majounie E, Escott-Price V, et al. The correlation between inflammatory biomarkers and polygenic risk score in Alzheimer’s disease. J Alzheimers Dis 2017; 56: 25–36. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Reitz C, Kunkle BW, Perry W, Park YS, et al. Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol 2014; 71: 1394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettiksimmons J, DeCarli C, Landau S, Beckett L, Alzheimer’s Disease Neuroimaging Initiative. Biological heterogeneity in ADNI amnestic mild cognitive impairment. Alzheimers Dement 2014; 10: 511–21.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh Y, Jeon S, Lee JM, Seo SW, Kim GH, Cho H, et al. Anatomical heterogeneity of Alzheimer disease: based on cortical thickness on MRIs. Neurology 2014; 83: 1936–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 1999; 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanrewaju O, Clare L, Barnes L, Brayne C. A multimodal approach to dementia prevention: A report from the Cambridge Institute of Public Health. Alzheimers Dement 2015; 1: 151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban P, Dansereau C, Desbois L, Mongeau-Pérusse V, Giguère C-É, Nguyen H, et al. Multisite generalizability of schizophrenia diagnosis classification based on functional brain connectivity. Schizophr Res 2017a; 192: 167–71. [DOI] [PubMed] [Google Scholar]
- Orban P, Tam A, Urchs S, Savard M, Madjar C, Badhwar A, et al. Subtypes of functional brain connectivity as early markers of neurodegeneration in Alzheimer’s disease. bioRxiv 2017b:195164. [Google Scholar]
- Orešič M, Hyötyläinen T, Herukka S-K, Sysi-Aho M, Mattila I, Seppänan-Laakso T, et al. Metabolome in progression to Alzheimer’s disease. Transl Psychiatry 2011; 1: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenkoppele R, Cohn-Sheehy BI, La Joie R, Vogel JW, Möller C, Lehmann M, et al. Atrophy patterns in early clinical stages across distinct phenotypes of Alzheimer’s disease. Hum Brain Mapp 2015; 36: 4421–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-Y, Na HK, Kim S, Kim H,, Kim HJ, Seo SW, et al. Robust identification of Alzheimer’s disease subtypes based on cortical atrophy patterns. Sci Rep 2017; 7: 43270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Park MTMAlzheimer’s Disease Neuroimaging InitiativeChakravarty MM, Knight J. Gene prioritization for imaging genetics studies using gene ontology and a stratified false discovery rate approach. Front Neuroinform 2016; 10: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HTT, Hata T, Morita M, Yoda T, Hamada T, Vestergaard MC, et al. The effect of oxysterols on the interaction of Alzheimer’s amyloid beta with model membranes. Biochim Biophys Acta 2013; 1828: 2487–95. [DOI] [PubMed] [Google Scholar]
- Poulakis K, Pereira JB, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, et al. Heterogeneous patterns of brain atrophy in Alzheimer’s disease. Neurobiol Aging 2018; 65: 98–108. [DOI] [PubMed] [Google Scholar]
- Raj T, Shulman JM, Keenan BT, Chibnik LB, Evans DA, Bennett DA, et al. Alzheimer disease susceptibility loci: evidence for a protein network under natural selection. Am J Hum Genet 2012; 90: 720–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore S, Habes M, Iftikhar MA, Shacklett A, Davatzikos C. A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages. Neuroimage 2017; 155: 530–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Lee EB, Xie SX, Rennert L, Suh E,, Bredenberg C, et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain 2018; 141: 2181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal SL, Barmada MM, Wang X, Demirci FY, Kamboh MI. Connecting the dots: potential of data integration to identify regulatory SNPs in late-onset Alzheimer’s disease GWAS findings. PLoS One 2014; 9: e95152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota S, Dixon RA. A network of genetic effects on non-demented cognitive aging: Alzheimer’s genetic risk (CLU + CR1 + PICALM) intensifies cognitive aging genetic risk (COMT + BDNF) selectively for APOEɛ4 carriers. J Alzheimers Dis 2018; 62: 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens NME, Galindo-Garre F, Pijnenburg YAL, van der Vlies AE, Smits LL, Koene T, et al. The identification of cognitive subtypes in Alzheimer’s disease dementia using latent class analysis. J Neurol Neurosurg Psychiatry 2016; 87: 235–43. [DOI] [PubMed] [Google Scholar]
- Seghier ML. Clustering of fMRI data: the elusive optimal number of clusters. PeerJ 2018; 6: e5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K, Bettens K, De Roeck A, Van Cauwenberghe C, Cuyvers E, Verheijen J, et al. A 22-single nucleotide polymorphism Alzheimer’s disease risk score correlates with family history, onset age, and cerebrospinal fluid Aβ42. Alzheimers Dement 2015; 11: 1452–60. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011; 7: 280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam A, Dansereau C, Iturria-Medina Y, Urchs S, Orban P, Sharmarke H, et al. A highly predictive signature of cognition and brain atrophy for progression to Alzheimer’s dementia. Gigascience 2019; 8 Available from: 10.1093/gigascience/giz055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Kate M, Dicks E, Visser PJ, van der Flier WM, Teunissen CE, Barkhof F, et al. Atrophy subtypes in prodromal Alzheimer’s disease are associated with cognitive decline. Brain 2018; 141: 3443–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa G, Staurenghi E, Zerbinati C, Gargiulo S, Iuliano L, Giaccone G, et al. Changes in brain oxysterols at different stages of Alzheimer’s disease: Their involvement in neuroinflammation. Redox Biol 2016; 10: 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet 2018; 19: 581–90. [DOI] [PubMed] [Google Scholar]
- Tosto G, Bird TD, Tsuang D, Bennett DA, Boeve BF, Cruchaga C, et al. Polygenic risk scores in familial Alzheimer disease. Neurology 2017; 88: 1180–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushina E, Dutta T, Persson X-MT, Mielke MM, Petersen RC. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS One 2013; 8: e63644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol E, Sotiras A, Davatzikos C, Alzheimer’s Disease Neuroimaging Initiative. HYDRA: revealing heterogeneity of imaging and genetic patterns through a multiple max-margin discriminative analysis framework. Neuroimage 2017; 145: 346–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux G. Cross-validation failure: Small sample sizes lead to large error bars. Neuroimage 2018; 180: 68–77. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhou Y, Huang F-J, Tang H-D, Xu X-H, Liu J-J, et al. Plasma metabolite profiles of Alzheimer’s disease and mild cognitive impairment. J Proteome Res 2014; 13: 2649–58. [DOI] [PubMed] [Google Scholar]
- Wilkins JM, Trushina E. Application of metabolomics in Alzheimer’s disease. Front Neurol 2017; 8: 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Liu Z-J, Tao S, Sun Y-M, Jiang D, Li H-L, et al. Risk prediction for sporadic Alzheimer’s disease using genetic risk score in the Han Chinese population. Oncotarget 2015; 6: 36955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Mormino EC, Sun N, Sperling RA, Sabuncu MR, Yeo BTT, et al. Bayesian model reveals latent atrophy factors with dissociable cognitive trajectories in Alzheimer’s disease. Proc Natl Acad Sci U S A 2016; 113: E6535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.