Abstract
Phototropism represents a simple physiological mechanism—differential growth across the growing organ of a plant—to respond to gradients of light and maximize photosynthetic light capture (in aerial tissues) and water/nutrient acquisition (in roots). The phototropin blue light receptors, phot1 and phot2, have been identified as the essential sensors for phototropism. Additionally, several downstream signal/response components have been identified, including the phot-interacting proteins NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3) and PHYTOCHROME SUBSTRATE 4 (PKS4). While the structural and photochemical properties of the phots are quite well understood, much less is known about how the phots signal through downstream regulators. Recent advances have, however, provided some intriguing clues. It appears that inactive receptor phot1 is found dispersed in a monomeric form at the plasma membrane in darkness. Upon light absorption dimerizes and clusters in sterol-rich microdomains where it is signal active. Additional studies showed that the phot-regulated phosphorylation status of both NPH3 and PKS4 is linked to phototropic responsiveness. While PKS4 can function as both a positive (in low light) and a negative (in high light) regulator of phototropism, NPH3 appears to function solely as a key positive regulator. Ultimately, it is the subcellular localization of NPH3 that appears crucial, an aspect regulated by its phosphorylation status. While phot1 activation promotes dephosphorylation of NPH3 and its movement from the plasma membrane to cytoplasmic foci, phot2 appears to modulate relocalization back to the plasma membrane. Together these findings are beginning to illuminate the complex biochemical and cellular events, involved in adaptively modifying phototropic responsiveness under a wide varying range of light conditions.
Keywords: NPH3, phosphorylation, phot1, phot2, phototropin, phototropism
Recent advances in the biochemical and cellular mechanisms of phototropin-dependent signaling are discussed.
Introduction
Lacking locomotion to seek out food and safety, plants instead rely on a variety of responses that allow them to fulfill these needs and adapt to changes in their environment (Huang, 2006; Mizutani and Kanaoka, 2018). Responses to alterations in the local light environment include phototropism, changes in leaf orientation, and chloroplast movements (de Witt et al., 2016; Fiorucci and Fankhauser, 2017). Each of the aforementioned responses requires perception of blue light (BL) by a class of photoreceptors known as the phototropins (phots) (Galvao and Fankhauser, 2015; de Witt et al., 2016). Though gene copy number can vary depending upon plant species, there are two phot isoforms, phot1 and phot2; both associate primarily with the plasma membrane (Fig. 1A), and can exhibit shared and unique functions (Morrow et al., 2018). Bimodular in structure, both phots contain a pair of LOV (light, oxygen, and voltage) domains that form an N-terminal sensory region, and a protein kinase domain (PKD) of the AGCVII subfamily of protein kinases as a C-terminal output domain (Morrow et al., 2018). One of the key molecules involved in the transduction of phot signals is NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3) (Liscum and Briggs, 1995, 1996; Motchoulski and Liscum, 1999; Inada et al., 2004; Inoue et al., 2008a; de Carbonnel et al., 2010; Harada et al., 2013), a founding member of the plant-specific NRL (NPH3/RPT2-Like) family of proteins (Pedmale et al., 2010; Liscum et al., 2014; Christie et al., 2018). NPH3 and NRL family members are generally trimodular in structure (Pedmale et al., 2010), with an N-terminal BTB (broad complex, tramtrack, and bric-a-brac) protein–protein interaction domain (Stogios et al., 2005), a central ‘NPH3 domain’ (PFam, PF0300) of conserved amino acid sequence across the NRL family, and a C-terminal coiled-coil (CC) protein–protein interaction domain (Motchoulski and Liscum, 1999). The CC and BTB domains of NPH3 have been shown to mediate direct interaction with phot1 (Fig. 1A) and CULLIN3 (CUL3), respectively (Motchoulski and Liscum, 1999; Roberts et al., 2011). These protein interactions facilitate ubiquitination (both mono- and polyubiquitination) of phot1 by a CULLIN3-Ring E3 ubiquitin ligase complex, CRL3NPH3, in which NPH3 functions as a substrate adaptor (Roberts et al., 2011). The phot signaling capacity of NPH3 appears to be linked to its phosphorylation status, which is itself modulated by light condition and phot activity (Motchoulski and Liscum, 1999; Pedmale and Liscum, 2007;Tsuchita-Mayama et al., 2008; Haga et al., 2015). Here we will discuss recent studies (see Box 1) that are beginning to address some of the most pressing questions, such as: what proteins are substrates for the phot PKD and how do these proteins influence phot signaling; how do post-translational modifications of phots and NPH3 alter signaling; and how does light-dependent re-localization of NPH3 influence signaling?
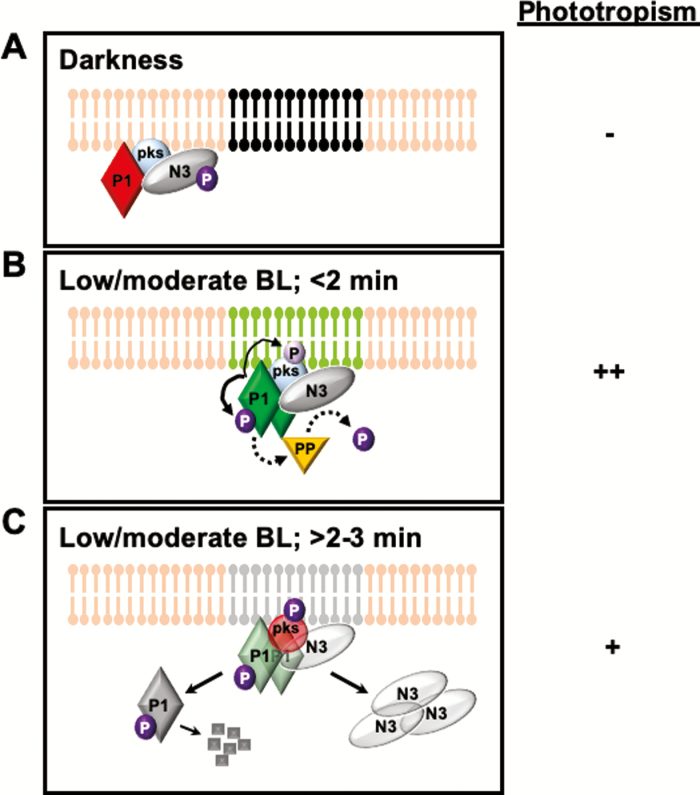
Fig. 1.
Model for phot1-associated early events in phototropic responsiveness under low to moderate light in etiolated seedlings. The plasma membrane lipid bilayer is shown in pink, black, light green, and gray; where the black and green areas designate sterol-rich microdomains that are phot inactive and active, respectively. Gray areas of the membrane are presumed to represent phot-active regions. Solid arrows represent known and characterized events, whereas dashed lines represent experimentally inferred events without a characterized mechanism. The relative phototropic responsiveness is given to the right for each of the light conditions: –, no or very little response; +, weak response; ++, moderate to strong response. (A) The basal phot1 signaling complex in etiolated seedlings kept in darkness. The complex is found dispersed on the inner face of the plasma membrane outside of sterol-rich microdomains. Phot1 (P1) is inactive (solid red); PKS4 (pks) is unphosphorylated and inactive; NPH3 (N3; solid gray) is fully phosphorylated (deep purple P) and presumed inactive. (B) The phot1 complex in etiolated seedlings exposed to low or moderate BL for a short duration (<2 min). Phot1 is activated (green) and moves into clusters within sterol-rich regions of the membrane (presumably with its interacting proteins), where it dimerizes and trans-autophosphorylates (purple P) and phosphorylates PKS4. Low levels of PKS4 phosphorylation (light purple P) are associated with positive regulation of phototropism. Phot1 activation also leads to the activation of an as yet identified protein phosphatase (PP) that dephosphorylates NPH3. (C) The phot1 complex in etiolated seedlings that received low or moderate BL continuously for a moderate time (>2–3 min). While some phot1 is retained in an active state (transparent green) at the plasma membrane, and can continue to phosphorylate PKS4 (represented by the deep purple P), some phot1 (gray, and presumed inactive) is translocated to the cytoplasm. Some of the cytoplasmic phot1 is degraded via a 26S proteasome-dependent process (Roberts et al., 2011). Enhanced phosphorylation of PKS4 results in the conversion of this molecule to a repressor of phot1-dependent phototropism (pks in red with attached deep purple P). Dephosphorylated NPH3 (transparent gray) translocates from the plasma membrane to cytoplasmic aggregates. Each of these events results in down-regulation of phot1 signaling and response desensitization. While the phot1 complex is depicted as being localized to a sterol-rich region of the plasma membrane in (C), this is color-coded gray because it has not been experimentally determined.
Box 1. Key developments in understanding the phototropin-dependent signaling leading to phototropism.
PKS4 phosphorylation status regulates phot1 signaling strength
Schumacker et al. (2018) showed that phot1-dependent phosphorylation of PKS4 leads to a negative feedback regulation of phot1-induced phototropism. Under low blue light (BL), PKS4 exists largely in an unphosphorylated state and functions as a positive regulator of phot1-dependent phototropism. As the BL intensity increases, phot1 directly phosphorylates more and more PKS4, promoting a shift in PKS4 action from positive to negative regulator. The greater the proportion of PKS4 phosphorylated, the more phototropism is diminished.
A gatekeeper thiophosphorylation method to identify phototropin kinase substrates
Schnabel et al. (2018) have developed mutant phototropins, called phot-Cerberus, in which the PKD is mutated to allow N6-benzyl-APγãS to be utilized as an ATP source for subsequent thiophosphorylation of phot substrates. Both phot1- and phot2-Cerberus can thio-autophosphorylate in a BL-dependent fashion; and both can thiophosphorylate BLUS1, a known in planta substrate of the phototropins. Moreover, phot1-Cerberus can utilize endogenous ATP and functionally complement the aphototropic phenotype of a phot1phot2 double mutant. This represents a powerful tool for substrate discovery, in vitro and in vivo.
Sterol-rich plasma membrane microdomains as the site of phot1 activation
Xue et al. (2018) have utilized single particle FRET-FLIM/VA-TIRFM microscopic analysis to characterize the mono-/dimeric state, and intra-plasma membrane dynamics, of phot1 in response to BL exposure. It was found that phot1 exists predominantly as dispersed monomers at the inner surface of the plasma membrane in darkness, but rapidly dimerizes and forms aggregate clusters with sterol-rich microdomains. It appears that these clusters are the site of BL-induced trans-autophosphorylation, rather than the phosphorylation being a driver for intracellular movement. It is presumed that these sterol-rich phot1-containing microdomains are the sites of initial signal transduction/integration.
An NPH3 phosphorylation and localization ‘rheostat’ that regulates phot1-dependent phototropism
Sullivan et al. (2019) demonstrated that enhanced phototropic responsiveness observed in de-etiolated seedlings results from retention of phosphorylated NPH3 at the plasma membrane. In etiolated seedlings, and dark-adapted de-etiolated seedlings, NPH3 is found predominantly in its phosphorylated form at the inner surface of the plasma membrane, where it interacts with phot1. In response to directional BL, NPH3 is rapidly dephosphorylated and moves to the cytoplasm where it forms aggregates. De-etiolation (the shift from heterotrophy to autotrophy) results in a much higher proportion of phosphorylated NPH3 being present and retained at the plasma membrane, even after exposure to directional BL, and is positively correlated with enhanced phototropic responsiveness in de-etiolated seedlings. Treatment of etiolated seedlings with the protein phosphatase inhibitor OKA also reduces BL-induced dephosphorylation of NPH3 and its movement to the cytoplasm, suggesting that the phosphorylation status of NPH3 is a key determinant of phot1-dependent phototropic responsiveness.
High BL-induced phototropism requires phot2-dependent relocation of NPH3
Zhao et al. (2018) identified NPH3 as a crucial component of the phot2-regulated phototropism and demonstrated that phot2, like phot1, modulates the localization of NPH3. Unlike phot1, which promotes the dephosphorylation of NPH3 and its subsequent movement to cytoplasmic foci in response to BL, phot2 appears to modulate the relocalization of cytoplasmic NPH3 to the plasma membrane in response to high BL. RPT2, an NPH3 paralog, whose expression is light induced also appears to serve a similar function, though the mechanism is probably distinct. Differing regulation of the localization of NPH3, a critical phototropic signaling component, by the phototropins provides a dynamic means for adaptation and acclimation under varying light conditions.
Regulation of phot signaling via phot-dependent phosphorylation
It has been well established that BL-activated trans-autophosphorylation of phots (Fig. 1B) is critical for signaling function (Inoue et al., 2008b, 2011; Tseng and Briggs, 2010; Peterson et al., 2017). Yet much less is known about the downstream targets of phots (Christie et al., 2011; Demarsy et al., 2012; Takemiya et al., 2013; Hiyama et al., 2017), or how phosphorylation of such proteins regulates phot-dependent processes. One such target protein is PHYTOCHROME KINASE SUBSTRATE 4 (PKS4). It was previously determined that PKS4 acts as a positive regulator of phototropism under low light conditions (Lariguet et al., 2006; Demarsy et al., 2012; Kami et al., 2014; Fig. 1B), though phosphorylation of PKS4 is not essential for phototropic responsiveness (Demarsy et al., 2012). Interestingly, it was found that PKS4 phosphorylation may play a role in a negative feedback control, though the mechanism remained unknown (Demasry et al., 2012). Schumacher et al. (2018) have now identified a potential mechanism (see Box 1).
By mutating the critical residue (Ser299) in PKS4 that is phosphorylated by phot1, to either prevent phosphorylation (PKS4-S299A) or generate a constitutive phosphorylation mimic (PKS4-S299D), Schumacher et al. (2018) found that the phosphorylation status of PKS4 acts as a ‘rheostat’ to mediate phototropic responsiveness under varying light intensities (Figs 1 and 2). Specifically, they found that under low light conditions, the PKS4-S299A mutant lines displayed phototropic curvatures that were temporally and quantitatively similar to those of the wild type and transgenic pks4 mutants expressing a PKS4-WT protein (Schumacher et al., 2018). However, in comparison with the response under low light, phototropism is both delayed and repressed under high light conditions in the wild type and pks4 PKS4-WT transgenics, but neither of these alterations is observed in the PKS4-S299A mutants, suggesting that phosphorylated PKS4 may act as an inhibitor of the phototropism (Schumacher et al., 2018). Consistent with this conclusion, the authors found that PKS4-S299D mutants exhibited slower and dampened phototropic responses under low light conditions, much like those observed in wild-type plants exposed to high light (Schumacher et al., 2018). Thus, as the light intensity increases, so does the amount of phosphorylated PKS4, resulting in a progressively slower and weaker phototropic response. Schumacher et al. (2018) hypothesize that this sequence of events allows the plant to adapt appropriately to ever-changing light conditions in the natural diurnal environment. For instance, when a plant experiences a gradient of directional light but is in non-photosynthetically limiting light conditions, there would be no advantage to expending energy to exhibit a phototropic response. By examining the phototropic responses under conditions where plants are exposed to the same relative directional gradient of light but given at different light intensities, it was indeed found that wild-type seedlings exhibited ever decreasing phototropic curvature with increasing intensity (Schumacker et al., 2018). Strikingly, this response is dramatically diminished in the PKS4-S299A mutant (Schumacker et al., 2018).
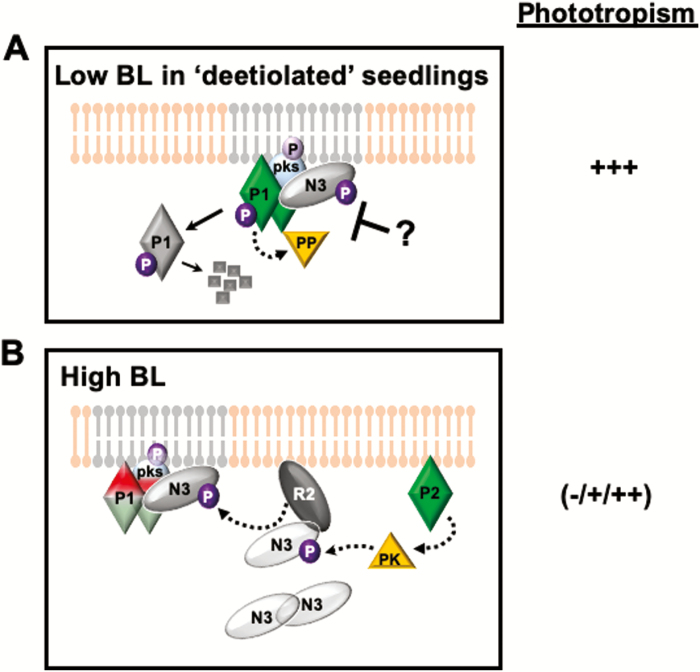
Fig. 2.
Model for phot1-associated early events in phototropic responsiveness in de-etiolated seedlings (A) or etiolated seedlings exposed to high intensity light for extended periods. The plasma membrane lipid bilayer is shown in pink and gray; where the gray areas designate presumed sterol-rich microdomains that are phot active. Solid arrows represent known and characterized events, whereas dashed lines represent experimentally inferred events without a characterized mechanism. The relative phototropic responsiveness is given to the right for each of the light conditions: –, no or very little response; +, weak response; +++, strong enhanced response. (A) The phot1 complex in de-etiolated seedlings exposed to low to moderate BL. De-etiolation results in the persistence of phosphophorylated NPH3 and its retention at the plasma membrane. Though the mechanism(s) for this is currently unknown, it would appear to influence either the protein phosphatase that dephosphorylates NPH3 in response to light, or the protein kinase that phosphorylates it. Phot1 is shown as an active (green) dimer associated with a presumed sterol-rich plasma membrane microdomain, and as a presumed inactive (green) cytoplasmic monomer which can be degraded by a 26S proteasome (Roberts et al., 2011). PKS4 is presumed to be in its active form as the seedlings are highly phototropic. (B) The phot1 complex in etiolated seedlings exposed to high BL. Short-term exposure to high BL results in PKS4-dependent (red pks; deep purple P) suppression of phot1 (red) activity and thus weak phototropism. Exposure to high BL for extended periods (>30 min) results in the expression and activity of phot2 (P2) and RPT2 (R2), both of which stimulate the relocalization of NPH3 from the cytoplasm to the plasma membrane. Though the mechanism by which RPT2 does this is unknown, NPH3 relocalization does require phosphorylation (deep purple P) by an unknown protein kinase (PK) that appears to be activated in response to phot2 activity. Because phototropism is stronger in extended versus short-term high BL, but similar to that observed in etiolated seedlings exposed to low/moderate BL for short periods, it is presumed that both PKS4 (light blue pks; light purple P) and phot1 (light green) activity are moderate. Phot2 and RPT2 relocalization of NPH3 in etiolated plants results in resensitization of phot1 signaling that allows for adaptive responses in high light conditions.
Despite advances like that just discussed, the dearth of identified phot kinase substrates severely limits our understanding of phot-dependent signal transduction. To date, just five substrates have been reported: the phots themselves (Deng et al., 2014; Inoue et al., 2008b, 2011; Sullivan et al., 2008); PKS4 (Demarsy et al., 2012); BLUS1 (Takemiya et al., 2013); and CBC1 (Hiyama et al., 2017). In a recent report, Schnabel et al. (2018) described the development of a method to identify potential phot kinase substrates via thiophosphorylation (see Box 1). In brief, the authors generated mutant ‘gatekeeper’ (Emrick et al., 2006; Allen et al., 2007) versions of the phots by substituting a glycine for a conserved threonine within the β5 strand in the N lobe of the kinase domain (Kornev and Taylor, 2015), thus allowing the accommodation of a bulky ATP analog, N6-benzyl-ATPγS (Schnabel et al., 2018). The phot gatekeeper mutants, called Cerberus for the ‘keeper’ of the underworld in Greek mythology, were shown to undergo trans-auto-thiophosphorylation in vitro (Schnabel et al., 2018). Moreover, both phot1- and phot2-Cerberus were found to thiophosphorylate BLUS1 (Schnabel et al., 2018), a known phot1 target (Takemiya et al., 2013). Interestingly, phot1-Cerberus expressed transgenically in Arabidopsis could still utilize native ATP and was capable of functionally complementing both loss of phototropism and leaf flattening in a phot1phot2 double mutant (Schnabel et al., 2018). This chemical genetic approach therefore represents a powerful tool to identify potential direct substrates of phot kinases, a potential game-changer indeed.
Activation of phot1 in membrane microdomains
In darkness, phots associate with the cytoplasmic face of the plasma membrane (Fig. 1A) via the C-terminal portion of the PKD through an as yet unknown mechanism (Kong et al., 2007, 2013a, b). Upon BL absorption, phots are autophosphorylated (Fig. 1B), probably in trans through BL-induced dimerization of the receptor (Kaiserli et al., 2009; Sullivan et al., 2010; Petersen et al., 2017). With increasing exposure time, some portion of phot is internalized to the cytoplasm (Sakamoto and Briggs, 2002; Kong et al., 2006; Han et al., 2008; Wan et al., 2008; Kaiserli et al., 2009; Sullivan et al., 2010; Fig. 1C). Although Preuten et al. (2015) have shown that internalization of phot1 is not necessary for primary signaling, it had also been reported that phot1 exhibits mosaic patterns and forms punctate aggregates at the surface of the membrane in response to BL exposure (Liscum, 2016), leaving open the question of whether dynamic BL-induced phot movements at the membrane might be involved in signal propagation.
Xue et al. (2018) have provided the first steps toward addressing this aforementioned question by utilizing single particle imaging (FRET-FLIM, Li et al., 2013; Wang et al., 2015; VA-TIRFM, Wan et al., 2011) to assess the dynamic movement of phot1–green fluorescent protein (GFP) within the plasma membrane (see Box 1). The authors found that in darkness phot1 is broadly distributed at the inner surface of the membrane as monomeric units (Fig. 1A). Upon BL exposure, phot1 dimerizes and aggregates into clusters on the inner surface of the plasma membrane (Xue et al., 2018; Fig. 1B), consistent with previous observation by spinning-disc confocal microscopy (Liscum, 2016). Moreover, it was found that phot1–GFP co-localizes with AtRem1.3, a marker for sterol-rich lipid environments (Demir et al., 2013), indicating that these phot1-containing foci are found within sterol-rich microdomains (Xue et al., 2018; Fig. 1B). Interestingly, while BL-induced dimerization appears to be essential for movement of phot1 to the sterol-rich microdomains, trans-autophosphorylation is not (Xue et al., 2018). Rather, trans-autophosphorylation appears to occur within the membrane microdomains containing dimeric phot1 (Xue et al., 2018; Fig. 1B). Findings that sterol depletion compromises phototropic responsiveness (Xue et al., 2018), together with previous results indicating that phot1 autophosphorylation is prerequisite for phototropism (Inoue et al., 2008b), strongly suggest that phot1 signaling occurs from these sterol-rich foci (Fig. 1B).
An NPH3 rheostat: phosphorylation status and localization regulate phot1-dependent phototropism
As discussed earlier, NPH3 is a phot1-interacting protein that is part of a CRL3NPH3 ubiquitin ligase complex necessary for phototropism (Liscum and Briggs, 1995, 1996; Motchoulski and Liscum, 1999; Inada et al., 2004; Roberts et al., 2011). Though reversible phosphorylation of NPH3 has been shown to be important for phototropic responsiveness (Motchoulski and Liscum, 1999; Pedmale and Liscum, 2007; Tsuchita-Mayama et al., 2008; Haga et al., 2015), the mechanistic impact of this post-translational modification has remained unknown. A recent study has, however, begun to shed some light on this open question (Sullivan et al., 2019; see Box 1).
Previous studies have shown that phototropism is enhanced in de-etiolated as compared with etiolated seedlings of several species (Hart and Macdonald, 1981; Gordon et al., 1982; Ellis, 1987), including Arabidopsis (Jin et al., 2001). Sullivan et al. (2019) found that a reduction in dephosphorylation of NPH3, as well as its retention at the plasma membrane, underpins this phytochrome- and cryptochrome-dependent enhancement of phototropism (Liscum et al., 2014) in de-etiolated Arabidopsis seedlings. In etiolated (or dark-adapted) seedlings, NPH3 exists predominantly in a phosphorylated state, and associates with the plasma membrane (Motchoulski and Liscum, 1999; Haga et al., 2015; Fig. 1A). In response to BL exposure, the dephosphorylated isoform accumulates (Motchoulski and Liscum, 1999; Pedmale and Liscum, 2007; Fig. 1B) and moves from the plasma membrane to form cytoplasmic aggregates in a phot1-dependent fashion (Haga et al., 2015; Fig. 1C). Surprisingly, given these previous findings, Sullivan et al. (2019) found that the phosphorylated form of NPH3 also predominates in de-etiolated seedlings. More surprising still, unlike what occurs in etiolated seedlings, much less NPH3 is dephosphorylated in de-etiolated seedlings exposed to unilateral BL (Sullivan et al., 2019). Interestingly, the higher levels of phosphorylated NPH3 in de-etiolated as compared with etiolated seedlings exposed to low to moderate intensity phototropic stimuli (Fig. 1B) also resulted in higher retention of NPH3 at the plasma membrane (Sullivan et al., 2019) where it interacts with phot1 (Lariguet et al., 2006; Roberts et al., 2011; Haga et al., 2015). Based on these observations, Sullivan et al. (2019) hypothesize that it is the proportion of phosphorylated NPH3 interacting with phot1 at the plasma membrane that determines the phototropic responsiveness.
NPH3 as a regulator of high light-induced, phot2-dependent phototropism
Under conditions where phot1 function is redundant to phot2, such as during the induction of phototropism by high intensity BL (Sakai et al., 2001; Inada et al., 2004), it can be difficult to isolate the phot2-dependent properties of signaling from those of phot1. Zhao et al. (2018) recently tackled this problem by screening for mutants in a phot1-null background that had impaired phot2-dependent phototropism under high BL. Perhaps not surprisingly, given its apparent essential role for phototropic signaling under a wide range of light intensities (Liscum et al., 2014; Fankhauser and Christie, 2015), one of these mutants turned out to be a new allele of NPH3 (Zhao et al., 2018; see Box 1). The surprises came when NPH3 localization was examined under high BL in various genetic backgrounds (wild type, phot1, phot2, phot1phot2, rpt2, or pks1pks2pks4; Zhao et al., 2018). First, as observed previously for etiolated seedlings exposed to low BL (Haga et al., 2015), NPH3 was found in clusters at the plasma membrane in darkness independent of genotype (Fig. 1A), and was relocalized into cytoplasmic aggregates in wild-type seedlings in a phot1-dependent fashion in response to high BL exposure (Zhao et al., 2018; Fig. 2B). However, high BL-induced cytoplasmic aggregation of NPH3 was found to be enhanced in the phot2 background (Zhao et al., 2018). Secondly, again similarly to what occurs in low BL (Haga et al., 2015), prolonged irradiation with high BL results in the RPT2-dependent relocation of cytoplasmic NPH3 back to the plasma membrane in etiolated wild-type seedlings, but this relocalization did not occur in the phot2 or rpt2 backgrounds (Zhao et al., 2018). Together these findings indicate that phot1 regulates the dissociation of NPH3 from the plasma membrane into cytoplasmic aggregates in high BL as a means of sensory response desensitization (Zhao et al., 2018), as occurs in low BL (Haga et al., 2015) (see Fig. 1C); whereas, both phot2 and RPT2 regulate the relocation of NPH3 from cytosol to the plasma membrane, and thus ‘reconstruction’ of a phototropically active phot1–NPH3 complex, as a means to acclimate to prolonged high BL exposure (Zhao et al., 2018) (see Fig. 2B).
Concluding remarks and future directions
While the recent findings highlighted here are both exciting and enlightening, like any good science they beg more questions. For example, how do phosphorylated isoforms of PKS4 inhibit phot1-dependent phototropism? Is it through a direct interaction with, and modulation of, phot1? Or does it require other yet identified components? Given that PKS proteins are intrinsically disordered proteins (Schumacker et al., 2018), answers to these questions may provide insight into the function of other disordered proteins (Wright and Dyson, 2015) in phot-dependent signaling, such as NPH3 (Sullivan et al., 2019). How does phot1 partitioning to sterol-rich membrane microdomains in response to phototropic stimulation link phot signaling to alterations in auxin transport that ultimately drive differential growth (Morrow et al., 2018; Zhang et al., 2017)? Does PKS4 phosphorylation status influence phot1 partitioning in these microdomains? How does the ‘NPH3 phosphorylation rheostat’ impact this cell biology? What protein phosphatase(s) and protein kinase(s) regulate the phot1- and phot2-induced changes in NPH3 phosphorylation and localization? How does NPH3 phosphorylation state impact its function within the CRL3NPH3 ubiquitin ligase, and how does that alter phot1 ubiquitination and function? Also, how are the PKS4-dependent inhibition of phot1 function and phot2-dependent re-sensitization of phot1 signaling capacity integrated in natural environments to allow for enhanced phototropism in de-etiolated seedlings? These and many other questions remain, but, thanks to the studies discussed here, paths forward have been illuminated.
Acknowledgements
The authors are grateful to the Bond Life Sciences Center and the Interdisciplinary Plant Group for financial support that made this work possible. EL wishes to thank H. Gov-Ari, S. Harris, B. Dickinson, A. Smith, D. Murray, J. Gers, N. McBrain, T. Araya, K. King, J. Hanneman, G. Holt, P. Bostaph, and D. Lombardo for ongoing inspiration during the writing.
References
- Allen JJ, Li M, Brinkworth CS, et al. 2007. A semisynthetic epitope for kinase substrates. Nature Methods 4, 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Suetsugu N, Sullivan S, Wada M. 2018. Shining light on the function of NPH3/RPT2-like proteins in phototropin signaling. Plant Physiology 176, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Yang H, Richter GL, et al. 2011. Phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biology 9, e1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carbonnel M, Davis P, Roelfsema MR, Inoue S, Schepens I, Lariguet P, Geisler M, Shimazaki K, Hangarter R, Fankhauser C. 2010. The Arabidopsis Phytochrome Kinase Substrate2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiology 152, 1391–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarsy E, Schepens I, Okajima K, Hersch M, Bergmann S, Christie J, Shimazaki K, Tokutomi S, Fankhauser C. 2012. Phytochrome Kinase Substrate 4 is phosphorylated by the phototropin 1 photoreceptor. The EMBO Journal 31, 3457–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir F, Horntrich C, Blachutzik JO, et al. 2013. Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proceedings of the National Academy of Sciences, USA 110, 8296–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Oses-Prieto JA, Kutschera U, Tseng TS, Hao L, Burlingame AL, Wang ZY, Briggs WR. 2014. Blue light-induced proteomic changes in etiolated Arabidopsis seedlings. Journal of Proteome Research 13, 2524–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M, Galvão VC, Fankhauser C. 2016. Light-mediated hormonal regulation of plant growth and development. Annual Review of Plant Biology 67, 513–537. [DOI] [PubMed] [Google Scholar]
- Ellis RJ. 1987. Comparison of fluence–response relationships of phototropism in light- and dark-grown buckwheat. Plant Physiology 85, 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrick MA, Lee T, Starkey PJ, Mumby MC, Resing KA, Ahn NG. 2006. The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proceedings of the National Academy of Sciences, USA 103, 18101–18106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Christie JM. 2015. Plant phototropic growth. Current Biology 25, R384–R389. [DOI] [PubMed] [Google Scholar]
- Fiorucci AS, Fankhauser C. 2017. Plant strategies for enhancing access to sunlight. Current Biology 27, R931–R940. [DOI] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C. 2015. Sensing the light environment in plants: photoreceptors and early signaling steps. Current Opinion in Neurobiology 34, 46–53. [DOI] [PubMed] [Google Scholar]
- Gordon DC, Macdonald IR, Hart JW. 1982. Regional growth-patterns in the hypocotyls of etiolated and green cress seedlings in light and darkness. Plant, Cell & Environment 5, 347–353. [Google Scholar]
- Haga K, Tsuchida-Mayama T, Yamada M, Sakai T. 2015. Arabidopsis ROOT PHOTOTROPISM2 contributes to the adaptation to high-intensity light in phototropic responses. The Plant Cell 27, 1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han IS, Tseng TS, Eisinger W, Briggs WR. 2008. Phytochrome A regulates the intracellular distribution of phototropin 1–green fluorescent protein in Arabidopsis thaliana. The Plant Cell 20, 2835–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Takemiya A, Inoue S, Sakai T, Shimazaki K. 2013. Role of RPT2 in leaf positioning and flattening and a possible inhibition of phot2 signaling by phot1. Plant & Cell Physiology 54, 36–47. [DOI] [PubMed] [Google Scholar]
- Hart JW, Macdonald IR. 1981. Phototropi responses of hypocotyls of etiolated and green seedlings. Plant Science 21, 151–158. [Google Scholar]
- Hiyama A, Takemiya A, Munemasa S, Okuma E, Sugiyama N, Tada Y, Murata Y, Shimazaki KI. 2017. Blue light and CO2 signals converge to regulate light-induced stomatal opening. Nature Communications 8, 1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B. ed. 2006. Plant–environment interactions, 3rd edn. Boca Raton, FL: CRC Press, Taylor & Francis Group. [Google Scholar]
- Inada S, Ohgishi M, Mayama T, Okada K, Sakai T. 2004. RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. The Plant Cell 16, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Kinoshita T, Matsumoto M, Nakayama KI, Doi M, Shimazaki K. 2008b Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proceedings of the National Academy of Sciences, USA 105, 5626–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Kinoshita T, Takemiya A, Doi M, Shimazaki K. 2008a Leaf positioning of Arabidopsis in response to blue light. Molecular Plant 1, 15–26. [DOI] [PubMed] [Google Scholar]
- Inoue S, Matsushita T, Tomokiyo Y, Matsumoto M, Nakayama KI, Kinoshita T, Shimazaki K. 2011. Functional analyses of the activation loop of phototropin2 in Arabidopsis. Plant Physiology 156, 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Zhu J, Zeiger E. 2001. The hypocotyl chloroplast plays a role in phototropic bending of Arabidopsis seedlings: developmental and genetic evidence. Journal of Experimental Botany 52, 91–97. [PubMed] [Google Scholar]
- Kaiserli E, Sullivan S, Jones MA, Feeney KA, Christie JM. 2009. Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation and endocytosis by blue light. The Plant Cell 21, 3226–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Allenbach L, Zourelidou M, Ljung K, Schütz F, Isono E, Watahiki MK, Yamamoto KT, Schwechheimer C, Fankhauser C. 2014. Reduced phototropism in pks mutants may be due to altered auxin-regulated gene expression or reduced lateral auxin transport. The Plant Journal 77, 393–403. [DOI] [PubMed] [Google Scholar]
- Kong SG, Kagawa T, Wada M, Nagatani A. 2013a A C-terminal membrane association domain of phototropin 2 is necessary for chloroplast movement. Plant & Cell Physiology 54, 57–68. [DOI] [PubMed] [Google Scholar]
- Kong SG, Kinoshita T, Shimazaki K, Mochizuki N, Suzuki T, Nagatani A. 2007. The C-terminal kinase fragment of Arabidopsis phototropin 2 triggers constitutive phototropin responses. The Plant Journal 51, 862–873. [DOI] [PubMed] [Google Scholar]
- Kong SG, Suetsugu N, Kikuchi S, Nakai M, Nagatani A, Wada M. 2013b Both phototropin 1 and 2 localize on the chloroplast outer membrane with distinct localization activity. Plant & Cell Physiology 54, 80–92. [DOI] [PubMed] [Google Scholar]
- Kong SG, Suzuki T, Tamura K, Mochizuki N, Hara-Nishimura I, Nagatani A. 2006. Blue light-induced association of phototropin 2 with the Golgi apparatus. The Plant Journal 45, 994–1005. [DOI] [PubMed] [Google Scholar]
- Kornev AP, Taylor SS. 2015. Dynamics-driven allostery in protein kinases. Trends in Biochemical Sciences 40, 628–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariguet P, Schepens I, Hodgson D, et al. 2006. PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proceedings of the National Academy of Sciences, USA 103, 10134–10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Luu DT, Maurel C, Lin J. 2013. Probing plasma membrane dynamics at the single-molecule level. Trends in Plant Science 18, 617–624. [DOI] [PubMed] [Google Scholar]
- Liscum E. 2016. Blue light-induced intracellular movement of phototropins: functional relevance or red herring? Frontiers in Plant Science 7, 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Askinosie SK, Leuchtman DL, Morrow J, Willenburg KT, Coats DR. 2014. Phototropism: growing towards an understanding of plant movement. The Plant Cell 26, 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Briggs WR. 1995. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. The Plant Cell 7, 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Briggs WR. 1996. Mutations of Arabidopsis in potential transduction and response components of the phototropic signaling pathway. Plant Physiology 112, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Kanaoka MM. 2018. Environmental sensing and morphological plasticity in plants. Seminars in Cell & Developmental Biology 83, 69–77. [DOI] [PubMed] [Google Scholar]
- Morrow J, Willenburg KT, Liscum E. 2018. Phototropism in land plants: molecules and mechanism from light perception to response. Frontiers in Biology 13, 342–356. [Google Scholar]
- Motchoulski A, Liscum E. 1999. Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science 286, 961–964. [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Celaya RB, Liscum E. 2010. Phototropism: mechanism and outcomes. The Arabidopsis Book 8, e0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Liscum E. 2007. Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. Journal of Biological Chemistry 282, 19992–20001. [DOI] [PubMed] [Google Scholar]
- Peterson J, Inoue S-i, Kelly SM, Sullivan S, Kinoshita T, Christie JM. 2017. Functional characterizatio of a constitutively active kinase variant of Arabidopsis phototropin 1. Journal of Biological Chemistry 292, 13843–13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuten T, Blackwood L, Christie JM, Fankhauser C. 2015. Lipid anchoring of Arabidopsis phototropin 1 to assess the functional significance of receptor internalization: should I stay or should I go? New Phytologist 206, 1038–1050. [DOI] [PubMed] [Google Scholar]
- Roberts D, Pedmale UV, Morrow J, Sachdev S, Lechner E, Tang X, Zheng N, Hannink M, Genschik P, Liscum E. 2011. Modulation of phototropic responsiveness in Arabidopsis through ubiquitination of phototropin 1 by the CUL3-Ring E3 ubiquitin ligase CRL3(NPH3). The Plant Cell 23, 3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. 2001. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proceedings of the National Academy of Sciences, USA 98, 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR. 2002. Cellular and subcellular localization of phototropin 1. The Plant Cell 14, 1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel J, Hombach P, Waksman T, Giuriani G, Petersen J, Christie JM. 2018. A chemical genetic approach to engineer phototropin kinases for substrate labeling. Journal of Biological Chemistry 293, 5613–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher P, Demarsy E, Waridel P, Petrolati LA, Trevisan M, Fankhauser C. 2018. A phosphorylation switch turns a positive regulator of phototropism into an inhibitor of the process. Nature Communications 9, 2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Privé GG. 2005. Sequence and structural analysis of BTB domain proteins. Genome Biology 6, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Kaiserli E, Tseng TS, Christie JM. 2010. Subcellular localization and turnover of Arabidopsis phototropin 1. Plant Signaling & Behavior 5, 184–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Kharshiing E, Laird J, Sakai T, Christie JM. 2019. Deetiolation enhances phototropism by modulating NON-PHOTOTROPIC HYPOCOTYL3 phosphorylation status. Plant Physiology 180, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Thomson CE, Lamont DJ, Jones MA, Christie JM. 2008. In vivo phosphorylation site mapping and functional characterization of Arabidopsis phototropin 1. Molecular Plant 1, 178–194. [DOI] [PubMed] [Google Scholar]
- Takemiya A, Sugiyama N, Fujimoto H, Tsutsumi T, Yamauchi S, Hiyama A, Tada Y, Christie JM, Shimazaki K. 2013. Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nature Communications 4, 2094. [DOI] [PubMed] [Google Scholar]
- Tseng TS, Briggs WR. 2010. The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses. The Plant Cell 22, 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida-Mayama T, Nakano M, Uehara Y, Sano M, Fujisawa N, Okada K, Sakai T. 2008. Mapping of the phosphorylation sites on the phototropic signal transducer, NPH3. Plant Science 174, 626–633. [Google Scholar]
- Wan Y, Ash WM 3rd, Fan L, Hao H, Kim MK, Lin J. 2011. Variable-angle total internal reflection fluorescence microscopy of intact cells of Arabidopsis thaliana. Plant Methods 7, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YL, Eisinger W, Ehrhardt D, Kubitscheck U, Baluska F, Briggs W. 2008. The subcellular localization and blue-light-induced movement of phototropin 1–GFP in etiolated seedlings of Arabidopsis thaliana. Molecular Plant 1, 103–117. [DOI] [PubMed] [Google Scholar]
- Wang X, Li X, Deng X, Luu DT, Maurel C, Lin J. 2015. Single-molecule fluorescence imaging to quantify membrane protein dynamics and oligomerization in living plant cells. Nature Protocols 10, 2054–2063. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. 2015. Intrinsically disordered proteins in cellular signalling and regulation. Nature Reviews. Molecular Cell Biology 16, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Xing J, Wan Y, et al. 2018. Arabidopsis blue light receptor phototropin 1 undergoes blue light-induced activation in membrane microdomains. Molecular Plant 11, 846–859. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yu Q, Jiang N, et al. 2017. Clathrin regulates blue light-triggered lateral auxin distribution and hypocotyl phototropism in Arabidopsis. Plant, Cell & Environment 40, 165–176. [DOI] [PubMed] [Google Scholar]
- Zhao X, Zhao Q, Xu C, Wang J, Zhu J, Shang B, Zhang X. 2018. Phot2-regulated relocation of NPH3 mediates phototropic response to high-intensity blue light in Arabidopsis thaliana. Journal of Integrative Plant Biology 60, 562–577. [DOI] [PubMed] [Google Scholar]