NbCycB2 is specifically expressed in trichomes of Nicotiana benthamiana and inhibits their formation by repressing Nbwo activity via a negative feedback loop.
Keywords: Feedback loop, L1-like box, NbCycB2, Nbwo, Nicotiana benthamiana, trichome formation, woolly motif
Abstract
The transcription factor Woolly (Wo) and its downstream gene CycB2 have been shown to regulate trichome development in tomato (Solanum lycopersicum). It has been demonstrated that only the gain-of-function allele of Slwo (SlWoV, the Slwo woolly motif mutant allele) can increase the trichome density; however, it remains unclear why the two alleles function differently in trichome development. In this study, we used Nicotiana benthamiana as a model and cloned the homologues of Slwo and SlCycB2 (named Nbwo and NbCycB2). We also constructed a Nbwo gain-of-function allele with the same mutation site as SlWoV (named NbWoV). We found that both Nbwo and NbWoV directly regulate NbCycB2 and their own expression by binding to the promoter of NbCycB2 and their own genomic sequences. As form of a feedback regulation, NbCycB2 negatively regulates trichome formation by repressing Nbwo activity at the protein level. We also found that mutations in the Nbwo woolly motif can prevent repression of NbWoV by NbCycB2, which results in a significant increase in the amount of active Nbwo proteins and in increases in trichome density and the number of branches. Our results reveal a novel reciprocal regulation mechanism between NbCycB2 and Nbwo during trichome formation in N. benthamiana.
Introduction
Trichomes are specialized epidermal protuberances that are found on aerial parts of nearly all terrestrial plants. They can be classified into different types according to cell numbers and shapes, namely unicellular/multicellular, and glandular/non-glandular. It has been demonstrated that the development of trichomes in Arabidopsis (which are unicellular and non-glandular) is regulated by the trimeric MYB-bHLH-WDR complex of protein activators GL1-GL3/EGL-TTG13 (Oppenheimer et al., 1991; Walker et al., 1999; Payne et al., 2000). This transcriptional complex activates the expression of the homeodomain protein GLABROUS2 (GL2) to induce the formation of trichomes (Rerie et al., 1994; Grebe, 2012). It also triggers the expression of single-repeat R3 MYBs including TRY (Schnittger et al., 1999), CPC (Wada et al., 1997), ETC1, ETC2, ETC3 (Kirik et al., 2004, Wester et al., 2009), and TCL2 (Gan et al., 2011), and these act as negative regulators of GL3 or EGL3 in trichome development by forming a repressor complex, GL3/EGL3-TRY/CPC-TTG1 (Wang et al., 2008; Wester et al., 2009). Thus, the control of trichome development in Arabidopsis requires a regulatory loop that includes both activators and repressors (Grebe, 2012; Pattanaik et al., 2014).
Multicellular glandular secreting trichomes (GSTs) are present in ~30% of all vascular plants (Glas et al., 2012). Since many useful phytochemical compounds are synthesized and secreted by such GSTs (Mauricio and Rausher, 1997; Hollósy, 2002; Valkama et al., 2003; Freeman and Beattie, 2008), they have considerable potential economic potential (Sallets et al., 2014; Huchelmann et al., 2017). However, it has been shown that the networks that regulate unicellular trichomes do not function in the development of multicellular trichomes (Serna and Martin, 2006; Yang et al., 2011; Kang et al., 2016; Yan et al., 2017).
In tomato (Solanum lycopersicum), a HD-ZIP IV transcription factor, Slwo, has been shown to regulate trichome initiation (Yang et al., 2011). This contains four conserved domains, namely a homeodomain domain (HD), a leucine zipper (LZ) domain, a steroidogenic acute regulatory protein-related lipid transfer (START) domain, and a START-adjacent domain (SAD). However, overexpression of Slwo fails to induce a change of trichome density, and only ectopic expression of its gain-of-function mutant allele, SlWoV, can cause a higher density of trichomes in tomato and tobacco (Nicotiana tabacum) (Yang et al., 2011, 2015). The SlWoV allele has two point-mutations at the C-terminal domain (since this motif is conserved in most Slwo homologous genes, we name it as the ‘woolly motif’ in this study). Sequence analysis in Arabidopsis has shown that the Slwo protein is more similar to PROTODERMAL FACTOR2 (PDF2) and the PDF2 redundant protein MERISTEM L1 (ML1), both of which are involved in the differentiation of shoot epidermal cells (Abe et al., 2001, Ogawa et al., 2015), than to GL2.
The ectopic expression of a constitutive active B-type cyclin in Arabidopsis induces mitotic divisions and results in an increase in the number of multicellular trichomes (Schnittger et al., 2005). SlCycB2, a hypothetical B-type cyclin, has been reported to directly interact with Slwo to promote the development of type I trichomes (Yang et al., 2011, 2015). Its homologous protein in Arabidopsis (AtGIR1, AT5G06270) has also been found to interact with GL2 and co-repressor TOPLESS proteins (Wu and Citovsky, 2017a, 2017b). However, overexpression of SlCycB2 results in a non-trichome phenotype, while suppression of SlCycB2 promotes trichomes formation in tomato (Gao et al., 2017). These inconsistent results raise important questions about the function of SlCycB2 in trichome formation and why the mutation of the woolly motif can promote formation.
Similar to tomato, trichomes in Nicotiana benthamiana are typically multicellular structures, and almost all of them are glandular (Supplementary Fig. S1 at JXB online), making it a better system for their study than tomato. In addition, the genome map of N. benthamiana has been constructed (Bombarely et al., 2012), and thus it represents an excellent model to study the molecular mechanisms of multicellular trichome formation (Goodin et al., 2008). Here, we cloned the homologues of Slwo and SlCycB2 in N. benthamiana (named Nbwo and NbCycB2, respectively) and constructed a previously identified two-point mutation of the Nbwo allele in the woolly motif, NbWoV (Yang et al., 2015). To investigate their biological functions in trichome development, we constructed transgenic lines with over- and underexpression of the genes. Our results demonstrate that Nbwo and NbWoV can positively regulate the expression of NbCycB2 through targeting to the cis-element in the promoter sequence. In contrast, NbCycB2 can act as a negative regulator of multicellular trichome formation by directly binding to and inhibiting the activity of Nbwo. The mutation of woolly motif blocked the interaction between NbCycB2 and Nbwo, thus removing the repression of Nbwo by NbCycB2 and resulting in increased trichome density. Our results reveal the mechanisms of the interaction between Nbwo and NbCycB2 in regulating the development of glandular trichomes.
Materials and methods
Plant materials and growth conditions
Sterilized seeds of Nicotiana benthamiana were germinated and grown to seedlings under a photoperiod of 14/10 h light/dark (120 μmol m–2 s–1) at 26 °C on MS medium that was solidified with 0.8% (w/v) gellan gum. At 2 weeks old the plants were transferred to either sterilized bottles (for genetic transformation) or to soil in pots to grow to maturity. All wild-type and transgenic plants were grown in a greenhouse under a photoperiod of 14/10 h light/dark (120 μmol m–2 s–1) at 26 °C.
Sequence analysis
The sequences of the similar proteins Slwo and SlCycB2 were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/) and the Sol Genomics Network (https://solgenomics.net/;Fernandez-Pozo et al., 2015). Details of these proteins are given in Supplementary Tables S1 and S2. The aligned sequences were used to construct phylogenetic trees in MEGA 5 by using the maximum-likelihood (ML) criterion with 100 bootstraps. In addition, the relative conservation for each amino acid position in the protein sequences of Nbwo and NbCycB2 were evaluated using WebLoGo (https://weblogo.berkeley.edu/;Crooks et al., 2004), followed by predictions of their conserved domains using SMART (https://smart.embl.de;Letunic and Bork, 2018).
RNA extraction and real-time PCR
Total RNA was extracted from leaves of ~3-week-old plants by using an Eastep® Super Total RNA Extraction Kit (Promega). The cDNA was synthesized using a M-MLV 1st Strand Kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) was carried out using SYBR Premix Ex Taq II (TaKaRa). L25 ribosomal protein (L18908) was used as an endogenous control (Schmidt and Delaney, 2010). Primers are listed in Supplementary Table S3.
Plasmid construction and transformation of N. benthamiana
The full-length coding sequences of Nbwo and NbCycB2 were amplified from the general cDNA of N. benthamiana leaves. The NbWoV allele with two point-mutations at loci 2084 (T replaced with G) and 2092 (G replaced with T) of Nbwo was generated by using a KOD -Plus- Mutagenesis Kit (Toyobo). To construct the overexpression lines of Nbwo, NbWoV, and NbCycB2, these genes were inserted into the pCXSN-HA (Nbwo and NbWoV fused to the HA tag) and pCXSN-FLAG (NbCycB2 fused to the Flag tag) vectors under the control of the CaMV 35S promoter (Chen et al., 2009). The underexpression vectors of Nbwo and NbCycB2 were constructed by recombination with the RNAi vector pH7GWIWGII with the LR Clonase II enzyme (Invitrogen). Approximately 2800 bp of the upstream promoter sequences of NbCycB2 and Nbwo were inserted into the pH2GW7 vector to create the promoter-driven GFP-GUS constructs (Cui et al., 2015).
All of these constructs were transferred into Agrobacterium tumefaciens strain GV3101 to generate transgenic lines via Agrobacterium-mediated transformation. All the primers used are listed in Supplementary Table S3.
Analysis of subcellular localization and tissue distribution
To analyse the subcellular localization of Nbwo and NbWoV, these genes were fused to green fluorescent protein (GFP) driven by the CaMV 35S promoter (p35S::GFP-Nbwo, p35S::GFP-NbWoV). The constructs were transferred into A. tumefaciens strain GV3101 and then transiently transformed into leaves of 4-week-old N. benthamiana. After cultivation under low light conditions for 48–72 h, GFP was observed using confocal microscopy (LSM 780, Carl Zeiss) with staining in DAPI solution (1 mg ml–1) for 15 min before observation. The subcellular localization of NbCycB2 was observed in leaves of NbCycB2-overexpressing (-OE) transgenic plants (p35S::GFP-NbCycB2).
Tissue distribution assays were performed as described previously by Jefferson et al. (1987). GUS staining was repeated in at least three independent transgenic lines.
Yeast hybrid assays
For yeast one-hybrid (Y1H) assays, the promoter of NbCycB2, separated into five fragments (E, –1027 to –831 bp; D, –830 to –631 bp; C, –630 to –411 bp; B, –410 to –201 bp; A, –200 to –1 bp), was amplified and inserted into the pHIS 2 vector (NbCycB2proE, NbCycB2proD, NbCycB2proC, NbCycB2proB, NbCycB2proA). Further investigations of the targeted sequences in the NbCycB2 promoter were conducted by point-mutations in the two L1-like boxes in the D fragment: proD-m1, mutant one L1-like box, with 5´-GCAAATATTTACTC-3´ changed to 5´-GCGGGTGACTC-3´; and proD-m2, mutant two L1-like boxes, with 5´-GCAAATATTTACTC-3´ to 5´-GCGGGTGACTC-3´, and 5´-ATTTACTC-3´ changed to 5´-GGGACTCC-3´. To test the specific region of the Nbwo genomic sequence that binds with the Nbwo protein, four genomic fragments of Nbwo (G1, –8 to 251 bp including the T3 fragment; G2, 2169 to 2522 bp including the T4 fragment; G3, 3485 to 3780 bp including the T5 fragment; G4, 4333 to 4660 bp including the T6 fragment) were amplified and inserted into the pHIS 2 vector (Nbwo-G1, Nbwo-G2, Nbwo-G3, Nbwo-G4). The coding sequences (CDSs) of Nbwo and NbWoV were fused to the GAL4 activation domain in pGADT7 vectors (AD-Nbwo and AD-NbWoV). The bait and prey constructs were co-transformed into Saccharomyces cerevisiae Y187 to test the DNA–protein interactions. The empty pGADT7 vector (AD) served as the negative control, and was cultivated on SD/–Leu/–Trp (–L–W) medium and tested on SD/–Leu/–His/–Trp (–L–-W–--H) medium with 60 mM 3-amino-1,2,4-triazole (Sangon Biotech Co., Ltd).
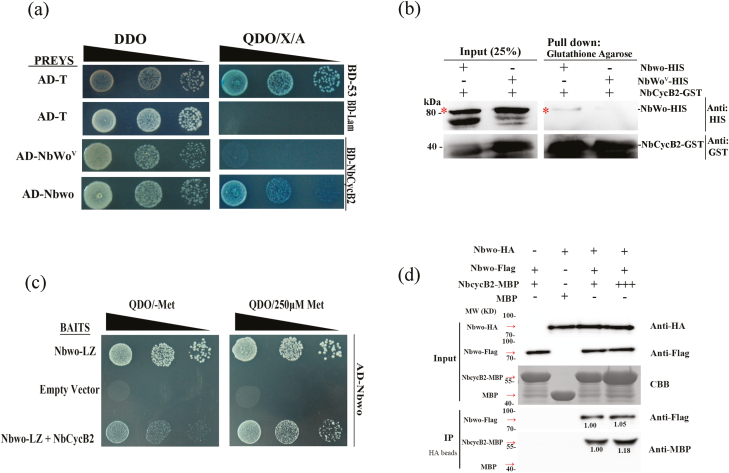
For yeast two-hybrid (Y2H) assays, four truncated Nbwo segments (including the HD, LZ domain, START domain, and SAD) and NbCycB2 were fused to the GAL4 binding domain (BD-Nbwo-HD, BD-Nbwo-LZ, BD-Nbwo-START, BD-Nbwo-SAD, BD-NbCycB2) to verify the interactions with Nbwo or NbWoV. The Nbwo and NbWoV genes fused to the GAL4 binding domain were used to test for auto-activation. Each pair of AD and BD plasmids were co-transformed into the Y2HGold yeast strain (Clontech). Clones containing the BD-53 and AD-T vectors served as positive controls, and BD-Lam and AD-T served as negative controls. The transformants were then cultivated on SD/–Leu/–Trp medium (DDO) and tested on SD/–Ade/–Leu/–His/–Trp medium with 40 mg l–1 X-a-Gal and 400 µg l–1 aureobasidin A (QDO/X/A).
For yeast three-hybrid (Y3H) assays, Nbwo-LZ was fused to the pBridge GAL4 binding domain, and NbCycB2 was inserted downstream of the pBridge vector methionine repressible promoter (Nbwo-LZ+NbCycB2). The BD-Nbwo-LZ plasmid was transferred with AD-Nbwo as a positive control, and the empty pBridge vector was transferred with AD-Nbwo into Y2HGold strain as a negative control. The transformants were then tested on SD/–Ade/–Leu/–His/–Trp media with different concentrations of methionine (0 μM, 250 μM).
Bimolecular fluorescence complementation assays
To examine the interactions of NbCycB2 and Nbwo (or NbWoV) in N. benthamiana protoplasts bimolecular fluorescence complementation (BiFC) assays were performed. The protoplasm was extracted from the leaves according to the method described by Yoo et al. (2007). The CDSs of Nbwo, NbWoV, and NbCycB2 were each inserted into the pSAT6-cEYFP-C1-B vector (2x35S::YFPc-Nbwo, 2x35S::YFPc-NbWoV) and the pSAT6-n(1–174)EYFP-C1 vector (2x35S::YFPn-NbCycB2) (Citovsky et al., 2006). Pairs of the two plasmids were then transiently transformed into the protoplasts using the PEG–calcium transfection method as described by Yoo et al. (2007).
To determine the interactions between the Nbwo LZ domain and Nbwo itself, the CDSs of Nbwo, NbWoV, and NbCycB2 were each fused to the C-terminal fragment of YFP in the p2YC vector; Nbwo-LZ and NbWoV were also fused to the N-terminal fragment of YFP in the p2YN vector. Different plasmid combinations were co-infiltrated into leaves of N. benthamiana as described by (Shen et al., 2011).
Fluorescence of the yellow fluorescent protein (YFP) was observed using confocal microscopy (LSM 780, Carl Zeiss). Three biological replicates were observed independently for each sample.
Dual-luciferase assays
The regulatory effectors of 2x35S::HA-Nbwo, 2x35s::HA-NbWoV, and 2x35s::Flag-NbCycB2 were generated. The firefly luciferase reporters were driven by the B or D fragments of the NbCycB2 promoter, and the Renilla luciferase was driven by the 35S promoter in the pGreen-0800-II report vector (35S::REN-proB::LUC, 35S::REN-proD::LUC). Mutants of the NbCycB2 promoter D fragments were also constructed using a mutagenesis kit (35S::REN-proD-m2::LUC; KOD -Plus- Mutagenesis Kit, Toyobo). The regulatory effector and reporter were used at a ratio of 5:1 or 5:5:1 for the expression tests of two or three plasmids.
Immunoblotting and pull-down assays
Leaves of ~4-week-old N. benthamiana (~0.5 g) were homogenized in liquid nitrogen and then solubilized in 0.4 ml of lysis buffer (25 mM Tris-HCl, 2.5 mM EDTA, pH 8.0, 0.05% v/v NP-40, 5% glycerol, 150 mM NaCl, 1 mM phenylmethylsulphonyl fluoride, 20 µM MG132) for 30 min at 4 °C. Total protein (~80 µg) was then used for immunoblotting assays. After SDS-PAGE separation, the proteins were electrophoretically transferred to a PVDF membrane for immunodetection.
For pull-down assays of the interaction of NbCycB2 with Nbwo and NbWoV, the CDSs of Nbwo, NbWoV, and NbCycB2 were inserted into the pET22b and PGEX-4T-1 vectors to create the fusion proteins (His-Nbwo, His-NbWoV, and GST-NbCycB2), and were then transformed into the Escherichia coli BL21 strain. The purified recombinant bait proteins (2 mg His-Nbwo or His-NbWoV) and prey proteins (2 mg GST-NbCycB2) were then mixed with 1 ml of binding buffer (50 mM Tris-HCl, pH 7.5, 0.6% Triton and X-100, 100 mM NaCl). After incubation at 4 °C for 2 h, 50 µl of glutathione agarose was added to the mixtures, followed by incubation for an additional 1 h. The immunoprecipitates were washed five times with binding buffer. The isolated proteins were detected by immunoblotting with anti-His or anti-GST antibodies.
The interaction of the NbCycB2 and Nbwo dimers was also determined by pull-down assays. NbCycB2 was fused to the MBP tag and transformed into the E. coli BL21 strain. The MBP and NbCycB2-MBP proteins were purified using amylose resin (NEB). The Nbwo overexpression plasmids (35S::HA-Nbwo and 35S::Flag-Nbwo) were transformed into N. benthamiana protoplasts according to the method described by Yoo et al. (2007). After transformation for 12 h, the HA-Nbwo and Flag-Nbwo proteins were each extracted from the protoplasts. Total protein was then uniformly mixed and incubated with purified MBP (2 mg) or NbCycB2-MBP (2 mg or 6 mg) proteins. Next, 5 µl of anti-HA antibody (Sigma) was added to each reaction tube. After incubation at 4 °C for 3 h, 10 µl of protein A magnetic beads (ThermoFisher Scientific) was added to the mixtures and they were further incubated for 1 h. The immunoprecipitates were washed five times with lysis buffer. The isolated proteins were detected by immunoblotting with anti-Flag or anti-MBP antibodies.
Chromatin immunoprecipitation assays
NbWo V-OE plants at 4 weeks old were used for the chromatin immunoprecipitation (ChIP) assay as described previously by (Gendrel et al., 2005). The HA-NbWoV proteins were precipitated using anti-HA antibody (Santa Cruz Biotechnology). Primers were designed to amplify three fragments (length ~120–210 bp) within the 1.7-kbp upstream sequence of the NbCycB2 transcription start site, and seven fragments within ~8.7 kbp of the genomic DNA sequence of Nbwo. After immunoprecipitation, the purified DNA was analysed using real-time PCR. Enrichment was calculated from the ratio of the immunoprecipitated sequences.
Observation of phenotypes
Leaves from 10-d-old plants of the transgenic lines and wild-type were fixed with 2% glutaraldehyde (0.1M phosphate buffer, PH 7.4) at 4 °C for 12 h for SEM analysis. The samples were then dehydrated with an ethanol series of 10–100% in steps of 10% for 20 min each time. Finally, the samples were dried in a critical-point drying device (Leica EMCPD030) and coated with gold particles before being observed using a JSM-6390/LV SEM.
Accession numbers
The accession numbers of sequences reported here are as follows: Nbwo (Niben101Scf07790g01007.1), Nbwo-allele (Niben101 Scf00176g11005.1), NbCycB2 (Niben101Scf10299g00003.1), NbCycB2- allele (Niben101Scf10396g00002.1), NbML1 (Niben101Scf00703g 00003.1), NbML1-allele (Niben101Scf01158g03010.1).
Results
Expression and cellular analysis of Nbwo and NbCycB2
We obtained 14 protein sequences similar to Nbwo and eight similar to NbCycB2 from the NCBI databases. Phylogenetic analyses showed that Nbwo and NbCycB2 had the highest similarity with Slwo and SlCycB2, respectively (Supplementary Fig. S2a, b). One allele of Nbwo was identified via BLAST with 95.82% identity and one allele of NbCycB2 was identified with 95.20% identity. We introduced two mutation sites in Nbwo (at loci 2084 and 2092) that were identical to SlWoV and named the mutant as NbWoV. These two point-mutations caused two amino acid substitutions in the woolly motif (Ile to Arg, Asp to Tyr; Supplementary Fig. S2c). Conserved domain analysis indicated that NbCycB2 contained a WD40-like domain in the N-terminus (NbCycB2-WD40, including an EAR-like motif) and a RING-like domain in the C-terminus (NbCycB2-RING) (Supplementary Fig. S2d). However, no conserved domain of B-type cyclin protein was found in the NbCycB2 sequences.
Subcellular examination indicated that both Nbwo and NbWoV were localized in the nucleus, and that NbcycB2 was localized in the nucleus and cytoplasm (Supplementary Fig. S3a). Y2H assays showed that compared to the positive control containing the BD-53 and AD-T vectors, clones of BD-Nbwo and BD-NbWoV with empty AD combinations could grow on QDO/X/A medium (Supplementary Fig. S3b), while AD-Nbwo and AD-NbWoV with empty BD combinations and the negative control containing the BD-Lam and AD-T vectors failed to grow. This suggested that Nbwo and NbWoV had strong auto-activating ability and that the mutations of the woolly motif did not affect the transactivation ability of Nbwo.
Examination of the spatial expression of NbCycB2 and Nbwo indicated that they were expressed at low levels in the roots but at high levels in organs containing trichomes (Supplementary Fig. S4a). Further investigation showed that GUS was only detected in the leaf and stem trichomes of the GFP-GUS transgenic lines driven by the NbCycB2 promoter (Supplementary Fig. S4b–c). In transgenic lines driven by the Nbwo promoter, although GUS was strongly expressed in the basal and venous regions of young leaves, it was also expressed in the trichomes of cotyledons and young leaves. (Supplementary Fig. S4d).
NbCycB2 negatively regulates trichome initiation
Most NbCycB2-overexpression (-OE) transgenic T1 lines showed a dramatic reduction in the density of trichomes on leaves and stems (Supplementary Fig. S5a, Fig. 1c, f), whilst root length and the number of branch roots significantly increased (Supplementary Fig. S6). Western blotting and qRT-PCR analyses showed that NbCycB2 significantly accumulated in the NbCycB2-OE lines, while the expression levels of Nbwo and endogenous NbCycB2 were significantly reduced (Supplementary Fig. S5b, c).
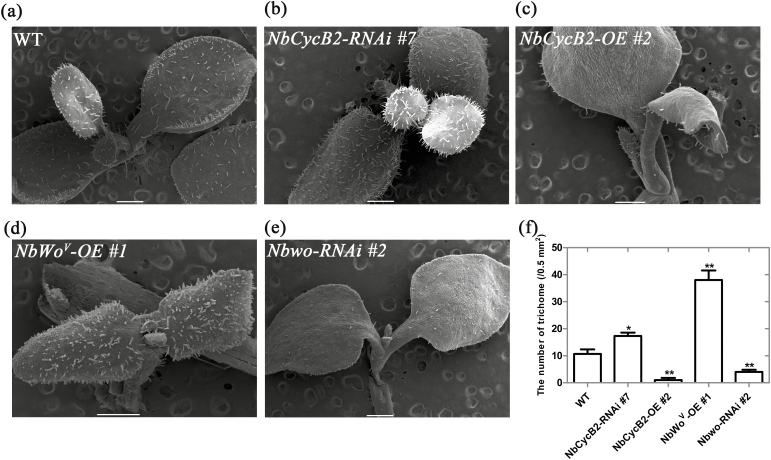
Fig. 1.
The trichome phenotypes of N. benthamiana NbCycB2, Nbwo, and NbWoV transgenic seedlings. SEM images of the trichomes of 10-d-old seedlings of (a) the wild-type (WT), (b) NbCycB2-RNAi #7 T1, (c) NbCycB2-overexpressing (-OE) #2 T1, (d) NbWoV-OE #1 T1, and (e) Nbwo-RNAi #2 T1. Scale bars are 500 μm. (f) The trichome densities of the plants shown in (a–e). Data are means (±SD), n=3. Significant differences compared to the wild-type were determined using Student’s t-test: *P<0.05, **P<0.01.
In contrast, the density of trichomes increased significantly on the leaves and stems of 16 NbCycB2-RNAi T1 lines (Supplementary Fig. S5d, Fig. 1b, f) whilst qRT-PCR analysis showed that the expression level of NbCycB2 decreased (Supplementary Fig. S5e). This suggested that NbCycB2 might play a negative role in trichome initiation.
NbWoV positively regulates trichome initiation
To confirm the function of Nbwo, we generated 22 Nbwo-knockdown transgenic T1 plants (Nbwo-RNAi). Compared with the wild-type, the trichome densities were clearly reduced on the leaves and stems of most Nbwo-RNAi plants (Supplementary Fig. S7a, e). The efficiency of the RNAi-mediated knockdown was confirmed by qRT-PCR in two independent lines in which the expression of Nbwo and NbCycB2 was significantly reduced (Supplementary Fig. S7b).
In common with previous results for tomato (Yang et al., 2011), we found significant increases in the density and branching of trichomes on the leaves and stems of NbWoV-OE plants (Fig. 1d, f, Supplementary Fig. S7d, h). qRT-PCR assays showed significantly up-regulated expression levels of NbWoV, endogenous Nbwo, and NbCycB2 in the transgenic lines (Supplementary Fig. S7f, g). Compared to the wild-type, the density of glandular trichomes increased significantly in NbWoV-OE #1, but root hairs were almost absent (Fig. 2d, f). A dwarfism phenotype observed in the T1 plants of NbWoV-OE (Supplementary Fig. S8).
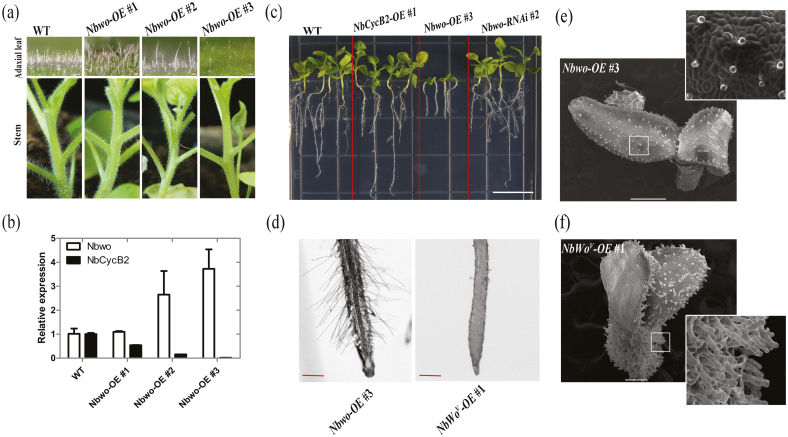
Fig. 2.
Overexpression of N. benthamiana Nbwo causes dwarfism. (a) Phenotypes of Nbwo-overexpressing (-OE) transgenic plants. Compared to wild-type (WT) plants, the density of the trichomes in the stems and leaves of the transgenic lines decreased. (b) Relative expression levels of Nbwo and NbCycB2 in the transgenic lines as measured by qRT-PCR. Compared with the wild-type plants, the expression level of NbCycB2 decreased with the increase in total expression of Nbwo in the transgenic plants. Data are means (±SD), n=3. Expression is relative to that of the WT, the value of which was set as 1. (c) Root lengths in 2-week-old seedlings of the WT, NbCycB2-OE #1 T1, Nbwo-OE #3 T1, and Nbwo-RNAi #2 T1. The scale bar is 1 cm. (d) Root hairs of 2-week-old seedlings of Nbwo-OE #3 T1 and NbWoV-OE #1 T1. The root hairs were normal in the Nbwo-OE line. The scale bars are 1 mm. (e, f) SEM images of 10-d-old seedlings of (e) Nbwo-OE #3 T1 and (f) NbWoV-OE #1 T1. The trichome density of Nbwo-OE plants was normal, whilst the NbWoV-OE plants showed a significant increase. The scale bars are 500 μm. (This figure is available in colour at JXB online.)
Overexpression of Nbwo does not increase trichome density
A total of 20 Nbwo-OE plants were generated. Interestingly, the trichome density was found to be negatively related to the expression level of Nbwo in T0Nbwo-OE plants (Fig. 2a, b). The expression levels of NbCycB2 were significantly reduced in T0 plants with reduced trichomes (Fig. 2b). However, the trichome density was decreased in T1 plants (Fig. 2e). Compared to the NbWoV-OE #1, the development of trichomes and root hair appeared to be unaffected in the Nbwo-OE plants (Fig. 2d-f). These results indicated that the functions of Nbwo in trichome and root hair development were quite different to those of NbWoV. However, the higher expression level of exogenous Nbwo (e.g. Nbwo-OE #3) resulted in dwarfism (Fig. 2c), which was similar to the NbWoV-OE lines (Supplementary Fig. S8).
Nbwo and NbWoV directly target the L1-like box of the NbCycB2 promoter
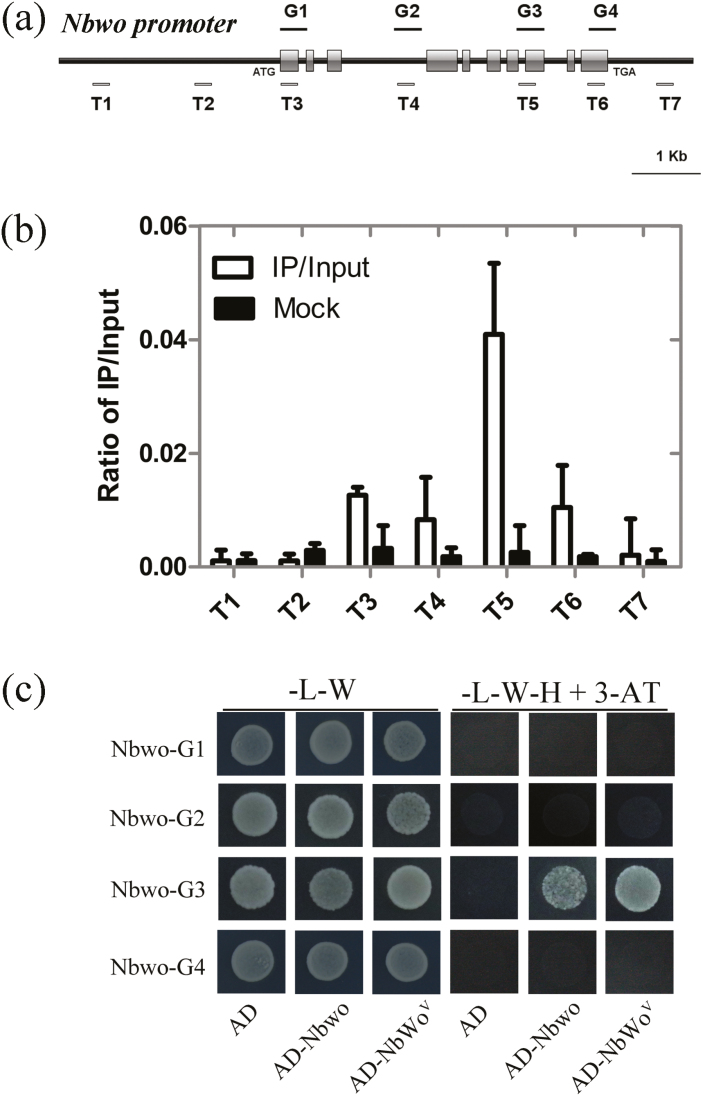
The expression of NbCycB2 was significantly up-regulated in the NbWoV-OE lines and decreased in the Nbwo-RNAi lines (Supplementary Fig. S7b, f), which indicated that NbCycB2 was positively regulated by Nbwo and NbWoV. To test whether Nbwo directly binds to the promoter of NbCycB2, NbWoV-OE plants with NbWoV fused to the HA tag were analysed using ChIP qRT-PCR assays with a HA antibody. Strong enrichment of NbWoV was observed in the P2 region of the NbCycB2 promoter in the NbWoV-OE plants (Fig. 3a, b).
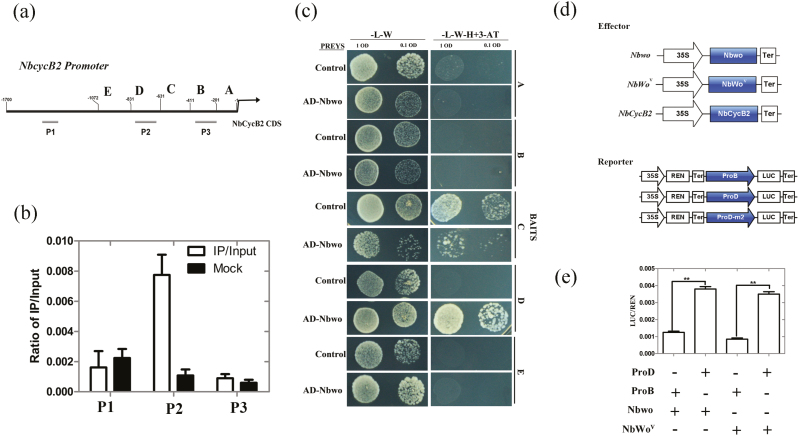
Fig. 3.
Nicotiana benthamiana Nbwo and NbWoV can bind to the NbCycB2 promoter in vitro and in vivo. (a) Fragments of the NbCycB2 promoter were used for ChIP (P1–P3) and yeast one-hybrid (Y1H) assays (A–E). The numbers indicate the positions of the truncations. (b) The ratio of bound promoter fragments versus total input detected by qRT-PCR after immuno-precipitation of HA-NbWoV by HA antibodies. Data are means (±SE), n=3. (c) Y1H assays to determine the interactions of the NbCycB2 promoter fragments and AD-Nbwo or the negative control (AD) in the Y187 yeast strain. (d) Schematic diagram of the effector and reporter constructs used in the LUC assays. (e) Relative reporter activities in N. benthamiana protoplasts after transient transformation of the effector and reporter constructs. The relative LUC activity normalized to REN activity are shown (LUC/REN). Data are means (±SD), n=3. Significant differences were determined using Student’s t-test: **P<0.01. (This figure is available in colour at JXB online.)
To further determine the specific area of binding of Nbwo, we performed Y1H assays. The five truncated fragments of NbCycB2 promoter that were used are shown in Fig. 3a. Yeast colonies containing the AD-Nbwo and NbCycB2-proC-pHIS2 or NbCycB2-proD-pHIS 2 constructs were grown on selection medium with 60 mM 3-aminotriazole (Fig. 3c). Colonies containing NbCycB2-proC-pHIS2 and the empty vector pGADT7 (AD) grew normally on the medium, which indicated that the NbCycB2 promoter proC fragment possessed high autoactivation activity. This suggested that the D fragment of the NbCycB2 promoter sequence was the binding targe of the Nbwo protein.
To determine whether Nbwo and NbWoV could directly affect the expression of the D fragment in vivo, we performed dual-luciferase assays. The reporters 35S::REN-NbCycB2proD::LUC and 35S::REN-NbCycB2proB::LUC and the effectors are shown in Fig. 3d. Each reporter and effector pair was transiently co-expressed in N. benthamiana protoplasts (Fig. 3e). Compared to the B fragment, when Nbwo or NbWoV were transiently co-expressed, LUC expression driven by the D fragment was significantly higher in protoplasts. This indicated that the D fragment of the NbCycB2 promoter appeared to be a specific site for binding of Nbwo and NbWoV.
Further analysis of the targeting sequence of NbCycB2proD revealed that this sequence contained two L1-like boxes (5´-ATTTACTC-3´) (Supplementary Fig. S9a). When two L1-like boxes were mutated (proD-m1, proD-m2), Y1H and in vivo LUC assays showed that the interaction with the Nbwo protein was abolished (Supplementary Fig. S9b, c). Based on these results, we inferred that the L1-like boxes might be the binding targets of the Nbwo and NbWoV proteins.
NbCycB2 represses the activity of Nbwo rather than NbWoV
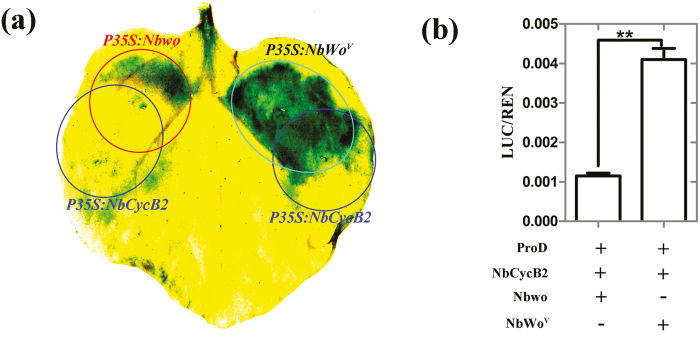
Since the NbCycB2-OE and Nbwo-RNAi transgenic lines shared the non-trichome phenotype and the expression of Nbwo did not increase in the NbCycB2-RNAi lines (Supplementary Fig. S5e), we suspected that NbCycB2 could affect the transactivation ability of Nbwo at the protein levels. To test this hypothesis, Nbwo, NbWoV, and NbCycB2 were transiently co-overexpressed under the control of the 35S promoter in the leaves of the proNbCycB2::GFP-GUS transgenic line using Agrobacterium-mediated transformation (Fig. 4a). We found that co-expression of NbCycB2 could inhibit the expression of GUS induced by overexpression of Nbwo in the leaves of proNbCycB2::GFP-GUS plants. However, GUS expression induced by overexpression of NbWoV was not affected by NbCycB2 expression. These results were further supported by LUC assays, in which co-expression with NbCycB2 clearly repressed the activity of Nbwo, but did not affect NbWoV (Fig. 4b). These results suggested that NbCycB2 may act as a negative regulator of Nbwo rather than NbWoV.
Fig. 4.
Nicotiana benthamiana NbCycB2 can suppress the function of Nbwo, but has no effect on NbWoV. (a) Co-expression of NbCycB2 in leaves of proNbCycB2-GFP-GUS transgenic plants inhibits the expression of GUS induced by overexpression of Nbwo, but it has no effect on the expression of GUS induced by NbWoV. All the constructs were expressed under the control of the 35S promoter. The areas injected with P35S::Nbwo, P35S::NbWoV, and P35S::NbCycB2 GV3101 strain are indicated. At 72 h after injection, the leaves were stained with GUS substrate. (b) Dual-LUC activity test, confirming that co-expression with NbCycB2 decreased the transactivation activity of Nbwo compared with NbWoV. Data are means (±SD), n=3. Significant differences were determined using Student’s t-test: **P<0.01.
The interaction between NbCycB2 and Nbwo can be reduced by mutations in the woolly motif
An interaction between SlCycB2 and Slwo has been reported previously (Yang et al., 2011). To explore the domain involved in the physical interaction between NbCycB2 and Nbwo, four truncated fragments of Nbwo containing the HD, LZ domain, START domain, and SAD were used (Supplementary Fig. S10a). Y2H assays suggested that the LZ domain of Nbwo interacted with NbCycB2 (Supplementary Fig. S10b). Bimolecular fluorescence complementation (BiFC) assays were further used to verify the interaction between the Nbwo LZ domain and NbCycB2 in vivo (Supplementary Fig. S10c).
Additional Y2H assays were performed to determine whether NbCycB2 also interacted with NbWoV. Only the clones of BD-NbCycB2 with AD-Nbwo combinations could grow on the selection medium, while BD-NbCycB2 with the AD-NbWoV combination and the negative control failed to grow (Fig. 5a). These results indicated that NbCycB2 physically interacted with Nbwo but not with NbWoV. We then used pull-down and BiFC assays to confirm the lack of interaction between NbCycB2 and NbWoV. Purified Nbwo-HIS and NbWoV-HIS were incubated with equal amounts of NbCycB2-GST, and immunoblotting showed that only Nbwo-HIS could retain NbCycB2-GST, whereas NbWoV-HIS could not (Fig. 5b). For the BiFC analysis, Nbwo and NbWoV were individually fused to the C-terminal part of yellow fluorescent protein (YFPC) to generate YFPC-Nbwo and YFPC-NbWoV, while NbCycB2 was ligated to the N-terminal fragment of YFP (YFPN) to generate YFPN-NbCycB2. We found that co-expression of YFPC-Nbwo with YFPN-NbCycB2 in N. benthamiana protoplasts resulted in strong YFP fluorescence in the nucleus, whereas no YFP signal was observed in the combinations of YFPC-NbWoV and YFPN-NbCycB2, or in the negative controls (Supplementary Fig. S10e). These results suggested that the interaction between NbCycB2 and Nbwo could be removed by mutations in the woolly motif.
Fig. 5.
Mutation of the woolly motif attenuates the interaction between Nbwo and NbCycB2 in N. benthamiana. (a) Yeast two-hybrid (Y2H) assays of the interactions between NbCycB2 and Nbwo or NbWoV using QDO/X/A medium. Clones with pGADT7-53 (BD-53) and pGADT7-T (AD-T) served as positive controls, and clones with pGBKT7-Lam (BD-Lam) and pGADT7-T (AD-T) served as negative controls. Only the positive control and AD-Nbwo and BD-NbcycB2 could grow on the medium. (b) Pull-down assays between Nbwo or NbWoV with NbCycB2 proteins. The NbCycB2-GST protein was immunoprecipitated with glutathione agarose, and the immunoblots were probed with anti-HIS and anti-GST antibodies. Only the recombinant HIS-Nbwo protein could co-precipitate with GST-NbCycB2. The asterisks indicate the bands for Nbwo or NbWoV. (c) Y3H assays to determine the competition between NbCycB2 and the LZ domain of Nbwo for binding to Nbwo (fused to GAL4 DNA-AD). The methionine-repressible promoter in the pBridge vector controlled the expression of NbCycB2 in the presence of the Nbwo LZ domain (fused to GAL4 DNA-BD). (d) Pull-down assays to determine whether NbCycB2 could compete for binding to Nbwo. The total protein from P35S::HA-Nbwo protoplasts was immunoprecipitated with anti-HA beads, and the immunoblots were probed with anti-Flag and anti-MBP antibodies. (This figure is available in colour at JXB online.)
NbCycB2 does not competitively bind to Nbwo homodimers
It is known that HD-Zip proteins bind to DNA as dimers via the LZ domain (Ariel et al., 2007). To determine whether Nbwo could be dimerized via the LZ domain, we first used Y2H assays to demonstrate that the LZ domain could bind to the Nbwo protein (Supplementary Fig. S10f). BiFC assays were also used to confirm the interaction between the LZ domain and Nbwo (or NbWoV) proteins in vivo (Supplementary Fig. S10c).
Y3H assays were used to determine whether NbCycB2 could competitively bind to the LZ domain of Nbwo homodimers. The methionine-repressible promoter in the pBridge vector was used to control the expression of NbCycB2 in the presence of the Nbwo LZ domain (Nbwo LZ domain fused to GAL4 DNA-BD, Nbwo-LZ+NbCycB2), and the pBridge vector containing only the Nbwo LZ domain served as a positive control (Nbwo LZ domain fused to GAL4 DNA-BD, Nbwo-LZ). The methionine promoter is inactive on media with high concentrations of methionine. As shown in Fig. 5c, the clone containing Nbwo-LZ+NbCycB2 was able to grow normally in comparison with the positive control, whether in medium containing methionine (250 μM) or lacking methionine. This suggested that NbCycB2 might not competitively bind to the LZ domain of Nbwo homodimers.
To verify this, we further performed a pull-down assay. Nbwo-HA and Nbwo-Flag proteins were extracted after expression in N. benthamiana protoplasts. The total proteins were then uniformly mixed and incubated with the purified NbCycB2-MBP protein. Immunoblot analysis showed that the increased NbCycB2-MBP protein did not affect the amount of Nbwo-Flag retained by Nbwo-HA (Fig. 5d).
Nbwo can bind to its own genomic DNA
The endogenous expression level of Nbwo was reduced in the NbCycB2-OE lines (Supplementary Fig. S5b) and increased in the NbWoV-OE plants (Supplementary Fig. S7f), which indicated that Nbwo might be able to regulate its self-expression. To test this hypothesis, ChIP assays were carried out to check whether Nbwo could bind to its genomic DNA sequence in the leaves of the NbWoV-OE transgenic line. Interestingly, an enrichment of NbWoV was detected in the T5 fragment in NbWoV-OE plants (Fig. 6a, b). This result was further demonstrated by Y1H assays, in which only the clones with AD-Nbwo (or AD-NbWoV) and Nbwo-G3-pHIS 2 constructs could grow on the resistant medium (Fig. 6c). This suggested that Nbwo and NbWoV could bind to the G3 fragments (including the T5 fragment, Fig. 6a) of its own genomic DNA sequence.
Fig. 6.
Nicotiana benthamiana Nbwo and NbWoV can bind to their own genomic DNA sequences. (a) The fragments of the Nbwo genomic sequences used in the ChIP (T1–T7) and yeast one-hybrid (Y1H) assays (G1–G4). (b) Ratio of bound genomic fragments versus total input detected by real-time PCR after immuno-precipitation from the NbWoV-overexpressing (-OE) lines by HA antibodies. Data means (±SE), n=3. (c) Y1H assays to determine the interaction of Nbwo genomic sequence fragment bait constructs and AD-Nbwo, AD-NbWoV, or empty-vector pGADT7 constructs in the Y187 yeast strain. The clones were grown on SD/–Leu/–His/–Trp (–L–W–H) with 60 mM 3-AT medium. (This figure is available in colour at JXB online.)
Overexpression of NbCycB2 can reduce the dwarf phenotype of Nbwo-OE plants
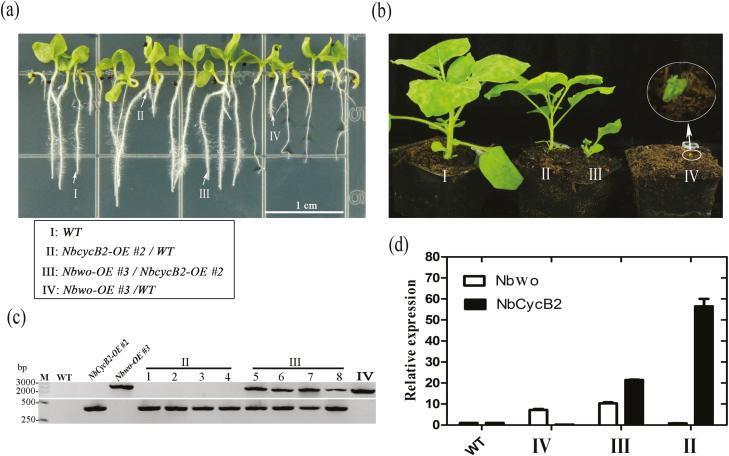
To determine whether NbCycB2 could inhibit the activity of Nbwo in vivo, we crossed NbCycB2-OE #2 T1 with Nbwo-OE #3 T0 plants. We found that the dwarf and short-root phenotypes of Nbwo-OE #3 were indeed reduced by NbCycB2-OE #2 (Fig. 7a, b). The crossed F1 plants were tested using PCR (Fig. 7c), and the expression of NbCycB2 and Nbwo was also verified by qRT-PCR. Compared with T1Nbwo-OE #3 plants, the balance between NbCycB2 and Nbwo expression was restored in the NbCycB2-OE #2 × Nbwo-OE #3 crossed F1 plants (Fig. 7d).
Fig. 7.
Hybridization between Nbwo-overexpressing (-OE) and NbCycB2-OE plants of N. benthamiana. (a, b) Phenotypes of NbCycB2-OE #2 and Nbwo-OE #3 hybridization F1 plants at (a) 2-weeks-old and (b) at maturity. I, wild-type (WT); II, NbCycB2-OE/WT hybrid T1; III, NbCycB2-OE #2 and Nbwo-OE #3 hybridization F1; IV, Nbwo-OE #3/WT hybrid T1. (c) F1 plants were examined using PCR. The reverse primer for the overexpression vector served as the 3´-terminal detection primer, and the forward primer for the detected genes served as the 5´-terminal primer. The WT served as a negative control, and Nbwo-OE #3 and NbCycB2-OE #2 served as positive controls. No DNA bands were detected in the WT. A ~330-bp DNA band was detected in the NbCycB2-OE #2 lines and a ~2199-bp band was detected in the Nbwo-OE #3 lines. In contrast, two bands were detected in NbCycB2-OE #2 and Nbwo-OE #3 hybridization F1 plants. (d) Relative expression levels of Nbwo and NbCycB2 in F1 plants as determined by qRT-PCR. Expression is relative to that of the WT, the value of which was set as 1. Data are means (±SD), n=3. (This figure is available in colour at JXB online.)
Discussion
Trichomes play important roles in plants. They participate in resistance mechanisms to a variety of abiotic stresses such as UV radiation and dehydration (Mauricio and Rausher, 1997; Werker, 2000), and biotic stresses such as pathogenic bacteria and insect herbivores (Freeman and Beattie, 2008; Tian et al., 2012). In tobacco, glandular trichomes function as synthesis and secretion sites of sucrose and diterpenoids (Tissier, 2012), so they are important targets of biotechnological engineering for quality improvement of varieties (Glas et al., 2012). However, few studies so far have examined the development of tobacco trichomes.
Slwo is a key gene in tomato and functions to regulate the initiation of multicellular trichomes, with multiple gain-of-function mutant alleles that are capable of increasing the density of trichomes (Among these alleles, the mutation sites of SlWo and SlWoV are both within the woolly motif, Yang et al., 2011). Of these mutants, SlWoV shows one of the most obvious increases in density (Yang et al., 2015); however, little is known about the mechanisms involved. Expression of SlCycB2 is significantly increased in SlWo and SlWoV lines, but it is significantly decreased in Slwo-RNAi plants (Yang et al., 2011). Because SlCycB2 is similar to AT5G06270.1, a hypothetical B-type cyclin (now renamed as AtGIR1) in Arabidopsis, it was previously thought to have a similar function to that in Arabidopsis, where it promotes the differentiation of unicellular to multicellular trichomes (Schnittger et al., 2005). SlCycB2 was considered to promote the development of type I trichomes in tomato (Yang et al., 2011); however, the detailed mechanism has not been fully studied.
In our current study, we cloned the homologues of SlCycB2 and Slwo in N. benthamiana (named Nbwo and NbCycB2), and constructed a mutant Nbwo allele, NbWoV. We found that overexpression of NbCycB2 lead to suppression rather than promotion of trichome development on stems and leaves (Fig.1b, Supplementary Fig. S5a). Consistent with qRT-PCR results (Supplementary Fig. S4a), GUS staining assays indicated that NbCycB2 was specifically expressed in the trichomes of leaves and stems (Supplementary Fig. S4b, c), and that Nbwo was also expressed in the trichomes of cotyledons and young leaves (Supplementary Fig. S4d). These results suggested that NbCycB2 may serves as a negative regulator of trichome initiation. None of the B-type cyclin conserved domains were found in the SlCycB2 and NbCycB2 protein sequences (Supplementary Fig. S2d). Hence, whether SlCycB2 can function as a B-type cyclin protein requires further study.
NbWoV and Nbwo directly regulate the expression of NbCycB2 through binding to the L1-like boxes in its promoter
SlCycB2 has been reported to be indirectly regulated by Slwo (Yang et al., 2011, 2015). However, we found that the expression level of NbCycB2 was up-regulated in NbWoV-OE plants and down-regulated in Nbwo-RNAi lines (Supplementary Fig. S7), indicating that NbCycB2 might be the downstream gene of Nbwo. ChIP, Y1H, and LUC assays confirmed that the D fragments of the NbCycB2 promoter were the binding target of Nbwo and NbWoV (Fig. 3). Mutation of the two L1-like box sequences in NbCycB2proD inhibited the binding of Nbwo and NbWoV both in vitro and in vivo (Supplementary Fig. S9 a–c). We thus demonstrated that the expression of NbCycB2 was directly regulated by Nbwo and NbWoV through the binding of L1-like boxes in the promoter. Using ChIP and YIH assays, we further demonstrated that Nbwo and NbWoV could self-regulate their own endogenous expression by binding to their own genomic DNA sequences (Fig. 6).
Increased trichome density is induced by mutation of the Nbwo woolly motif
NbWo V contained only two point-mutations (at loci 2084 and 2092 of the Nbwo CDS), which caused two amino acid replacements in the woolly motif of the Nbwo protein (Ile to Arg, Asp to Tyr; Supplementary Fig. S2c). However, these mutations caused a large difference between the functioning of Nbwo and NbWoV. Although overexpression of either Nbwo or NbWoV could cause a dwarf phenotype in offspring (Fig. 2c, Supplementary Fig. S6a), the trichome phenotypes of were completely different. Trichome density was decreased with the expression of Nbwo in the T0 generation of the Nbwo-OE lines (Fig.2a, b). In contrast, as NbWoV expression increased in the NbWoV-OE lines, the density and branching of the trichomes on the leaves were also significantly increased (Fig. 2f, Supplementary Fig. S7d, f). These results implied that the Nbwo SAD (including the woolly motif) might itself possess repression activity. However, compared to wild-type, there was no difference in trichome density in the Nbwo-OE lines where the SAD had been deleted (Supplementary Fig. S11). This suggested that deletion of the SAD might disrupt the function of Nbwo, and that mutation of the woolly motif in the SAD was important for enhancing the functioning of Nbwo.
NbCycB2 represses the transactivation activity of Nbwo at the protein level
Our results demonstrated that NbCycB2 was directly regulated by Nbwo; however, similar non-trichome phenotypes were found in the NbCycB2-OE and Nbwo-RNAi transgenic lines (Supplementary Figs S5a, S7a). In addition, the expression of Nbwo was not increased in NbCycB2-RNAi plants Supplementary (Fig. S5e). This suggested that NbCycB2 might repress the transactivation activity of Nbwo at the protein level. GUS expression was up-regulated by the expression of Nbwo and inhibited by the co-expression of NbCycB2 in the leaves of the NbCycB2pro::GFP-GUS transgenic line (Fig. 4a), and the same result was found in LUC assays (Fig. 4b). In addition, hybridization with NbCycB2-OE was able to attenuate the dwarf phenotype of T1Nbwo-OE (Fig. 7a, b). Further examination showed that the expression levels of endogenous NbCycB2 and Nbwo were reduced in the NbCycB2-OE lines (Supplementary Fig. S5b), and they were found to be downstream of the regulatory genes of Nbwo (Figs 3, 6). Taken together, these results supported our hypotheses that NbCycB2 may act as a negative regulator of Nbwo at the protein level.
In a previous study, SlCycB2 was reported to interact with Slwo (Yang et al., 2011). Further investigation of the interaction between Nbwo and NbCycB2 revealed that the dimerized LZ domain of Nbwo binds to NbCycB2 (Supplementary Fig. S10b, c). Using Y2H, BiFC, Y3H, and co-IP assays we also found that Nbwo could form a homodimer through the LZ domain, and the NbCycB2 protein did not competitively bind to the LZ domain of Nbwo (Fig. 5c, d, Supplementary S10b, c). These results indicated that NbCycB2 might bind to the Nbwo protein via its LZ domain to repress its transactivation ability by recruiting another inhibitor. However, further study is required to determine whether NbCycB2 functions similarly to its Arabidopsis homologues AtGIR1 and AtGIR2, which also interact with the co-repressor TOPLESS (Long et al., 2006, Szemenyei et al., 2008, Pauwels et al., 2010, Wu and Citovsky, 2017b).
The interaction between Nbwo and NbCycB2 is blocked by mutation in the Nbwo woolly motif
In Arabidopsis, feedback-loop regulation mechanisms of R3 MYBs (TRY, CPC, and others) occur through competitively binding to GL3/EGL3 to form a non-functional trimeric protein complex (MYB-bHLH-WDR) that inhibits the formation of trichomes (Wang et al., 2008, Wester et al., 2009). Feedback-loop regulation has been reported as an effective strategy for many HD-ZIP proteins to maintain normal organism development (Ohgishi et al., 2001, Williams and Fletcher, 2005, Kim et al., 2008, San-Bento et al., 2014). However, trichome formation was not repressed by the high expression level of NbCycB2 in NbWoV-OE plants (Supplementary Fig. S7d, f), which suggested that the negative effect of NbCycB2 could be eliminated by the mutation in NbWoV. This hypothesis was verified by LUC and GUS activity assays, which showed that the transactivation activity of NbWoV was not affected by the expression or non-expression of NbCycB2 (Fig. 4a, b).
Further investigation determined that the interaction between NbCycB2 and Nbwo could be blocked by mutation of the woolly motif in the NbWoV protein both in vitro and in vivo (Fig. 5a, b, Supplementary S10d). Elimination of the interaction prevented NbWoV from being inhibited by NbCycB2. The high expression levels of NbCycB2 and endogenous Nbwo in the NbWoV-OE lines further supported this hypothesis (Supplementary Fig. S7f).
Conclusions
In summary, we found that NbCycB2 was specifically expressed in the trichomes of N. benthamiana and negatively affected trichome formation. Further analysis revealed that Nbwo and NbWoV directly regulated the expressions of NbCycB2 and Nbwo by binding to the L1-like box in the NbCycB2 promoter and its own genomic DNA sequence. In addition, NbCycB2 may function via binding to the LZ domain of Nbwo, which represses the activity of Nbwo and reduces the expression of genes downstream of Nbwo , eventually leading to the inhibition of trichome initiation. The interaction between NbCycB2 and Nbwo could be blocked by a mutation in the woolly motif (the Nbwo gain-of-function mutation allele, NbWoV), which prevented repression by NbCycB2 and resulted in a dramatic increase in trichome density and branching. A model of the regulation network is given in Supplementary Fig. S12. Our study has determined the relationship between NbCycB2 and Nbwo, the key regulatory genes of multicellular trichomes in N. benthamiana, and hopefully will facilitate biotechnological engineering and research on multicellular trichomes more generally in plants.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. SEM images of trichomes in the leaves of N. benthamiana.
Fig. S2. Sequence analysis of Nbwo, NbCycB2, and their similar proteins.
Fig. S3. Subcellular localization and auto-activation of NbCycB2, Nbwo, and NbWoV.
Fig. S4. Expression patterns of NbCycB2 and Nbwo in N. benthamiana plants.
Fig. S5. Overexpression of and RNA-interference of NbCycB2 in N. benthamiana.
Fig. S6. Root phenotypes of wild-type, NbCycB2-RNAi #7 T1, NbCycB2-OE #2 T1, NbWoV-OE #1 T1, and Nbwo-RNAi #2 T1 seedlings.
Fig. S7. RNA-interference of Nbwo and overexpression of NbWoV in N. benthamiana.
Fig. S8. The phenotypes of NbWoV-OE lines.
Fig. S9. Nbwo and NbWoV bind directly to L1-like boxes of the NbCycB2 promoter.
Fig. S10. The interactions between Nbwo and NbCycB2, and Nbwo and the Nbwo LZ domain.
Fig. S11. The phenotype of the overexpressing Nbwo-SAD-mutant in N. benthamiana.
Fig. S12. A simplified model for regulation between Nbwo and NbCycB2.
Table S1. List of similar proteins for Nbwo.
Table S2. List of similar proteins for NbCycB2.
Table S3. Primers used in this study.
Supplementary Material
Acknowledgements
We would like to thank Dr. Yong-Jia Zhong (Fujian Agriculture and Forestry University), for kindly providing p2YN and p2YC vectors and thank Prof. Xi Huang (School of Life Sciences, XMU) for kindly providing pBridge and pGreen-0800-II report vectors. We thank Prof. Tao Huang, Prof. Yi Tao, Prof. Hong-Rui Wang (School of Life Sciences, XMU) and Xiao-Ling Guo (College of the Environment & Ecology, XMU) for their help in conducting the experiments. This study was supported by the State Tobacco Monopoly Administration of China (grant no. 110201401003 (JY-03)), the XMU Training Program of Innovation and Entrepreneurship for Undergraduates (2016Y0635) and theGuizhou Science and Technology Major Project (grant no. (2019)3001–2). The authors declare that they have no competing financial interests.
Author contributions
L. C, H. C, and ML. W conceived and designed the experiments; ML. W, YC, C, L. G, LP. C, ZC. X, HY. Z, ZJ. W, and D. Z performed the experiments and analysed the results; ML. W and S. W wrote the main manuscript; L. C supervised the project.
References
- Abe M, Takahashi T, Komeda Y. 2001. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. The Plant Journal 26, 487–494. [DOI] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. 2007. The true story of the HD-Zip family. Trends in Plant Science 12, 419–426. [DOI] [PubMed] [Google Scholar]
- Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB. 2012. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant–microbe biology research. Molecular Plant-Microbe Interactions 25, 1523–1530. [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. 2009. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiology 150, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T. 2006. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. Journal of Molecular Biology 362, 1120–1131. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Research 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Rao S, Chang B, et al. 2015. AtLa1 protein initiates IRES-dependent translation of WUSCHEL mRNA and regulates the stem cell homeostasis of Arabidopsis in response to environmental hazards. Plant, Cell & Environment 38, 2098–2114. [DOI] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Menda N, Edwards JD, et al. 2015. The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Research 43, D1036–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Beattie GA. 2008. An overview of plant defenses against pathogens and herbivores. The Plant Health Instructor. doi: 10.1094/PHI-I-2008-0226-01. [DOI] [Google Scholar]
- Gan L, Xia K, Chen JG, Wang S. 2011. Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biology 11, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Gao Y, Xiong C, Yu G, Chang J, Yang Q, Yang C, Ye Z. 2017. The tomato B-type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Science 262, 103–114. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V. 2005. Profiling histone modification patterns in plants using genomic tiling microarrays. Nature Methods 2, 213–218. [DOI] [PubMed] [Google Scholar]
- Glas JJ, Schimmel BC, Alba JM, Escobar-Bravo R, Schuurink RC, Kant MR. 2012. Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. International Journal of Molecular Sciences 13, 17077–17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Zaitlin D, Naidu RA, Lommel SA. 2008. Nicotiana benthamiana: its history and future as a model for plant–pathogen interactions. Molecular Plant-Microbe Interactions 21, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Grebe M. 2012. The patterning of epidermal hairs in Arabidopsis—updated. Current Opinion in Plant Biology 15, 31–37. [DOI] [PubMed] [Google Scholar]
- Hollósy F. 2002. Effects of ultraviolet radiation on plant cells. Micron 33, 179–197. [DOI] [PubMed] [Google Scholar]
- Huchelmann A, Boutry M, Hachez C. 2017. Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiology 175, 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Campos ML, Zemelis-Durfee S, Al-Haddad JM, Jones AD, Telewski FW, Brandizzi F, Howe GA. 2016. Molecular cloning of the tomato Hairless gene implicates actin dynamics in trichome-mediated defense and mechanical properties of stem tissue. Journal of Experimental Botany 67, 5313–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Lee M, et al. 2008. HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. The Plant Cell 20, 920–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J. 2004. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Developmental Biology 268, 506–513. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Research 46, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. 2006. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523. [DOI] [PubMed] [Google Scholar]
- Mauricio R, Rausher MD. 1997. Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution 51, 1435–1444. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Yamada Y, Sezaki N, Kosaka S, Kondo H, Kamata N, Abe M, Komeda Y, Takahashi T. 2015. ATML1 and PDF2 play a redundant and essential role in Arabidopsis embryo development. Plant & Cell Physiology 56, 1183–1192. [DOI] [PubMed] [Google Scholar]
- Ohgishi M, Oka A, Morelli G, Ruberti I, Aoyama T. 2001. Negative autoregulation of the Arabidopsis homeobox gene ATHB-2. The Plant Journal 25, 389–398. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. 1991. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. [DOI] [PubMed] [Google Scholar]
- Pattanaik S, Patra B, Singh SK, Yuan L. 2014. An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Frontiers in Plant Science 5, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, et al. 2010. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. 2000. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. 1994. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes & Development 8, 1388–1399. [DOI] [PubMed] [Google Scholar]
- Sallets A, Beyaert M, Boutry M, Champagne A. 2014. Comparative proteomics of short and tall glandular trichomes of Nicotiana tabacum reveals differential metabolic activities. Journal of Proteome Research 13, 3386–3396. [DOI] [PubMed] [Google Scholar]
- San-Bento R, Farcot E, Galletti R, Creff A, Ingram G. 2014. Epidermal identity is maintained by cell–cell communication via a universally active feedback loop in Arabidopsis thaliana. The Plant Journal 77, 46–58. [DOI] [PubMed] [Google Scholar]
- Schmidt GW, Delaney SK. 2010. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Molecular Genetics and Genomics 283, 233–241. [DOI] [PubMed] [Google Scholar]
- Schnittger A, Folkers U, Schwab B, Jürgens G, Hülskamp M. 1999. Generation of a spacing pattern: the role of triptychon in trichome patterning in Arabidopsis. The Plant Cell 11, 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Schöbinger U, Stierhof Y-D, Hülskamp M. 2005. Erratum. Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Current Biology 15, 980. [DOI] [PubMed] [Google Scholar]
- Serna L, Martin C. 2006. Trichomes: different regulatory networks lead to convergent structures. Trends in Plant Science 11, 274–280. [DOI] [PubMed] [Google Scholar]
- Shen Q, Liu Z, Song F, Xie Q, Hanley-Bowdoin L, Zhou X. 2011. Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus β-satellite. Plant Physiology 157, 1394–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386. [DOI] [PubMed] [Google Scholar]
- Tian D, Tooker J, Peiffer M, Chung SH, Felton GW. 2012. Role of trichomes in defense against herbivores: comparison of herbivore response to woolly and hairless trichome mutants in tomato (Solanum lycopersicum). Planta 236, 1053–1066. [DOI] [PubMed] [Google Scholar]
- Tissier A. 2012. Glandular trichomes: what comes after expressed sequence tags? The Plant Journal 70, 51–68. [DOI] [PubMed] [Google Scholar]
- Valkama E, Salminen JP, Koricheva J, Pihlaja K. 2003. Comparative analysis of leaf trichome structure and composition of epicuticular flavonoids in Finnish birch species. Annals of Botany 91, 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. 1997. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. 1999. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. The Plant Cell 11, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hubbard L, Chang Y, Guo J, Schiefelbein J, Chen JG. 2008. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biology 8, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werker E. 2000. Trichome diversity and development. Advances in Botanical Research 31, 1–35. [Google Scholar]
- Wester K, Digiuni S, Geier F, Timmer J, Fleck C, Hülskamp M. 2009. Functional diversity of R3 single-repeat genes in trichome development. Development 136, 1487–1496. [DOI] [PubMed] [Google Scholar]
- Williams L, Fletcher JC. 2005. Stem cell regulation in the Arabidopsis shoot apical meristem. Current Opinion in Plant Biology 8, 582–586. [DOI] [PubMed] [Google Scholar]
- Wu R, Citovsky V. 2017a Adaptor proteins GIR1 and GIR2. I. Interaction with the repressor GLABRA2 and regulation of root hair development. Biochemical and Biophysical Research Communications 488, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Citovsky V. 2017b Adaptor proteins GIR1 and GIR2. II. Interaction with the co-repressor TOPLESS and promotion of histone deacetylation of target chromatin. Biochemical and Biophysical Research Communications 488, 609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Chen M, Shen Q, et al. 2017. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytologist 213, 1145–1155. [DOI] [PubMed] [Google Scholar]
- Yang C, Gao Y, Gao S, Yu G, Xiong C, Chang J, Li H, Ye Z. 2015. Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato Wov gene in tobacco. BMC Genomics 16, 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Li H, Zhang J, et al. 2011. A regulatory gene induces trichome formation and embryo lethality in tomato. Proceedings of the National Academy of Sciences, USA 108, 11836–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.